Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
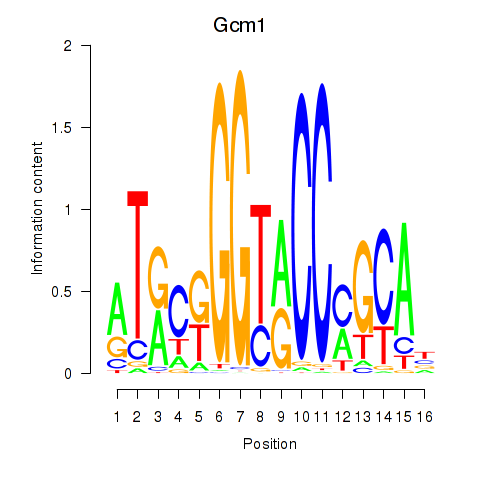
Results for Gcm1
Z-value: 0.52
Transcription factors associated with Gcm1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Gcm1
|
ENSMUSG00000023333.7 | glial cells missing homolog 1 |
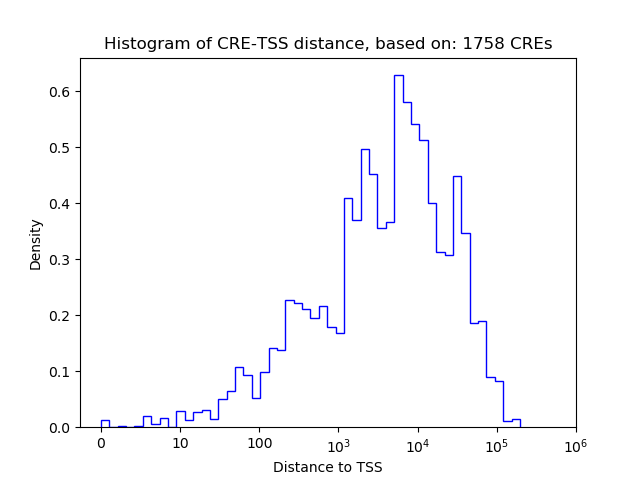
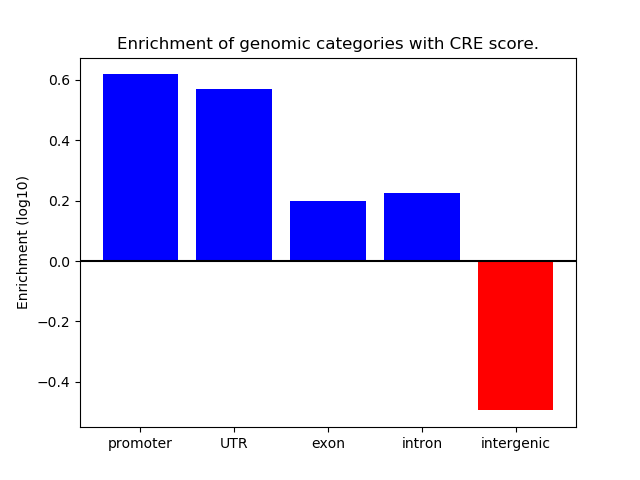
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr9_78050435_78050589 | Gcm1 | 1412 | 0.281499 | -0.38 | 4.5e-01 | Click! |
| chr9_78046870_78047029 | Gcm1 | 4975 | 0.124173 | 0.23 | 6.6e-01 | Click! |
Activity of the Gcm1 motif across conditions
Conditions sorted by the z-value of the Gcm1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
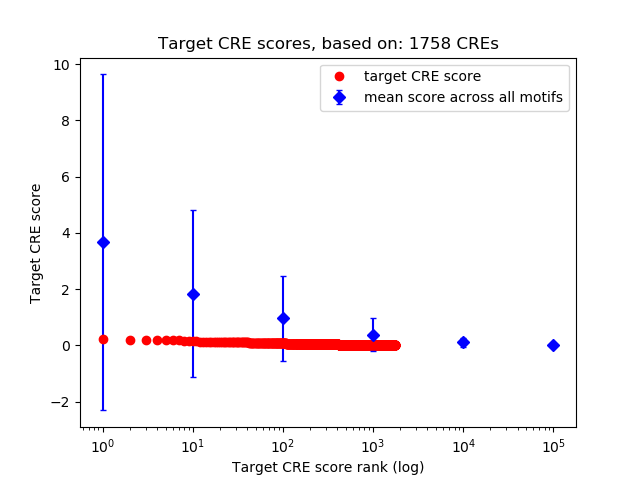
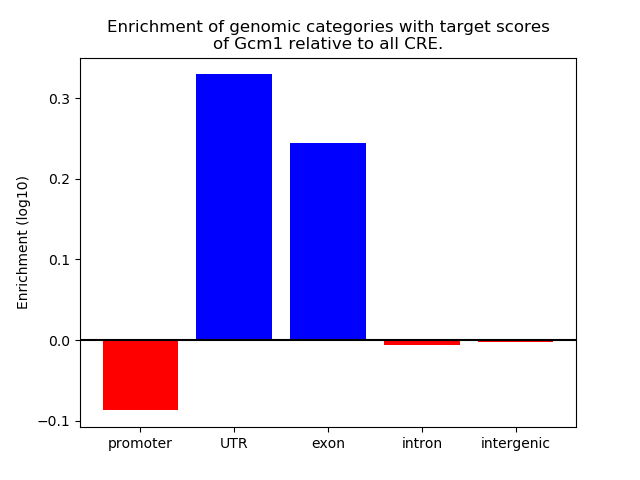
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr6_113669738_113669918 | 0.23 |
Irak2 |
interleukin-1 receptor-associated kinase 2 |
50 |
0.93 |
| chr4_141711616_141711801 | 0.20 |
Ddi2 |
DNA-damage inducible protein 2 |
11711 |
0.12 |
| chr10_83316057_83316237 | 0.20 |
Slc41a2 |
solute carrier family 41, member 2 |
6736 |
0.18 |
| chr11_101921459_101921610 | 0.19 |
Rpl27-ps2 |
ribosomal protein L27, pseudogene 2 |
17967 |
0.1 |
| chr9_98429359_98429510 | 0.19 |
Rbp1 |
retinol binding protein 1, cellular |
6473 |
0.22 |
| chr11_95387473_95387807 | 0.18 |
Slc35b1 |
solute carrier family 35, member B1 |
2345 |
0.19 |
| chr6_90796530_90796964 | 0.18 |
Gm44105 |
predicted gene, 44105 |
2272 |
0.27 |
| chr3_95693812_95693983 | 0.16 |
Adamtsl4 |
ADAMTS-like 4 |
5980 |
0.11 |
| chr8_123807438_123807645 | 0.15 |
Rab4a |
RAB4A, member RAS oncogene family |
1473 |
0.23 |
| chr12_82419285_82419473 | 0.15 |
Sipa1l1 |
signal-induced proliferation-associated 1 like 1 |
1236 |
0.58 |
| chr19_4194784_4194988 | 0.14 |
Ppp1ca |
protein phosphatase 1 catalytic subunit alpha |
237 |
0.73 |
| chr15_3368495_3368667 | 0.13 |
Ccdc152 |
coiled-coil domain containing 152 |
65055 |
0.12 |
| chr13_93627953_93628190 | 0.13 |
Gm15622 |
predicted gene 15622 |
2689 |
0.23 |
| chr1_154149857_154150038 | 0.13 |
A830008E24Rik |
RIKEN cDNA A830008E24 gene |
31996 |
0.15 |
| chr1_194977569_194977733 | 0.13 |
Gm16897 |
predicted gene, 16897 |
691 |
0.46 |
| chr11_117966799_117967405 | 0.13 |
Socs3 |
suppressor of cytokine signaling 3 |
2084 |
0.24 |
| chr15_96284208_96284391 | 0.12 |
2610037D02Rik |
RIKEN cDNA 2610037D02 gene |
196 |
0.94 |
| chr9_108090294_108090594 | 0.12 |
Apeh |
acylpeptide hydrolase |
1924 |
0.13 |
| chr5_112292069_112292220 | 0.12 |
Tpst2 |
protein-tyrosine sulfotransferase 2 |
3382 |
0.15 |
| chr7_98353281_98353473 | 0.12 |
Tsku |
tsukushi, small leucine rich proteoglycan |
6702 |
0.18 |
| chr3_82275838_82275989 | 0.11 |
Map9 |
microtubule-associated protein 9 |
82131 |
0.1 |
| chr2_32679057_32679208 | 0.11 |
Gm26236 |
predicted gene, 26236 |
4023 |
0.08 |
| chr7_63926888_63927066 | 0.11 |
Klf13 |
Kruppel-like factor 13 |
2107 |
0.23 |
| chr19_55779594_55779745 | 0.11 |
Tcf7l2 |
transcription factor 7 like 2, T cell specific, HMG box |
36824 |
0.2 |
| chr19_30120420_30120571 | 0.11 |
Gldc |
glycine decarboxylase |
24736 |
0.16 |
| chr9_57656306_57656491 | 0.11 |
Csk |
c-src tyrosine kinase |
2767 |
0.17 |
| chr17_56124312_56124572 | 0.11 |
Lrg1 |
leucine-rich alpha-2-glycoprotein 1 |
2441 |
0.13 |
| chr2_181522671_181522856 | 0.11 |
Dnajc5 |
DnaJ heat shock protein family (Hsp40) member C5 |
1816 |
0.2 |
| chr1_174765891_174766327 | 0.11 |
Fmn2 |
formin 2 |
51783 |
0.17 |
| chr15_79765471_79765632 | 0.11 |
Dnal4 |
dynein, axonemal, light chain 4 |
670 |
0.51 |
| chr14_61689110_61689310 | 0.11 |
Gm37820 |
predicted gene, 37820 |
5700 |
0.11 |
| chr11_106055229_106055380 | 0.11 |
Gm11646 |
predicted gene 11646 |
3452 |
0.13 |
| chr9_37636423_37636601 | 0.11 |
Siae |
sialic acid acetylesterase |
6168 |
0.12 |
| chr4_8910236_8910387 | 0.11 |
Rps18-ps2 |
ribosomal protein S18, pseudogene 2 |
30948 |
0.22 |
| chr7_98171190_98171341 | 0.11 |
Capn5 |
calpain 5 |
5824 |
0.15 |
| chr7_97630457_97630610 | 0.10 |
Rsf1os1 |
remodeling and spacing factor 1, opposite strand 1 |
14579 |
0.11 |
| chr17_32073432_32073589 | 0.10 |
Rrp1b |
ribosomal RNA processing 1 homolog B (S. cerevisiae) |
14342 |
0.13 |
| chr1_182585215_182585532 | 0.10 |
Capn8 |
calpain 8 |
20335 |
0.16 |
| chr3_94699549_94699901 | 0.10 |
Selenbp2 |
selenium binding protein 2 |
6066 |
0.12 |
| chr1_75652903_75653256 | 0.10 |
Gm5257 |
predicted gene 5257 |
16689 |
0.16 |
| chr17_64644726_64644989 | 0.10 |
Man2a1 |
mannosidase 2, alpha 1 |
44121 |
0.17 |
| chr19_37115604_37115755 | 0.10 |
Gm22714 |
predicted gene, 22714 |
33483 |
0.14 |
| chr4_117131790_117132241 | 0.10 |
Plk3 |
polo like kinase 3 |
1784 |
0.13 |
| chr1_121333853_121334026 | 0.10 |
Insig2 |
insulin induced gene 2 |
1350 |
0.38 |
| chr3_109307549_109307748 | 0.10 |
Vav3 |
vav 3 oncogene |
33005 |
0.21 |
| chr15_89378952_89379124 | 0.10 |
Odf3b |
outer dense fiber of sperm tails 3B |
35 |
0.93 |
| chr4_61401423_61401691 | 0.10 |
Mup15 |
major urinary protein 15 |
38186 |
0.14 |
| chr6_29444857_29445042 | 0.10 |
Flnc |
filamin C, gamma |
11673 |
0.09 |
| chr16_4355769_4355950 | 0.10 |
Gm6142 |
predicted pseudogene 6142 |
35179 |
0.15 |
| chr1_31051779_31051950 | 0.10 |
Gm28644 |
predicted gene 28644 |
9945 |
0.17 |
| chr14_76731472_76731694 | 0.09 |
4930431P22Rik |
RIKEN cDNA 4930431P22 gene |
28548 |
0.16 |
| chr19_46088425_46088600 | 0.09 |
Nolc1 |
nucleolar and coiled-body phosphoprotein 1 |
12458 |
0.12 |
| chr5_121329036_121329365 | 0.09 |
Hectd4 |
HECT domain E3 ubiquitin protein ligase 4 |
9177 |
0.13 |
| chr4_41064027_41064178 | 0.09 |
Gm25637 |
predicted gene, 25637 |
13882 |
0.11 |
| chr3_127895067_127895247 | 0.09 |
Fam241a |
family with sequence similarity 241, member A |
1131 |
0.39 |
| chr4_136178037_136178188 | 0.09 |
E2f2 |
E2F transcription factor 2 |
2671 |
0.2 |
| chr5_81686942_81687101 | 0.09 |
Adgrl3 |
adhesion G protein-coupled receptor L3 |
26718 |
0.21 |
| chr7_64501555_64501718 | 0.09 |
Apba2 |
amyloid beta (A4) precursor protein-binding, family A, member 2 |
70 |
0.97 |
| chr8_12896613_12896795 | 0.09 |
Gm15353 |
predicted gene 15353 |
12978 |
0.11 |
| chr14_65798900_65799063 | 0.09 |
Pbk |
PDZ binding kinase |
6856 |
0.2 |
| chr8_109999167_109999388 | 0.09 |
Tat |
tyrosine aminotransferase |
8771 |
0.12 |
| chr4_105225403_105225583 | 0.09 |
Plpp3 |
phospholipid phosphatase 3 |
68146 |
0.13 |
| chr4_140860963_140861126 | 0.08 |
4930515B02Rik |
RIKEN cDNA 4930515B02 gene |
11649 |
0.12 |
| chr19_23150997_23151148 | 0.08 |
Mir1192 |
microRNA 1192 |
1641 |
0.33 |
| chr15_99620057_99620228 | 0.08 |
Racgap1 |
Rac GTPase-activating protein 1 |
12662 |
0.1 |
| chr1_131286770_131286934 | 0.08 |
Ikbke |
inhibitor of kappaB kinase epsilon |
7246 |
0.13 |
| chr3_27788248_27788425 | 0.08 |
Fndc3b |
fibronectin type III domain containing 3B |
77029 |
0.1 |
| chr16_14940270_14940627 | 0.08 |
Efcab1 |
EF-hand calcium binding domain 1 |
33783 |
0.17 |
| chr15_85868388_85868539 | 0.08 |
Gtse1 |
G two S phase expressed protein 1 |
8517 |
0.12 |
| chr11_11945884_11946107 | 0.08 |
Grb10 |
growth factor receptor bound protein 10 |
335 |
0.89 |
| chr7_127262385_127262536 | 0.08 |
Dctpp1 |
dCTP pyrophosphatase 1 |
1751 |
0.15 |
| chr3_152763675_152763826 | 0.08 |
Pigk |
phosphatidylinositol glycan anchor biosynthesis, class K |
369 |
0.88 |
| chr4_109800666_109800954 | 0.08 |
Faf1 |
Fas-associated factor 1 |
39975 |
0.16 |
| chr9_48738508_48738659 | 0.08 |
Zbtb16 |
zinc finger and BTB domain containing 16 |
97362 |
0.07 |
| chr1_30947654_30948200 | 0.08 |
Ptp4a1 |
protein tyrosine phosphatase 4a1 |
1382 |
0.38 |
| chr18_39050085_39050237 | 0.08 |
Arhgap26 |
Rho GTPase activating protein 26 |
49150 |
0.14 |
| chr6_5045977_5046153 | 0.08 |
Ppp1r9a |
protein phosphatase 1, regulatory subunit 9A |
64594 |
0.1 |
| chr2_160767067_160767218 | 0.08 |
Plcg1 |
phospholipase C, gamma 1 |
7024 |
0.18 |
| chr19_3881058_3881209 | 0.07 |
Chka |
choline kinase alpha |
5728 |
0.09 |
| chr19_7160219_7160696 | 0.07 |
Otub1 |
OTU domain, ubiquitin aldehyde binding 1 |
40007 |
0.09 |
| chr5_32029942_32030169 | 0.07 |
Babam2 |
BRISC and BRCA1 A complex member 2 |
15923 |
0.18 |
| chr5_30835594_30835763 | 0.07 |
Gm15461 |
predicted gene 15461 |
13066 |
0.09 |
| chr19_58864470_58864651 | 0.07 |
Hspa12a |
heat shock protein 12A |
3548 |
0.23 |
| chr9_89827135_89827296 | 0.07 |
4930524O08Rik |
RIKEN cDNA 4930524O08 gene |
233 |
0.92 |
| chr13_111686251_111686408 | 0.07 |
Mier3 |
MIER family member 3 |
3 |
0.97 |
| chr3_51226339_51227024 | 0.07 |
Noct |
nocturnin |
2211 |
0.23 |
| chr7_101012379_101012581 | 0.07 |
P2ry2 |
purinergic receptor P2Y, G-protein coupled 2 |
386 |
0.81 |
| chr15_25188113_25188310 | 0.07 |
5830426I08Rik |
RIKEN cDNA 5830426I08 gene |
37643 |
0.13 |
| chr8_123978504_123978907 | 0.07 |
Abcb10 |
ATP-binding cassette, sub-family B (MDR/TAP), member 10 |
4417 |
0.12 |
| chr6_31146835_31147009 | 0.07 |
Gm37728 |
predicted gene, 37728 |
497 |
0.71 |
| chr5_89380116_89380277 | 0.07 |
Gc |
vitamin D binding protein |
55432 |
0.15 |
| chr16_43979287_43979438 | 0.07 |
Zdhhc23 |
zinc finger, DHHC domain containing 23 |
200 |
0.93 |
| chr1_60342961_60343669 | 0.07 |
Cyp20a1 |
cytochrome P450, family 20, subfamily a, polypeptide 1 |
8 |
0.98 |
| chr16_35485278_35485437 | 0.07 |
Pdia5 |
protein disulfide isomerase associated 5 |
5232 |
0.18 |
| chr14_31636168_31636433 | 0.07 |
Gm49387 |
predicted gene, 49387 |
4578 |
0.15 |
| chr10_60290245_60290396 | 0.07 |
Psap |
prosaposin |
5859 |
0.2 |
| chr9_70269742_70269893 | 0.07 |
Myo1e |
myosin IE |
62449 |
0.11 |
| chr4_19632365_19632516 | 0.07 |
Gm12353 |
predicted gene 12353 |
26989 |
0.17 |
| chr12_85879731_85879892 | 0.07 |
Ttll5 |
tubulin tyrosine ligase-like family, member 5 |
825 |
0.66 |
| chr3_83915891_83916061 | 0.07 |
Tmem131l |
transmembrane 131 like |
6432 |
0.26 |
| chr9_29661595_29661754 | 0.07 |
Gm15521 |
predicted gene 15521 |
69164 |
0.14 |
| chr19_46133855_46134398 | 0.07 |
Elovl3 |
elongation of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 3 |
2229 |
0.19 |
| chr1_36781526_36781677 | 0.07 |
Zap70 |
zeta-chain (TCR) associated protein kinase |
2617 |
0.18 |
| chr16_15890654_15890858 | 0.07 |
Cebpd |
CCAAT/enhancer binding protein (C/EBP), delta |
2340 |
0.29 |
| chr4_55959382_55959646 | 0.07 |
Gm12519 |
predicted gene 12519 |
34225 |
0.23 |
| chr4_62054018_62054169 | 0.07 |
Mup20 |
major urinary protein 20 |
65 |
0.96 |
| chr9_53771104_53771255 | 0.06 |
Slc35f2 |
solute carrier family 35, member F2 |
359 |
0.86 |
| chr2_91055387_91055557 | 0.06 |
Psmc3 |
proteasome (prosome, macropain) 26S subunit, ATPase 3 |
899 |
0.36 |
| chr1_134237854_134238025 | 0.06 |
Adora1 |
adenosine A1 receptor |
2508 |
0.21 |
| chr12_16717737_16717911 | 0.06 |
Greb1 |
gene regulated by estrogen in breast cancer protein |
6051 |
0.19 |
| chr13_73971445_73971616 | 0.06 |
Zdhhc11 |
zinc finger, DHHC domain containing 11 |
7668 |
0.1 |
| chr4_147194953_147195141 | 0.06 |
Gm13163 |
predicted gene 13163 |
164 |
0.92 |
| chr17_28886568_28886733 | 0.06 |
1700030A11Rik |
RIKEN cDNA 1700030A11 gene |
18615 |
0.08 |
| chr8_95349502_95349653 | 0.06 |
Mmp15 |
matrix metallopeptidase 15 |
2691 |
0.18 |
| chr18_50136754_50136905 | 0.06 |
Cd63-ps |
CD63 antigen, pseudogene |
2034 |
0.3 |
| chr11_103142469_103142688 | 0.06 |
Hexim2 |
hexamethylene bis-acetamide inducible 2 |
8713 |
0.12 |
| chr11_106785417_106785568 | 0.06 |
Ddx5 |
DEAD (Asp-Glu-Ala-Asp) box polypeptide 5 |
446 |
0.63 |
| chr9_15681946_15682110 | 0.06 |
Gm5119 |
predicted gene 5119 |
874 |
0.57 |
| chr2_163530402_163530553 | 0.06 |
Hnf4aos |
hepatic nuclear factor 4 alpha, opposite strand |
11215 |
0.12 |
| chr8_70799967_70800448 | 0.06 |
Mir7240 |
microRNA 7240 |
2072 |
0.12 |
| chr6_51267772_51267983 | 0.06 |
Mir148a |
microRNA 148a |
2033 |
0.36 |
| chr11_82856121_82856282 | 0.06 |
Rffl |
ring finger and FYVE like domain containing protein |
10109 |
0.11 |
| chr6_113469767_113469928 | 0.06 |
Il17rc |
interleukin 17 receptor C |
1580 |
0.17 |
| chr9_21229195_21229369 | 0.06 |
Gm16754 |
predicted gene, 16754 |
3879 |
0.11 |
| chr18_12682851_12683167 | 0.06 |
Ttc39c |
tetratricopeptide repeat domain 39C |
6719 |
0.16 |
| chr2_94405812_94406198 | 0.06 |
2810002D19Rik |
RIKEN cDNA 2810002D19 gene |
146 |
0.87 |
| chr1_95049965_95050120 | 0.06 |
Gm23181 |
predicted gene, 23181 |
9999 |
0.31 |
| chr4_125457322_125457488 | 0.06 |
Grik3 |
glutamate receptor, ionotropic, kainate 3 |
33295 |
0.17 |
| chr9_67815035_67815189 | 0.06 |
C2cd4a |
C2 calcium-dependent domain containing 4A |
17218 |
0.16 |
| chr8_119396900_119397055 | 0.06 |
Mlycd |
malonyl-CoA decarboxylase |
2079 |
0.26 |
| chr1_155797721_155797909 | 0.06 |
Qsox1 |
quiescin Q6 sulfhydryl oxidase 1 |
2398 |
0.2 |
| chr6_90363398_90363571 | 0.06 |
Zxdc |
ZXD family zinc finger C |
6008 |
0.12 |
| chr11_119489837_119489988 | 0.06 |
A930037H05Rik |
RIKEN cDNA A930037H05 gene |
1277 |
0.31 |
| chr19_58864190_58864413 | 0.06 |
Hspa12a |
heat shock protein 12A |
3289 |
0.24 |
| chr4_84619745_84620056 | 0.06 |
Bnc2 |
basonuclin 2 |
55096 |
0.15 |
| chr12_102650732_102650905 | 0.06 |
Itpk1 |
inositol 1,3,4-triphosphate 5/6 kinase |
9884 |
0.13 |
| chr2_165987296_165987478 | 0.06 |
Platr29 |
pluripotency associated transcript 29 |
1463 |
0.3 |
| chr1_40236545_40236756 | 0.06 |
Gm38070 |
predicted gene, 38070 |
1370 |
0.45 |
| chr6_91486783_91486970 | 0.06 |
Tmem43 |
transmembrane protein 43 |
10276 |
0.1 |
| chr8_33871610_33871761 | 0.06 |
Gm26978 |
predicted gene, 26978 |
14062 |
0.16 |
| chr2_30959100_30959284 | 0.06 |
Tor1b |
torsin family 1, member B |
3939 |
0.14 |
| chr15_9596456_9596681 | 0.06 |
Il7r |
interleukin 7 receptor |
66392 |
0.12 |
| chr8_72495274_72495445 | 0.06 |
Gm17435 |
predicted gene, 17435 |
2415 |
0.13 |
| chr8_13339386_13339572 | 0.06 |
Tfdp1 |
transcription factor Dp 1 |
153 |
0.93 |
| chr1_193172058_193172209 | 0.06 |
A130010J15Rik |
RIKEN cDNA A130010J15 gene |
1335 |
0.28 |
| chr5_89321713_89321897 | 0.06 |
Gc |
vitamin D binding protein |
113823 |
0.06 |
| chr1_186587150_186587347 | 0.06 |
A730004F24Rik |
RIKEN cDNA A730004F24 gene |
28567 |
0.18 |
| chr4_47039722_47039883 | 0.06 |
Anks6 |
ankyrin repeat and sterile alpha motif domain containing 6 |
341 |
0.81 |
| chr17_86697779_86697978 | 0.05 |
Gm22235 |
predicted gene, 22235 |
5529 |
0.21 |
| chr17_3246800_3247190 | 0.05 |
Gm49797 |
predicted gene, 49797 |
15724 |
0.16 |
| chr10_93494492_93494717 | 0.05 |
Hal |
histidine ammonia lyase |
1801 |
0.28 |
| chr3_10292843_10293025 | 0.05 |
Fabp12 |
fatty acid binding protein 12 |
8240 |
0.1 |
| chr5_117096808_117097327 | 0.05 |
Suds3 |
suppressor of defective silencing 3 homolog (S. cerevisiae) |
1744 |
0.29 |
| chr13_102646594_102646778 | 0.05 |
Gm47014 |
predicted gene, 47014 |
42153 |
0.14 |
| chr13_45965539_45965830 | 0.05 |
5033430I15Rik |
RIKEN cDNA 5033430I15 gene |
333 |
0.72 |
| chr18_12731812_12732024 | 0.05 |
Ttc39c |
tetratricopeptide repeat domain 39C |
2787 |
0.19 |
| chr12_41349181_41349335 | 0.05 |
Gm25497 |
predicted gene, 25497 |
686 |
0.57 |
| chr15_71996936_71997098 | 0.05 |
Col22a1 |
collagen, type XXII, alpha 1 |
37210 |
0.21 |
| chr4_146480712_146480882 | 0.05 |
Gm13245 |
predicted gene 13245 |
130 |
0.95 |
| chr6_8421479_8421656 | 0.05 |
Umad1 |
UMAP1-MVP12 associated (UMA) domain containing 1 |
1889 |
0.3 |
| chr1_89107539_89107745 | 0.05 |
Gm38312 |
predicted gene, 38312 |
10569 |
0.21 |
| chr2_113535511_113535708 | 0.05 |
Gm13964 |
predicted gene 13964 |
31575 |
0.17 |
| chr15_101236999_101237173 | 0.05 |
A330009N23Rik |
RIKEN cDNA A330009N23 gene |
11900 |
0.09 |
| chr11_94677695_94677908 | 0.05 |
Gm11542 |
predicted gene 11542 |
236 |
0.53 |
| chr1_131992475_131992670 | 0.05 |
Slc45a3 |
solute carrier family 45, member 3 |
13608 |
0.12 |
| chr18_25554784_25554942 | 0.05 |
Gm3227 |
predicted gene 3227 |
42627 |
0.17 |
| chr5_135022147_135022305 | 0.05 |
Stx1a |
syntaxin 1A (brain) |
1256 |
0.22 |
| chr1_194337170_194337321 | 0.05 |
4930503O07Rik |
RIKEN cDNA 4930503O07 gene |
114486 |
0.07 |
| chr14_33205736_33205895 | 0.05 |
Gm25498 |
predicted gene, 25498 |
1337 |
0.3 |
| chr3_27474078_27474242 | 0.05 |
Gm43344 |
predicted gene 43344 |
72481 |
0.1 |
| chr11_19924765_19925082 | 0.05 |
Spred2 |
sprouty-related EVH1 domain containing 2 |
481 |
0.87 |
| chr2_84408991_84409142 | 0.05 |
Calcrl |
calcitonin receptor-like |
16200 |
0.18 |
| chr9_43050755_43050915 | 0.05 |
Arhgef12 |
Rho guanine nucleotide exchange factor (GEF) 12 |
8203 |
0.22 |
| chr14_25493328_25493730 | 0.05 |
Zmiz1 |
zinc finger, MIZ-type containing 1 |
9126 |
0.14 |
| chr2_36091235_36091637 | 0.05 |
Lhx6 |
LIM homeobox protein 6 |
2837 |
0.18 |
| chr11_121354577_121354790 | 0.05 |
Wdr45b |
WD repeat domain 45B |
238 |
0.9 |
| chr12_104076433_104076597 | 0.05 |
Serpina4-ps1 |
serine (or cysteine) peptidase inhibitor, clade A, member 4, pseudogene 1 |
4134 |
0.12 |
| chr5_90447216_90447550 | 0.05 |
Alb |
albumin |
13514 |
0.15 |
| chr19_32573081_32573276 | 0.05 |
Papss2 |
3'-phosphoadenosine 5'-phosphosulfate synthase 2 |
22612 |
0.17 |
| chr13_52008098_52008268 | 0.05 |
Gm37872 |
predicted gene, 37872 |
13744 |
0.21 |
| chr4_61959781_61960042 | 0.05 |
Mup-ps19 |
major urinary protein, pseudogene 19 |
0 |
0.98 |
| chr17_84282731_84282889 | 0.05 |
Gm24492 |
predicted gene, 24492 |
4937 |
0.22 |
| chr13_101513249_101513636 | 0.05 |
Gm47533 |
predicted gene, 47533 |
31764 |
0.14 |
| chr15_10628189_10628357 | 0.05 |
Gm10389 |
predicted gene 10389 |
27936 |
0.14 |
| chr3_131653385_131653536 | 0.05 |
Gm42881 |
predicted gene 42881 |
39680 |
0.18 |
| chr3_38885313_38885466 | 0.05 |
Fat4 |
FAT atypical cadherin 4 |
1551 |
0.42 |
| chr5_75862712_75862874 | 0.05 |
Gm28865 |
predicted gene 28865 |
9835 |
0.16 |
| chr6_114931581_114931753 | 0.05 |
Vgll4 |
vestigial like family member 4 |
9846 |
0.23 |
| chr5_92726013_92726169 | 0.05 |
Gm20500 |
predicted gene 20500 |
10114 |
0.19 |
| chr5_92110647_92110859 | 0.05 |
Gm24931 |
predicted gene, 24931 |
7530 |
0.13 |
| chr6_121880801_121881009 | 0.05 |
Mug1 |
murinoglobulin 1 |
4672 |
0.22 |
| chr2_28544002_28544162 | 0.05 |
Ralgds |
ral guanine nucleotide dissociation stimulator |
1211 |
0.31 |
| chr11_4048460_4048709 | 0.05 |
Sec14l4 |
SEC14-like lipid binding 4 |
6847 |
0.11 |
| chr2_163480538_163480689 | 0.05 |
Fitm2 |
fat storage-inducing transmembrane protein 2 |
7984 |
0.11 |
| chr10_115778750_115778901 | 0.05 |
Tspan8 |
tetraspanin 8 |
38007 |
0.19 |
| chr5_52979532_52979683 | 0.05 |
Gm30301 |
predicted gene, 30301 |
2430 |
0.23 |
| chr10_27015331_27015545 | 0.05 |
Lama2 |
laminin, alpha 2 |
26924 |
0.19 |
| chr8_45358039_45358319 | 0.05 |
Fam149a |
family with sequence similarity 149, member A |
583 |
0.7 |
| chr1_93688304_93688651 | 0.05 |
Bok |
BCL2-related ovarian killer |
731 |
0.58 |
| chr9_57657975_57658137 | 0.05 |
Csk |
c-src tyrosine kinase |
4425 |
0.13 |
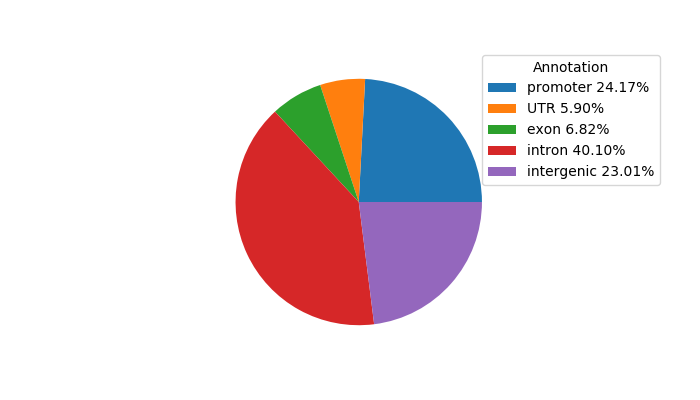
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0008355 | olfactory learning(GO:0008355) |
| 0.0 | 0.2 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.0 | 0.1 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.0 | 0.0 | GO:2000599 | regulation of cyclin catabolic process(GO:2000598) negative regulation of cyclin catabolic process(GO:2000599) |
| 0.0 | 0.1 | GO:0045719 | negative regulation of glycogen biosynthetic process(GO:0045719) |
| 0.0 | 0.1 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.0 | 0.0 | GO:0097195 | pilomotor reflex(GO:0097195) |
| 0.0 | 0.0 | GO:0019065 | receptor-mediated endocytosis of virus by host cell(GO:0019065) endocytosis involved in viral entry into host cell(GO:0075509) |
| 0.0 | 0.0 | GO:0072531 | pyrimidine-containing compound transmembrane transport(GO:0072531) |
| 0.0 | 0.0 | GO:0018894 | dibenzo-p-dioxin metabolic process(GO:0018894) |
| 0.0 | 0.0 | GO:2000622 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 0.1 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 0.0 | GO:1990597 | AIP1-IRE1 complex(GO:1990597) |
| 0.0 | 0.0 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0018588 | mono-butyltin dioxygenase activity(GO:0018586) tri-n-butyltin dioxygenase activity(GO:0018588) di-n-butyltin dioxygenase activity(GO:0018589) methylsilanetriol hydroxylase activity(GO:0018590) methyl tertiary butyl ether 3-monooxygenase activity(GO:0018591) 4-nitrocatechol 4-monooxygenase activity(GO:0018592) 4-chlorophenoxyacetate monooxygenase activity(GO:0018593) tert-butanol 2-monooxygenase activity(GO:0018594) alpha-pinene monooxygenase activity(GO:0018595) dimethylsilanediol hydroxylase activity(GO:0018596) ammonia monooxygenase activity(GO:0018597) hydroxymethylsilanetriol oxidase activity(GO:0018598) 2-hydroxyisobutyrate 3-monooxygenase activity(GO:0018599) alpha-pinene dehydrogenase activity(GO:0018600) bisphenol A hydroxylase B activity(GO:0034559) 2,2-bis(4-hydroxyphenyl)-1-propanol hydroxylase activity(GO:0034562) 9-fluorenone-3,4-dioxygenase activity(GO:0034786) anthracene 9,10-dioxygenase activity(GO:0034816) 2-(methylthio)benzothiazole monooxygenase activity(GO:0034857) 2-hydroxybenzothiazole monooxygenase activity(GO:0034858) benzothiazole monooxygenase activity(GO:0034859) 2,6-dihydroxybenzothiazole monooxygenase activity(GO:0034862) pinacolone 5-monooxygenase activity(GO:0034870) thioacetamide S-oxygenase activity(GO:0034873) thioacetamide S-oxide S-oxygenase activity(GO:0034874) endosulfan monooxygenase I activity(GO:0034888) N-nitrodimethylamine hydroxylase activity(GO:0034893) 4-(1-ethyl-1,4-dimethyl-pentyl)phenol monoxygenase activity(GO:0034897) endosulfan ether monooxygenase activity(GO:0034903) pyrene 4,5-monooxygenase activity(GO:0034925) pyrene 1,2-monooxygenase activity(GO:0034927) 1-hydroxypyrene 6,7-monooxygenase activity(GO:0034928) 1-hydroxypyrene 7,8-monooxygenase activity(GO:0034929) phenylboronic acid monooxygenase activity(GO:0034950) spheroidene monooxygenase activity(GO:0043823) |
| 0.0 | 0.1 | GO:0015065 | uridine nucleotide receptor activity(GO:0015065) G-protein coupled pyrimidinergic nucleotide receptor activity(GO:0071553) |
| 0.0 | 0.1 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.0 | 0.0 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |