Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
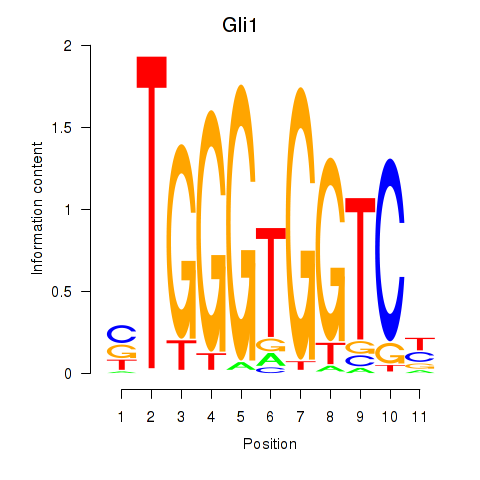
Results for Gli1
Z-value: 0.66
Transcription factors associated with Gli1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Gli1
|
ENSMUSG00000025407.6 | GLI-Kruppel family member GLI1 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr10_127342400_127342556 | Gli1 | 504 | 0.573543 | -0.36 | 4.9e-01 | Click! |
| chr10_127339210_127339386 | Gli1 | 661 | 0.452215 | 0.19 | 7.2e-01 | Click! |
| chr10_127338950_127339111 | Gli1 | 929 | 0.317120 | -0.17 | 7.5e-01 | Click! |
| chr10_127342165_127342330 | Gli1 | 273 | 0.786497 | 0.03 | 9.5e-01 | Click! |
| chr10_127341973_127342136 | Gli1 | 80 | 0.924080 | 0.00 | 1.0e+00 | Click! |
Activity of the Gli1 motif across conditions
Conditions sorted by the z-value of the Gli1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
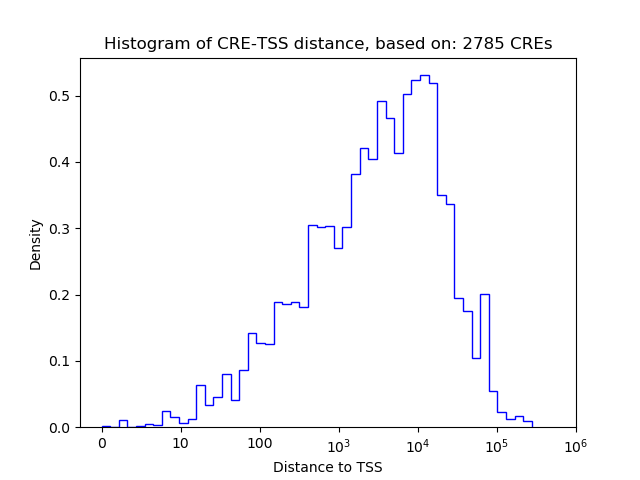
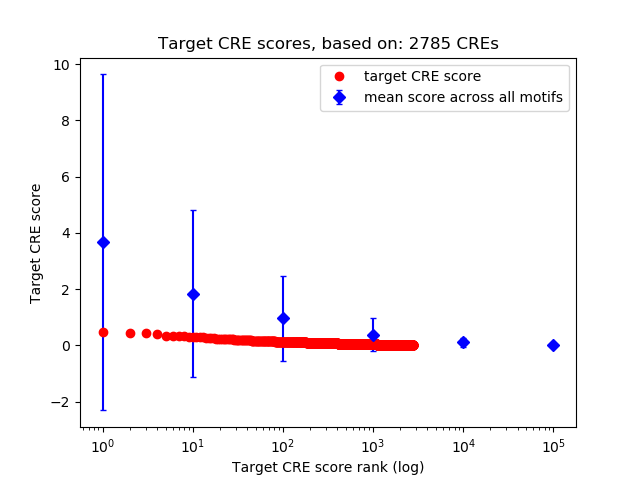
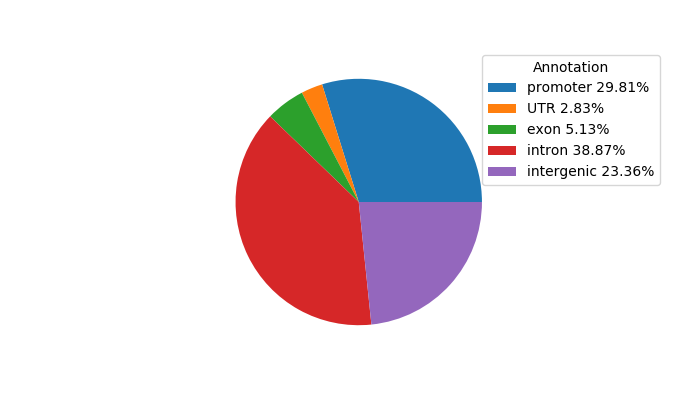
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr2_33430783_33430934 | 0.46 |
Zbtb34 |
zinc finger and BTB domain containing 34 |
459 |
0.78 |
| chr13_93627456_93627944 | 0.45 |
Gm15622 |
predicted gene 15622 |
2318 |
0.25 |
| chr19_3840572_3840723 | 0.45 |
Chka |
choline kinase alpha |
11126 |
0.09 |
| chr15_100625793_100625948 | 0.40 |
Dazap2 |
DAZ associated protein 2 |
9538 |
0.08 |
| chr2_103846113_103846314 | 0.34 |
Gm13879 |
predicted gene 13879 |
2557 |
0.13 |
| chr3_108011504_108011674 | 0.33 |
Gstm1 |
glutathione S-transferase, mu 1 |
1727 |
0.15 |
| chr11_86954758_86954966 | 0.33 |
Ypel2 |
yippee like 2 |
17162 |
0.18 |
| chr5_33726324_33726568 | 0.32 |
Fgfr3 |
fibroblast growth factor receptor 3 |
2674 |
0.15 |
| chr7_127804168_127804541 | 0.30 |
9430064I24Rik |
RIKEN cDNA 9430064I24 gene |
1592 |
0.16 |
| chr2_167260546_167260697 | 0.30 |
Ptgis |
prostaglandin I2 (prostacyclin) synthase |
20017 |
0.13 |
| chr15_27560020_27560329 | 0.30 |
Ank |
progressive ankylosis |
11273 |
0.16 |
| chr8_124843018_124843401 | 0.29 |
Gm16163 |
predicted gene 16163 |
11511 |
0.11 |
| chr11_119056746_119056911 | 0.28 |
Cbx8 |
chromobox 8 |
15859 |
0.14 |
| chr19_46133855_46134398 | 0.27 |
Elovl3 |
elongation of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 3 |
2229 |
0.19 |
| chr6_141875439_141875590 | 0.25 |
Gm30784 |
predicted gene, 30784 |
9156 |
0.21 |
| chr3_121168919_121169132 | 0.25 |
Gm10652 |
predicted gene 10652 |
369 |
0.83 |
| chr10_59978256_59978409 | 0.25 |
Anapc16 |
anaphase promoting complex subunit 16 |
12051 |
0.16 |
| chr8_13889986_13890137 | 0.24 |
Coprs |
coordinator of PRMT5, differentiation stimulator |
71 |
0.95 |
| chr10_76580112_76580438 | 0.22 |
Ftcd |
formiminotransferase cyclodeaminase |
4623 |
0.13 |
| chr1_72836877_72837059 | 0.22 |
Igfbp2 |
insulin-like growth factor binding protein 2 |
11646 |
0.21 |
| chr5_110809507_110809658 | 0.22 |
Ulk1 |
unc-51 like kinase 1 |
471 |
0.7 |
| chr11_60113222_60113373 | 0.21 |
4930412M03Rik |
RIKEN cDNA 4930412M03 gene |
1933 |
0.28 |
| chr15_89378952_89379124 | 0.21 |
Odf3b |
outer dense fiber of sperm tails 3B |
35 |
0.93 |
| chr6_117431300_117431501 | 0.21 |
Gm4640 |
predicted gene 4640 |
7783 |
0.22 |
| chr12_28898892_28899474 | 0.21 |
Gm31508 |
predicted gene, 31508 |
11046 |
0.17 |
| chr5_93282543_93282755 | 0.21 |
Ccng2 |
cyclin G2 |
13941 |
0.18 |
| chr19_4623588_4623859 | 0.21 |
Mir6986 |
microRNA 6986 |
232 |
0.84 |
| chr9_46230822_46231218 | 0.21 |
Apoa1 |
apolipoprotein A-I |
2299 |
0.14 |
| chr16_91379054_91379459 | 0.21 |
Ifnar2 |
interferon (alpha and beta) receptor 2 |
6357 |
0.12 |
| chr19_16437834_16438025 | 0.20 |
Gna14 |
guanine nucleotide binding protein, alpha 14 |
1371 |
0.4 |
| chr3_96451459_96451723 | 0.20 |
BC107364 |
cDNA sequence BC107364 |
715 |
0.31 |
| chr3_96458907_96459172 | 0.20 |
Gm10685 |
predicted gene 10685 |
830 |
0.26 |
| chr8_41041775_41041987 | 0.20 |
Mtus1 |
mitochondrial tumor suppressor 1 |
15 |
0.97 |
| chr8_46386589_46386784 | 0.19 |
Gm45253 |
predicted gene 45253 |
798 |
0.57 |
| chr19_61049084_61049293 | 0.19 |
Gm22520 |
predicted gene, 22520 |
35643 |
0.14 |
| chr3_96402109_96402496 | 0.19 |
Gm26654 |
predicted gene, 26654 |
79 |
0.72 |
| chr11_75196330_75196483 | 0.19 |
Rtn4rl1 |
reticulon 4 receptor-like 1 |
2623 |
0.15 |
| chr2_36092832_36093004 | 0.18 |
Lhx6 |
LIM homeobox protein 6 |
1355 |
0.33 |
| chr7_19955922_19956099 | 0.18 |
Rpl7a-ps8 |
ribosomal protein L7A, pseudogene 8 |
2227 |
0.14 |
| chr14_31427037_31427204 | 0.18 |
Sh3bp5 |
SH3-domain binding protein 5 (BTK-associated) |
8945 |
0.15 |
| chr1_153900403_153900624 | 0.18 |
Glul |
glutamate-ammonia ligase (glutamine synthetase) |
72 |
0.72 |
| chr9_48755281_48755902 | 0.17 |
Zbtb16 |
zinc finger and BTB domain containing 16 |
80354 |
0.09 |
| chr18_35831562_35831748 | 0.17 |
Gm29417 |
predicted gene 29417 |
562 |
0.55 |
| chr12_104414666_104414826 | 0.17 |
Gm24334 |
predicted gene, 24334 |
4654 |
0.15 |
| chr9_31851816_31851990 | 0.17 |
Gm31497 |
predicted gene, 31497 |
54090 |
0.1 |
| chr8_126774523_126774869 | 0.17 |
Gm45805 |
predicted gene 45805 |
16362 |
0.22 |
| chr4_33284555_33284721 | 0.17 |
Pnrc1 |
proline-rich nuclear receptor coactivator 1 |
5525 |
0.18 |
| chr17_25727890_25728059 | 0.17 |
Rpusd1 |
RNA pseudouridylate synthase domain containing 1 |
242 |
0.65 |
| chr5_120560470_120560640 | 0.17 |
2510016D11Rik |
RIKEN cDNA 2510016D11 gene |
7516 |
0.1 |
| chr8_126685189_126685340 | 0.16 |
Gm45805 |
predicted gene 45805 |
73070 |
0.1 |
| chr1_39205802_39205953 | 0.16 |
Npas2 |
neuronal PAS domain protein 2 |
11665 |
0.18 |
| chr2_94047308_94047485 | 0.16 |
Hsd17b12 |
hydroxysteroid (17-beta) dehydrogenase 12 |
19073 |
0.16 |
| chr8_119443770_119443946 | 0.16 |
Necab2 |
N-terminal EF-hand calcium binding protein 2 |
2861 |
0.21 |
| chr15_99716487_99716652 | 0.16 |
Gpd1 |
glycerol-3-phosphate dehydrogenase 1 (soluble) |
946 |
0.3 |
| chr15_79351473_79351624 | 0.16 |
Maff |
v-maf musculoaponeurotic fibrosarcoma oncogene family, protein F (avian) |
3146 |
0.14 |
| chr13_43493297_43493485 | 0.16 |
Gm47683 |
predicted gene, 47683 |
6776 |
0.14 |
| chr8_105759107_105759440 | 0.16 |
Gfod2 |
glucose-fructose oxidoreductase domain containing 2 |
609 |
0.58 |
| chr7_81541084_81541411 | 0.16 |
Gm37829 |
predicted gene, 37829 |
8726 |
0.1 |
| chr8_13350961_13351112 | 0.16 |
Tfdp1 |
transcription factor Dp 1 |
11362 |
0.12 |
| chr14_30916422_30916581 | 0.16 |
Itih3 |
inter-alpha trypsin inhibitor, heavy chain 3 |
1114 |
0.35 |
| chr1_39612023_39612366 | 0.16 |
Gm5100 |
predicted gene 5100 |
14189 |
0.12 |
| chr14_70338039_70338256 | 0.15 |
Slc39a14 |
solute carrier family 39 (zinc transporter), member 14 |
3949 |
0.15 |
| chr9_57283258_57283868 | 0.15 |
1700017B05Rik |
RIKEN cDNA 1700017B05 gene |
20951 |
0.14 |
| chr9_120731019_120731170 | 0.15 |
Gm47064 |
predicted gene, 47064 |
20049 |
0.12 |
| chr10_67536858_67537009 | 0.15 |
Egr2 |
early growth response 2 |
936 |
0.47 |
| chr1_134065116_134065322 | 0.15 |
Btg2 |
BTG anti-proliferation factor 2 |
13901 |
0.12 |
| chr4_56961559_56961774 | 0.15 |
Tmem245 |
transmembrane protein 245 |
14229 |
0.15 |
| chr5_9038233_9038384 | 0.15 |
Gm40264 |
predicted gene, 40264 |
3184 |
0.2 |
| chr16_97918472_97918638 | 0.14 |
C2cd2 |
C2 calcium-dependent domain containing 2 |
4051 |
0.21 |
| chr1_93686775_93686953 | 0.14 |
Bok |
BCL2-related ovarian killer |
601 |
0.66 |
| chr3_96646975_96647126 | 0.14 |
Itga10 |
integrin, alpha 10 |
1466 |
0.17 |
| chr10_121423941_121424099 | 0.14 |
Gm18730 |
predicted gene, 18730 |
23307 |
0.11 |
| chr10_58233103_58233399 | 0.14 |
Duxf3 |
double homeobox family member 3 |
576 |
0.63 |
| chr9_62344600_62344925 | 0.14 |
Anp32a |
acidic (leucine-rich) nuclear phosphoprotein 32 family, member A |
37 |
0.98 |
| chr7_110853928_110854130 | 0.14 |
Lyve1 |
lymphatic vessel endothelial hyaluronan receptor 1 |
8924 |
0.15 |
| chr11_51832795_51832987 | 0.14 |
Jade2 |
jade family PHD finger 2 |
24234 |
0.15 |
| chr13_45907083_45907460 | 0.14 |
4930453C13Rik |
RIKEN cDNA 4930453C13 gene |
28769 |
0.18 |
| chr7_141478122_141478322 | 0.14 |
Tspan4 |
tetraspanin 4 |
1822 |
0.13 |
| chr11_81374622_81374773 | 0.14 |
4930527B05Rik |
RIKEN cDNA 4930527B05 gene |
10316 |
0.3 |
| chr15_82668190_82668753 | 0.14 |
Cyp2d35-ps |
cytochrome P450, family 2, subfamily d, member 35, pseudogene |
13760 |
0.07 |
| chr8_35290423_35290789 | 0.14 |
Gm34597 |
predicted gene, 34597 |
45127 |
0.11 |
| chr8_126656995_126657172 | 0.13 |
Irf2bp2 |
interferon regulatory factor 2 binding protein 2 |
63097 |
0.12 |
| chr17_4199793_4199983 | 0.13 |
4930548J01Rik |
RIKEN cDNA 4930548J01 gene |
77775 |
0.11 |
| chr11_67535494_67535665 | 0.13 |
Gas7 |
growth arrest specific 7 |
11575 |
0.23 |
| chr3_36079982_36080141 | 0.13 |
Acad9 |
acyl-Coenzyme A dehydrogenase family, member 9 |
85 |
0.95 |
| chr7_24653592_24653770 | 0.13 |
Gm4598 |
predicted gene 4598 |
7613 |
0.08 |
| chr2_32259552_32259703 | 0.13 |
Uck1 |
uridine-cytidine kinase 1 |
6 |
0.94 |
| chr4_155028352_155028518 | 0.13 |
Plch2 |
phospholipase C, eta 2 |
9003 |
0.13 |
| chr8_116185346_116185497 | 0.13 |
4930422C21Rik |
RIKEN cDNA 4930422C21 gene |
1948 |
0.49 |
| chr15_36476548_36476699 | 0.13 |
Ankrd46 |
ankyrin repeat domain 46 |
20092 |
0.13 |
| chr16_24082765_24082916 | 0.13 |
Gm46545 |
predicted gene, 46545 |
150 |
0.95 |
| chr4_141536459_141536662 | 0.13 |
Spen |
spen family transcription repressor |
1693 |
0.27 |
| chr14_30939550_30939725 | 0.13 |
Itih1 |
inter-alpha trypsin inhibitor, heavy chain 1 |
3640 |
0.13 |
| chr11_75430733_75430997 | 0.13 |
Gm12337 |
predicted gene 12337 |
3112 |
0.11 |
| chr11_101728654_101728805 | 0.13 |
Dhx8 |
DEAH (Asp-Glu-Ala-His) box polypeptide 8 |
4190 |
0.16 |
| chr12_37906736_37906912 | 0.13 |
Dgkb |
diacylglycerol kinase, beta |
26108 |
0.21 |
| chr9_108083457_108083608 | 0.13 |
Rnf123 |
ring finger protein 123 |
186 |
0.77 |
| chr2_155272603_155272754 | 0.13 |
Map1lc3a |
microtubule-associated protein 1 light chain 3 alpha |
3619 |
0.2 |
| chr4_134399064_134399223 | 0.13 |
Pafah2 |
platelet-activating factor acetylhydrolase 2 |
1672 |
0.26 |
| chr18_67800429_67800656 | 0.13 |
Cep192 |
centrosomal protein 192 |
408 |
0.57 |
| chr4_49501233_49501521 | 0.13 |
Baat |
bile acid-Coenzyme A: amino acid N-acyltransferase |
1743 |
0.25 |
| chr4_53263517_53263892 | 0.13 |
AI427809 |
expressed sequence AI427809 |
127 |
0.96 |
| chr1_138322744_138322953 | 0.13 |
Atp6v1g3 |
ATPase, H+ transporting, lysosomal V1 subunit G3 |
49110 |
0.12 |
| chr1_131963395_131963600 | 0.13 |
Slc45a3 |
solute carrier family 45, member 3 |
530 |
0.42 |
| chr16_30550091_30550467 | 0.12 |
Tmem44 |
transmembrane protein 44 |
249 |
0.93 |
| chr5_9059465_9059641 | 0.12 |
Gm40264 |
predicted gene, 40264 |
24429 |
0.13 |
| chr6_48595408_48595586 | 0.12 |
Repin1 |
replication initiator 1 |
1178 |
0.2 |
| chr11_70489793_70489955 | 0.12 |
Tm4sf5 |
transmembrane 4 superfamily member 5 |
15370 |
0.06 |
| chr15_36933623_36933947 | 0.12 |
Gm41300 |
predicted gene, 41300 |
2099 |
0.27 |
| chr18_46713777_46714063 | 0.12 |
Cdo1 |
cysteine dioxygenase 1, cytosolic |
14109 |
0.13 |
| chr8_106936247_106936398 | 0.12 |
Sntb2 |
syntrophin, basic 2 |
173 |
0.92 |
| chr5_114581477_114581628 | 0.12 |
Fam222a |
family with sequence similarity 222, member A |
13535 |
0.15 |
| chr8_35410788_35410939 | 0.12 |
Gm45301 |
predicted gene 45301 |
1357 |
0.39 |
| chr17_36019112_36019264 | 0.12 |
H2-T24 |
histocompatibility 2, T region locus 24 |
1337 |
0.17 |
| chr12_105546152_105546368 | 0.12 |
Gm47796 |
predicted gene, 47796 |
15003 |
0.14 |
| chr15_100682914_100683075 | 0.12 |
Cela1 |
chymotrypsin-like elastase family, member 1 |
125 |
0.93 |
| chr1_133307113_133307383 | 0.12 |
Golt1a |
golgi transport 1A |
2552 |
0.17 |
| chr15_77160485_77160636 | 0.12 |
Rbfox2 |
RNA binding protein, fox-1 homolog (C. elegans) 2 |
6748 |
0.15 |
| chr11_109502246_109502399 | 0.12 |
Gm22378 |
predicted gene, 22378 |
2498 |
0.21 |
| chr6_72624193_72624362 | 0.12 |
Gm15401 |
predicted gene 15401 |
5011 |
0.09 |
| chr8_90371977_90372137 | 0.12 |
Tox3 |
TOX high mobility group box family member 3 |
23714 |
0.27 |
| chr8_126694488_126694675 | 0.12 |
Gm45805 |
predicted gene 45805 |
63753 |
0.12 |
| chr1_132318004_132318180 | 0.12 |
Nuak2 |
NUAK family, SNF1-like kinase, 2 |
1927 |
0.22 |
| chr7_143502927_143503132 | 0.12 |
Phlda2 |
pleckstrin homology like domain, family A, member 2 |
36 |
0.96 |
| chr11_58307244_58307445 | 0.12 |
Zfp692 |
zinc finger protein 692 |
172 |
0.9 |
| chr3_88406373_88406524 | 0.12 |
Pmf1 |
polyamine-modulated factor 1 |
3817 |
0.08 |
| chr1_182648871_182649054 | 0.12 |
Gm37516 |
predicted gene, 37516 |
30977 |
0.15 |
| chr5_124636798_124636967 | 0.12 |
Atp6v0a2 |
ATPase, H+ transporting, lysosomal V0 subunit A2 |
7713 |
0.14 |
| chr11_101733748_101733899 | 0.12 |
Dhx8 |
DEAH (Asp-Glu-Ala-His) box polypeptide 8 |
782 |
0.56 |
| chr7_141628129_141628296 | 0.11 |
Mir7063 |
microRNA 7063 |
7511 |
0.13 |
| chr10_22645135_22645353 | 0.11 |
Slc2a12 |
solute carrier family 2 (facilitated glucose transporter), member 12 |
233 |
0.94 |
| chr11_62487650_62487823 | 0.11 |
Gm12278 |
predicted gene 12278 |
4939 |
0.13 |
| chr1_180227522_180227852 | 0.11 |
Psen2 |
presenilin 2 |
1881 |
0.26 |
| chr15_86275887_86276051 | 0.11 |
Tbc1d22a |
TBC1 domain family, member 22a |
40464 |
0.16 |
| chr1_181210349_181210792 | 0.11 |
Wdr26 |
WD repeat domain 26 |
856 |
0.52 |
| chr15_97059709_97059903 | 0.11 |
Slc38a4 |
solute carrier family 38, member 4 |
3850 |
0.33 |
| chr10_127903210_127903408 | 0.11 |
Sdr9c7 |
4short chain dehydrogenase/reductase family 9C, member 7 |
105 |
0.93 |
| chr5_92110647_92110859 | 0.11 |
Gm24931 |
predicted gene, 24931 |
7530 |
0.13 |
| chr13_43525896_43526216 | 0.11 |
Gm32939 |
predicted gene, 32939 |
4164 |
0.16 |
| chr2_167944308_167944486 | 0.11 |
Ptpn1 |
protein tyrosine phosphatase, non-receptor type 1 |
6651 |
0.19 |
| chr6_38343853_38344004 | 0.11 |
Zc3hav1 |
zinc finger CCCH type, antiviral 1 |
10345 |
0.13 |
| chr12_25249300_25249688 | 0.11 |
Gm19340 |
predicted gene, 19340 |
33101 |
0.15 |
| chr2_26974148_26974379 | 0.11 |
Adamts13 |
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 13 |
770 |
0.43 |
| chr14_48138613_48138798 | 0.11 |
Gm49310 |
predicted gene, 49310 |
4481 |
0.16 |
| chr15_67020007_67020163 | 0.11 |
Gm31342 |
predicted gene, 31342 |
19973 |
0.18 |
| chr12_104798236_104798545 | 0.11 |
Clmn |
calmin |
17519 |
0.19 |
| chr11_98769689_98769972 | 0.11 |
Nr1d1 |
nuclear receptor subfamily 1, group D, member 1 |
596 |
0.57 |
| chr10_95625719_95626123 | 0.11 |
Gm33336 |
predicted gene, 33336 |
6318 |
0.14 |
| chr19_43898660_43898822 | 0.11 |
Dnmbp |
dynamin binding protein |
8050 |
0.14 |
| chr13_63562924_63563160 | 0.11 |
Ptch1 |
patched 1 |
773 |
0.56 |
| chr11_4119733_4119903 | 0.11 |
Sec14l2 |
SEC14-like lipid binding 2 |
987 |
0.33 |
| chr10_41959970_41960121 | 0.11 |
Armc2 |
armadillo repeat containing 2 |
16077 |
0.2 |
| chr8_34147011_34147181 | 0.11 |
Leprotl1 |
leptin receptor overlapping transcript-like 1 |
63 |
0.95 |
| chr5_102460519_102460675 | 0.11 |
1700013M08Rik |
RIKEN cDNA 1700013M08 gene |
19399 |
0.2 |
| chr8_11921702_11921858 | 0.11 |
Gm45680 |
predicted gene 45680 |
26517 |
0.14 |
| chr6_72218048_72218558 | 0.11 |
Atoh8 |
atonal bHLH transcription factor 8 |
16234 |
0.15 |
| chr10_77551087_77551238 | 0.11 |
Itgb2 |
integrin beta 2 |
20782 |
0.1 |
| chr3_104220440_104221564 | 0.11 |
Magi3 |
membrane associated guanylate kinase, WW and PDZ domain containing 3 |
628 |
0.6 |
| chr5_33218123_33218481 | 0.11 |
Spon2 |
spondin 2, extracellular matrix protein |
107 |
0.96 |
| chr10_28078586_28078966 | 0.11 |
Ptprk |
protein tyrosine phosphatase, receptor type, K |
3766 |
0.28 |
| chr4_123589098_123589491 | 0.11 |
Macf1 |
microtubule-actin crosslinking factor 1 |
16234 |
0.16 |
| chr15_98591099_98591252 | 0.10 |
Gm29331 |
predicted gene 29331 |
1185 |
0.26 |
| chr10_96231610_96231804 | 0.10 |
4930459C07Rik |
RIKEN cDNA 4930459C07 gene |
5500 |
0.22 |
| chr2_118726808_118726959 | 0.10 |
Plcb2 |
phospholipase C, beta 2 |
1476 |
0.29 |
| chr11_118215876_118216042 | 0.10 |
Cyth1 |
cytohesin 1 |
3066 |
0.2 |
| chr11_116435092_116435485 | 0.10 |
Ubald2 |
UBA-like domain containing 2 |
779 |
0.48 |
| chr2_144182222_144182616 | 0.10 |
Gm5535 |
predicted gene 5535 |
3333 |
0.2 |
| chr5_74304899_74305225 | 0.10 |
Gm15981 |
predicted gene 15981 |
11972 |
0.16 |
| chr17_5004485_5004673 | 0.10 |
Arid1b |
AT rich interactive domain 1B (SWI-like) |
8150 |
0.25 |
| chr15_10344412_10344563 | 0.10 |
Agxt2 |
alanine-glyoxylate aminotransferase 2 |
14045 |
0.18 |
| chr11_83944358_83944548 | 0.10 |
Ddx52 |
DEAD (Asp-Glu-Ala-Asp) box polypeptide 52 |
1802 |
0.33 |
| chr8_22124586_22124925 | 0.10 |
Nek5 |
NIMA (never in mitosis gene a)-related expressed kinase 5 |
283 |
0.87 |
| chr15_62063425_62063595 | 0.10 |
Pvt1 |
Pvt1 oncogene |
24253 |
0.23 |
| chr13_32970123_32970274 | 0.10 |
Serpinb6b |
serine (or cysteine) peptidase inhibitor, clade B, member 6b |
1300 |
0.35 |
| chr16_94545906_94546079 | 0.10 |
Vps26c |
VPS26 endosomal protein sorting factor C |
19162 |
0.16 |
| chr16_75345372_75345557 | 0.10 |
Gm49677 |
predicted gene, 49677 |
53810 |
0.16 |
| chr17_26647402_26647562 | 0.10 |
Ergic1 |
endoplasmic reticulum-golgi intermediate compartment (ERGIC) 1 |
11459 |
0.13 |
| chr8_90841291_90841442 | 0.10 |
Chd9 |
chromodomain helicase DNA binding protein 9 |
8301 |
0.13 |
| chr19_55754460_55754655 | 0.10 |
Tcf7l2 |
transcription factor 7 like 2, T cell specific, HMG box |
11712 |
0.28 |
| chr4_105225983_105226273 | 0.10 |
Plpp3 |
phospholipid phosphatase 3 |
68781 |
0.12 |
| chr11_120493774_120493943 | 0.10 |
Gm11788 |
predicted gene 11788 |
1464 |
0.16 |
| chr13_32821794_32821951 | 0.10 |
Wrnip1 |
Werner helicase interacting protein 1 |
1106 |
0.41 |
| chr9_73050700_73050869 | 0.10 |
Rab27a |
RAB27A, member RAS oncogene family |
5800 |
0.09 |
| chr3_122462488_122462639 | 0.10 |
Gm25153 |
predicted gene, 25153 |
2164 |
0.23 |
| chr2_156190488_156190836 | 0.10 |
Phf20 |
PHD finger protein 20 |
5804 |
0.13 |
| chr13_37452717_37452952 | 0.10 |
Gm29458 |
predicted gene 29458 |
374 |
0.73 |
| chr14_63133928_63134119 | 0.10 |
Ctsb |
cathepsin B |
1495 |
0.31 |
| chr8_119426295_119426446 | 0.10 |
Osgin1 |
oxidative stress induced growth inhibitor 1 |
7754 |
0.15 |
| chr18_60737126_60737516 | 0.10 |
Ndst1 |
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1 |
4171 |
0.18 |
| chr5_138823785_138824155 | 0.10 |
Gm5294 |
predicted gene 5294 |
3890 |
0.23 |
| chr9_123153278_123153441 | 0.10 |
Clec3b |
C-type lectin domain family 3, member b |
2413 |
0.18 |
| chr9_53355672_53355823 | 0.10 |
Exph5 |
exophilin 5 |
14558 |
0.16 |
| chr15_85710318_85710476 | 0.10 |
Mirlet7b |
microRNA let7b |
3078 |
0.17 |
| chr1_74190744_74190929 | 0.10 |
Cxcr1 |
chemokine (C-X-C motif) receptor 1 |
2399 |
0.18 |
| chr5_8971094_8971245 | 0.10 |
Gm15610 |
predicted gene 15610 |
779 |
0.53 |
| chr2_153214800_153214963 | 0.10 |
Tspyl3 |
TSPY-like 3 |
10560 |
0.13 |
| chr9_121388029_121388181 | 0.10 |
Trak1 |
trafficking protein, kinesin binding 1 |
3856 |
0.22 |
| chr2_129800360_129800704 | 0.10 |
Stk35 |
serine/threonine kinase 35 |
15 |
0.98 |
| chr2_75563293_75563456 | 0.10 |
Gm13655 |
predicted gene 13655 |
70008 |
0.08 |
| chr17_56609964_56610227 | 0.09 |
Micos13 |
mitochondrial contact site and cristae organizing system subunit 13 |
324 |
0.78 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0043985 | histone H4-R3 methylation(GO:0043985) |
| 0.0 | 0.1 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.0 | 0.1 | GO:0019676 | ammonia assimilation cycle(GO:0019676) |
| 0.0 | 0.2 | GO:0043097 | pyrimidine-containing compound salvage(GO:0008655) pyrimidine nucleoside salvage(GO:0043097) |
| 0.0 | 0.1 | GO:0051541 | elastin metabolic process(GO:0051541) |
| 0.0 | 0.1 | GO:0021593 | rhombomere morphogenesis(GO:0021593) |
| 0.0 | 0.1 | GO:1902065 | response to L-glutamate(GO:1902065) |
| 0.0 | 0.1 | GO:2000823 | regulation of androgen receptor activity(GO:2000823) |
| 0.0 | 0.1 | GO:0061144 | alveolar secondary septum development(GO:0061144) |
| 0.0 | 0.1 | GO:0015772 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.0 | 0.1 | GO:0006362 | transcription elongation from RNA polymerase I promoter(GO:0006362) |
| 0.0 | 0.1 | GO:0052490 | negative regulation by symbiont of host apoptotic process(GO:0033668) negative regulation by symbiont of host programmed cell death(GO:0052041) negative regulation by organism of programmed cell death in other organism involved in symbiotic interaction(GO:0052490) |
| 0.0 | 0.1 | GO:0045200 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.0 | 0.1 | GO:0071288 | cellular response to mercury ion(GO:0071288) |
| 0.0 | 0.1 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.0 | 0.1 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.0 | 0.2 | GO:0021684 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.0 | 0.1 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.0 | 0.1 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.0 | 0.1 | GO:0009436 | glyoxylate catabolic process(GO:0009436) |
| 0.0 | 0.1 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.0 | 0.1 | GO:0006216 | cytidine catabolic process(GO:0006216) cytidine deamination(GO:0009972) cytidine metabolic process(GO:0046087) |
| 0.0 | 0.1 | GO:0071879 | positive regulation of adrenergic receptor signaling pathway(GO:0071879) |
| 0.0 | 0.0 | GO:0032058 | positive regulation of translational initiation in response to stress(GO:0032058) |
| 0.0 | 0.1 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 | 0.1 | GO:1902744 | negative regulation of lamellipodium organization(GO:1902744) |
| 0.0 | 0.1 | GO:0070857 | regulation of bile acid biosynthetic process(GO:0070857) |
| 0.0 | 0.1 | GO:0021853 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.0 | 0.2 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.0 | 0.1 | GO:0006361 | transcription initiation from RNA polymerase I promoter(GO:0006361) |
| 0.0 | 0.0 | GO:0030070 | insulin processing(GO:0030070) |
| 0.0 | 0.1 | GO:0071265 | amino acid salvage(GO:0043102) L-methionine biosynthetic process(GO:0071265) L-methionine salvage(GO:0071267) |
| 0.0 | 0.0 | GO:0045896 | regulation of transcription during mitosis(GO:0045896) positive regulation of transcription during mitosis(GO:0045897) |
| 0.0 | 0.0 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.0 | 0.1 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.0 | 0.0 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.0 | 0.0 | GO:0010901 | regulation of very-low-density lipoprotein particle remodeling(GO:0010901) |
| 0.0 | 0.1 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.0 | 0.1 | GO:0035428 | hexose transmembrane transport(GO:0035428) |
| 0.0 | 0.0 | GO:0010908 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.0 | 0.0 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.0 | 0.0 | GO:1902564 | negative regulation of neutrophil activation(GO:1902564) |
| 0.0 | 0.0 | GO:1901844 | regulation of cell communication by electrical coupling involved in cardiac conduction(GO:1901844) |
| 0.0 | 0.0 | GO:0048022 | negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) |
| 0.0 | 0.1 | GO:1904587 | glycoprotein ERAD pathway(GO:0097466) response to glycoprotein(GO:1904587) |
| 0.0 | 0.0 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.1 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.0 | 0.0 | GO:0034165 | positive regulation of toll-like receptor 9 signaling pathway(GO:0034165) |
| 0.0 | 0.0 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.0 | 0.2 | GO:0010758 | regulation of macrophage chemotaxis(GO:0010758) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0097629 | extrinsic component of omegasome membrane(GO:0097629) |
| 0.0 | 0.1 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.1 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 0.0 | 0.1 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.0 | 0.1 | GO:0016939 | kinesin II complex(GO:0016939) |
| 0.0 | 0.0 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.0 | 0.0 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.0 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 0.0 | 0.1 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.0 | 0.0 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.0 | 0.1 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.0 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.0 | 0.0 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.0 | 0.1 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.0 | 0.0 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.0 | 0.0 | GO:0010009 | cytoplasmic side of endosome membrane(GO:0010009) |
| 0.0 | 0.1 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 0.0 | 0.0 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 0.1 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.0 | 0.0 | GO:1990696 | USH2 complex(GO:1990696) |
| 0.0 | 0.0 | GO:0097443 | sorting endosome(GO:0097443) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.0 | 0.2 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.1 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.1 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.0 | 0.1 | GO:0034845 | 4-methyloctanoyl-CoA dehydrogenase activity(GO:0034580) naphthyl-2-methyl-succinyl-CoA dehydrogenase activity(GO:0034845) 2-methylhexanoyl-CoA dehydrogenase activity(GO:0034916) propionyl-CoA dehydrogenase activity(GO:0043820) thiol-driven fumarate reductase activity(GO:0043830) coenzyme F420-dependent 2,4,6-trinitrophenol reductase activity(GO:0052758) coenzyme F420-dependent 2,4,6-trinitrophenol hydride reductase activity(GO:0052759) coenzyme F420-dependent 2,4-dinitrophenol reductase activity(GO:0052760) |
| 0.0 | 0.1 | GO:0008898 | S-adenosylmethionine-homocysteine S-methyltransferase activity(GO:0008898) |
| 0.0 | 0.1 | GO:0008515 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.0 | 0.1 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 0.0 | 0.1 | GO:0050252 | retinol O-fatty-acyltransferase activity(GO:0050252) |
| 0.0 | 0.1 | GO:0004103 | choline kinase activity(GO:0004103) |
| 0.0 | 0.0 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.0 | 0.1 | GO:0045340 | mercury ion binding(GO:0045340) |
| 0.0 | 0.1 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.0 | 0.1 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.0 | GO:0019961 | interferon binding(GO:0019961) |
| 0.0 | 0.1 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.0 | 0.0 | GO:0004920 | interleukin-10 receptor activity(GO:0004920) |
| 0.0 | 0.0 | GO:0000171 | ribonuclease MRP activity(GO:0000171) |
| 0.0 | 0.1 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 0.0 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.0 | 0.1 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 0.0 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.0 | 0.0 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.0 | 0.0 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 0.1 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.1 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.1 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |