Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
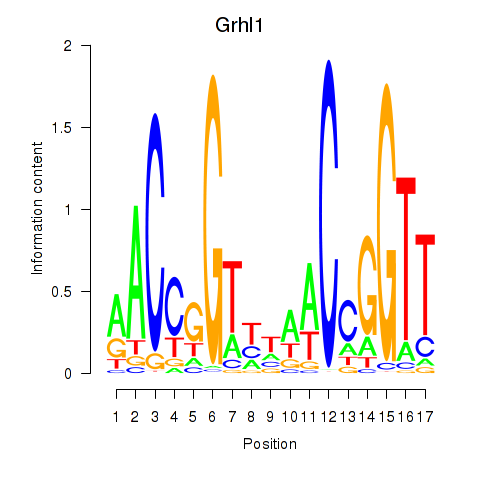
Results for Grhl1
Z-value: 1.75
Transcription factors associated with Grhl1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Grhl1
|
ENSMUSG00000020656.9 | grainyhead like transcription factor 1 |
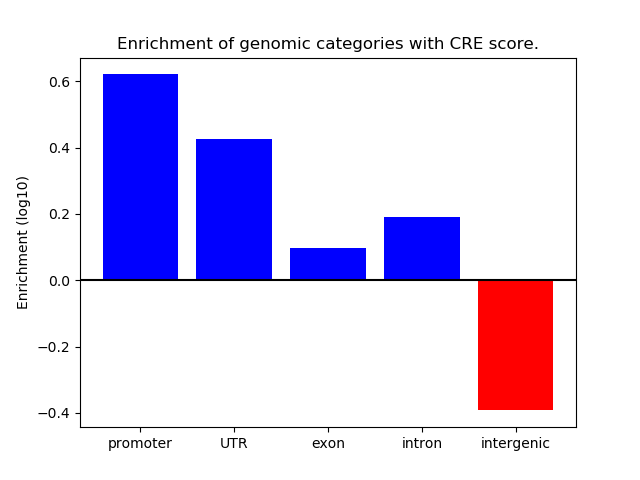
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
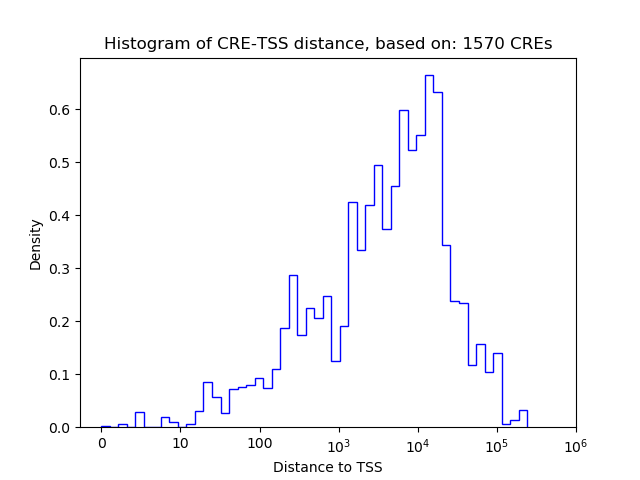
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr12_24571852_24572003 | Grhl1 | 356 | 0.868272 | 0.84 | 3.7e-02 | Click! |
| chr12_24600908_24601076 | Grhl1 | 20243 | 0.147325 | 0.76 | 7.9e-02 | Click! |
| chr12_24593470_24593854 | Grhl1 | 12913 | 0.164995 | 0.63 | 1.8e-01 | Click! |
| chr12_24602623_24602807 | Grhl1 | 21966 | 0.142995 | 0.59 | 2.2e-01 | Click! |
| chr12_24599648_24599816 | Grhl1 | 18983 | 0.150443 | -0.58 | 2.3e-01 | Click! |
Activity of the Grhl1 motif across conditions
Conditions sorted by the z-value of the Grhl1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
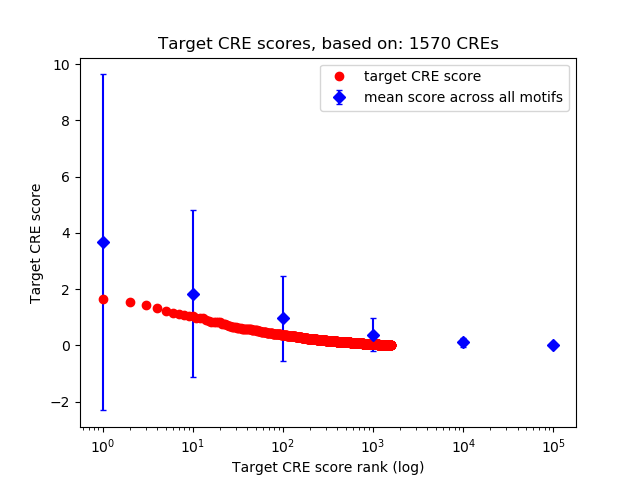
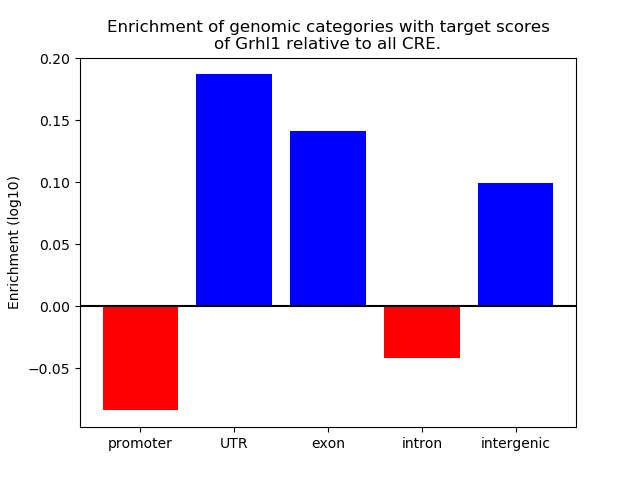
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr11_112219735_112219893 | 1.65 |
Gm11680 |
predicted gene 11680 |
112376 |
0.07 |
| chr1_39650381_39650532 | 1.52 |
D930019O06Rik |
RIKEN cDNA D930019O06 |
280 |
0.82 |
| chr7_113219987_113220140 | 1.45 |
Arntl |
aryl hydrocarbon receptor nuclear translocator-like |
12506 |
0.21 |
| chr15_89209095_89209277 | 1.32 |
Ppp6r2 |
protein phosphatase 6, regulatory subunit 2 |
2367 |
0.16 |
| chr19_12488282_12488454 | 1.22 |
Dtx4 |
deltex 4, E3 ubiquitin ligase |
13086 |
0.1 |
| chr5_52975314_52975474 | 1.16 |
Gm30301 |
predicted gene, 30301 |
6643 |
0.16 |
| chr19_30128513_30128664 | 1.10 |
Gldc |
glycine decarboxylase |
16643 |
0.18 |
| chr7_30361119_30361389 | 1.07 |
Lrfn3 |
leucine rich repeat and fibronectin type III domain containing 3 |
1518 |
0.17 |
| chr6_116119166_116119329 | 1.06 |
Tmcc1 |
transmembrane and coiled coil domains 1 |
8853 |
0.16 |
| chr5_9047464_9048021 | 1.05 |
Gm40264 |
predicted gene, 40264 |
12618 |
0.15 |
| chr7_68709416_68709593 | 0.98 |
Gm44692 |
predicted gene 44692 |
16963 |
0.21 |
| chr5_93250808_93250985 | 0.97 |
Ccng2 |
cyclin G2 |
16361 |
0.15 |
| chr2_156447550_156447712 | 0.96 |
Gm14225 |
predicted gene 14225 |
23 |
0.96 |
| chr8_106611072_106611260 | 0.90 |
Cdh1 |
cadherin 1 |
7023 |
0.18 |
| chr16_22958475_22958991 | 0.85 |
Hrg |
histidine-rich glycoprotein |
7633 |
0.12 |
| chr4_116176807_116176958 | 0.85 |
Gm12951 |
predicted gene 12951 |
636 |
0.51 |
| chr4_60658099_60658456 | 0.84 |
Mup11 |
major urinary protein 11 |
1461 |
0.38 |
| chr15_9361830_9361987 | 0.84 |
Ugt3a2 |
UDP glycosyltransferases 3 family, polypeptide A2 |
26319 |
0.17 |
| chr8_3252795_3252979 | 0.84 |
Gm16180 |
predicted gene 16180 |
6817 |
0.21 |
| chr4_124630137_124630316 | 0.83 |
Pou3f1 |
POU domain, class 3, transcription factor 1 |
26581 |
0.11 |
| chr4_61777961_61778274 | 0.77 |
Mup19 |
major urinary protein 19 |
4107 |
0.15 |
| chr8_119439834_119440348 | 0.77 |
Osgin1 |
oxidative stress induced growth inhibitor 1 |
2902 |
0.2 |
| chr6_5510024_5510175 | 0.77 |
Pdk4 |
pyruvate dehydrogenase kinase, isoenzyme 4 |
13790 |
0.27 |
| chr17_56112675_56112850 | 0.74 |
Plin4 |
perilipin 4 |
2959 |
0.12 |
| chr15_79741187_79741771 | 0.70 |
Sun2 |
Sad1 and UNC84 domain containing 2 |
613 |
0.45 |
| chr10_127591032_127591346 | 0.67 |
Gm16217 |
predicted gene 16217 |
5359 |
0.11 |
| chr7_138538849_138539011 | 0.66 |
4930543N07Rik |
RIKEN cDNA 4930543N07 gene |
15300 |
0.26 |
| chr1_52727224_52727404 | 0.66 |
Mfsd6 |
major facilitator superfamily domain containing 6 |
44 |
0.97 |
| chr18_20927286_20927596 | 0.65 |
Rnf125 |
ring finger protein 125 |
17184 |
0.19 |
| chr1_165667059_165667210 | 0.64 |
Gm18407 |
predicted gene, 18407 |
22446 |
0.1 |
| chrX_42112143_42112724 | 0.62 |
Gm14615 |
predicted gene 14615 |
18290 |
0.16 |
| chr15_31062841_31062999 | 0.62 |
4930430F21Rik |
RIKEN cDNA 4930430F21 gene |
23792 |
0.19 |
| chr11_119058051_119058489 | 0.61 |
Cbx8 |
chromobox 8 |
17301 |
0.14 |
| chr2_67976602_67976753 | 0.61 |
Gm37964 |
predicted gene, 37964 |
78081 |
0.1 |
| chr2_103801242_103801527 | 0.60 |
Caprin1 |
cell cycle associated protein 1 |
3735 |
0.12 |
| chr8_84130948_84131146 | 0.59 |
Podnl1 |
podocan-like 1 |
3229 |
0.09 |
| chr11_117176448_117176608 | 0.59 |
Sec14l1 |
SEC14-like lipid binding 1 |
21459 |
0.13 |
| chr19_3958627_3958778 | 0.59 |
Aldh3b3 |
aldehyde dehydrogenase 3 family, member B3 |
57 |
0.92 |
| chr2_69248790_69248965 | 0.58 |
Gm13613 |
predicted gene 13613 |
570 |
0.71 |
| chr10_43235483_43235634 | 0.58 |
Pdss2 |
prenyl (solanesyl) diphosphate synthase, subunit 2 |
13822 |
0.17 |
| chr2_126584131_126584290 | 0.57 |
Hdc |
histidine decarboxylase |
14051 |
0.16 |
| chr11_21235775_21236103 | 0.57 |
Vps54 |
VPS54 GARP complex subunit |
3342 |
0.17 |
| chr11_82858816_82858975 | 0.56 |
Rffl |
ring finger and FYVE like domain containing protein |
11849 |
0.1 |
| chr19_21038157_21038331 | 0.56 |
4930554I06Rik |
RIKEN cDNA 4930554I06 gene |
66258 |
0.11 |
| chr2_170287488_170287839 | 0.56 |
Gm14270 |
predicted gene 14270 |
2628 |
0.33 |
| chr9_115481085_115481255 | 0.56 |
Gm5921 |
predicted gene 5921 |
42589 |
0.13 |
| chr15_9306411_9306595 | 0.55 |
Ugt3a1 |
UDP glycosyltransferases 3 family, polypeptide A1 |
23002 |
0.17 |
| chr2_166674892_166675063 | 0.55 |
Gm23152 |
predicted gene, 23152 |
19835 |
0.18 |
| chr4_125116849_125117000 | 0.55 |
Zc3h12a |
zinc finger CCCH type containing 12A |
5596 |
0.15 |
| chr15_94591469_94591640 | 0.54 |
Twf1 |
twinfilin actin binding protein 1 |
1665 |
0.39 |
| chr13_45475320_45475484 | 0.54 |
Gm23387 |
predicted gene, 23387 |
10928 |
0.21 |
| chr10_93491843_93492023 | 0.51 |
Hal |
histidine ammonia lyase |
3130 |
0.19 |
| chr16_21332789_21332974 | 0.51 |
Magef1 |
melanoma antigen family F, 1 |
248 |
0.93 |
| chr5_128806405_128806573 | 0.50 |
Rimbp2 |
RIMS binding protein 2 |
13533 |
0.17 |
| chr14_58032344_58032511 | 0.50 |
Gm9012 |
predicted gene 9012 |
17396 |
0.15 |
| chr10_43723444_43723691 | 0.49 |
F930017D23Rik |
RIKEN cDNA F930017D23 gene |
16669 |
0.12 |
| chr4_57954096_57954315 | 0.49 |
Txn1 |
thioredoxin 1 |
2206 |
0.32 |
| chr17_48002012_48002364 | 0.48 |
Gm14871 |
predicted gene 14871 |
1384 |
0.35 |
| chr2_93445693_93445960 | 0.48 |
Cd82 |
CD82 antigen |
6853 |
0.17 |
| chr5_9045029_9045180 | 0.48 |
Gm40264 |
predicted gene, 40264 |
9980 |
0.15 |
| chr3_19986524_19986675 | 0.48 |
Cp |
ceruloplasmin |
946 |
0.55 |
| chr13_112071667_112072207 | 0.47 |
Gm31104 |
predicted gene, 31104 |
66179 |
0.09 |
| chr19_7310938_7311109 | 0.47 |
Gm17227 |
predicted gene 17227 |
13289 |
0.11 |
| chr11_7197846_7198073 | 0.47 |
Igfbp1 |
insulin-like growth factor binding protein 1 |
177 |
0.94 |
| chr4_86658429_86658687 | 0.46 |
Plin2 |
perilipin 2 |
86 |
0.97 |
| chr10_61427386_61427579 | 0.45 |
Nodal |
nodal |
9510 |
0.11 |
| chr2_145855907_145856101 | 0.45 |
Rin2 |
Ras and Rab interactor 2 |
6640 |
0.18 |
| chr8_12800767_12800949 | 0.45 |
Atp11a |
ATPase, class VI, type 11A |
10558 |
0.16 |
| chr13_46630953_46631134 | 0.45 |
Cap2 |
CAP, adenylate cyclase-associated protein, 2 (yeast) |
9015 |
0.17 |
| chr7_118534971_118535395 | 0.44 |
Coq7 |
demethyl-Q 7 |
1827 |
0.29 |
| chr7_65692899_65693066 | 0.43 |
Tm2d3 |
TM2 domain containing 3 |
426 |
0.84 |
| chr11_22814100_22814280 | 0.42 |
Gm20456 |
predicted gene 20456 |
25109 |
0.1 |
| chr3_82291765_82291930 | 0.42 |
Map9 |
microtubule-associated protein 9 |
66197 |
0.13 |
| chr10_81088340_81088663 | 0.42 |
Creb3l3 |
cAMP responsive element binding protein 3-like 3 |
3405 |
0.1 |
| chr14_29996020_29996356 | 0.42 |
Il17rb |
interleukin 17 receptor B |
2271 |
0.17 |
| chr17_13714881_13715054 | 0.42 |
Tcte2 |
t-complex-associated testis expressed 2 |
17931 |
0.13 |
| chr16_24085290_24085914 | 0.42 |
Gm46545 |
predicted gene, 46545 |
2912 |
0.23 |
| chr11_101729161_101729323 | 0.42 |
Dhx8 |
DEAH (Asp-Glu-Ala-His) box polypeptide 8 |
3677 |
0.17 |
| chr5_122131253_122131408 | 0.41 |
Ccdc63 |
coiled-coil domain containing 63 |
673 |
0.61 |
| chr4_60066252_60066431 | 0.41 |
Mup7 |
major urinary protein 7 |
4070 |
0.21 |
| chr2_69418752_69418922 | 0.41 |
Dhrs9 |
dehydrogenase/reductase (SDR family) member 9 |
38392 |
0.15 |
| chr4_61435652_61435957 | 0.40 |
Mup15 |
major urinary protein 15 |
3939 |
0.23 |
| chr1_182495453_182495613 | 0.40 |
Gm37069 |
predicted gene, 37069 |
1377 |
0.32 |
| chr16_35742175_35742539 | 0.40 |
Gm25967 |
predicted gene, 25967 |
20124 |
0.13 |
| chr14_120534643_120535027 | 0.40 |
Rap2a |
RAS related protein 2a |
56391 |
0.14 |
| chr14_69728721_69728885 | 0.40 |
Chmp7 |
charged multivesicular body protein 7 |
2072 |
0.22 |
| chr2_69340135_69340332 | 0.39 |
Abcb11 |
ATP-binding cassette, sub-family B (MDR/TAP), member 11 |
2367 |
0.28 |
| chr2_32252717_32252887 | 0.39 |
Pomt1 |
protein-O-mannosyltransferase 1 |
1529 |
0.19 |
| chr19_4326385_4326550 | 0.39 |
Kdm2a |
lysine (K)-specific demethylase 2A |
1602 |
0.23 |
| chr4_61948678_61948918 | 0.39 |
Mup-ps19 |
major urinary protein, pseudogene 19 |
11113 |
0.17 |
| chr11_97126973_97127136 | 0.39 |
Gm11572 |
predicted gene 11572 |
10139 |
0.09 |
| chr10_67792831_67793043 | 0.39 |
Gm29774 |
predicted gene, 29774 |
265 |
0.9 |
| chr11_67516215_67516384 | 0.39 |
Gas7 |
growth arrest specific 7 |
30855 |
0.19 |
| chr11_119191869_119192119 | 0.38 |
Gm11753 |
predicted gene 11753 |
7478 |
0.13 |
| chr18_60848442_60848618 | 0.38 |
Tcof1 |
treacle ribosome biogenesis factor 1 |
84 |
0.96 |
| chr13_20007652_20007822 | 0.38 |
Gm5446 |
predicted gene 5446 |
11933 |
0.27 |
| chr9_44268479_44268675 | 0.38 |
Nlrx1 |
NLR family member X1 |
22 |
0.93 |
| chr10_63160777_63160928 | 0.38 |
Mypn |
myopalladin |
38168 |
0.1 |
| chr15_67434225_67434396 | 0.37 |
1700012I11Rik |
RIKEN cDNA 1700012I11 gene |
207541 |
0.02 |
| chr11_48819928_48820131 | 0.37 |
Trim41 |
tripartite motif-containing 41 |
2676 |
0.13 |
| chr15_62017519_62017928 | 0.37 |
Pvt1 |
Pvt1 oncogene |
20263 |
0.19 |
| chr8_40896761_40897111 | 0.37 |
Slc7a2 |
solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 |
1502 |
0.36 |
| chr11_75653315_75653490 | 0.36 |
Myo1c |
myosin IC |
1326 |
0.31 |
| chr7_44496721_44496957 | 0.36 |
Fam71e1 |
family with sequence similarity 71, member E1 |
145 |
0.66 |
| chr12_80107353_80107544 | 0.35 |
Zfp36l1 |
zinc finger protein 36, C3H type-like 1 |
5546 |
0.13 |
| chr3_58683500_58683862 | 0.35 |
Siah2 |
siah E3 ubiquitin protein ligase 2 |
2356 |
0.2 |
| chr10_8066302_8066722 | 0.35 |
Gm48614 |
predicted gene, 48614 |
45220 |
0.16 |
| chr19_44985600_44985928 | 0.35 |
4930414N06Rik |
RIKEN cDNA 4930414N06 gene |
1214 |
0.29 |
| chr13_41249141_41249488 | 0.35 |
Smim13 |
small integral membrane protein 13 |
530 |
0.68 |
| chr17_35667833_35668010 | 0.35 |
Vars2 |
valyl-tRNA synthetase 2, mitochondrial |
329 |
0.65 |
| chr11_34072364_34072537 | 0.35 |
Lcp2 |
lymphocyte cytosolic protein 2 |
5727 |
0.22 |
| chr4_62049824_62050253 | 0.35 |
Mup20 |
major urinary protein 20 |
4120 |
0.16 |
| chr17_29049918_29050142 | 0.35 |
Gm41556 |
predicted gene, 41556 |
3306 |
0.11 |
| chr19_3609825_3610173 | 0.35 |
Lrp5 |
low density lipoprotein receptor-related protein 5 |
5793 |
0.17 |
| chr19_24225293_24225657 | 0.35 |
Tjp2 |
tight junction protein 2 |
445 |
0.83 |
| chr17_48324406_48324576 | 0.35 |
B430306N03Rik |
RIKEN cDNA B430306N03 gene |
8067 |
0.12 |
| chr16_32008396_32008555 | 0.34 |
Senp5 |
SUMO/sentrin specific peptidase 5 |
5188 |
0.1 |
| chr1_160982820_160983305 | 0.34 |
Serpinc1 |
serine (or cysteine) peptidase inhibitor, clade C (antithrombin), member 1 |
1706 |
0.15 |
| chr12_100477440_100477617 | 0.34 |
Ttc7b |
tetratricopeptide repeat domain 7B |
11230 |
0.16 |
| chr18_65037226_65037468 | 0.34 |
Nedd4l |
neural precursor cell expressed, developmentally down-regulated gene 4-like |
11545 |
0.22 |
| chr4_86656497_86656682 | 0.34 |
Plin2 |
perilipin 2 |
2055 |
0.29 |
| chr6_136469183_136469418 | 0.33 |
Gm6728 |
predicted gene 6728 |
17886 |
0.12 |
| chr17_24236940_24237256 | 0.33 |
Mir5134 |
microRNA 5134 |
2494 |
0.12 |
| chr14_73135659_73135810 | 0.33 |
Rcbtb2 |
regulator of chromosome condensation (RCC1) and BTB (POZ) domain containing protein 2 |
3034 |
0.27 |
| chr9_43151950_43152122 | 0.33 |
Pou2f3 |
POU domain, class 2, transcription factor 3 |
8832 |
0.18 |
| chr11_96860496_96860647 | 0.33 |
Copz2 |
coatomer protein complex, subunit zeta 2 |
6440 |
0.09 |
| chr13_34154759_34154927 | 0.32 |
Psmg4 |
proteasome (prosome, macropain) assembly chaperone 4 |
8121 |
0.12 |
| chr8_126588770_126588952 | 0.32 |
Irf2bp2 |
interferon regulatory factor 2 binding protein 2 |
5125 |
0.24 |
| chr10_99384294_99384806 | 0.32 |
B530045E10Rik |
RIKEN cDNA B530045E10 gene |
18240 |
0.17 |
| chr10_75741017_75741184 | 0.32 |
Cabin1 |
calcineurin binding protein 1 |
7329 |
0.12 |
| chr7_140899673_140899825 | 0.32 |
Cox8b |
cytochrome c oxidase subunit 8B |
697 |
0.38 |
| chr5_89018649_89018800 | 0.32 |
Slc4a4 |
solute carrier family 4 (anion exchanger), member 4 |
9242 |
0.29 |
| chr1_88211821_88211976 | 0.32 |
Ugt1a1 |
UDP glucuronosyltransferase 1 family, polypeptide A1 |
61 |
0.93 |
| chr2_32453958_32454145 | 0.32 |
Slc25a25 |
solute carrier family 25 (mitochondrial carrier, phosphate carrier), member 25 |
2606 |
0.15 |
| chr19_47402965_47403381 | 0.32 |
Sh3pxd2a |
SH3 and PX domains 2A |
7186 |
0.23 |
| chr4_60497810_60497972 | 0.31 |
Mup1 |
major urinary protein 1 |
1441 |
0.3 |
| chr4_126097782_126097933 | 0.31 |
Lsm10 |
U7 snRNP-specific Sm-like protein LSM10 |
1197 |
0.32 |
| chr7_14536045_14536258 | 0.31 |
Obox4-ps2 |
oocyte specific homeobox 4, pseudogene 2 |
6085 |
0.12 |
| chr2_52298963_52299373 | 0.31 |
Gm22095 |
predicted gene, 22095 |
16091 |
0.19 |
| chr4_60737134_60737406 | 0.31 |
Mup12 |
major urinary protein 12 |
4056 |
0.21 |
| chr19_55744520_55744671 | 0.31 |
Tcf7l2 |
transcription factor 7 like 2, T cell specific, HMG box |
1750 |
0.5 |
| chr2_38378853_38379004 | 0.31 |
Gm13584 |
predicted gene 13584 |
14948 |
0.14 |
| chr9_110361719_110362313 | 0.31 |
Scap |
SREBF chaperone |
399 |
0.76 |
| chr3_30593178_30593362 | 0.31 |
1600017P15Rik |
RIKEN cDNA 1600017P15 gene |
2463 |
0.17 |
| chr1_36466108_36466267 | 0.30 |
Cnnm4 |
cyclin M4 |
5433 |
0.11 |
| chr8_126595451_126595638 | 0.30 |
Irf2bp2 |
interferon regulatory factor 2 binding protein 2 |
1558 |
0.45 |
| chr6_108163692_108163904 | 0.30 |
Sumf1 |
sulfatase modifying factor 1 |
11512 |
0.21 |
| chr11_119976291_119976457 | 0.30 |
Baiap2 |
brain-specific angiogenesis inhibitor 1-associated protein 2 |
16353 |
0.1 |
| chr8_69920774_69920972 | 0.29 |
Gatad2a |
GATA zinc finger domain containing 2A |
15653 |
0.1 |
| chr10_13965620_13965780 | 0.29 |
Hivep2 |
human immunodeficiency virus type I enhancer binding protein 2 |
375 |
0.87 |
| chr15_61942121_61942278 | 0.29 |
Gm37947 |
predicted gene, 37947 |
41103 |
0.16 |
| chr3_138527126_138527297 | 0.29 |
Eif4e |
eukaryotic translation initiation factor 4E |
74 |
0.92 |
| chr5_66214590_66214851 | 0.29 |
Gm3822 |
predicted gene 3822 |
27 |
0.96 |
| chr1_192094576_192094741 | 0.28 |
Traf5 |
TNF receptor-associated factor 5 |
2099 |
0.2 |
| chr5_140797604_140797806 | 0.28 |
Gna12 |
guanine nucleotide binding protein, alpha 12 |
32452 |
0.15 |
| chr16_24489944_24490119 | 0.28 |
Morf4l1-ps1 |
mortality factor 4 like 1, pseudogene 1 |
39363 |
0.15 |
| chr5_64805521_64805672 | 0.28 |
Gm20033 |
predicted gene, 20033 |
1479 |
0.26 |
| chr13_93814179_93814589 | 0.28 |
Mir5624 |
microRNA 5624 |
23607 |
0.15 |
| chr18_69355911_69356077 | 0.28 |
Tcf4 |
transcription factor 4 |
7050 |
0.28 |
| chr2_103454731_103454882 | 0.28 |
Elf5 |
E74-like factor 5 |
6068 |
0.19 |
| chr5_88918674_88918881 | 0.28 |
Slc4a4 |
solute carrier family 4 (anion exchanger), member 4 |
15755 |
0.23 |
| chr2_167118045_167118226 | 0.28 |
Kcnb1 |
potassium voltage gated channel, Shab-related subfamily, member 1 |
16309 |
0.1 |
| chr9_116101907_116102117 | 0.27 |
Gm9385 |
predicted pseudogene 9385 |
60948 |
0.11 |
| chr12_54738742_54738899 | 0.27 |
Gm22317 |
predicted gene, 22317 |
198 |
0.84 |
| chr12_57231180_57231526 | 0.27 |
Mipol1 |
mirror-image polydactyly 1 |
862 |
0.42 |
| chr2_30967908_30968194 | 0.27 |
Tor1a |
torsin family 1, member A (torsin A) |
118 |
0.94 |
| chr4_133756501_133756653 | 0.27 |
Arid1a |
AT rich interactive domain 1A (SWI-like) |
192 |
0.93 |
| chr10_128303547_128303722 | 0.27 |
Pan2 |
PAN2 poly(A) specific ribonuclease subunit |
236 |
0.8 |
| chr16_37503864_37504075 | 0.26 |
Gtf2e1 |
general transcription factor II E, polypeptide 1 (alpha subunit) |
35807 |
0.13 |
| chr11_103113058_103113588 | 0.26 |
Hexim1 |
hexamethylene bis-acetamide inducible 1 |
2908 |
0.16 |
| chr2_103760559_103761137 | 0.26 |
Nat10 |
N-acetyltransferase 10 |
78 |
0.96 |
| chr6_48395304_48395463 | 0.26 |
Gm44696 |
predicted gene 44696 |
25 |
0.71 |
| chr4_61223975_61224206 | 0.26 |
Mup13 |
major urinary protein 13 |
4181 |
0.23 |
| chr4_53120634_53120947 | 0.26 |
Abca1 |
ATP-binding cassette, sub-family A (ABC1), member 1 |
39105 |
0.15 |
| chr17_8652292_8652725 | 0.26 |
Gm15425 |
predicted gene 15425 |
83648 |
0.08 |
| chr17_36836564_36836799 | 0.26 |
Trim26 |
tripartite motif-containing 26 |
453 |
0.65 |
| chr13_52828377_52828538 | 0.26 |
BB123696 |
expressed sequence BB123696 |
71252 |
0.11 |
| chr19_61048615_61048813 | 0.26 |
Gm22520 |
predicted gene, 22520 |
35169 |
0.14 |
| chr11_4123188_4123541 | 0.25 |
Sec14l2 |
SEC14-like lipid binding 2 |
51 |
0.94 |
| chr4_105196590_105196741 | 0.25 |
Plpp3 |
phospholipid phosphatase 3 |
39318 |
0.2 |
| chr11_115619159_115619461 | 0.25 |
Slc25a19 |
solute carrier family 25 (mitochondrial thiamine pyrophosphate carrier), member 19 |
1405 |
0.23 |
| chr8_84989857_84990140 | 0.25 |
Hook2 |
hook microtubule tethering protein 2 |
605 |
0.43 |
| chr11_75439481_75439671 | 0.25 |
Serpinf2 |
serine (or cysteine) peptidase inhibitor, clade F, member 2 |
2 |
0.94 |
| chr4_134796652_134796803 | 0.24 |
Maco1 |
macoilin 1 |
14486 |
0.18 |
| chr4_118410618_118411038 | 0.24 |
Med8 |
mediator complex subunit 8 |
480 |
0.64 |
| chr10_72310190_72310549 | 0.24 |
Gm9923 |
predicted pseudogene 9923 |
1048 |
0.69 |
| chr3_58334130_58334299 | 0.24 |
Gm43730 |
predicted gene 43730 |
18904 |
0.18 |
| chr2_180118773_180118959 | 0.24 |
Osbpl2 |
oxysterol binding protein-like 2 |
440 |
0.73 |
| chr17_88597845_88598024 | 0.24 |
Ston1 |
stonin 1 |
238 |
0.91 |
| chr2_50813860_50814047 | 0.24 |
Gm13498 |
predicted gene 13498 |
95731 |
0.08 |
| chr5_30231710_30232189 | 0.24 |
Selenoi |
selenoprotein I |
632 |
0.63 |
| chr11_115831081_115831263 | 0.24 |
Llgl2 |
LLGL2 scribble cell polarity complex component |
796 |
0.45 |
| chr7_109526487_109526662 | 0.24 |
Gm24888 |
predicted gene, 24888 |
5294 |
0.12 |
| chr3_98296908_98297093 | 0.24 |
Gm43189 |
predicted gene 43189 |
10012 |
0.15 |
| chr5_136170628_136170833 | 0.24 |
Orai2 |
ORAI calcium release-activated calcium modulator 2 |
17 |
0.96 |
| chr3_87009765_87009927 | 0.24 |
Cd1d1 |
CD1d1 antigen |
10405 |
0.13 |
| chr16_23081291_23081442 | 0.23 |
Gm8134 |
predicted gene 8134 |
11577 |
0.06 |
| chr6_86609579_86609730 | 0.23 |
Gm44369 |
predicted gene, 44369 |
18224 |
0.1 |
| chr10_8657884_8658275 | 0.23 |
Gm24374 |
predicted gene, 24374 |
15145 |
0.23 |
| chr18_13671248_13671407 | 0.23 |
AC103362.1 |
novel transcript |
69927 |
0.13 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0060137 | maternal process involved in parturition(GO:0060137) |
| 0.1 | 0.3 | GO:0043578 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.1 | 0.4 | GO:1900242 | regulation of synaptic vesicle endocytosis(GO:1900242) |
| 0.1 | 0.3 | GO:0039536 | negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.1 | 0.2 | GO:0018879 | biphenyl metabolic process(GO:0018879) |
| 0.1 | 0.2 | GO:0006438 | valyl-tRNA aminoacylation(GO:0006438) |
| 0.1 | 0.3 | GO:1903011 | negative regulation of bone development(GO:1903011) |
| 0.1 | 0.2 | GO:0006546 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.1 | 0.3 | GO:0006344 | maintenance of chromatin silencing(GO:0006344) |
| 0.1 | 0.2 | GO:0045472 | response to ether(GO:0045472) |
| 0.1 | 0.2 | GO:0060066 | oviduct development(GO:0060066) |
| 0.1 | 0.2 | GO:0045041 | protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.1 | 0.2 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.1 | 0.2 | GO:0001306 | age-dependent response to oxidative stress(GO:0001306) age-dependent general metabolic decline(GO:0007571) |
| 0.0 | 0.3 | GO:0051918 | negative regulation of fibrinolysis(GO:0051918) |
| 0.0 | 0.1 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.0 | 0.1 | GO:0010908 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.0 | 0.1 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.0 | 0.2 | GO:0072367 | regulation of lipid transport by regulation of transcription from RNA polymerase II promoter(GO:0072367) |
| 0.0 | 0.1 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) |
| 0.0 | 0.2 | GO:0006548 | histidine catabolic process(GO:0006548) imidazole-containing compound catabolic process(GO:0052805) |
| 0.0 | 0.1 | GO:1903660 | regulation of complement-dependent cytotoxicity(GO:1903659) negative regulation of complement-dependent cytotoxicity(GO:1903660) |
| 0.0 | 0.2 | GO:0001712 | ectodermal cell fate commitment(GO:0001712) |
| 0.0 | 0.2 | GO:0019087 | transformation of host cell by virus(GO:0019087) |
| 0.0 | 0.1 | GO:0044533 | induction of programmed cell death(GO:0012502) positive regulation of apoptotic process in other organism(GO:0044533) positive regulation by symbiont of host programmed cell death(GO:0052042) positive regulation by symbiont of host apoptotic process(GO:0052151) positive regulation by organism of programmed cell death in other organism involved in symbiotic interaction(GO:0052330) positive regulation by organism of apoptotic process in other organism involved in symbiotic interaction(GO:0052501) positive regulation of apoptotic process by virus(GO:0060139) |
| 0.0 | 0.1 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.0 | 0.1 | GO:1902837 | amino acid import into cell(GO:1902837) |
| 0.0 | 0.4 | GO:0042790 | transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:0042790) |
| 0.0 | 0.2 | GO:0046618 | drug export(GO:0046618) |
| 0.0 | 0.3 | GO:0032211 | negative regulation of telomere maintenance via telomerase(GO:0032211) |
| 0.0 | 0.2 | GO:2001286 | regulation of caveolin-mediated endocytosis(GO:2001286) |
| 0.0 | 0.1 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.0 | 0.1 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.2 | GO:0015760 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.0 | 0.1 | GO:0051791 | medium-chain fatty acid metabolic process(GO:0051791) |
| 0.0 | 0.1 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.0 | 0.5 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.0 | 0.1 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.0 | 0.1 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.0 | 0.1 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.0 | 0.1 | GO:1901727 | positive regulation of histone deacetylase activity(GO:1901727) |
| 0.0 | 0.2 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.0 | 0.1 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.0 | 0.2 | GO:0046826 | negative regulation of protein export from nucleus(GO:0046826) |
| 0.0 | 0.2 | GO:0071501 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.0 | 0.1 | GO:0008355 | olfactory learning(GO:0008355) |
| 0.0 | 0.0 | GO:0002752 | cell surface pattern recognition receptor signaling pathway(GO:0002752) |
| 0.0 | 0.1 | GO:0043097 | pyrimidine-containing compound salvage(GO:0008655) pyrimidine nucleoside salvage(GO:0043097) |
| 0.0 | 0.1 | GO:0051562 | negative regulation of mitochondrial calcium ion concentration(GO:0051562) |
| 0.0 | 0.1 | GO:0050689 | negative regulation of defense response to virus by host(GO:0050689) |
| 0.0 | 0.5 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.0 | 0.1 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.0 | 0.0 | GO:0032489 | regulation of Cdc42 protein signal transduction(GO:0032489) |
| 0.0 | 0.1 | GO:0070889 | platelet alpha granule organization(GO:0070889) |
| 0.0 | 0.1 | GO:0002447 | eosinophil activation involved in immune response(GO:0002278) eosinophil mediated immunity(GO:0002447) eosinophil degranulation(GO:0043308) |
| 0.0 | 0.1 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.0 | 0.0 | GO:2001268 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001268) |
| 0.0 | 0.1 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.0 | 0.2 | GO:0090308 | regulation of methylation-dependent chromatin silencing(GO:0090308) |
| 0.0 | 0.1 | GO:0070544 | histone H3-K36 demethylation(GO:0070544) |
| 0.0 | 0.0 | GO:0006578 | amino-acid betaine biosynthetic process(GO:0006578) |
| 0.0 | 0.1 | GO:0071442 | positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.0 | 0.1 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.0 | 0.1 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.8 | GO:0015909 | long-chain fatty acid transport(GO:0015909) |
| 0.0 | 0.1 | GO:2000667 | positive regulation of interleukin-5 secretion(GO:2000664) positive regulation of interleukin-13 secretion(GO:2000667) |
| 0.0 | 0.1 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.1 | GO:0006057 | cell wall mannoprotein biosynthetic process(GO:0000032) mannoprotein metabolic process(GO:0006056) mannoprotein biosynthetic process(GO:0006057) cell wall glycoprotein biosynthetic process(GO:0031506) cell wall biogenesis(GO:0042546) cell wall macromolecule biosynthetic process(GO:0044038) chain elongation of O-linked mannose residue(GO:0044845) cellular component macromolecule biosynthetic process(GO:0070589) |
| 0.0 | 0.1 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.1 | GO:0015766 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.0 | 0.0 | GO:0014858 | positive regulation of skeletal muscle cell proliferation(GO:0014858) |
| 0.0 | 0.1 | GO:1903977 | positive regulation of glial cell migration(GO:1903977) |
| 0.0 | 0.2 | GO:0009396 | folic acid-containing compound biosynthetic process(GO:0009396) |
| 0.0 | 0.2 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.0 | GO:2000065 | negative regulation of aldosterone metabolic process(GO:0032345) negative regulation of aldosterone biosynthetic process(GO:0032348) negative regulation of cortisol biosynthetic process(GO:2000065) |
| 0.0 | 0.0 | GO:0003289 | atrial septum primum morphogenesis(GO:0003289) |
| 0.0 | 0.2 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.0 | 0.0 | GO:2000586 | regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000586) negative regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000587) |
| 0.0 | 0.1 | GO:0072095 | regulation of branch elongation involved in ureteric bud branching(GO:0072095) |
| 0.0 | 0.0 | GO:0045869 | negative regulation of single stranded viral RNA replication via double stranded DNA intermediate(GO:0045869) |
| 0.0 | 0.0 | GO:1903298 | regulation of hypoxia-induced intrinsic apoptotic signaling pathway(GO:1903297) negative regulation of hypoxia-induced intrinsic apoptotic signaling pathway(GO:1903298) |
| 0.0 | 0.1 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.0 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.0 | 0.1 | GO:0042904 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.0 | 0.1 | GO:0072675 | osteoclast fusion(GO:0072675) |
| 0.0 | 0.0 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) platelet activating factor metabolic process(GO:0046469) |
| 0.0 | 0.1 | GO:0044791 | modulation by host of viral release from host cell(GO:0044789) positive regulation by host of viral release from host cell(GO:0044791) |
| 0.0 | 0.1 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.0 | 0.1 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.0 | 0.1 | GO:1901628 | positive regulation of postsynaptic membrane organization(GO:1901628) |
| 0.0 | 0.1 | GO:0035745 | T-helper 2 cell cytokine production(GO:0035745) |
| 0.0 | 0.0 | GO:1902071 | regulation of hypoxia-inducible factor-1alpha signaling pathway(GO:1902071) |
| 0.0 | 0.0 | GO:0060399 | positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.0 | 0.3 | GO:0071549 | cellular response to dexamethasone stimulus(GO:0071549) |
| 0.0 | 0.1 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 0.0 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.0 | 0.1 | GO:0032497 | detection of lipopolysaccharide(GO:0032497) |
| 0.0 | 0.0 | GO:0042998 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) |
| 0.0 | 0.0 | GO:1903596 | regulation of gap junction assembly(GO:1903596) positive regulation of gap junction assembly(GO:1903598) |
| 0.0 | 0.1 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.0 | 0.0 | GO:0010989 | negative regulation of low-density lipoprotein particle clearance(GO:0010989) |
| 0.0 | 0.1 | GO:0045404 | positive regulation of interleukin-4 biosynthetic process(GO:0045404) |
| 0.0 | 0.1 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.0 | 0.1 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 0.0 | 0.1 | GO:2001184 | positive regulation of interleukin-12 secretion(GO:2001184) |
| 0.0 | 0.1 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 | 0.0 | GO:0009174 | UMP biosynthetic process(GO:0006222) pyrimidine ribonucleoside monophosphate metabolic process(GO:0009173) pyrimidine ribonucleoside monophosphate biosynthetic process(GO:0009174) UMP metabolic process(GO:0046049) |
| 0.0 | 0.0 | GO:0030997 | regulation of centriole-centriole cohesion(GO:0030997) |
| 0.0 | 0.0 | GO:0070268 | cornification(GO:0070268) |
| 0.0 | 0.1 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.0 | 0.0 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.0 | 0.0 | GO:0014745 | negative regulation of muscle adaptation(GO:0014745) |
| 0.0 | 0.0 | GO:0051599 | response to hydrostatic pressure(GO:0051599) |
| 0.0 | 0.1 | GO:0032484 | Ral protein signal transduction(GO:0032484) |
| 0.0 | 0.1 | GO:2001214 | positive regulation of vasculogenesis(GO:2001214) |
| 0.0 | 0.0 | GO:2000169 | regulation of peptidyl-cysteine S-nitrosylation(GO:2000169) |
| 0.0 | 0.0 | GO:1904469 | positive regulation of tumor necrosis factor secretion(GO:1904469) |
| 0.0 | 0.0 | GO:1901668 | regulation of superoxide dismutase activity(GO:1901668) |
| 0.0 | 0.0 | GO:1900825 | regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900825) |
| 0.0 | 0.1 | GO:0090160 | Golgi to lysosome transport(GO:0090160) |
| 0.0 | 0.0 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.0 | 0.1 | GO:1902715 | positive regulation of interferon-gamma secretion(GO:1902715) |
| 0.0 | 0.0 | GO:0035771 | interleukin-4-mediated signaling pathway(GO:0035771) |
| 0.0 | 0.2 | GO:0032878 | regulation of establishment or maintenance of cell polarity(GO:0032878) |
| 0.0 | 0.0 | GO:0032066 | nucleolus to nucleoplasm transport(GO:0032066) |
| 0.0 | 0.0 | GO:0060671 | epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 0.0 | 0.0 | GO:0046098 | guanine metabolic process(GO:0046098) |
| 0.0 | 0.1 | GO:2001185 | regulation of CD8-positive, alpha-beta T cell activation(GO:2001185) |
| 0.0 | 0.1 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 0.1 | GO:0043922 | negative regulation by host of viral transcription(GO:0043922) |
| 0.0 | 0.0 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.0 | 0.0 | GO:2000646 | positive regulation of receptor catabolic process(GO:2000646) |
| 0.0 | 0.2 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.0 | 0.2 | GO:0009813 | flavonoid biosynthetic process(GO:0009813) flavonoid glucuronidation(GO:0052696) |
| 0.0 | 0.1 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) determination of liver left/right asymmetry(GO:0071910) |
| 0.0 | 0.0 | GO:0003415 | chondrocyte hypertrophy(GO:0003415) |
| 0.0 | 0.2 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 | 0.0 | GO:0097283 | keratinocyte apoptotic process(GO:0097283) regulation of keratinocyte apoptotic process(GO:1902172) |
| 0.0 | 0.0 | GO:0060283 | negative regulation of oocyte development(GO:0060283) negative regulation of oocyte maturation(GO:1900194) |
| 0.0 | 0.1 | GO:1902287 | semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 0.0 | 0.1 | GO:0090267 | positive regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090267) |
| 0.0 | 0.0 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.0 | 0.0 | GO:0035425 | autocrine signaling(GO:0035425) |
| 0.0 | 0.1 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) |
| 0.0 | 0.1 | GO:0031998 | regulation of fatty acid beta-oxidation(GO:0031998) |
| 0.0 | 0.1 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.0 | 0.0 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.1 | 0.2 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.1 | 0.2 | GO:0005745 | m-AAA complex(GO:0005745) |
| 0.0 | 0.1 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 0.1 | GO:0032010 | phagolysosome(GO:0032010) |
| 0.0 | 0.6 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.0 | 0.3 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.1 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.2 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.1 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.0 | 0.2 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.2 | GO:0070695 | FHF complex(GO:0070695) |
| 0.0 | 0.1 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.0 | 0.1 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.0 | 0.3 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.1 | GO:0031502 | dolichyl-phosphate-mannose-protein mannosyltransferase complex(GO:0031502) |
| 0.0 | 0.1 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.0 | 0.1 | GO:0000938 | GARP complex(GO:0000938) |
| 0.0 | 0.1 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.0 | 0.2 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.1 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.0 | 0.1 | GO:0005683 | U7 snRNP(GO:0005683) |
| 0.0 | 0.2 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.1 | GO:0033503 | HULC complex(GO:0033503) |
| 0.0 | 0.3 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.2 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.0 | GO:0032127 | dense core granule membrane(GO:0032127) |
| 0.0 | 0.1 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.0 | 0.1 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 0.0 | GO:0097433 | dense body(GO:0097433) |
| 0.0 | 0.1 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 0.1 | GO:0070033 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) |
| 0.0 | 0.2 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 0.0 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.0 | 0.9 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.1 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.0 | 0.0 | GO:0097057 | TRAF2-GSTP1 complex(GO:0097057) |
| 0.0 | 0.1 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.2 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 0.0 | GO:0020018 | ciliary pocket(GO:0020016) ciliary pocket membrane(GO:0020018) |
| 0.0 | 0.1 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.0 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.0 | 0.2 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.1 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 0.1 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.0 | 0.1 | GO:0000235 | astral microtubule(GO:0000235) |
| 0.0 | 0.0 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.0 | 0.0 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.0 | 0.1 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.0 | 0.0 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.0 | 0.0 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.0 | 0.1 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 0.0 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 0.1 | GO:0005579 | membrane attack complex(GO:0005579) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.1 | 0.2 | GO:0004832 | valine-tRNA ligase activity(GO:0004832) |
| 0.1 | 0.6 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.1 | 0.6 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.1 | 0.2 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.1 | GO:0015526 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.0 | 0.3 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.1 | GO:0017098 | sulfonylurea receptor binding(GO:0017098) |
| 0.0 | 0.1 | GO:0071208 | histone pre-mRNA DCP binding(GO:0071208) |
| 0.0 | 0.3 | GO:0019113 | 3-(3-hydroxyphenyl)propionate hydroxylase activity(GO:0008688) 4-chlorobenzaldehyde oxidase activity(GO:0018471) 3,5-xylenol methylhydroxylase activity(GO:0018630) phenylacetate hydroxylase activity(GO:0018631) 4-nitrophenol 4-monooxygenase activity(GO:0018632) dimethyl sulfide monooxygenase activity(GO:0018633) alpha-pinene monooxygenase [NADH] activity(GO:0018634) 1-hydroxy-2-naphthoate hydroxylase activity(GO:0018637) toluene 4-monooxygenase activity(GO:0018638) xylene monooxygenase activity(GO:0018639) dibenzothiophene monooxygenase activity(GO:0018640) 6-hydroxy-3-methyl-2-oxo-1,2-dihydroquinoline 6-monooxygenase activity(GO:0018641) chlorophenol 4-monooxygenase activity(GO:0018642) carbon disulfide oxygenase activity(GO:0018643) toluene 2-monooxygenase activity(GO:0018644) 1-hydroxy-2-oxolimonene 1,2-monooxygenase activity(GO:0018646) phenanthrene 1,2-monooxygenase activity(GO:0018647) tetrahydrofuran hydroxylase activity(GO:0018649) styrene monooxygenase activity(GO:0018650) toluene-4-sulfonate monooxygenase activity(GO:0018651) toluene-sulfonate methyl-monooxygenase activity(GO:0018652) 3-methyl-2-oxo-1,2-dihydroquinoline 6-monooxygenase activity(GO:0018653) 2-hydroxy-phenylacetate hydroxylase activity(GO:0018654) 2-oxo-delta3-4,5,5-trimethylcyclopentenylacetyl-CoA 1,2-monooxygenase activity(GO:0018655) phenanthrene 3,4-monooxygenase activity(GO:0018656) toluene 3-monooxygenase activity(GO:0018657) 4-hydroxyphenylacetate,NADH:oxygen oxidoreductase (3-hydroxylating) activity(GO:0018660) limonene monooxygenase activity(GO:0019113) 2-methylnaphthalene hydroxylase activity(GO:0034526) 1-methylnaphthalene hydroxylase activity(GO:0034534) bisphenol A hydroxylase A activity(GO:0034560) salicylate 5-hydroxylase activity(GO:0034785) isobutylamine N-hydroxylase activity(GO:0034791) branched-chain dodecylbenzene sulfonate monooxygenase activity(GO:0034802) 3-HSA hydroxylase activity(GO:0034819) 4-hydroxypyridine-3-hydroxylase activity(GO:0034894) 2-octaprenyl-3-methyl-6-methoxy-1,4-benzoquinol hydroxylase activity(GO:0043719) 6-hydroxynicotinate 3-monooxygenase activity(GO:0043731) thalianol hydroxylase activity(GO:0080014) |
| 0.0 | 0.2 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.0 | 0.2 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.1 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.0 | 0.5 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.2 | GO:0035473 | lipase binding(GO:0035473) |
| 0.0 | 0.1 | GO:0015087 | cobalt ion transmembrane transporter activity(GO:0015087) |
| 0.0 | 0.1 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.1 | GO:1990239 | steroid hormone binding(GO:1990239) |
| 0.0 | 0.1 | GO:0051870 | methotrexate binding(GO:0051870) |
| 0.0 | 0.1 | GO:0015181 | arginine transmembrane transporter activity(GO:0015181) L-lysine transmembrane transporter activity(GO:0015189) |
| 0.0 | 0.1 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.1 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.0 | 0.1 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.0 | 0.3 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 0.1 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.0 | 0.1 | GO:0015037 | peptide disulfide oxidoreductase activity(GO:0015037) |
| 0.0 | 0.1 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.1 | GO:0032767 | copper-dependent protein binding(GO:0032767) |
| 0.0 | 0.1 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.0 | 0.1 | GO:0004096 | catalase activity(GO:0004096) |
| 0.0 | 0.2 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.1 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 0.2 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.1 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 0.1 | GO:0050309 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.0 | 0.1 | GO:0004305 | ethanolamine kinase activity(GO:0004305) |
| 0.0 | 0.1 | GO:0052658 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) |
| 0.0 | 0.1 | GO:0070698 | type I activin receptor binding(GO:0070698) |
| 0.0 | 0.1 | GO:0008506 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.0 | 0.1 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 0.1 | GO:0071723 | lipopeptide binding(GO:0071723) |
| 0.0 | 0.2 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.0 | 0.2 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.1 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.0 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.0 | 0.0 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.0 | 0.1 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.0 | 0.1 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.0 | 0.1 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.0 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 0.0 | 0.2 | GO:0016594 | glycine binding(GO:0016594) |
| 0.0 | 0.1 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.2 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.0 | GO:0004920 | interleukin-10 receptor activity(GO:0004920) |
| 0.0 | 0.1 | GO:0009374 | biotin binding(GO:0009374) |
| 0.0 | 0.2 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 0.0 | GO:0016508 | long-chain-enoyl-CoA hydratase activity(GO:0016508) |
| 0.0 | 0.1 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.0 | 0.1 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.0 | 0.0 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.0 | 0.1 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 0.1 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 0.0 | GO:0070139 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.0 | 0.1 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.0 | 0.2 | GO:0005186 | pheromone activity(GO:0005186) |
| 0.0 | 0.1 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.0 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.0 | 0.1 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.0 | 0.3 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.0 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.1 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.0 | 0.0 | GO:0036374 | gamma-glutamyltransferase activity(GO:0003840) glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.1 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.0 | 0.0 | GO:0034584 | piRNA binding(GO:0034584) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.1 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.0 | 0.0 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 1.1 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.0 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.0 | 0.2 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.6 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 0.3 | ST P38 MAPK PATHWAY | p38 MAPK Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.3 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.2 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 0.5 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.1 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.0 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 0.6 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.2 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.2 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.2 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.5 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.1 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.3 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 1.1 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.1 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 0.4 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.3 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 0.1 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.2 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.0 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 0.1 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |