Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
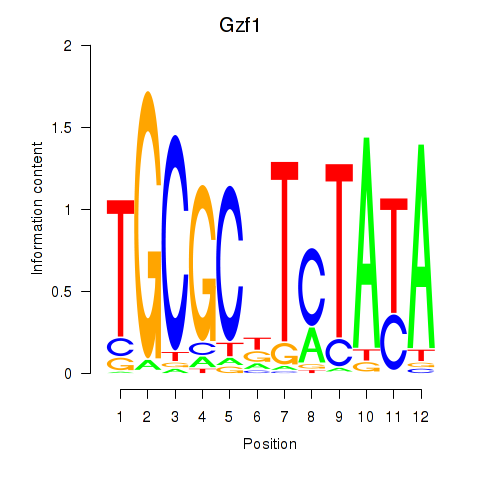
Results for Gzf1
Z-value: 1.16
Transcription factors associated with Gzf1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Gzf1
|
ENSMUSG00000027439.9 | GDNF-inducible zinc finger protein 1 |
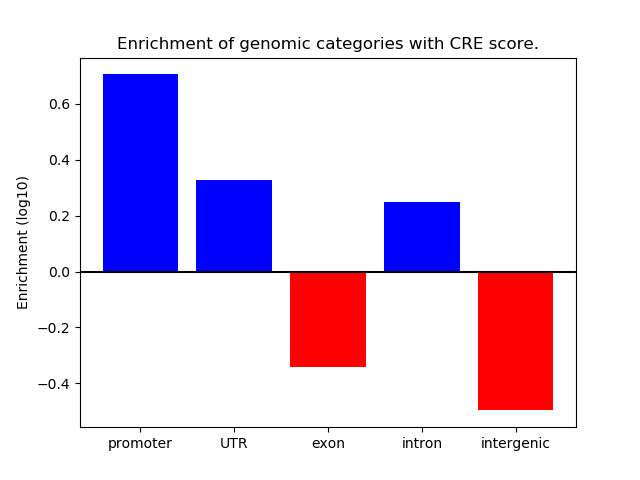
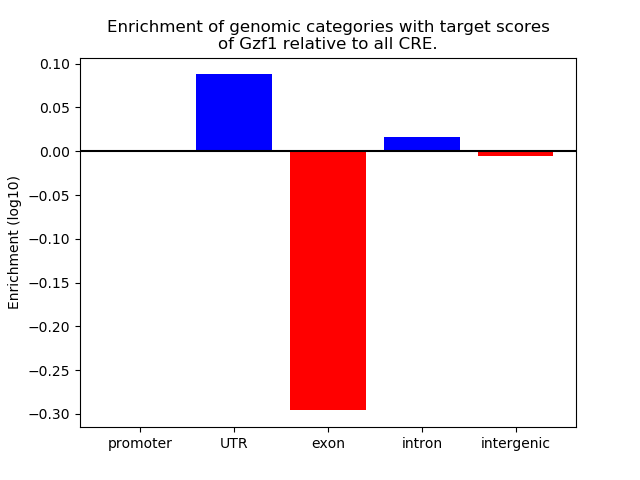
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr2_148680889_148681282 | Gzf1 | 62 | 0.967076 | -0.82 | 4.4e-02 | Click! |
| chr2_148693353_148693640 | Gzf1 | 8884 | 0.145886 | 0.82 | 4.7e-02 | Click! |
| chr2_148684656_148684837 | Gzf1 | 134 | 0.950211 | -0.80 | 5.4e-02 | Click! |
| chr2_148680577_148680742 | Gzf1 | 364 | 0.837005 | 0.80 | 5.8e-02 | Click! |
| chr2_148690686_148690855 | Gzf1 | 6158 | 0.156373 | 0.77 | 7.4e-02 | Click! |
Activity of the Gzf1 motif across conditions
Conditions sorted by the z-value of the Gzf1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
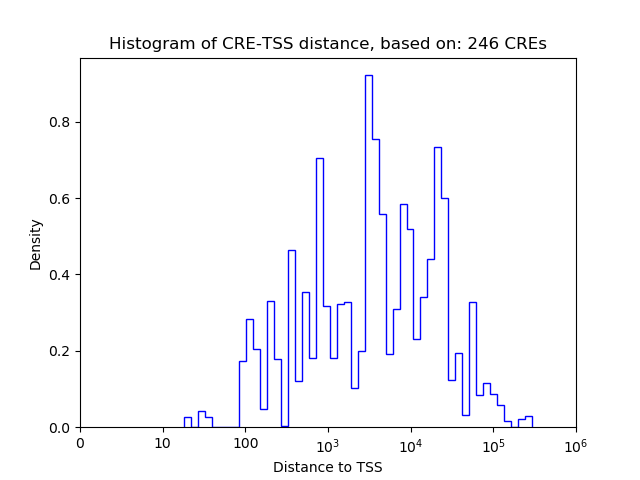
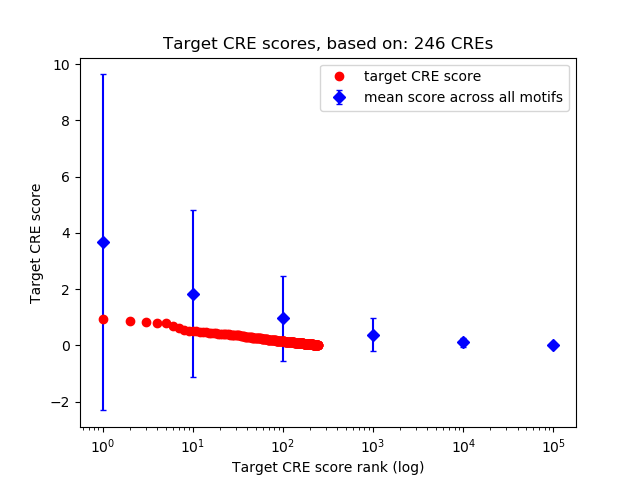
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr6_8209868_8210031 | 0.93 |
Mios |
meiosis regulator for oocyte development |
727 |
0.65 |
| chr4_133451667_133451825 | 0.88 |
Tent5b |
terminal nucleotidyltransferase 5B |
28386 |
0.1 |
| chr1_88908948_88909111 | 0.82 |
Gm4753 |
predicted gene 4753 |
23290 |
0.18 |
| chr7_63921305_63921456 | 0.81 |
Klf13 |
Kruppel-like factor 13 |
3490 |
0.16 |
| chr18_38370754_38370932 | 0.80 |
Gm4949 |
predicted gene 4949 |
10305 |
0.12 |
| chr8_40922878_40923029 | 0.67 |
Pdgfrl |
platelet-derived growth factor receptor-like |
3259 |
0.22 |
| chr8_13109068_13109219 | 0.61 |
Cul4a |
cullin 4A |
3207 |
0.13 |
| chr9_55279082_55279254 | 0.53 |
Nrg4 |
neuregulin 4 |
4404 |
0.21 |
| chr2_71256092_71256345 | 0.52 |
Dync1i2 |
dynein cytoplasmic 1 intermediate chain 2 |
4329 |
0.23 |
| chr6_116240202_116240376 | 0.52 |
Washc2 |
WASH complex subunit 2` |
14317 |
0.11 |
| chr18_12793031_12793182 | 0.51 |
Osbpl1a |
oxysterol binding protein-like 1A |
25969 |
0.14 |
| chr2_116983968_116984177 | 0.49 |
Gm29340 |
predicted gene 29340 |
7644 |
0.2 |
| chr17_43138649_43138810 | 0.47 |
E130008D07Rik |
RIKEN cDNA E130008D07 gene |
19467 |
0.26 |
| chr6_91703122_91703273 | 0.47 |
Slc6a6 |
solute carrier family 6 (neurotransmitter transporter, taurine), member 6 |
1540 |
0.28 |
| chr9_116174746_116175061 | 0.43 |
Tgfbr2 |
transforming growth factor, beta receptor II |
362 |
0.86 |
| chr2_120404351_120404527 | 0.42 |
Ganc |
glucosidase, alpha; neutral C |
101 |
0.77 |
| chr11_90627862_90628033 | 0.42 |
Stxbp4 |
syntaxin binding protein 4 |
2970 |
0.25 |
| chr12_84163524_84163675 | 0.42 |
Elmsan1 |
ELM2 and Myb/SANT-like domain containing 1 |
3461 |
0.13 |
| chr6_47792412_47792563 | 0.42 |
Rpl31-ps7 |
ribosomal protein L31, pseudogene 7 |
6113 |
0.13 |
| chr2_90561333_90561502 | 0.41 |
Ptprj |
protein tyrosine phosphatase, receptor type, J |
19230 |
0.2 |
| chr8_25753638_25753875 | 0.41 |
Ddhd2 |
DDHD domain containing 2 |
524 |
0.47 |
| chr1_152399824_152400209 | 0.40 |
Colgalt2 |
collagen beta(1-O)galactosyltransferase 2 |
186 |
0.95 |
| chr10_77898372_77898523 | 0.39 |
Lrrc3 |
leucine rich repeat containing 3 |
4089 |
0.09 |
| chr13_45863946_45864126 | 0.39 |
Atxn1 |
ataxin 1 |
8252 |
0.26 |
| chr1_151154930_151155204 | 0.39 |
C730036E19Rik |
RIKEN cDNA C730036E19 gene |
17033 |
0.11 |
| chr1_67142809_67143033 | 0.38 |
Cps1 |
carbamoyl-phosphate synthetase 1 |
19895 |
0.22 |
| chr6_7983342_7983669 | 0.37 |
Rnps1-ps |
RNA binding protein with serine rich domain 1, pseudogene |
122 |
0.97 |
| chr7_97415846_97416028 | 0.37 |
Thrsp |
thyroid hormone responsive |
1582 |
0.28 |
| chr4_141546149_141546719 | 0.37 |
B330016D10Rik |
RIKEN cDNA B330016D10 gene |
245 |
0.89 |
| chrX_140540020_140540246 | 0.36 |
Tsc22d3 |
TSC22 domain family, member 3 |
2535 |
0.29 |
| chr4_155848027_155848340 | 0.36 |
Dvl1 |
dishevelled segment polarity protein 1 |
512 |
0.52 |
| chr6_6885058_6885209 | 0.36 |
Dlx5 |
distal-less homeobox 5 |
3048 |
0.17 |
| chr6_129183742_129184088 | 0.35 |
Clec2d |
C-type lectin domain family 2, member d |
3300 |
0.15 |
| chr1_155808676_155809110 | 0.33 |
Qsox1 |
quiescin Q6 sulfhydryl oxidase 1 |
3927 |
0.15 |
| chr12_26182684_26183045 | 0.33 |
Gm29687 |
predicted gene, 29687 |
27641 |
0.15 |
| chr18_12827399_12827550 | 0.33 |
Osbpl1a |
oxysterol binding protein-like 1A |
7567 |
0.19 |
| chr2_32524561_32524721 | 0.31 |
Gm13412 |
predicted gene 13412 |
390 |
0.73 |
| chr15_97048387_97048745 | 0.31 |
Slc38a4 |
solute carrier family 38, member 4 |
1521 |
0.52 |
| chrX_140957060_140957250 | 0.31 |
Atg4a |
autophagy related 4A, cysteine peptidase |
205 |
0.73 |
| chr8_105268773_105269119 | 0.31 |
Hsf4 |
heat shock transcription factor 4 |
855 |
0.29 |
| chr16_23056944_23057241 | 0.30 |
Kng1 |
kininogen 1 |
773 |
0.39 |
| chr10_82699263_82699421 | 0.30 |
Hcfc2 |
host cell factor C2 |
131 |
0.95 |
| chr3_135485099_135485441 | 0.29 |
Manba |
mannosidase, beta A, lysosomal |
341 |
0.77 |
| chr12_73545525_73545713 | 0.28 |
Tmem30b |
transmembrane protein 30B |
773 |
0.58 |
| chr19_41418415_41418566 | 0.28 |
Pik3ap1 |
phosphoinositide-3-kinase adaptor protein 1 |
33388 |
0.16 |
| chr15_103273560_103273712 | 0.28 |
Copz1 |
coatomer protein complex, subunit zeta 1 |
724 |
0.49 |
| chr12_99429400_99429551 | 0.28 |
Foxn3 |
forkhead box N3 |
16870 |
0.17 |
| chr19_45805094_45805257 | 0.28 |
Kcnip2 |
Kv channel-interacting protein 2 |
1601 |
0.31 |
| chr1_156741988_156742155 | 0.27 |
Fam20b |
family with sequence similarity 20, member B |
22985 |
0.14 |
| chr7_90158580_90158753 | 0.27 |
Picalm |
phosphatidylinositol binding clathrin assembly protein |
6847 |
0.12 |
| chr4_135221177_135221356 | 0.26 |
Clic4 |
chloride intracellular channel 4 (mitochondrial) |
51548 |
0.09 |
| chr17_65594339_65595183 | 0.26 |
Vapa |
vesicle-associated membrane protein, associated protein A |
209 |
0.93 |
| chr7_120176026_120176177 | 0.26 |
Anks4b |
ankyrin repeat and sterile alpha motif domain containing 4B |
2243 |
0.22 |
| chr3_40746359_40746539 | 0.26 |
Hspa4l |
heat shock protein 4 like |
581 |
0.72 |
| chr3_138222614_138222780 | 0.26 |
Adh7 |
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
1226 |
0.36 |
| chrX_103288811_103289364 | 0.26 |
Gm19200 |
predicted gene, 19200 |
117 |
0.9 |
| chr11_83834151_83834516 | 0.25 |
Gm12576 |
predicted gene 12576 |
15160 |
0.12 |
| chr7_99837512_99837672 | 0.24 |
Neu3 |
neuraminidase 3 |
9175 |
0.12 |
| chr2_73056230_73056536 | 0.23 |
Gm13665 |
predicted gene 13665 |
60365 |
0.09 |
| chr18_44540954_44541143 | 0.22 |
Mcc |
mutated in colorectal cancers |
21532 |
0.24 |
| chr9_48757158_48757309 | 0.22 |
Zbtb16 |
zinc finger and BTB domain containing 16 |
78712 |
0.09 |
| chr16_13276512_13276712 | 0.22 |
Mrtfb |
myocardin related transcription factor B |
11536 |
0.26 |
| chr15_82244141_82244292 | 0.22 |
Cenpm |
centromere protein M |
116 |
0.91 |
| chr17_87143110_87143261 | 0.22 |
Socs5 |
suppressor of cytokine signaling 5 |
35327 |
0.11 |
| chr2_52476715_52476893 | 0.21 |
A430018G15Rik |
RIKEN cDNA A430018G15 gene |
51792 |
0.11 |
| chr10_69106905_69107081 | 0.21 |
Gm47107 |
predicted gene, 47107 |
9127 |
0.18 |
| chr7_24564744_24564916 | 0.21 |
Xrcc1 |
X-ray repair complementing defective repair in Chinese hamster cells 1 |
739 |
0.44 |
| chr10_89502299_89502578 | 0.21 |
Nr1h4 |
nuclear receptor subfamily 1, group H, member 4 |
4211 |
0.26 |
| chr15_28387409_28387579 | 0.20 |
Gm49278 |
predicted gene, 49278 |
117697 |
0.06 |
| chr11_98588166_98588531 | 0.20 |
Ormdl3 |
ORM1-like 3 (S. cerevisiae) |
980 |
0.38 |
| chr19_5294343_5294670 | 0.20 |
Sf3b2 |
splicing factor 3b, subunit 2 |
128 |
0.83 |
| chr8_33991786_33992077 | 0.19 |
Gm45817 |
predicted gene 45817 |
4731 |
0.17 |
| chr18_16608064_16608271 | 0.19 |
Cdh2 |
cadherin 2 |
61902 |
0.14 |
| chr13_8649005_8649216 | 0.19 |
Gm48262 |
predicted gene, 48262 |
96090 |
0.07 |
| chr11_120005928_120006087 | 0.19 |
Aatk |
apoptosis-associated tyrosine kinase |
6401 |
0.11 |
| chr16_14152987_14153329 | 0.19 |
Marf1 |
meiosis regulator and mRNA stability 1 |
6109 |
0.14 |
| chr9_74993629_74993780 | 0.18 |
Fam214a |
family with sequence similarity 214, member A |
17593 |
0.17 |
| chr11_86907144_86907318 | 0.18 |
Ypel2 |
yippee like 2 |
64793 |
0.09 |
| chr10_89491324_89491638 | 0.18 |
Nr1h4 |
nuclear receptor subfamily 1, group H, member 4 |
15168 |
0.21 |
| chr11_115857814_115857965 | 0.18 |
Myo15b |
myosin XVB |
517 |
0.62 |
| chr7_72519858_72520162 | 0.18 |
Gm37620 |
predicted gene, 37620 |
81979 |
0.09 |
| chr18_39426891_39427064 | 0.17 |
Gm15337 |
predicted gene 15337 |
37552 |
0.16 |
| chr15_41162949_41163116 | 0.17 |
4930555K19Rik |
RIKEN cDNA 4930555K19 gene |
10455 |
0.28 |
| chr9_42138118_42138269 | 0.17 |
Gm39321 |
predicted gene, 39321 |
3800 |
0.24 |
| chr1_138044670_138044827 | 0.17 |
Gm23782 |
predicted gene, 23782 |
56972 |
0.07 |
| chr17_34911940_34912115 | 0.17 |
Ehmt2 |
euchromatic histone lysine N-methyltransferase 2 |
1052 |
0.2 |
| chr14_74741561_74741891 | 0.17 |
Esd |
esterase D/formylglutathione hydrolase |
3220 |
0.27 |
| chr7_88294280_88294445 | 0.17 |
Ctsc |
cathepsin C |
2490 |
0.3 |
| chr19_55126042_55126256 | 0.16 |
Gpam |
glycerol-3-phosphate acyltransferase, mitochondrial |
1054 |
0.54 |
| chr14_41109398_41109729 | 0.16 |
Mat1a |
methionine adenosyltransferase I, alpha |
4182 |
0.14 |
| chr8_94874425_94874581 | 0.16 |
Dok4 |
docking protein 4 |
1593 |
0.24 |
| chr9_48612421_48612590 | 0.16 |
Nnmt |
nicotinamide N-methyltransferase |
7352 |
0.25 |
| chr15_36275360_36275511 | 0.16 |
Rnf19a |
ring finger protein 19A |
7663 |
0.12 |
| chr5_8947767_8947928 | 0.16 |
Abcb4 |
ATP-binding cassette, sub-family B (MDR/TAP), member 4 |
10705 |
0.13 |
| chr1_74061759_74062050 | 0.16 |
Tns1 |
tensin 1 |
24747 |
0.17 |
| chr8_69790804_69791266 | 0.16 |
Atp13a1 |
ATPase type 13A1 |
128 |
0.51 |
| chr18_40255569_40255732 | 0.16 |
Kctd16 |
potassium channel tetramerisation domain containing 16 |
1312 |
0.41 |
| chr5_140794441_140794653 | 0.16 |
Gna12 |
guanine nucleotide binding protein, alpha 12 |
35610 |
0.14 |
| chr10_69352418_69352569 | 0.16 |
Cdk1 |
cyclin-dependent kinase 1 |
410 |
0.83 |
| chr13_23518940_23519232 | 0.15 |
n-TStga1 |
nuclear encoded tRNA serine 1 (anticodon TGA) |
944 |
0.24 |
| chr5_9101079_9101755 | 0.15 |
Tmem243 |
transmembrane protein 243, mitochondrial |
670 |
0.68 |
| chr4_11239151_11239302 | 0.14 |
Ints8 |
integrator complex subunit 8 |
3258 |
0.21 |
| chr15_82793132_82793326 | 0.14 |
Cyp2d26 |
cytochrome P450, family 2, subfamily d, polypeptide 26 |
993 |
0.37 |
| chr12_82190257_82190917 | 0.14 |
Sipa1l1 |
signal-induced proliferation-associated 1 like 1 |
20561 |
0.18 |
| chr8_117725888_117726161 | 0.13 |
Hsd17b2 |
hydroxysteroid (17-beta) dehydrogenase 2 |
5088 |
0.16 |
| chr2_90868308_90868585 | 0.13 |
Mtch2 |
mitochondrial carrier 2 |
4987 |
0.12 |
| chr11_101278340_101278806 | 0.13 |
Coa3 |
cytochrome C oxidase assembly factor 3 |
370 |
0.44 |
| chr5_44234175_44234326 | 0.13 |
Tapt1 |
transmembrane anterior posterior transformation 1 |
7624 |
0.12 |
| chr14_66118821_66119188 | 0.13 |
Ephx2 |
epoxide hydrolase 2, cytoplasmic |
5496 |
0.18 |
| chr10_100589197_100589356 | 0.13 |
4930430F08Rik |
RIKEN cDNA 4930430F08 gene |
28 |
0.95 |
| chr10_127066593_127066777 | 0.13 |
Cdk4 |
cyclin-dependent kinase 4 |
609 |
0.45 |
| chr9_21836570_21836722 | 0.13 |
Angptl8 |
angiopoietin-like 8 |
1136 |
0.32 |
| chr10_42254686_42254909 | 0.12 |
Foxo3 |
forkhead box O3 |
3569 |
0.32 |
| chr16_72663718_72663897 | 0.12 |
Robo1 |
roundabout guidance receptor 1 |
603 |
0.86 |
| chr4_35156757_35157337 | 0.12 |
Mob3b |
MOB kinase activator 3B |
437 |
0.81 |
| chr11_50037172_50037336 | 0.12 |
Rnf130 |
ring finger protein 130 |
11906 |
0.16 |
| chr8_104442536_104442722 | 0.12 |
Dync1li2 |
dynein, cytoplasmic 1 light intermediate chain 2 |
115 |
0.77 |
| chr15_85806972_85807337 | 0.12 |
Cdpf1 |
cysteine rich, DPF motif domain containing 1 |
3936 |
0.16 |
| chr6_137754996_137755471 | 0.12 |
Dera |
deoxyribose-phosphate aldolase (putative) |
618 |
0.77 |
| chr7_78910767_78910959 | 0.12 |
Isg20 |
interferon-stimulated protein |
2561 |
0.19 |
| chr4_88034364_88034770 | 0.12 |
Gm12631 |
predicted gene 12631 |
1066 |
0.37 |
| chr7_44816822_44816988 | 0.12 |
Atf5 |
activating transcription factor 5 |
247 |
0.55 |
| chr10_127888444_127888627 | 0.12 |
Rdh7 |
retinol dehydrogenase 7 |
205 |
0.88 |
| chr14_79627613_79627788 | 0.12 |
Sugt1 |
SGT1, suppressor of G2 allele of SKP1 (S. cerevisiae) |
3061 |
0.22 |
| chr10_117494201_117494365 | 0.11 |
Gm32141 |
predicted gene, 32141 |
46790 |
0.1 |
| chr2_80580802_80580972 | 0.11 |
Nckap1 |
NCK-associated protein 1 |
377 |
0.79 |
| chr13_6830007_6830186 | 0.11 |
2900024D18Rik |
RIKEN cDNA 2900024D18 gene |
95755 |
0.08 |
| chr12_76177934_76178097 | 0.11 |
Esr2 |
estrogen receptor 2 (beta) |
756 |
0.62 |
| chr1_164450614_164451017 | 0.11 |
Atp1b1 |
ATPase, Na+/K+ transporting, beta 1 polypeptide |
2962 |
0.2 |
| chr13_93996913_93997222 | 0.10 |
Gm47216 |
predicted gene, 47216 |
5299 |
0.2 |
| chr6_74797061_74797232 | 0.10 |
Gm22254 |
predicted gene, 22254 |
291982 |
0.01 |
| chr8_123191935_123192230 | 0.10 |
Dpep1 |
dipeptidase 1 |
1412 |
0.19 |
| chr4_150660019_150660333 | 0.10 |
Slc45a1 |
solute carrier family 45, member 1 |
8002 |
0.19 |
| chr19_4508485_4508947 | 0.10 |
Pcx |
pyruvate carboxylase |
1756 |
0.26 |
| chr11_78202734_78203046 | 0.10 |
Proca1 |
protein interacting with cyclin A1 |
8144 |
0.06 |
| chr18_35134558_35134709 | 0.10 |
Ctnna1 |
catenin (cadherin associated protein), alpha 1 |
15191 |
0.19 |
| chr12_4455773_4455955 | 0.10 |
Ncoa1 |
nuclear receptor coactivator 1 |
21318 |
0.16 |
| chr7_116039868_116040228 | 0.09 |
1110004F10Rik |
RIKEN cDNA 1110004F10 gene |
336 |
0.79 |
| chr17_80172841_80173158 | 0.09 |
Galm |
galactose mutarotase |
27847 |
0.12 |
| chr15_3396811_3397215 | 0.09 |
Ghr |
growth hormone receptor |
74631 |
0.1 |
| chr5_35028540_35028744 | 0.09 |
Rgs12 |
regulator of G-protein signaling 12 |
1672 |
0.27 |
| chr12_59218810_59218961 | 0.09 |
Fbxo33 |
F-box protein 33 |
84 |
0.96 |
| chr2_167337145_167337487 | 0.09 |
B4galt5 |
UDP-Gal:betaGlcNAc beta 1,4-galactosyltransferase, polypeptide 5 |
11867 |
0.18 |
| chr1_180726272_180726587 | 0.09 |
Acbd3 |
acyl-Coenzyme A binding domain containing 3 |
386 |
0.77 |
| chr6_54042034_54042185 | 0.09 |
Chn2 |
chimerin 2 |
2023 |
0.35 |
| chr16_15637762_15637937 | 0.09 |
Prkdc |
protein kinase, DNA activated, catalytic polypeptide |
17 |
0.89 |
| chr11_50388012_50388306 | 0.09 |
Hnrnph1 |
heterogeneous nuclear ribonucleoprotein H1 |
3196 |
0.18 |
| chr9_50848931_50849288 | 0.09 |
Ppp2r1b |
protein phosphatase 2, regulatory subunit A, beta |
3808 |
0.18 |
| chr6_82647878_82648248 | 0.08 |
Pole4 |
polymerase (DNA-directed), epsilon 4 (p12 subunit) |
4455 |
0.23 |
| chr4_63236027_63236196 | 0.08 |
Col27a1 |
collagen, type XXVII, alpha 1 |
1825 |
0.32 |
| chr4_120387588_120387746 | 0.08 |
Scmh1 |
sex comb on midleg homolog 1 |
17614 |
0.22 |
| chr3_110250026_110250177 | 0.08 |
C130013H08Rik |
RIKEN cDNA C130013H08 gene |
86 |
0.93 |
| chr2_93456519_93456670 | 0.08 |
Gm10804 |
predicted gene 10804 |
3773 |
0.2 |
| chr14_73369940_73370196 | 0.08 |
Itm2b |
integral membrane protein 2B |
944 |
0.58 |
| chr14_66268304_66268471 | 0.08 |
Ptk2b |
PTK2 protein tyrosine kinase 2 beta |
12595 |
0.17 |
| chr1_36465535_36465686 | 0.07 |
Cnnm4 |
cyclin M4 |
6010 |
0.11 |
| chr7_120679030_120679297 | 0.07 |
Mosmo |
modulator of smoothened |
1483 |
0.21 |
| chr14_74865691_74866187 | 0.07 |
Lrch1 |
leucine-rich repeats and calponin homology (CH) domain containing 1 |
19316 |
0.21 |
| chr6_34718943_34719094 | 0.07 |
Npn2 |
neoplastic progression 2 |
7887 |
0.14 |
| chr15_39006699_39006925 | 0.07 |
Fzd6 |
frizzled class receptor 6 |
479 |
0.5 |
| chr5_90492199_90492408 | 0.07 |
Afp |
alpha fetoprotein |
1065 |
0.43 |
| chr4_85831605_85831761 | 0.07 |
Gm25811 |
predicted gene, 25811 |
211811 |
0.02 |
| chr3_7649491_7649673 | 0.07 |
Il7 |
interleukin 7 |
35822 |
0.17 |
| chr6_108282461_108282737 | 0.07 |
Itpr1 |
inositol 1,4,5-trisphosphate receptor 1 |
29641 |
0.21 |
| chr5_107961440_107961605 | 0.07 |
Dipk1a |
divergent protein kinase domain 1A |
11476 |
0.13 |
| chr2_164911992_164912374 | 0.07 |
Zfp335 |
zinc finger protein 335 |
426 |
0.6 |
| chr17_56686614_56686939 | 0.07 |
Ranbp3 |
RAN binding protein 3 |
9959 |
0.11 |
| chr10_112916902_112917062 | 0.07 |
Gm26596 |
predicted gene, 26596 |
11519 |
0.16 |
| chr8_84810711_84810864 | 0.07 |
Nfix |
nuclear factor I/X |
10443 |
0.08 |
| chr11_106842172_106842332 | 0.06 |
Smurf2 |
SMAD specific E3 ubiquitin protein ligase 2 |
2954 |
0.17 |
| chr10_127643746_127644134 | 0.06 |
Stat6 |
signal transducer and activator of transcription 6 |
938 |
0.35 |
| chr7_45124707_45124921 | 0.06 |
Rps11 |
ribosomal protein S11 |
375 |
0.45 |
| chr4_59000010_59000161 | 0.06 |
Dnajc25 |
DnaJ heat shock protein family (Hsp40) member C25 |
3087 |
0.17 |
| chr18_20673992_20674203 | 0.06 |
Ttr |
transthyretin |
8817 |
0.16 |
| chr19_44544853_44545006 | 0.06 |
Sec31b |
Sec31 homolog B (S. cerevisiae) |
918 |
0.45 |
| chr8_3392646_3392851 | 0.06 |
Arhgef18 |
rho/rac guanine nucleotide exchange factor (GEF) 18 |
258 |
0.91 |
| chr1_34842105_34842416 | 0.06 |
Fam168b |
family with sequence similarity 168, member B |
704 |
0.58 |
| chr13_13994363_13994818 | 0.06 |
B3galnt2 |
UDP-GalNAc:betaGlcNAc beta 1,3-galactosaminyltransferase, polypeptide 2 |
170 |
0.92 |
| chr5_32282757_32282925 | 0.06 |
Gm15615 |
predicted gene 15615 |
29278 |
0.14 |
| chr17_32787291_32787473 | 0.05 |
Zfp871 |
zinc finger protein 871 |
165 |
0.84 |
| chr3_104218844_104219050 | 0.05 |
Magi3 |
membrane associated guanylate kinase, WW and PDZ domain containing 3 |
1156 |
0.35 |
| chr2_26118492_26118757 | 0.05 |
Nacc2 |
nucleus accumbens associated 2, BEN and BTB (POZ) domain containing |
4159 |
0.15 |
| chr1_194362178_194362358 | 0.05 |
4930503O07Rik |
RIKEN cDNA 4930503O07 gene |
139509 |
0.05 |
| chr13_109343509_109343689 | 0.05 |
Mir582 |
microRNA 582 |
18855 |
0.28 |
| chr8_120281580_120281753 | 0.05 |
Gse1 |
genetic suppressor element 1, coiled-coil protein |
53210 |
0.11 |
| chr16_70535063_70535483 | 0.05 |
Gbe1 |
glucan (1,4-alpha-), branching enzyme 1 |
25721 |
0.23 |
| chr12_55447086_55447286 | 0.05 |
Psma6 |
proteasome subunit alpha 6 |
39803 |
0.11 |
| chr12_91709555_91709810 | 0.05 |
Ston2 |
stonin 2 |
4292 |
0.19 |
| chr3_50381633_50381790 | 0.05 |
Slc7a11 |
solute carrier family 7 (cationic amino acid transporter, y+ system), member 11 |
10912 |
0.23 |
| chr9_71163682_71163840 | 0.05 |
Gm32511 |
predicted gene, 32511 |
33 |
0.89 |
| chr16_85085948_85086156 | 0.05 |
Gm49227 |
predicted gene, 49227 |
5941 |
0.22 |
| chr4_101250409_101250587 | 0.05 |
Jak1 |
Janus kinase 1 |
14725 |
0.13 |
| chr2_73910509_73910709 | 0.05 |
Atp5g3 |
ATP synthase, H+ transporting, mitochondrial F0 complex, subunit C3 (subunit 9) |
150 |
0.96 |
| chr17_10496047_10496417 | 0.05 |
Gm16168 |
predicted gene 16168 |
214 |
0.96 |
| chr15_77330063_77330442 | 0.04 |
Rbfox2 |
RNA binding protein, fox-1 homolog (C. elegans) 2 |
23248 |
0.11 |
| chr18_35703072_35703241 | 0.04 |
Dnajc18 |
DnaJ heat shock protein family (Hsp40) member C18 |
35 |
0.52 |
| chr19_38364433_38364731 | 0.04 |
Gm50155 |
predicted gene, 50155 |
20932 |
0.12 |
| chr17_13654745_13655106 | 0.04 |
2700054A10Rik |
RIKEN cDNA 2700054A10 gene |
13966 |
0.15 |
| chr14_41120025_41120433 | 0.04 |
Sftpa1 |
surfactant associated protein A1 |
11553 |
0.11 |
| chr11_98031142_98031301 | 0.04 |
Gm27597 |
predicted gene, 27597 |
1808 |
0.17 |
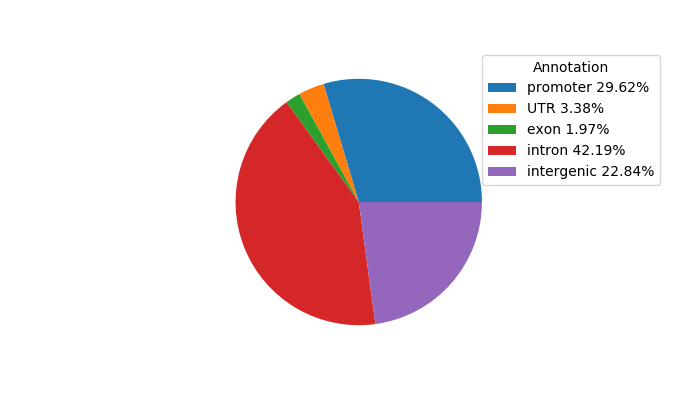
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0003431 | growth plate cartilage chondrocyte development(GO:0003431) |
| 0.1 | 0.2 | GO:0051697 | protein delipidation(GO:0051697) |
| 0.1 | 0.3 | GO:0044789 | modulation by host of viral release from host cell(GO:0044789) positive regulation by host of viral release from host cell(GO:0044791) |
| 0.0 | 0.2 | GO:0022007 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.0 | 0.2 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.0 | 0.1 | GO:0021827 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) postnatal olfactory bulb interneuron migration(GO:0021827) |
| 0.0 | 0.1 | GO:1903279 | regulation of calcium:sodium antiporter activity(GO:1903279) |
| 0.0 | 0.1 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.0 | 0.1 | GO:1904179 | positive regulation of adipose tissue development(GO:1904179) |
| 0.0 | 0.1 | GO:0007354 | zygotic determination of anterior/posterior axis, embryo(GO:0007354) establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.0 | 0.1 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.0 | 0.1 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.0 | 0.6 | GO:0034198 | cellular response to amino acid starvation(GO:0034198) |
| 0.0 | 0.1 | GO:1900060 | negative regulation of ceramide biosynthetic process(GO:1900060) |
| 0.0 | 0.1 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.0 | 0.1 | GO:0006046 | N-acetylglucosamine catabolic process(GO:0006046) |
| 0.0 | 0.1 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.0 | 0.1 | GO:0090166 | Golgi disassembly(GO:0090166) |
| 0.0 | 0.2 | GO:0070863 | positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.0 | 0.1 | GO:0015819 | lysine transport(GO:0015819) |
| 0.0 | 0.1 | GO:0060166 | olfactory pit development(GO:0060166) |
| 0.0 | 0.1 | GO:0048294 | negative regulation of isotype switching to IgE isotypes(GO:0048294) |
| 0.0 | 0.1 | GO:0010482 | epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 0.0 | 0.2 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.0 | 0.1 | GO:0044849 | estrous cycle(GO:0044849) |
| 0.0 | 0.0 | GO:0046292 | formaldehyde metabolic process(GO:0046292) |
| 0.0 | 0.1 | GO:0002326 | B cell lineage commitment(GO:0002326) |
| 0.0 | 0.1 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.0 | 0.1 | GO:0010288 | response to lead ion(GO:0010288) |
| 0.0 | 0.1 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.0 | 0.1 | GO:1903300 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.0 | 0.1 | GO:0010572 | positive regulation of platelet activation(GO:0010572) |
| 0.0 | 0.0 | GO:1903334 | positive regulation of protein folding(GO:1903334) |
| 0.0 | 0.1 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.0 | 0.1 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.0 | 0.2 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.0 | 0.0 | GO:0035880 | embryonic nail plate morphogenesis(GO:0035880) |
| 0.0 | 0.1 | GO:0090267 | positive regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090267) |
| 0.0 | 0.1 | GO:0010838 | positive regulation of keratinocyte proliferation(GO:0010838) |
| 0.0 | 0.1 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 0.1 | GO:1904970 | brush border assembly(GO:1904970) |
| 0.0 | 0.1 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.0 | 0.0 | GO:0071314 | cellular response to cocaine(GO:0071314) |
| 0.0 | 0.0 | GO:1905049 | negative regulation of metalloendopeptidase activity(GO:1904684) negative regulation of metallopeptidase activity(GO:1905049) |
| 0.0 | 0.1 | GO:0021891 | olfactory bulb interneuron development(GO:0021891) |
| 0.0 | 0.0 | GO:0097252 | oligodendrocyte apoptotic process(GO:0097252) |
| 0.0 | 0.0 | GO:1900211 | mesenchymal cell apoptotic process involved in metanephros development(GO:1900200) regulation of mesenchymal cell apoptotic process involved in metanephros development(GO:1900211) negative regulation of mesenchymal cell apoptotic process involved in metanephros development(GO:1900212) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.0 | 0.1 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 0.0 | 0.2 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.0 | 0.1 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.0 | 0.1 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.2 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.0 | GO:0097418 | neurofibrillary tangle(GO:0097418) |
| 0.0 | 0.1 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.1 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.0 | 0.1 | GO:0090533 | cation-transporting ATPase complex(GO:0090533) |
| 0.0 | 0.1 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 0.0 | GO:0032010 | phagolysosome(GO:0032010) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.1 | 0.3 | GO:0004558 | alpha-1,4-glucosidase activity(GO:0004558) |
| 0.0 | 0.1 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.0 | 0.1 | GO:0038181 | bile acid receptor activity(GO:0038181) |
| 0.0 | 0.1 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.1 | GO:0016743 | carboxyl- or carbamoyltransferase activity(GO:0016743) |
| 0.0 | 0.1 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.1 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.0 | 0.1 | GO:0043426 | MRF binding(GO:0043426) |
| 0.0 | 0.1 | GO:0015189 | L-ornithine transmembrane transporter activity(GO:0000064) arginine transmembrane transporter activity(GO:0015181) L-lysine transmembrane transporter activity(GO:0015189) |
| 0.0 | 0.1 | GO:1990239 | steroid hormone binding(GO:1990239) |
| 0.0 | 0.0 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.0 | 0.1 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.1 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.0 | 0.2 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 0.1 | GO:0103116 | alpha-D-galactofuranose transporter activity(GO:0103116) |
| 0.0 | 0.0 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.0 | 0.1 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.1 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.4 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.2 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.1 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.1 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 0.1 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |