Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
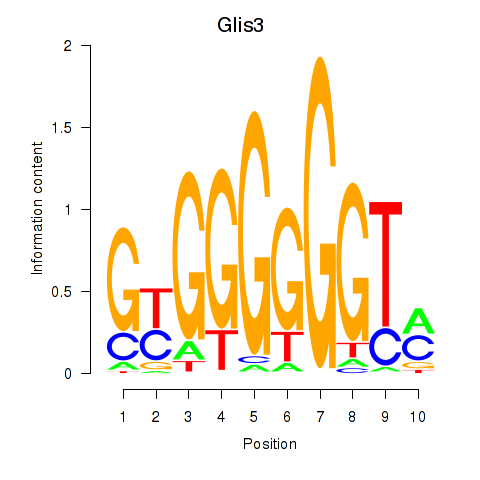
Results for Glis3
Z-value: 0.28
Transcription factors associated with Glis3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Glis3
|
ENSMUSG00000052942.7 | GLIS family zinc finger 3 |
|
Glis3
|
ENSMUSG00000091294.1 | GLIS family zinc finger 3 |
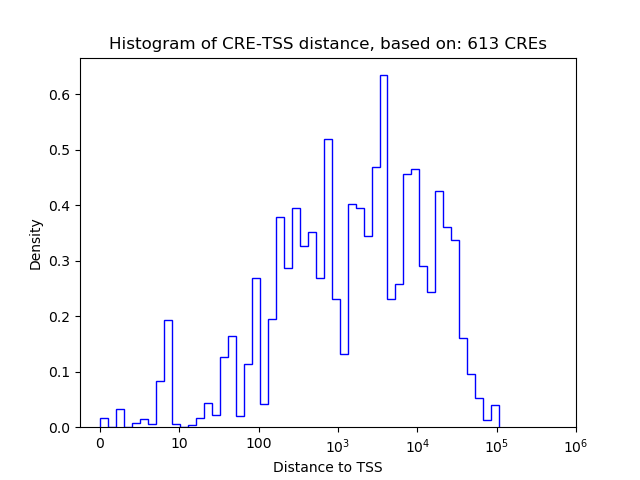
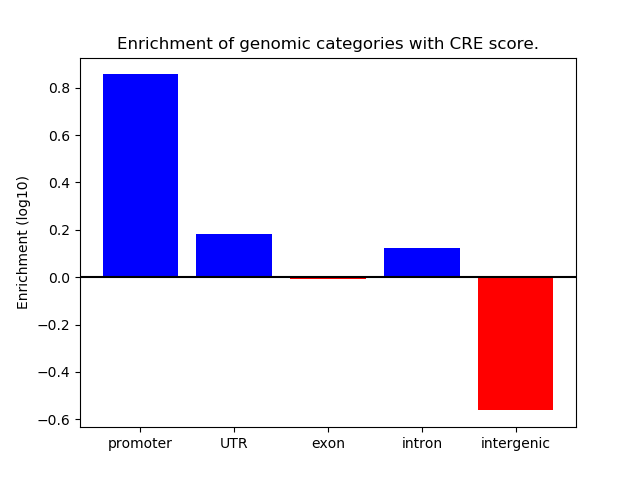
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr19_28520868_28521034 | Glis3 | 19469 | 0.235136 | -0.70 | 1.2e-01 | Click! |
| chr19_28656986_28657167 | Glis3 | 9609 | 0.196316 | -0.66 | 1.5e-01 | Click! |
| chr19_28530943_28531108 | Glis3 | 9395 | 0.254385 | -0.50 | 3.1e-01 | Click! |
| chr19_28347532_28347702 | Glis3 | 21611 | 0.263215 | -0.16 | 7.6e-01 | Click! |
| chr19_28678405_28678738 | Glis3 | 119 | 0.771688 | -0.14 | 8.0e-01 | Click! |
Activity of the Glis3 motif across conditions
Conditions sorted by the z-value of the Glis3 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
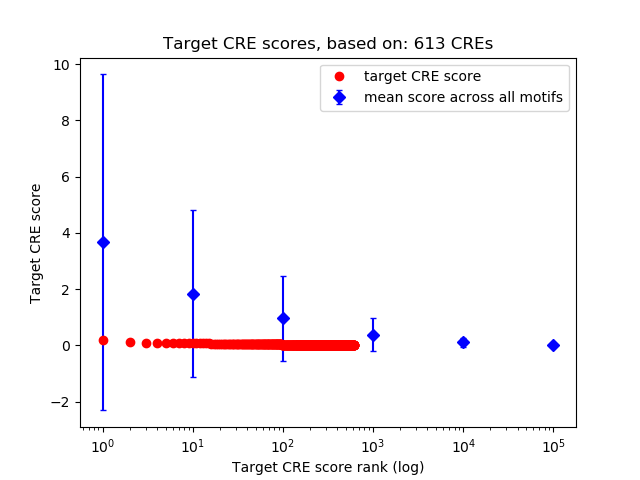
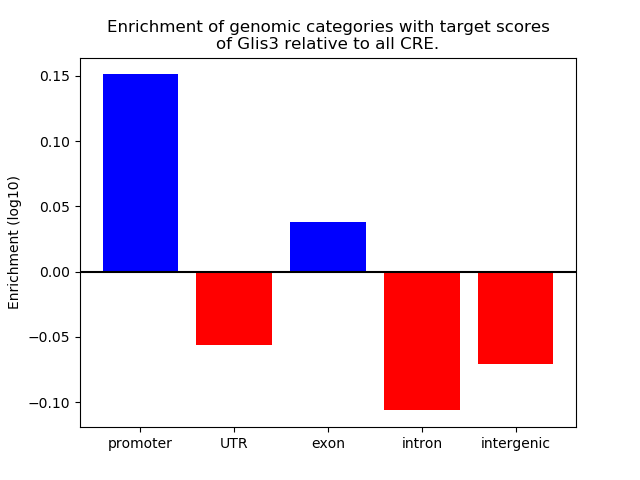
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr1_131276568_131276872 | 0.18 |
Ikbke |
inhibitor of kappaB kinase epsilon |
7 |
0.96 |
| chr10_83324464_83324615 | 0.11 |
Slc41a2 |
solute carrier family 41, member 2 |
1656 |
0.34 |
| chr13_44640104_44640258 | 0.10 |
Gm47805 |
predicted gene, 47805 |
17999 |
0.21 |
| chr10_21689999_21690150 | 0.09 |
Gm5420 |
predicted gene 5420 |
771 |
0.7 |
| chr17_12402301_12402715 | 0.09 |
Plg |
plasminogen |
23849 |
0.15 |
| chr7_16793142_16793312 | 0.09 |
Slc1a5 |
solute carrier family 1 (neutral amino acid transporter), member 5 |
50 |
0.95 |
| chr1_121474052_121474217 | 0.08 |
Ccdc93 |
coiled-coil domain containing 93 |
17083 |
0.14 |
| chr12_104153598_104153749 | 0.08 |
Serpina3c |
serine (or cysteine) peptidase inhibitor, clade A, member 3C |
197 |
0.89 |
| chr17_12751352_12751573 | 0.07 |
Igf2r |
insulin-like growth factor 2 receptor |
662 |
0.61 |
| chr6_89644800_89644964 | 0.07 |
Txnrd3 |
thioredoxin reductase 3 |
894 |
0.58 |
| chr3_19987089_19987240 | 0.07 |
Cp |
ceruloplasmin |
381 |
0.85 |
| chr5_135695656_135695889 | 0.07 |
Gm16061 |
predicted gene 16061 |
3247 |
0.14 |
| chr4_33243933_33244238 | 0.07 |
Pnrc1 |
proline-rich nuclear receptor coactivator 1 |
3490 |
0.21 |
| chr17_6106744_6106904 | 0.07 |
Tulp4 |
tubby like protein 4 |
34 |
0.96 |
| chr10_31553345_31553496 | 0.07 |
Gm47693 |
predicted gene, 47693 |
4020 |
0.21 |
| chr8_120533900_120534066 | 0.06 |
Gse1 |
genetic suppressor element 1, coiled-coil protein |
1874 |
0.21 |
| chr4_122455220_122455373 | 0.06 |
Gm12895 |
predicted gene 12895 |
86 |
0.98 |
| chr4_122122855_122123016 | 0.06 |
Gm12896 |
predicted gene 12896 |
88 |
0.98 |
| chr19_61052885_61053046 | 0.06 |
Gm22520 |
predicted gene, 22520 |
39420 |
0.13 |
| chr3_95687775_95688070 | 0.06 |
Adamtsl4 |
ADAMTS-like 4 |
5 |
0.96 |
| chr1_120083236_120083428 | 0.06 |
Tmem37 |
transmembrane protein 37 |
9258 |
0.18 |
| chr6_28845646_28845808 | 0.05 |
Snd1 |
staphylococcal nuclease and tudor domain containing 1 |
3180 |
0.25 |
| chr4_3682969_3683158 | 0.05 |
Lyn |
LYN proto-oncogene, Src family tyrosine kinase |
4933 |
0.18 |
| chr17_36231281_36231449 | 0.05 |
Gm18604 |
predicted gene, 18604 |
1896 |
0.09 |
| chr3_101918798_101918949 | 0.05 |
Slc22a15 |
solute carrier family 22 (organic anion/cation transporter), member 15 |
5506 |
0.25 |
| chr2_167886467_167886618 | 0.05 |
Gm14319 |
predicted gene 14319 |
27957 |
0.14 |
| chr13_45963217_45963398 | 0.05 |
Atxn1 |
ataxin 1 |
1650 |
0.33 |
| chr8_25529060_25529214 | 0.05 |
Fgfr1 |
fibroblast growth factor receptor 1 |
2988 |
0.17 |
| chr4_61830694_61830880 | 0.05 |
Mup5 |
major urinary protein 5 |
4446 |
0.15 |
| chr9_21149629_21149877 | 0.05 |
Cdc37 |
cell division cycle 37 |
189 |
0.89 |
| chr1_92474055_92474238 | 0.05 |
Ndufa10 |
NADH:ubiquinone oxidoreductase subunit A10 |
286 |
0.84 |
| chr17_33932704_33933052 | 0.05 |
Rgl2 |
ral guanine nucleotide dissociation stimulator-like 2 |
747 |
0.27 |
| chr11_67722605_67722756 | 0.05 |
Rcvrn |
recoverin |
27354 |
0.14 |
| chr11_21550683_21550834 | 0.05 |
Mdh1 |
malate dehydrogenase 1, NAD (soluble) |
8971 |
0.14 |
| chr1_107638144_107638321 | 0.05 |
Serpinb8 |
serine (or cysteine) peptidase inhibitor, clade B, member 8 |
32352 |
0.17 |
| chr11_120627879_120628086 | 0.05 |
Mafg |
v-maf musculoaponeurotic fibrosarcoma oncogene family, protein G (avian) |
2166 |
0.09 |
| chr6_113866107_113866258 | 0.05 |
Atp2b2 |
ATPase, Ca++ transporting, plasma membrane 2 |
25188 |
0.15 |
| chr11_101370588_101371223 | 0.05 |
G6pc |
glucose-6-phosphatase, catalytic |
3344 |
0.08 |
| chr2_158095379_158095534 | 0.05 |
2010009K17Rik |
RIKEN cDNA 2010009K17 gene |
3659 |
0.19 |
| chr11_97659776_97659960 | 0.05 |
Gm24158 |
predicted gene, 24158 |
459 |
0.41 |
| chr11_3266649_3266801 | 0.04 |
Drg1 |
developmentally regulated GTP binding protein 1 |
310 |
0.84 |
| chr11_60074241_60074392 | 0.04 |
Pemt |
phosphatidylethanolamine N-methyltransferase |
27827 |
0.12 |
| chr12_16734524_16734675 | 0.04 |
Greb1 |
gene regulated by estrogen in breast cancer protein |
5529 |
0.19 |
| chr14_66009383_66009534 | 0.04 |
Gulo |
gulonolactone (L-) oxidase |
251 |
0.91 |
| chr13_52928944_52929562 | 0.04 |
Auh |
AU RNA binding protein/enoyl-coenzyme A hydratase |
205 |
0.95 |
| chr1_155112239_155112390 | 0.04 |
Ier5 |
immediate early response 5 |
12678 |
0.14 |
| chr17_31872001_31872152 | 0.04 |
Sik1 |
salt inducible kinase 1 |
16272 |
0.13 |
| chr14_67631840_67632009 | 0.04 |
Gm47010 |
predicted gene, 47010 |
43132 |
0.12 |
| chr7_143502466_143502878 | 0.04 |
Phlda2 |
pleckstrin homology like domain, family A, member 2 |
131 |
0.93 |
| chr2_163301297_163301733 | 0.04 |
Tox2 |
TOX high mobility group box family member 2 |
18863 |
0.19 |
| chr14_122105762_122105950 | 0.04 |
A330035P11Rik |
RIKEN cDNA A330035P11 gene |
1097 |
0.31 |
| chr2_32420040_32420228 | 0.04 |
Slc25a25 |
solute carrier family 25 (mitochondrial carrier, phosphate carrier), member 25 |
316 |
0.79 |
| chr2_25453352_25453548 | 0.04 |
Clic3 |
chloride intracellular channel 3 |
3388 |
0.09 |
| chr11_43950012_43950201 | 0.04 |
Gm12153 |
predicted gene 12153 |
8983 |
0.26 |
| chr6_121878379_121878709 | 0.04 |
Mug1 |
murinoglobulin 1 |
7033 |
0.2 |
| chr11_78498755_78498922 | 0.04 |
Vtn |
vitronectin |
253 |
0.76 |
| chr6_125352883_125353034 | 0.04 |
Tnfrsf1a |
tumor necrosis factor receptor superfamily, member 1a |
3235 |
0.16 |
| chr13_80885021_80885448 | 0.04 |
1700023H06Rik |
RIKEN cDNA 1700023H06 gene |
290 |
0.8 |
| chr3_88492692_88492952 | 0.04 |
Lmna |
lamin A |
490 |
0.6 |
| chr14_31394009_31394182 | 0.04 |
Sh3bp5 |
SH3-domain binding protein 5 (BTK-associated) |
13290 |
0.14 |
| chr12_101075488_101075655 | 0.04 |
D130020L05Rik |
RIKEN cDNA D130020L05 gene |
6880 |
0.13 |
| chr7_80077642_80077793 | 0.04 |
Gm21057 |
predicted gene, 21057 |
2450 |
0.18 |
| chr11_18900383_18900558 | 0.03 |
2900018N21Rik |
RIKEN cDNA 2900018N21 gene |
10630 |
0.16 |
| chr1_157283514_157283666 | 0.03 |
Rasal2 |
RAS protein activator like 2 |
26908 |
0.17 |
| chr2_174021375_174021526 | 0.03 |
Vamp7-ps |
vesicle-associated membrane protein 7, pseudogene |
25823 |
0.15 |
| chr11_4108452_4108659 | 0.03 |
Sec14l2 |
SEC14-like lipid binding 2 |
456 |
0.54 |
| chr2_167753904_167754135 | 0.03 |
Gm14321 |
predicted gene 14321 |
23448 |
0.12 |
| chr7_110853928_110854130 | 0.03 |
Lyve1 |
lymphatic vessel endothelial hyaluronan receptor 1 |
8924 |
0.15 |
| chr7_30412740_30412933 | 0.03 |
Tyrobp |
TYRO protein tyrosine kinase binding protein |
924 |
0.3 |
| chr12_103897314_103897939 | 0.03 |
B430119L08Rik |
RIKEN cDNA B430119L08 gene |
1729 |
0.21 |
| chr9_46216986_46217137 | 0.03 |
Sik3 |
SIK family kinase 3 |
2510 |
0.15 |
| chr9_108083457_108083608 | 0.03 |
Rnf123 |
ring finger protein 123 |
186 |
0.77 |
| chr9_48595528_48595693 | 0.03 |
Nnmt |
nicotinamide N-methyltransferase |
1765 |
0.42 |
| chr5_33726324_33726568 | 0.03 |
Fgfr3 |
fibroblast growth factor receptor 3 |
2674 |
0.15 |
| chr1_93128050_93128201 | 0.03 |
Gm28086 |
predicted gene 28086 |
4022 |
0.15 |
| chr9_120933701_120933852 | 0.03 |
Ctnnb1 |
catenin (cadherin associated protein), beta 1 |
42 |
0.95 |
| chr5_122936823_122936974 | 0.03 |
Kdm2b |
lysine (K)-specific demethylase 2B |
11096 |
0.13 |
| chr5_142920701_142920990 | 0.03 |
Actb |
actin, beta |
14091 |
0.14 |
| chr5_110809507_110809658 | 0.03 |
Ulk1 |
unc-51 like kinase 1 |
471 |
0.7 |
| chr1_13373035_13373207 | 0.03 |
Ncoa2 |
nuclear receptor coactivator 2 |
682 |
0.56 |
| chr15_91049265_91049450 | 0.03 |
Kif21a |
kinesin family member 21A |
461 |
0.83 |
| chr8_108584590_108585054 | 0.03 |
Lncbate1 |
brown adipose tissue enriched long non-coding RNA 1 |
227 |
0.95 |
| chr15_81888131_81888282 | 0.03 |
Aco2 |
aconitase 2, mitochondrial |
7100 |
0.09 |
| chr19_3852307_3852467 | 0.03 |
Chka |
choline kinase alpha |
24 |
0.49 |
| chr15_77160485_77160636 | 0.03 |
Rbfox2 |
RNA binding protein, fox-1 homolog (C. elegans) 2 |
6748 |
0.15 |
| chr19_29252075_29252226 | 0.03 |
Jak2 |
Janus kinase 2 |
291 |
0.91 |
| chr1_190126920_190127155 | 0.03 |
Gm28172 |
predicted gene 28172 |
41633 |
0.14 |
| chr1_156073065_156073216 | 0.03 |
Tor1aip2 |
torsin A interacting protein 2 |
9927 |
0.17 |
| chr11_101783694_101783858 | 0.03 |
Etv4 |
ets variant 4 |
612 |
0.68 |
| chr14_30923762_30923926 | 0.03 |
Itih3 |
inter-alpha trypsin inhibitor, heavy chain 3 |
84 |
0.95 |
| chr9_121906088_121906239 | 0.03 |
Ackr2 |
atypical chemokine receptor 2 |
1 |
0.95 |
| chr19_46630947_46631098 | 0.03 |
Wbp1l |
WW domain binding protein 1 like |
7621 |
0.14 |
| chr17_46455115_46455282 | 0.03 |
Gm5093 |
predicted gene 5093 |
15101 |
0.09 |
| chr13_52771924_52772129 | 0.03 |
BB123696 |
expressed sequence BB123696 |
14821 |
0.27 |
| chr14_60737068_60737230 | 0.03 |
Spata13 |
spermatogenesis associated 13 |
4243 |
0.23 |
| chr13_109633045_109633234 | 0.03 |
Pde4d |
phosphodiesterase 4D, cAMP specific |
359 |
0.94 |
| chr14_66129671_66129842 | 0.03 |
Ephx2 |
epoxide hydrolase 2, cytoplasmic |
5256 |
0.17 |
| chr5_36697435_36697602 | 0.03 |
D5Ertd579e |
DNA segment, Chr 5, ERATO Doi 579, expressed |
1494 |
0.31 |
| chr17_26576750_26576907 | 0.03 |
Gm22420 |
predicted gene, 22420 |
4099 |
0.13 |
| chr2_90941045_90941219 | 0.03 |
Celf1 |
CUGBP, Elav-like family member 1 |
589 |
0.61 |
| chr10_96548942_96549289 | 0.03 |
Gm48505 |
predicted gene, 48505 |
25133 |
0.17 |
| chr4_49459901_49460052 | 0.03 |
Acnat1 |
acyl-coenzyme A amino acid N-acyltransferase 1 |
8839 |
0.13 |
| chr7_27163021_27163193 | 0.03 |
Egln2 |
egl-9 family hypoxia-inducible factor 2 |
2236 |
0.12 |
| chr4_130787632_130787854 | 0.03 |
Gm12972 |
predicted gene 12972 |
1674 |
0.23 |
| chr12_84221379_84221549 | 0.03 |
Elmsan1 |
ELM2 and Myb/SANT-like domain containing 1 |
2583 |
0.16 |
| chr1_153900403_153900624 | 0.03 |
Glul |
glutamate-ammonia ligase (glutamine synthetase) |
72 |
0.72 |
| chr5_124167184_124167456 | 0.03 |
Pitpnm2 |
phosphatidylinositol transfer protein, membrane-associated 2 |
17798 |
0.11 |
| chr4_126173656_126173812 | 0.03 |
Thrap3 |
thyroid hormone receptor associated protein 3 |
207 |
0.89 |
| chr6_89239871_89240105 | 0.03 |
Gm25961 |
predicted gene, 25961 |
9614 |
0.13 |
| chr4_135406321_135406484 | 0.03 |
Gm12990 |
predicted gene 12990 |
2746 |
0.14 |
| chr2_39014404_39014555 | 0.03 |
Golga1 |
golgi autoantigen, golgin subfamily a, 1 |
3927 |
0.11 |
| chr13_48625877_48626046 | 0.03 |
Ptpdc1 |
protein tyrosine phosphatase domain containing 1 |
297 |
0.87 |
| chr6_87810683_87810855 | 0.03 |
Rab43 |
RAB43, member RAS oncogene family |
1010 |
0.28 |
| chr15_81938386_81938558 | 0.03 |
Csdc2 |
cold shock domain containing C2, RNA binding |
1490 |
0.21 |
| chr8_46908369_46908545 | 0.03 |
1700011L03Rik |
RIKEN cDNA 1700011L03 gene |
40342 |
0.14 |
| chr10_111597521_111597672 | 0.03 |
Gm30262 |
predicted gene, 30262 |
1581 |
0.33 |
| chr14_70058075_70058262 | 0.03 |
Gm33524 |
predicted gene, 33524 |
801 |
0.6 |
| chr4_137653092_137653286 | 0.03 |
Usp48 |
ubiquitin specific peptidase 48 |
2888 |
0.23 |
| chr18_60736799_60737000 | 0.02 |
Ndst1 |
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1 |
3749 |
0.19 |
| chr19_30157135_30157327 | 0.02 |
Rpl31-ps20 |
ribosomal protein L31, pseudogene 20 |
3389 |
0.24 |
| chr11_101715500_101715663 | 0.02 |
Dhx8 |
DEAH (Asp-Glu-Ala-His) box polypeptide 8 |
17338 |
0.12 |
| chr12_80437192_80437401 | 0.02 |
Dcaf5 |
DDB1 and CUL4 associated factor 5 |
695 |
0.54 |
| chr11_57963469_57963736 | 0.02 |
Gm12249 |
predicted gene 12249 |
9532 |
0.16 |
| chr16_32202814_32202973 | 0.02 |
Bex6 |
brain expressed family member 6 |
16707 |
0.11 |
| chr7_145047841_145048023 | 0.02 |
Ccnd1 |
cyclin D1 |
108007 |
0.05 |
| chr6_39782094_39782403 | 0.02 |
Mrps33 |
mitochondrial ribosomal protein S33 |
23794 |
0.15 |
| chr14_122082959_122083124 | 0.02 |
Gm49236 |
predicted gene, 49236 |
8185 |
0.14 |
| chr17_14201623_14201787 | 0.02 |
Gm34510 |
predicted gene, 34510 |
2023 |
0.27 |
| chr5_123395676_123395864 | 0.02 |
Gm15747 |
predicted gene 15747 |
706 |
0.34 |
| chr2_93459658_93459954 | 0.02 |
Cd82 |
CD82 antigen |
2106 |
0.29 |
| chr1_133278341_133278519 | 0.02 |
Plekha6 |
pleckstrin homology domain containing, family A member 6 |
4176 |
0.16 |
| chr15_75909227_75909395 | 0.02 |
Eef1d |
eukaryotic translation elongation factor 1 delta (guanine nucleotide exchange protein) |
5 |
0.85 |
| chr4_119226096_119226268 | 0.02 |
Gm12898 |
predicted gene 12898 |
3008 |
0.11 |
| chr4_139683984_139684159 | 0.02 |
Tas1r2 |
taste receptor, type 1, member 2 |
30522 |
0.14 |
| chr2_75704785_75705107 | 0.02 |
E030042O20Rik |
RIKEN cDNA E030042O20 gene |
176 |
0.65 |
| chr9_78223304_78223524 | 0.02 |
Gsta1 |
glutathione S-transferase, alpha 1 (Ya) |
7242 |
0.1 |
| chr4_144895074_144895225 | 0.02 |
Dhrs3 |
dehydrogenase/reductase (SDR family) member 3 |
1930 |
0.37 |
| chr17_14990062_14990517 | 0.02 |
9030025P20Rik |
RIKEN cDNA 9030025P20 gene |
10790 |
0.1 |
| chr10_95716074_95716270 | 0.02 |
Gm47671 |
predicted gene, 47671 |
1335 |
0.29 |
| chr15_100681158_100681447 | 0.02 |
Cela1 |
chymotrypsin-like elastase family, member 1 |
281 |
0.82 |
| chr11_101668758_101669043 | 0.02 |
Arl4d |
ADP-ribosylation factor-like 4D |
3359 |
0.12 |
| chr8_70716563_70716714 | 0.02 |
Gm3336 |
predicted gene 3336 |
1905 |
0.14 |
| chr2_103867069_103867263 | 0.02 |
Gm13876 |
predicted gene 13876 |
21158 |
0.08 |
| chr10_41491217_41491378 | 0.02 |
Ppil6 |
peptidylprolyl isomerase (cyclophilin)-like 6 |
820 |
0.36 |
| chr1_67195986_67196354 | 0.02 |
Gm15668 |
predicted gene 15668 |
53030 |
0.13 |
| chr5_113735843_113736024 | 0.02 |
Ficd |
FIC domain containing |
130 |
0.93 |
| chr4_134186730_134186892 | 0.02 |
Cep85 |
centrosomal protein 85 |
170 |
0.89 |
| chr12_99417868_99418197 | 0.02 |
Foxn3 |
forkhead box N3 |
5427 |
0.19 |
| chr6_86209454_86209634 | 0.02 |
Tgfa |
transforming growth factor alpha |
2050 |
0.28 |
| chr10_58233103_58233399 | 0.02 |
Duxf3 |
double homeobox family member 3 |
576 |
0.63 |
| chr19_42518403_42518575 | 0.02 |
R3hcc1l |
R3H domain and coiled-coil containing 1 like |
270 |
0.92 |
| chr3_69778568_69778734 | 0.02 |
Nmd3 |
NMD3 ribosome export adaptor |
32219 |
0.16 |
| chr13_75976970_75977121 | 0.02 |
Spata9 |
spermatogenesis associated 9 |
9298 |
0.12 |
| chr5_122131727_122131989 | 0.02 |
Ccdc63 |
coiled-coil domain containing 63 |
145 |
0.94 |
| chr19_48700604_48700763 | 0.02 |
Sorcs3 |
sortilin-related VPS10 domain containing receptor 3 |
3533 |
0.37 |
| chr8_105207006_105207175 | 0.02 |
D230025D16Rik |
RIKEN cDNA D230025D16 gene |
18055 |
0.08 |
| chr2_157556257_157556418 | 0.02 |
Blcap |
bladder cancer associated protein |
3670 |
0.14 |
| chr13_93648931_93649116 | 0.02 |
Bhmt |
betaine-homocysteine methyltransferase |
11062 |
0.14 |
| chr6_72233913_72234064 | 0.02 |
Atoh8 |
atonal bHLH transcription factor 8 |
549 |
0.74 |
| chr1_74284201_74284570 | 0.02 |
Aamp |
angio-associated migratory protein |
36 |
0.87 |
| chr13_8996579_8996771 | 0.02 |
Gtpbp4 |
GTP binding protein 4 |
592 |
0.58 |
| chr11_79081331_79081482 | 0.02 |
Ksr1 |
kinase suppressor of ras 1 |
6920 |
0.23 |
| chr2_167336792_167337118 | 0.02 |
B4galt5 |
UDP-Gal:betaGlcNAc beta 1,4-galactosyltransferase, polypeptide 5 |
12228 |
0.18 |
| chr4_43415734_43415920 | 0.02 |
Rusc2 |
RUN and SH3 domain containing 2 |
611 |
0.6 |
| chr11_116165413_116165564 | 0.02 |
Fbf1 |
Fas (TNFRSF6) binding factor 1 |
395 |
0.67 |
| chr8_111625803_111625975 | 0.02 |
Ldhd |
lactate dehydrogenase D |
2698 |
0.22 |
| chr4_45496371_45496660 | 0.02 |
Shb |
src homology 2 domain-containing transforming protein B |
3674 |
0.19 |
| chr8_84706735_84707039 | 0.02 |
Nfix |
nuclear factor I/X |
829 |
0.42 |
| chr17_24959424_24959602 | 0.02 |
Jpt2 |
Jupiter microtubule associated homolog 2 |
1123 |
0.35 |
| chr17_5730727_5730878 | 0.02 |
3300005D01Rik |
RIKEN cDNA 3300005D01 gene |
67855 |
0.09 |
| chr8_105304667_105304842 | 0.02 |
E2f4 |
E2F transcription factor 4 |
829 |
0.23 |
| chr4_125122748_125123219 | 0.02 |
Zc3h12a |
zinc finger CCCH type containing 12A |
463 |
0.76 |
| chr9_20898316_20898471 | 0.02 |
Eif3g |
eukaryotic translation initiation factor 3, subunit G |
189 |
0.85 |
| chr6_138075559_138075718 | 0.02 |
Slc15a5 |
solute carrier family 15, member 5 |
2442 |
0.4 |
| chr17_34591378_34591776 | 0.02 |
Pbx2 |
pre B cell leukemia homeobox 2 |
311 |
0.65 |
| chr9_43232699_43232996 | 0.02 |
Oaf |
out at first homolog |
7064 |
0.15 |
| chr10_63247362_63247596 | 0.02 |
Herc4 |
hect domain and RLD 4 |
1657 |
0.24 |
| chr8_83895995_83896156 | 0.02 |
Adgrl1 |
adhesion G protein-coupled receptor L1 |
4030 |
0.14 |
| chr17_56673508_56673661 | 0.02 |
Ranbp3 |
RAN binding protein 3 |
200 |
0.9 |
| chr1_86441574_86441779 | 0.02 |
Tex44 |
testis expressed 44 |
15347 |
0.11 |
| chr11_4115259_4115410 | 0.02 |
Sec14l2 |
SEC14-like lipid binding 2 |
3362 |
0.11 |
| chr17_62883043_62883201 | 0.02 |
Efna5 |
ephrin A5 |
1805 |
0.52 |
| chr12_3850885_3851080 | 0.02 |
Dnmt3a |
DNA methyltransferase 3A |
1347 |
0.41 |
| chr10_80870759_80871034 | 0.02 |
Tmprss9 |
transmembrane protease, serine 9 |
952 |
0.32 |
| chr13_41220287_41220454 | 0.02 |
Elovl2 |
elongation of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 2 |
35 |
0.96 |
| chr18_31951040_31951212 | 0.02 |
Gpr17 |
G protein-coupled receptor 17 |
1490 |
0.29 |
| chr4_117124519_117125009 | 0.02 |
Btbd19 |
BTB (POZ) domain containing 19 |
402 |
0.61 |
| chr7_28811272_28811515 | 0.02 |
Hnrnpl |
heterogeneous nuclear ribonucleoprotein L |
453 |
0.54 |
| chr15_90679599_90679931 | 0.02 |
Cpne8 |
copine VIII |
333 |
0.89 |
| chr12_102284053_102284212 | 0.02 |
Rin3 |
Ras and Rab interactor 3 |
491 |
0.83 |
| chr12_24680312_24680550 | 0.02 |
Cys1 |
cystin 1 |
288 |
0.87 |
| chr6_91752698_91752849 | 0.02 |
Slc6a6 |
solute carrier family 6 (neurotransmitter transporter, taurine), member 6 |
2589 |
0.22 |
| chr3_60510914_60511240 | 0.02 |
Mbnl1 |
muscleblind like splicing factor 1 |
9799 |
0.23 |
| chr18_84084397_84084569 | 0.02 |
Tshz1 |
teashirt zinc finger family member 1 |
592 |
0.69 |
| chr13_93190090_93190414 | 0.02 |
Tent2 |
terminal nucleotidyltransferase 2 |
1859 |
0.36 |
| chr19_53892382_53892590 | 0.02 |
Pdcd4 |
programmed cell death 4 |
250 |
0.9 |
| chr1_179836734_179836979 | 0.02 |
Ahctf1 |
AT hook containing transcription factor 1 |
33176 |
0.15 |
| chr2_93045265_93045449 | 0.02 |
Prdm11 |
PR domain containing 11 |
214 |
0.94 |
| chr8_106149050_106149358 | 0.02 |
Pla2g15 |
phospholipase A2, group XV |
1195 |
0.33 |
| chr11_113632167_113632467 | 0.02 |
Sstr2 |
somatostatin receptor 2 |
8060 |
0.14 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | GO:0060399 | positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.0 | 0.1 | GO:0015825 | L-serine transport(GO:0015825) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |