Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
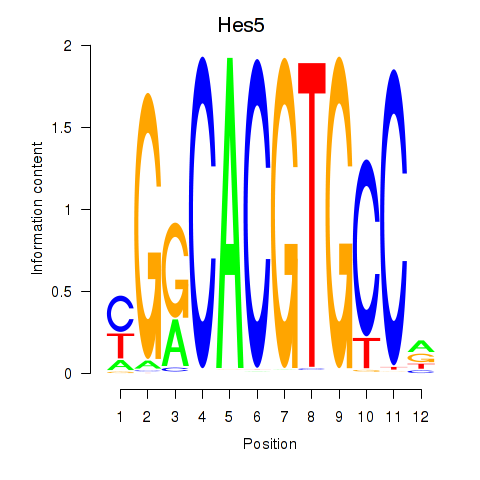
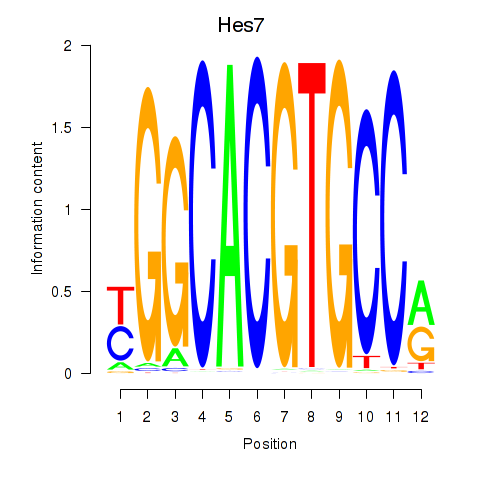
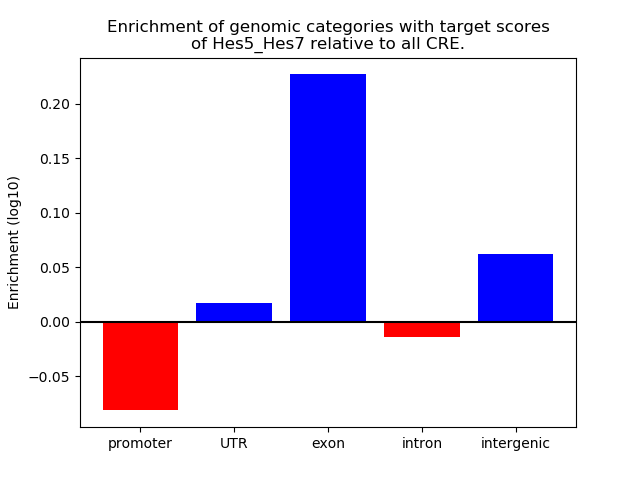
Results for Hes5_Hes7
Z-value: 2.64
Transcription factors associated with Hes5_Hes7
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hes5
|
ENSMUSG00000048001.7 | hes family bHLH transcription factor 5 |
|
Hes7
|
ENSMUSG00000023781.2 | hes family bHLH transcription factor 7 |
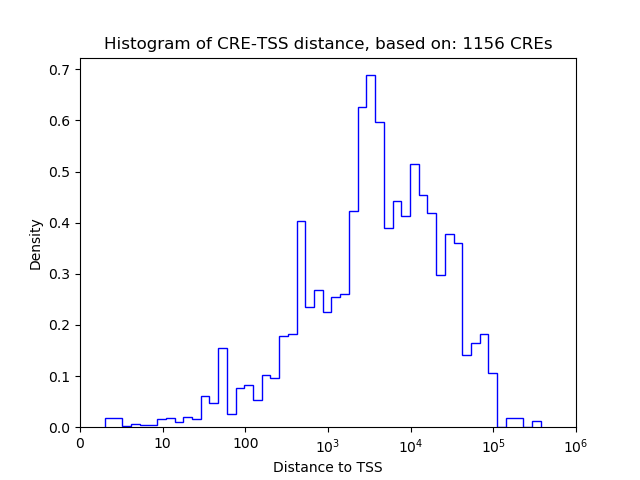
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr4_154946325_154946641 | Hes5 | 14440 | 0.105527 | 0.47 | 3.5e-01 | Click! |
| chr4_154946732_154946894 | Hes5 | 14110 | 0.105940 | 0.45 | 3.8e-01 | Click! |
| chr11_69114771_69114928 | Hes7 | 5555 | 0.080045 | 0.91 | 1.2e-02 | Click! |
| chr11_69110217_69110963 | Hes7 | 9814 | 0.070555 | 0.70 | 1.2e-01 | Click! |
| chr11_69115062_69115266 | Hes7 | 5240 | 0.081385 | -0.64 | 1.7e-01 | Click! |
| chr11_69111108_69111280 | Hes7 | 9210 | 0.071467 | -0.53 | 2.8e-01 | Click! |
| chr11_69118830_69118983 | Hes7 | 1498 | 0.185923 | -0.52 | 2.9e-01 | Click! |
Activity of the Hes5_Hes7 motif across conditions
Conditions sorted by the z-value of the Hes5_Hes7 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
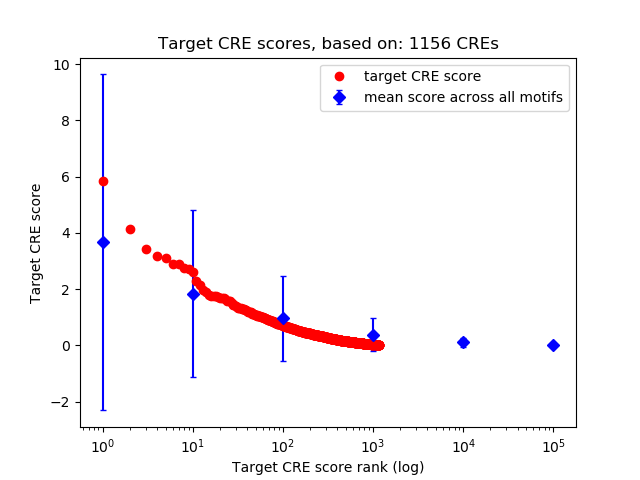
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr17_47329074_47329231 | 5.83 |
Trerf1 |
transcriptional regulating factor 1 |
35083 |
0.1 |
| chr6_116653898_116654072 | 4.13 |
Depp1 |
DEPP1 autophagy regulator |
3289 |
0.14 |
| chr12_85748786_85748937 | 3.43 |
Flvcr2 |
feline leukemia virus subgroup C cellular receptor 2 |
2322 |
0.22 |
| chr4_124005727_124005878 | 3.19 |
Gm12902 |
predicted gene 12902 |
79568 |
0.07 |
| chr16_17802864_17803015 | 3.10 |
Scarf2 |
scavenger receptor class F, member 2 |
463 |
0.67 |
| chr3_90473370_90473521 | 2.90 |
Gm43595 |
predicted gene 43595 |
2486 |
0.12 |
| chr5_113204337_113204488 | 2.89 |
2900026A02Rik |
RIKEN cDNA 2900026A02 gene |
16824 |
0.12 |
| chr11_55126402_55126555 | 2.73 |
Mup-ps22 |
major urinary protein, pseudogene 22 |
2783 |
0.16 |
| chr1_40217644_40217810 | 2.72 |
Il1r1 |
interleukin 1 receptor, type I |
7353 |
0.21 |
| chr12_82743501_82743658 | 2.62 |
Gm22149 |
predicted gene, 22149 |
28877 |
0.22 |
| chr16_93793312_93793530 | 2.28 |
Dop1b |
DOP1 leucine zipper like protein B |
632 |
0.59 |
| chr7_64186009_64186191 | 2.13 |
Trpm1 |
transient receptor potential cation channel, subfamily M, member 1 |
499 |
0.73 |
| chr13_73720273_73720547 | 1.98 |
Slc12a7 |
solute carrier family 12, member 7 |
12684 |
0.14 |
| chr12_110688126_110688290 | 1.89 |
Hsp90aa1 |
heat shock protein 90, alpha (cytosolic), class A member 1 |
4972 |
0.13 |
| chr16_23293902_23294209 | 1.79 |
St6gal1 |
beta galactoside alpha 2,6 sialyltransferase 1 |
3585 |
0.22 |
| chr11_95223583_95223772 | 1.76 |
Gm11515 |
predicted gene 11515 |
1955 |
0.26 |
| chr7_110969269_110969430 | 1.75 |
Mrvi1 |
MRV integration site 1 |
12709 |
0.18 |
| chr10_19443595_19443775 | 1.74 |
Gm33104 |
predicted gene, 33104 |
43030 |
0.15 |
| chr14_63479379_63479556 | 1.73 |
Gm47074 |
predicted gene, 47074 |
4091 |
0.16 |
| chr8_69822995_69823178 | 1.69 |
Lpar2 |
lysophosphatidic acid receptor 2 |
502 |
0.65 |
| chr13_59767243_59767411 | 1.69 |
Isca1 |
iron-sulfur cluster assembly 1 |
2150 |
0.13 |
| chr4_45487423_45487587 | 1.67 |
Shb |
src homology 2 domain-containing transforming protein B |
1898 |
0.29 |
| chr9_58112850_58113072 | 1.66 |
Ccdc33 |
coiled-coil domain containing 33 |
1399 |
0.31 |
| chr11_115249882_115250043 | 1.59 |
Gm25837 |
predicted gene, 25837 |
3316 |
0.14 |
| chr10_85829073_85829241 | 1.56 |
Pwp1 |
PWP1 homolog, endonuclein |
337 |
0.6 |
| chr1_74121947_74122123 | 1.56 |
Tns1 |
tensin 1 |
2412 |
0.22 |
| chr12_98919687_98919838 | 1.51 |
Ttc8 |
tetratricopeptide repeat domain 8 |
812 |
0.61 |
| chr11_118265609_118265912 | 1.45 |
Usp36 |
ubiquitin specific peptidase 36 |
288 |
0.89 |
| chr6_54760026_54760215 | 1.42 |
Znrf2 |
zinc and ring finger 2 |
56796 |
0.1 |
| chr2_119322511_119322662 | 1.40 |
Gm14207 |
predicted gene 14207 |
2957 |
0.15 |
| chr11_88849470_88849637 | 1.37 |
Akap1 |
A kinase (PRKA) anchor protein 1 |
1950 |
0.25 |
| chr8_119901427_119901603 | 1.33 |
Usp10 |
ubiquitin specific peptidase 10 |
8845 |
0.16 |
| chr6_112939448_112939605 | 1.32 |
Srgap3 |
SLIT-ROBO Rho GTPase activating protein 3 |
7228 |
0.15 |
| chr1_157594710_157594868 | 1.31 |
Gm37682 |
predicted gene, 37682 |
1517 |
0.33 |
| chr17_56780692_56780856 | 1.31 |
Rfx2 |
regulatory factor X, 2 (influences HLA class II expression) |
2930 |
0.15 |
| chrX_12103317_12103582 | 1.29 |
Bcor |
BCL6 interacting corepressor |
22896 |
0.23 |
| chr13_113032070_113032240 | 1.28 |
Cdc20b |
cell division cycle 20B |
2956 |
0.12 |
| chr2_172580659_172580839 | 1.27 |
Tfap2c |
transcription factor AP-2, gamma |
28633 |
0.18 |
| chr11_75213340_75213711 | 1.27 |
Rtn4rl1 |
reticulon 4 receptor-like 1 |
19742 |
0.1 |
| chr9_59664998_59665149 | 1.23 |
Pkm |
pyruvate kinase, muscle |
271 |
0.87 |
| chr6_56708894_56709045 | 1.20 |
Lsm5 |
LSM5 homolog, U6 small nuclear RNA and mRNA degradation associated |
4259 |
0.2 |
| chr5_51391034_51391201 | 1.19 |
Gm42614 |
predicted gene 42614 |
93188 |
0.07 |
| chr2_146247712_146247863 | 1.19 |
A930019D19Rik |
RIKEN cDNA A930019D19 gene |
11499 |
0.16 |
| chr16_97912494_97912647 | 1.17 |
C2cd2 |
C2 calcium-dependent domain containing 2 |
10036 |
0.18 |
| chr6_141943721_141944090 | 1.14 |
Slco1a1 |
solute carrier organic anion transporter family, member 1a1 |
2905 |
0.31 |
| chr19_44881162_44881335 | 1.12 |
Gm5246 |
predicted gene 5246 |
30239 |
0.11 |
| chr11_100830367_100830521 | 1.11 |
Stat5b |
signal transducer and activator of transcription 5B |
7916 |
0.13 |
| chr5_103976460_103976626 | 1.10 |
Hsd17b13 |
hydroxysteroid (17-beta) dehydrogenase 13 |
783 |
0.55 |
| chr13_51918889_51919046 | 1.09 |
Gm26651 |
predicted gene, 26651 |
54247 |
0.13 |
| chr1_180757970_180758155 | 1.07 |
Gm37768 |
predicted gene, 37768 |
2226 |
0.19 |
| chr8_88361045_88361206 | 1.07 |
Brd7 |
bromodomain containing 7 |
889 |
0.61 |
| chr16_31404159_31404311 | 1.06 |
Gm49736 |
predicted gene, 49736 |
8232 |
0.12 |
| chr1_22645397_22645563 | 1.06 |
Gm26866 |
predicted gene, 26866 |
95284 |
0.09 |
| chr17_56408931_56409082 | 1.04 |
Ptprs |
protein tyrosine phosphatase, receptor type, S |
5424 |
0.15 |
| chr7_127922875_127923045 | 1.03 |
Kat8 |
K(lysine) acetyltransferase 8 |
3172 |
0.09 |
| chr10_115586794_115587149 | 1.03 |
Lgr5 |
leucine rich repeat containing G protein coupled receptor 5 |
522 |
0.79 |
| chr9_107655076_107655227 | 1.01 |
Slc38a3 |
solute carrier family 38, member 3 |
869 |
0.34 |
| chr7_142081836_142082027 | 1.00 |
Gm25416 |
predicted gene, 25416 |
3494 |
0.11 |
| chr2_172511032_172511203 | 1.00 |
Gm14303 |
predicted gene 14303 |
1426 |
0.37 |
| chr13_43212570_43212901 | 1.00 |
Tbc1d7 |
TBC1 domain family, member 7 |
41234 |
0.15 |
| chr7_127930738_127930911 | 0.98 |
Prss8 |
protease, serine 8 (prostasin) |
720 |
0.39 |
| chr6_86129871_86130022 | 0.98 |
Gm19596 |
predicted gene, 19596 |
17206 |
0.14 |
| chr14_13356089_13356268 | 0.97 |
Gm15913 |
predicted gene 15913 |
2098 |
0.41 |
| chr18_38992299_38992593 | 0.96 |
Arhgap26 |
Rho GTPase activating protein 26 |
699 |
0.63 |
| chr11_107043437_107043599 | 0.95 |
Bptf |
bromodomain PHD finger transcription factor |
188 |
0.93 |
| chr1_74396566_74396720 | 0.94 |
Mir26b |
microRNA 26b |
2333 |
0.16 |
| chr1_168459622_168459790 | 0.94 |
Pbx1 |
pre B cell leukemia homeobox 1 |
27436 |
0.2 |
| chr4_148074902_148075079 | 0.91 |
Gm13201 |
predicted gene 13201 |
430 |
0.69 |
| chr4_72270584_72270739 | 0.91 |
C630043F03Rik |
RIKEN cDNA C630043F03 gene |
69313 |
0.11 |
| chr6_84583452_84583643 | 0.91 |
Cyp26b1 |
cytochrome P450, family 26, subfamily b, polypeptide 1 |
4133 |
0.29 |
| chr11_55117495_55117662 | 0.89 |
Atp5pb-ps |
ATP synthase peripheral stalk-membrane subunit b, pseudogene |
1217 |
0.33 |
| chr11_97958258_97958415 | 0.89 |
Gm11633 |
predicted gene 11633 |
10456 |
0.1 |
| chr10_21046859_21047052 | 0.87 |
Gm20149 |
predicted gene, 20149 |
1944 |
0.28 |
| chr5_76063442_76063602 | 0.87 |
Gm6051 |
predicted gene 6051 |
2759 |
0.24 |
| chr2_168003546_168003752 | 0.86 |
Gm14236 |
predicted gene 14236 |
6880 |
0.16 |
| chr4_126602196_126602347 | 0.85 |
5730409E04Rik |
RIKEN cDNA 5730409E04Rik gene |
7547 |
0.12 |
| chr3_104523724_104523899 | 0.85 |
Lrig2 |
leucine-rich repeats and immunoglobulin-like domains 2 |
11893 |
0.13 |
| chr17_25274284_25274449 | 0.84 |
Ube2i |
ubiquitin-conjugating enzyme E2I |
56 |
0.93 |
| chr14_61125286_61125469 | 0.83 |
Sacs |
sacsin |
13080 |
0.21 |
| chr10_17653872_17654032 | 0.82 |
Gm47771 |
predicted gene, 47771 |
4561 |
0.27 |
| chr17_5926825_5927009 | 0.82 |
Gm8376 |
predicted gene 8376 |
12170 |
0.17 |
| chr5_103776042_103776193 | 0.80 |
Aff1 |
AF4/FMR2 family, member 1 |
8242 |
0.22 |
| chr16_4415377_4415550 | 0.80 |
Adcy9 |
adenylate cyclase 9 |
4124 |
0.25 |
| chr6_32528153_32528323 | 0.80 |
Plxna4os1 |
plexin A4, opposite strand 1 |
17165 |
0.22 |
| chr3_85829172_85829346 | 0.79 |
Fam160a1 |
family with sequence similarity 160, member A1 |
11968 |
0.16 |
| chr4_37093076_37093227 | 0.79 |
Taf9-ps |
TATA-box binding protein associated factor 9, pseudogene |
46723 |
0.17 |
| chr5_139359350_139359501 | 0.77 |
Cyp2w1 |
cytochrome P450, family 2, subfamily w, polypeptide 1 |
4849 |
0.12 |
| chr11_59809129_59809311 | 0.77 |
Flcn |
folliculin |
368 |
0.65 |
| chr10_86685214_86685383 | 0.77 |
1810014B01Rik |
RIKEN cDNA 1810014B01 gene |
227 |
0.82 |
| chr18_38177426_38177598 | 0.77 |
Pcdh1 |
protocadherin 1 |
25651 |
0.12 |
| chr8_123046808_123046972 | 0.76 |
2810013P06Rik |
RIKEN cDNA 2810013P06 gene |
4424 |
0.12 |
| chr7_25220574_25220762 | 0.76 |
Dedd2 |
death effector domain-containing DNA binding protein 2 |
53 |
0.89 |
| chr3_94654096_94654253 | 0.76 |
Tuft1 |
tuftelin 1 |
4626 |
0.11 |
| chr19_3840572_3840723 | 0.75 |
Chka |
choline kinase alpha |
11126 |
0.09 |
| chr12_82856018_82856227 | 0.75 |
1700085C21Rik |
RIKEN cDNA 1700085C21 gene |
83033 |
0.09 |
| chr7_80636985_80637158 | 0.74 |
Gm15880 |
predicted gene 15880 |
1054 |
0.48 |
| chr3_85777976_85778136 | 0.73 |
Fam160a1 |
family with sequence similarity 160, member A1 |
31790 |
0.16 |
| chr14_45983632_45983829 | 0.73 |
Gm49192 |
predicted gene, 49192 |
7307 |
0.19 |
| chr9_65289050_65289226 | 0.72 |
Clpx |
caseinolytic mitochondrial matrix peptidase chaperone subunit |
5122 |
0.12 |
| chr13_69002748_69002930 | 0.70 |
Gm35161 |
predicted gene, 35161 |
2417 |
0.22 |
| chr14_25506802_25506986 | 0.70 |
Zmiz1 |
zinc finger, MIZ-type containing 1 |
4239 |
0.18 |
| chr14_31128387_31128552 | 0.70 |
Smim4 |
small integral membrane protein 4 |
369 |
0.76 |
| chr6_7882729_7882890 | 0.70 |
Gm22276 |
predicted gene, 22276 |
3246 |
0.2 |
| chr19_7180817_7180970 | 0.70 |
Otub1 |
OTU domain, ubiquitin aldehyde binding 1 |
19571 |
0.12 |
| chr15_99716487_99716652 | 0.69 |
Gpd1 |
glycerol-3-phosphate dehydrogenase 1 (soluble) |
946 |
0.3 |
| chr16_77551147_77551350 | 0.68 |
Gm37606 |
predicted gene, 37606 |
10402 |
0.12 |
| chr5_39219850_39220015 | 0.67 |
Gm40293 |
predicted gene, 40293 |
87152 |
0.08 |
| chr15_88850581_88850743 | 0.67 |
Pim3 |
proviral integration site 3 |
11524 |
0.13 |
| chr10_81057523_81057689 | 0.67 |
Sgta |
small glutamine-rich tetratricopeptide repeat (TPR)-containing, alpha |
2509 |
0.12 |
| chr15_81523272_81523474 | 0.66 |
Gm5218 |
predicted gene 5218 |
23828 |
0.11 |
| chr7_67709950_67710121 | 0.66 |
Ttc23 |
tetratricopeptide repeat domain 23 |
15226 |
0.13 |
| chr11_59787617_59787784 | 0.66 |
Pld6 |
phospholipase D family, member 6 |
55 |
0.95 |
| chr4_148868111_148868290 | 0.65 |
Casz1 |
castor zinc finger 1 |
21181 |
0.18 |
| chr9_26924278_26924451 | 0.65 |
Gm1110 |
predicted gene 1110 |
1253 |
0.44 |
| chr11_101066041_101066195 | 0.65 |
Naglu |
alpha-N-acetylglucosaminidase (Sanfilippo disease IIIB) |
3894 |
0.1 |
| chr4_136168101_136168252 | 0.64 |
E2f2 |
E2F transcription factor 2 |
4218 |
0.16 |
| chr11_59802025_59802176 | 0.63 |
Flcn |
folliculin |
7488 |
0.1 |
| chr15_38569454_38569605 | 0.63 |
Gm29697 |
predicted gene, 29697 |
4467 |
0.14 |
| chr2_132639484_132639653 | 0.63 |
AU019990 |
expressed sequence AU019990 |
6313 |
0.14 |
| chr7_49351868_49352264 | 0.63 |
Gm44913 |
predicted gene 44913 |
5972 |
0.24 |
| chr13_92846534_92846735 | 0.62 |
Mtx3 |
metaxin 3 |
1436 |
0.47 |
| chr2_148022052_148022231 | 0.62 |
9030622O22Rik |
RIKEN cDNA 9030622O22 gene |
16129 |
0.17 |
| chr8_125931903_125932065 | 0.62 |
Map3k21 |
mitogen-activated protein kinase kinase kinase 21 |
21534 |
0.17 |
| chr11_60015685_60015836 | 0.62 |
Pemt |
phosphatidylethanolamine N-methyltransferase |
5466 |
0.18 |
| chr10_80256725_80256907 | 0.60 |
Gamt |
guanidinoacetate methyltransferase |
2473 |
0.11 |
| chr6_29650247_29650407 | 0.60 |
Tnpo3 |
transportin 3 |
40440 |
0.11 |
| chr15_90992378_90992671 | 0.59 |
Kif21a |
kinesin family member 21A |
26122 |
0.16 |
| chr14_76812275_76812477 | 0.59 |
Gm48968 |
predicted gene, 48968 |
20445 |
0.17 |
| chr9_107289417_107289626 | 0.59 |
Mapkapk3 |
mitogen-activated protein kinase-activated protein kinase 3 |
177 |
0.89 |
| chr6_124813558_124813734 | 0.58 |
Tpi1 |
triosephosphate isomerase 1 |
508 |
0.53 |
| chr16_30064275_30064431 | 0.58 |
Hes1 |
hes family bHLH transcription factor 1 |
31 |
0.97 |
| chr13_113168060_113168232 | 0.58 |
Gzmk |
granzyme K |
12751 |
0.12 |
| chr18_11050629_11050793 | 0.57 |
Gata6os |
GATA binding protein 6, opposite strand |
776 |
0.58 |
| chr8_111148892_111149043 | 0.57 |
9430091E24Rik |
RIKEN cDNA 9430091E24 gene |
3487 |
0.18 |
| chr12_3862523_3862680 | 0.57 |
Dnmt3aos |
DNA methyltransferase 3A, opposite strand |
357 |
0.82 |
| chr3_9599812_9599994 | 0.57 |
Zfp704 |
zinc finger protein 704 |
10182 |
0.23 |
| chr11_3931044_3931195 | 0.56 |
Tcn2 |
transcobalamin 2 |
670 |
0.52 |
| chr5_122239684_122239867 | 0.56 |
Tctn1 |
tectonic family member 1 |
11903 |
0.12 |
| chr17_47463129_47463289 | 0.55 |
1700001C19Rik |
RIKEN cDNA 1700001C19 gene |
25833 |
0.1 |
| chr11_60932286_60932492 | 0.55 |
Map2k3 |
mitogen-activated protein kinase kinase 3 |
323 |
0.62 |
| chr18_61647742_61648023 | 0.55 |
Mir145a |
microRNA 145a |
12 |
0.95 |
| chr15_74704343_74704514 | 0.54 |
4933427E11Rik |
RIKEN cDNA 4933427E11 gene |
4733 |
0.09 |
| chr7_15980408_15980576 | 0.54 |
Bicra |
BRD4 interacting chromatin remodeling complex associated protein |
1767 |
0.22 |
| chr2_114775396_114775571 | 0.53 |
Gm13975 |
predicted gene 13975 |
67371 |
0.11 |
| chr9_46112348_46112526 | 0.53 |
Sik3 |
SIK family kinase 3 |
10693 |
0.18 |
| chr5_8992782_8992933 | 0.53 |
Crot |
carnitine O-octanoyltransferase |
4185 |
0.14 |
| chr18_84376797_84376986 | 0.53 |
Gm37216 |
predicted gene, 37216 |
745 |
0.73 |
| chr6_126106995_126107182 | 0.52 |
Ntf3 |
neurotrophin 3 |
57872 |
0.15 |
| chr7_141456887_141457063 | 0.52 |
Pnpla2 |
patatin-like phospholipase domain containing 2 |
43 |
0.92 |
| chr19_37583528_37583701 | 0.52 |
Exoc6 |
exocyst complex component 6 |
33155 |
0.16 |
| chr9_20730600_20730850 | 0.52 |
Olfm2 |
olfactomedin 2 |
2496 |
0.23 |
| chr1_84758953_84759122 | 0.51 |
Trip12 |
thyroid hormone receptor interactor 12 |
1147 |
0.47 |
| chr16_31996383_31996610 | 0.51 |
Hmgb1-ps6 |
high mobility group box 1, pseudogene 6 |
4628 |
0.09 |
| chr6_140674472_140674632 | 0.51 |
Aebp2 |
AE binding protein 2 |
27735 |
0.16 |
| chr16_87710092_87710297 | 0.51 |
Bach1 |
BTB and CNC homology 1, basic leucine zipper transcription factor 1 |
11190 |
0.21 |
| chr13_28507894_28508045 | 0.51 |
Gm6081 |
predicted gene 6081 |
3263 |
0.28 |
| chr12_112229300_112229492 | 0.50 |
Gm20368 |
predicted gene, 20368 |
24243 |
0.16 |
| chr4_133563360_133563542 | 0.50 |
Gm23158 |
predicted gene, 23158 |
4502 |
0.11 |
| chr4_150371789_150371980 | 0.50 |
Rere |
arginine glutamic acid dipeptide (RE) repeats |
34510 |
0.15 |
| chr6_71270702_71270888 | 0.49 |
Mir8112 |
microRNA 8112 |
876 |
0.31 |
| chr13_119791388_119791687 | 0.49 |
Zfp131 |
zinc finger protein 131 |
648 |
0.58 |
| chr9_56937902_56938069 | 0.49 |
Imp3 |
IMP3, U3 small nucleolar ribonucleoprotein |
510 |
0.64 |
| chr8_111726628_111726784 | 0.48 |
Bcar1 |
breast cancer anti-estrogen resistance 1 |
5399 |
0.19 |
| chr19_5038738_5038898 | 0.48 |
B4gat1 |
beta-1,4-glucuronyltransferase 1 |
8 |
0.53 |
| chr10_7268902_7269053 | 0.47 |
Cnksr3 |
Cnksr family member 3 |
56740 |
0.14 |
| chr11_67024896_67025068 | 0.47 |
Tmem220 |
transmembrane protein 220 |
172 |
0.93 |
| chrX_7679840_7680006 | 0.47 |
Magix |
MAGI family member, X-linked |
1166 |
0.23 |
| chr2_35233951_35234102 | 0.47 |
Gm13605 |
predicted gene 13605 |
9493 |
0.14 |
| chr1_120061492_120061672 | 0.47 |
Tmem37 |
transmembrane protein 37 |
12492 |
0.17 |
| chr10_44474418_44474575 | 0.47 |
Prdm1 |
PR domain containing 1, with ZNF domain |
15748 |
0.18 |
| chr17_25231680_25231997 | 0.46 |
Gnptg |
N-acetylglucosamine-1-phosphotransferase, gamma subunit |
8278 |
0.08 |
| chr7_142622273_142622480 | 0.46 |
Gm33148 |
predicted gene, 33148 |
30632 |
0.08 |
| chr2_26576085_26576254 | 0.46 |
Egfl7 |
EGF-like domain 7 |
3845 |
0.11 |
| chr13_45570061_45570212 | 0.46 |
Gmpr |
guanosine monophosphate reductase |
23983 |
0.23 |
| chr15_83772573_83772737 | 0.46 |
Mpped1 |
metallophosphoesterase domain containing 1 |
6812 |
0.24 |
| chr13_45406872_45407023 | 0.46 |
Mylip |
myosin regulatory light chain interacting protein |
16020 |
0.2 |
| chr12_75473113_75473300 | 0.45 |
Gm47690 |
predicted gene, 47690 |
35539 |
0.17 |
| chr17_29058068_29058237 | 0.45 |
Gm41556 |
predicted gene, 41556 |
77 |
0.94 |
| chr6_91424316_91424467 | 0.45 |
Gm4575 |
predicted gene 4575 |
4492 |
0.12 |
| chr8_71306147_71306308 | 0.45 |
Myo9b |
myosin IXb |
27590 |
0.11 |
| chr4_8400514_8400698 | 0.44 |
Gm37386 |
predicted gene, 37386 |
53641 |
0.14 |
| chr11_100819720_100819912 | 0.44 |
Stat5b |
signal transducer and activator of transcription 5B |
2712 |
0.19 |
| chr6_31081370_31081547 | 0.44 |
Lncpint |
long non-protein coding RNA, Trp53 induced transcript |
6146 |
0.12 |
| chr18_46455190_46455491 | 0.44 |
Mospd4 |
motile sperm domain containing 4 |
10450 |
0.14 |
| chr18_12869001_12869319 | 0.44 |
Osbpl1a |
oxysterol binding protein-like 1A |
53 |
0.97 |
| chr17_85040351_85040697 | 0.44 |
Slc3a1 |
solute carrier family 3, member 1 |
12148 |
0.18 |
| chr5_121775382_121775533 | 0.44 |
Atxn2 |
ataxin 2 |
2472 |
0.19 |
| chr8_124750214_124750687 | 0.44 |
Fam89a |
family with sequence similarity 89, member A |
1382 |
0.3 |
| chr7_79466451_79467019 | 0.44 |
Gm10616 |
predicted gene 10616 |
326 |
0.49 |
| chr13_45917431_45917595 | 0.44 |
4930453C13Rik |
RIKEN cDNA 4930453C13 gene |
18527 |
0.2 |
| chr10_120391230_120391401 | 0.43 |
9230105E05Rik |
RIKEN cDNA 9230105E05 gene |
1792 |
0.33 |
| chr15_67405630_67405995 | 0.43 |
1700012I11Rik |
RIKEN cDNA 1700012I11 gene |
179043 |
0.03 |
| chr15_84121144_84121320 | 0.43 |
Pnpla5 |
patatin-like phospholipase domain containing 5 |
1943 |
0.2 |
| chr9_71663328_71663759 | 0.43 |
Cgnl1 |
cingulin-like 1 |
15193 |
0.21 |
| chr12_109501140_109501291 | 0.43 |
Gm34081 |
predicted gene, 34081 |
15525 |
0.06 |
| chr7_80202018_80202199 | 0.43 |
Sema4b |
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4B |
3572 |
0.12 |
| chr16_18877584_18877767 | 0.42 |
Hira |
histone cell cycle regulator |
263 |
0.79 |
| chr9_23030248_23030409 | 0.42 |
Bmper |
BMP-binding endothelial regulator |
192748 |
0.03 |
| chr11_61974443_61974594 | 0.42 |
Gm12274 |
predicted gene 12274 |
168 |
0.92 |
| chr10_75937006_75937161 | 0.42 |
Chchd10 |
coiled-coil-helix-coiled-coil-helix domain containing 10 |
137 |
0.81 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:1903261 | regulation of serine phosphorylation of STAT3 protein(GO:1903261) |
| 0.2 | 0.6 | GO:0045608 | negative regulation of auditory receptor cell differentiation(GO:0045608) |
| 0.2 | 0.6 | GO:1901856 | negative regulation of cellular respiration(GO:1901856) |
| 0.2 | 0.7 | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.2 | 1.1 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 0.2 | 0.6 | GO:2000659 | regulation of interleukin-1-mediated signaling pathway(GO:2000659) |
| 0.1 | 0.3 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.1 | 1.8 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.1 | 0.4 | GO:0046544 | development of secondary male sexual characteristics(GO:0046544) |
| 0.1 | 0.4 | GO:0097503 | sialylation(GO:0097503) |
| 0.1 | 0.3 | GO:2001016 | positive regulation of skeletal muscle cell differentiation(GO:2001016) |
| 0.1 | 0.3 | GO:0006868 | glutamine transport(GO:0006868) |
| 0.1 | 0.7 | GO:0010635 | regulation of mitochondrial fusion(GO:0010635) |
| 0.1 | 0.4 | GO:1900194 | negative regulation of oocyte maturation(GO:1900194) |
| 0.1 | 0.5 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.1 | 0.6 | GO:0044351 | macropinocytosis(GO:0044351) |
| 0.1 | 0.7 | GO:0031282 | regulation of guanylate cyclase activity(GO:0031282) |
| 0.1 | 0.3 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.1 | 0.4 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.1 | 0.3 | GO:1900045 | negative regulation of protein K63-linked ubiquitination(GO:1900045) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.1 | 0.3 | GO:0021586 | pons maturation(GO:0021586) |
| 0.1 | 0.5 | GO:0032792 | negative regulation of CREB transcription factor activity(GO:0032792) |
| 0.1 | 0.4 | GO:1902570 | protein localization to nucleolus(GO:1902570) |
| 0.1 | 0.2 | GO:0045583 | regulation of cytotoxic T cell differentiation(GO:0045583) positive regulation of cytotoxic T cell differentiation(GO:0045585) |
| 0.1 | 0.3 | GO:0061074 | regulation of neural retina development(GO:0061074) |
| 0.1 | 0.2 | GO:0036515 | serotonergic neuron axon guidance(GO:0036515) |
| 0.1 | 0.3 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.1 | 0.3 | GO:0065001 | specification of axis polarity(GO:0065001) |
| 0.1 | 0.2 | GO:0021555 | midbrain-hindbrain boundary morphogenesis(GO:0021555) |
| 0.1 | 0.3 | GO:0060839 | endothelial cell fate commitment(GO:0060839) |
| 0.1 | 0.9 | GO:0033235 | positive regulation of protein sumoylation(GO:0033235) |
| 0.1 | 0.9 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.1 | 0.3 | GO:0006538 | glutamate catabolic process(GO:0006538) |
| 0.1 | 0.2 | GO:0060178 | regulation of exocyst localization(GO:0060178) |
| 0.1 | 0.3 | GO:0060750 | epithelial cell proliferation involved in mammary gland duct elongation(GO:0060750) |
| 0.1 | 0.1 | GO:0006533 | aspartate catabolic process(GO:0006533) |
| 0.1 | 0.4 | GO:0034626 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.1 | 0.5 | GO:0010898 | positive regulation of triglyceride catabolic process(GO:0010898) |
| 0.1 | 0.2 | GO:0046864 | retinol transport(GO:0034633) isoprenoid transport(GO:0046864) terpenoid transport(GO:0046865) |
| 0.1 | 0.2 | GO:0061589 | calcium activated phosphatidylserine scrambling(GO:0061589) |
| 0.1 | 0.2 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.1 | 0.3 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.1 | 0.3 | GO:1902774 | late endosome to lysosome transport(GO:1902774) |
| 0.1 | 0.2 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.1 | 0.1 | GO:0061419 | positive regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061419) |
| 0.1 | 0.2 | GO:0006116 | NADH oxidation(GO:0006116) |
| 0.1 | 0.4 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.1 | 0.2 | GO:0043985 | histone H4-R3 methylation(GO:0043985) |
| 0.1 | 0.2 | GO:0003358 | noradrenergic neuron development(GO:0003358) |
| 0.1 | 0.2 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.1 | 0.2 | GO:0006566 | threonine metabolic process(GO:0006566) |
| 0.1 | 0.3 | GO:0060087 | relaxation of vascular smooth muscle(GO:0060087) |
| 0.0 | 0.1 | GO:0060486 | Clara cell differentiation(GO:0060486) |
| 0.0 | 0.1 | GO:0034729 | histone H3-K79 methylation(GO:0034729) |
| 0.0 | 0.1 | GO:0040031 | snRNA modification(GO:0040031) |
| 0.0 | 0.2 | GO:0015808 | L-alanine transport(GO:0015808) |
| 0.0 | 0.1 | GO:0033058 | directional locomotion(GO:0033058) |
| 0.0 | 0.2 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.0 | 0.9 | GO:0060219 | camera-type eye photoreceptor cell differentiation(GO:0060219) |
| 0.0 | 0.2 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.0 | 0.1 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.0 | 0.3 | GO:0043045 | DNA methylation involved in embryo development(GO:0043045) changes to DNA methylation involved in embryo development(GO:1901538) |
| 0.0 | 0.2 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 | 1.9 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.1 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 0.0 | 0.3 | GO:0046498 | S-adenosylhomocysteine metabolic process(GO:0046498) |
| 0.0 | 0.3 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.0 | 1.4 | GO:1903146 | regulation of mitophagy(GO:1903146) |
| 0.0 | 0.1 | GO:0033030 | negative regulation of neutrophil apoptotic process(GO:0033030) |
| 0.0 | 0.0 | GO:0060137 | maternal process involved in parturition(GO:0060137) |
| 0.0 | 0.1 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.0 | 0.1 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.0 | 0.1 | GO:1990086 | lens fiber cell apoptotic process(GO:1990086) |
| 0.0 | 0.1 | GO:0045347 | negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 0.0 | 0.1 | GO:0061366 | behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 0.0 | 0.3 | GO:0015816 | glycine transport(GO:0015816) |
| 0.0 | 0.5 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.0 | 0.2 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.0 | 0.2 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.0 | 0.2 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.0 | 0.1 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.0 | 0.3 | GO:0030853 | negative regulation of granulocyte differentiation(GO:0030853) |
| 0.0 | 0.1 | GO:0046878 | positive regulation of saliva secretion(GO:0046878) |
| 0.0 | 0.0 | GO:1903431 | positive regulation of cell maturation(GO:1903431) |
| 0.0 | 0.3 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.1 | GO:0002309 | T cell proliferation involved in immune response(GO:0002309) |
| 0.0 | 0.1 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.0 | 0.1 | GO:0022009 | central nervous system vasculogenesis(GO:0022009) |
| 0.0 | 0.1 | GO:0097476 | motor neuron migration(GO:0097475) spinal cord motor neuron migration(GO:0097476) |
| 0.0 | 0.4 | GO:0006098 | pentose-phosphate shunt(GO:0006098) |
| 0.0 | 0.1 | GO:0070544 | histone H3-K36 demethylation(GO:0070544) |
| 0.0 | 0.1 | GO:0034047 | regulation of protein phosphatase type 2A activity(GO:0034047) |
| 0.0 | 0.1 | GO:2000302 | positive regulation of synaptic vesicle exocytosis(GO:2000302) |
| 0.0 | 0.1 | GO:1902262 | apoptotic process involved in patterning of blood vessels(GO:1902262) |
| 0.0 | 0.1 | GO:0007403 | glial cell fate determination(GO:0007403) |
| 0.0 | 0.1 | GO:0071847 | TNFSF11-mediated signaling pathway(GO:0071847) |
| 0.0 | 0.1 | GO:0010989 | negative regulation of low-density lipoprotein particle clearance(GO:0010989) |
| 0.0 | 0.2 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.0 | 0.0 | GO:0044778 | meiotic DNA integrity checkpoint(GO:0044778) |
| 0.0 | 0.1 | GO:1903898 | negative regulation of PERK-mediated unfolded protein response(GO:1903898) |
| 0.0 | 0.1 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.1 | GO:0017187 | peptidyl-glutamic acid carboxylation(GO:0017187) |
| 0.0 | 0.1 | GO:1902202 | regulation of hepatocyte growth factor receptor signaling pathway(GO:1902202) |
| 0.0 | 0.1 | GO:0050703 | interleukin-1 alpha secretion(GO:0050703) |
| 0.0 | 0.1 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.0 | 0.1 | GO:0017182 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.0 | 0.0 | GO:0035771 | interleukin-4-mediated signaling pathway(GO:0035771) |
| 0.0 | 0.2 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.0 | 0.1 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.0 | 0.1 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.0 | 0.1 | GO:0030311 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.0 | 0.0 | GO:0061205 | paramesonephric duct development(GO:0061205) |
| 0.0 | 0.1 | GO:1901475 | pyruvate transmembrane transport(GO:1901475) |
| 0.0 | 0.1 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.0 | 0.2 | GO:0061418 | regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061418) |
| 0.0 | 0.0 | GO:0090283 | regulation of protein glycosylation in Golgi(GO:0090283) |
| 0.0 | 0.3 | GO:0010592 | positive regulation of lamellipodium assembly(GO:0010592) |
| 0.0 | 0.1 | GO:0072675 | osteoclast fusion(GO:0072675) |
| 0.0 | 0.2 | GO:0072540 | T-helper 17 cell lineage commitment(GO:0072540) |
| 0.0 | 0.1 | GO:0051791 | medium-chain fatty acid metabolic process(GO:0051791) |
| 0.0 | 0.1 | GO:0031443 | fast-twitch skeletal muscle fiber contraction(GO:0031443) |
| 0.0 | 0.0 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.0 | 0.1 | GO:0035630 | bone mineralization involved in bone maturation(GO:0035630) |
| 0.0 | 0.5 | GO:0006921 | cellular component disassembly involved in execution phase of apoptosis(GO:0006921) apoptotic nuclear changes(GO:0030262) |
| 0.0 | 0.1 | GO:0032074 | negative regulation of nuclease activity(GO:0032074) |
| 0.0 | 0.2 | GO:0009992 | cellular water homeostasis(GO:0009992) |
| 0.0 | 0.1 | GO:0060298 | positive regulation of sarcomere organization(GO:0060298) |
| 0.0 | 0.2 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.1 | GO:0071895 | odontoblast differentiation(GO:0071895) |
| 0.0 | 0.0 | GO:0071288 | cellular response to mercury ion(GO:0071288) |
| 0.0 | 0.1 | GO:0008228 | opsonization(GO:0008228) |
| 0.0 | 0.3 | GO:0006336 | DNA replication-independent nucleosome assembly(GO:0006336) DNA replication-independent nucleosome organization(GO:0034724) |
| 0.0 | 0.1 | GO:2000418 | positive regulation of eosinophil migration(GO:2000418) |
| 0.0 | 0.3 | GO:0006206 | pyrimidine nucleobase metabolic process(GO:0006206) |
| 0.0 | 0.3 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.1 | GO:1904219 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) regulation of serine C-palmitoyltransferase activity(GO:1904220) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.0 | 0.2 | GO:0014041 | regulation of neuron maturation(GO:0014041) |
| 0.0 | 0.1 | GO:1903300 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.0 | 0.1 | GO:0045006 | DNA deamination(GO:0045006) |
| 0.0 | 0.1 | GO:0046078 | dUMP metabolic process(GO:0046078) |
| 0.0 | 0.1 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.0 | 0.1 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.0 | 0.3 | GO:0070633 | transepithelial transport(GO:0070633) |
| 0.0 | 0.2 | GO:0060576 | intestinal epithelial cell development(GO:0060576) |
| 0.0 | 0.4 | GO:0035066 | positive regulation of histone acetylation(GO:0035066) |
| 0.0 | 0.1 | GO:0019695 | choline metabolic process(GO:0019695) |
| 0.0 | 0.1 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.0 | 0.1 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) |
| 0.0 | 0.1 | GO:0060948 | cardiac vascular smooth muscle cell development(GO:0060948) |
| 0.0 | 0.1 | GO:0010693 | negative regulation of alkaline phosphatase activity(GO:0010693) |
| 0.0 | 0.1 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.0 | 0.2 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.0 | 0.2 | GO:1903405 | protein localization to nuclear body(GO:1903405) protein localization to Cajal body(GO:1904867) regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) protein localization to nucleoplasm(GO:1990173) |
| 0.0 | 0.1 | GO:0010481 | epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 0.0 | 0.1 | GO:0071351 | cellular response to interleukin-18(GO:0071351) |
| 0.0 | 0.1 | GO:0032782 | bile acid secretion(GO:0032782) |
| 0.0 | 0.1 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 0.0 | GO:0097491 | trunk segmentation(GO:0035290) trunk neural crest cell migration(GO:0036484) ventral trunk neural crest cell migration(GO:0036486) sympathetic neuron projection extension(GO:0097490) sympathetic neuron projection guidance(GO:0097491) neural crest cell migration involved in autonomic nervous system development(GO:1901166) |
| 0.0 | 0.1 | GO:0051409 | response to nitrosative stress(GO:0051409) |
| 0.0 | 0.1 | GO:0003383 | apical constriction(GO:0003383) |
| 0.0 | 0.1 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.0 | 0.1 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 0.3 | GO:0072337 | modified amino acid transport(GO:0072337) |
| 0.0 | 0.0 | GO:0007228 | positive regulation of hh target transcription factor activity(GO:0007228) |
| 0.0 | 0.1 | GO:0035728 | response to hepatocyte growth factor(GO:0035728) cellular response to hepatocyte growth factor stimulus(GO:0035729) |
| 0.0 | 0.1 | GO:1900364 | negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.0 | 0.2 | GO:0052697 | xenobiotic glucuronidation(GO:0052697) |
| 0.0 | 0.1 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 | 0.1 | GO:1903071 | positive regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903071) |
| 0.0 | 0.2 | GO:0097034 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 | 0.0 | GO:0035609 | C-terminal protein deglutamylation(GO:0035609) |
| 0.0 | 0.0 | GO:0032349 | positive regulation of aldosterone metabolic process(GO:0032346) positive regulation of aldosterone biosynthetic process(GO:0032349) |
| 0.0 | 0.0 | GO:0090258 | negative regulation of mitochondrial fission(GO:0090258) |
| 0.0 | 0.1 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.0 | 0.1 | GO:1904354 | negative regulation of telomere capping(GO:1904354) |
| 0.0 | 0.5 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.0 | GO:0034442 | regulation of lipoprotein oxidation(GO:0034442) |
| 0.0 | 0.2 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.0 | 0.0 | GO:0036016 | response to interleukin-3(GO:0036015) cellular response to interleukin-3(GO:0036016) |
| 0.0 | 0.1 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.0 | 0.0 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.0 | 0.0 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.0 | 0.0 | GO:0036151 | phosphatidylcholine acyl-chain remodeling(GO:0036151) |
| 0.0 | 0.0 | GO:0030174 | regulation of DNA-dependent DNA replication initiation(GO:0030174) |
| 0.0 | 0.0 | GO:0035509 | negative regulation of myosin-light-chain-phosphatase activity(GO:0035509) |
| 0.0 | 0.1 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.0 | 0.0 | GO:2000020 | positive regulation of male gonad development(GO:2000020) |
| 0.0 | 0.0 | GO:0046684 | response to pyrethroid(GO:0046684) |
| 0.0 | 0.0 | GO:0043309 | regulation of eosinophil degranulation(GO:0043309) |
| 0.0 | 0.1 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.0 | 0.0 | GO:0071335 | hair follicle cell proliferation(GO:0071335) |
| 0.0 | 0.1 | GO:0006362 | transcription elongation from RNA polymerase I promoter(GO:0006362) |
| 0.0 | 0.3 | GO:1903077 | negative regulation of protein localization to plasma membrane(GO:1903077) negative regulation of protein localization to cell periphery(GO:1904376) |
| 0.0 | 0.1 | GO:1904424 | regulation of GTP binding(GO:1904424) |
| 0.0 | 0.0 | GO:2000969 | positive regulation of glutamate receptor signaling pathway(GO:1900451) positive regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000969) |
| 0.0 | 0.1 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 0.0 | 0.2 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 0.0 | 0.0 | GO:0090154 | positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 0.0 | 0.0 | GO:2000847 | negative regulation of steroid hormone secretion(GO:2000832) negative regulation of corticosteroid hormone secretion(GO:2000847) negative regulation of glucocorticoid secretion(GO:2000850) |
| 0.0 | 0.0 | GO:0003223 | ventricular compact myocardium morphogenesis(GO:0003223) |
| 0.0 | 0.1 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.0 | 0.0 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.0 | 0.0 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.0 | 0.0 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.0 | 0.1 | GO:1901387 | positive regulation of voltage-gated calcium channel activity(GO:1901387) |
| 0.0 | 0.0 | GO:0060710 | chorio-allantoic fusion(GO:0060710) |
| 0.0 | 0.0 | GO:0043369 | CD4-positive or CD8-positive, alpha-beta T cell lineage commitment(GO:0043369) |
| 0.0 | 0.0 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.0 | GO:0034653 | diterpenoid catabolic process(GO:0016103) retinoic acid catabolic process(GO:0034653) |
| 0.0 | 0.0 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.0 | 0.1 | GO:0010993 | regulation of ubiquitin homeostasis(GO:0010993) free ubiquitin chain polymerization(GO:0010994) |
| 0.0 | 0.2 | GO:0006264 | mitochondrial DNA replication(GO:0006264) |
| 0.0 | 0.0 | GO:0046439 | cysteine catabolic process(GO:0009093) L-cysteine catabolic process(GO:0019448) L-cysteine metabolic process(GO:0046439) |
| 0.0 | 0.1 | GO:0061370 | testosterone biosynthetic process(GO:0061370) |
| 0.0 | 0.1 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.0 | 0.0 | GO:0060300 | regulation of cytokine activity(GO:0060300) |
| 0.0 | 0.1 | GO:0044818 | mitotic G2 DNA damage checkpoint(GO:0007095) mitotic G2/M transition checkpoint(GO:0044818) |
| 0.0 | 0.1 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.0 | 0.1 | GO:1904321 | response to forskolin(GO:1904321) cellular response to forskolin(GO:1904322) |
| 0.0 | 0.1 | GO:0071493 | cellular response to UV-B(GO:0071493) |
| 0.0 | 0.1 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.0 | 0.1 | GO:0036010 | protein localization to endosome(GO:0036010) |
| 0.0 | 0.2 | GO:0071880 | adenylate cyclase-activating adrenergic receptor signaling pathway(GO:0071880) |
| 0.0 | 0.0 | GO:0060988 | lipid tube assembly(GO:0060988) |
| 0.0 | 0.4 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.0 | 0.1 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.0 | 0.0 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.0 | 0.1 | GO:1901533 | negative regulation of hematopoietic progenitor cell differentiation(GO:1901533) |
| 0.0 | 0.0 | GO:0048208 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.0 | 0.0 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.0 | 0.0 | GO:1902548 | negative regulation of cellular response to vascular endothelial growth factor stimulus(GO:1902548) |
| 0.0 | 0.0 | GO:2000467 | positive regulation of glycogen (starch) synthase activity(GO:2000467) |
| 0.0 | 0.1 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.0 | GO:0009642 | response to light intensity(GO:0009642) |
| 0.0 | 0.0 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.0 | 0.0 | GO:0014873 | response to muscle activity involved in regulation of muscle adaptation(GO:0014873) |
| 0.0 | 0.1 | GO:0071294 | cellular response to zinc ion(GO:0071294) |
| 0.0 | 0.0 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.0 | 0.0 | GO:0090170 | regulation of Golgi inheritance(GO:0090170) |
| 0.0 | 0.1 | GO:0002934 | desmosome organization(GO:0002934) |
| 0.0 | 0.2 | GO:0034198 | cellular response to amino acid starvation(GO:0034198) |
| 0.0 | 0.1 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.0 | 0.0 | GO:0048170 | positive regulation of long-term neuronal synaptic plasticity(GO:0048170) |
| 0.0 | 0.0 | GO:0006203 | dGTP catabolic process(GO:0006203) |
| 0.0 | 0.1 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.0 | 0.0 | GO:0007039 | protein catabolic process in the vacuole(GO:0007039) |
| 0.0 | 0.1 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.0 | 0.1 | GO:0045075 | interleukin-12 biosynthetic process(GO:0042090) regulation of interleukin-12 biosynthetic process(GO:0045075) |
| 0.0 | 0.1 | GO:0043981 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.0 | 0.0 | GO:0072710 | response to hydroxyurea(GO:0072710) cellular response to hydroxyurea(GO:0072711) |
| 0.0 | 0.1 | GO:0060075 | regulation of resting membrane potential(GO:0060075) |
| 0.0 | 0.0 | GO:0016081 | synaptic vesicle docking(GO:0016081) |
| 0.0 | 0.1 | GO:0071670 | smooth muscle cell chemotaxis(GO:0071670) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.5 | GO:0035838 | growing cell tip(GO:0035838) |
| 0.1 | 0.9 | GO:0016589 | NURF complex(GO:0016589) |
| 0.1 | 0.8 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.1 | 0.9 | GO:0034464 | BBSome(GO:0034464) |
| 0.1 | 0.3 | GO:0031074 | nucleocytoplasmic shuttling complex(GO:0031074) |
| 0.1 | 0.3 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.1 | 0.8 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.1 | 0.3 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.1 | 0.3 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.1 | 0.2 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.1 | 0.2 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.1 | 0.2 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.1 | 0.2 | GO:1990131 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.1 | 0.2 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.0 | 0.2 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.0 | 0.2 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.0 | 0.2 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.0 | 0.1 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.7 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.2 | GO:0000235 | astral microtubule(GO:0000235) |
| 0.0 | 0.2 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.0 | 0.1 | GO:0090661 | box H/ACA telomerase RNP complex(GO:0090661) |
| 0.0 | 0.1 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.0 | 0.4 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 0.2 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.1 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.2 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.0 | 0.1 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.0 | 0.2 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.0 | 0.3 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 0.3 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.9 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 0.1 | GO:0005745 | m-AAA complex(GO:0005745) |
| 0.0 | 0.6 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.1 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.0 | 0.1 | GO:0044462 | cell outer membrane(GO:0009279) cell envelope(GO:0030313) external encapsulating structure part(GO:0044462) |
| 0.0 | 0.1 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.1 | GO:0032127 | dense core granule membrane(GO:0032127) |
| 0.0 | 0.1 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 0.1 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 1.1 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.4 | GO:0090544 | BAF-type complex(GO:0090544) |
| 0.0 | 0.3 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.1 | GO:0001674 | female germ cell nucleus(GO:0001674) |
| 0.0 | 0.2 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.0 | 0.1 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 0.2 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.2 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 0.1 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.1 | GO:0005767 | secondary lysosome(GO:0005767) |
| 0.0 | 0.0 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 0.1 | GO:0070419 | nonhomologous end joining complex(GO:0070419) |
| 0.0 | 0.0 | GO:0038037 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor complex(GO:0097648) |
| 0.0 | 0.1 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.0 | 0.3 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.6 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.0 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.0 | 0.2 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.2 | GO:0001741 | XY body(GO:0001741) |
| 0.0 | 0.0 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 0.1 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.0 | 0.0 | GO:0072558 | NLRP1 inflammasome complex(GO:0072558) |
| 0.0 | 0.1 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.1 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.3 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.0 | GO:0036195 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.0 | 0.0 | GO:0097629 | extrinsic component of omegasome membrane(GO:0097629) |
| 0.0 | 0.1 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.6 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.0 | 0.1 | GO:0005915 | zonula adherens(GO:0005915) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0043398 | HLH domain binding(GO:0043398) |
| 0.2 | 0.7 | GO:0030572 | cardiolipin synthase activity(GO:0008808) phosphatidyltransferase activity(GO:0030572) |
| 0.2 | 0.8 | GO:0010853 | cyclase activator activity(GO:0010853) guanylate cyclase activator activity(GO:0030250) |
| 0.1 | 0.8 | GO:0022858 | L-alanine transmembrane transporter activity(GO:0015180) alanine transmembrane transporter activity(GO:0022858) |
| 0.1 | 0.8 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.1 | 0.3 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.1 | 0.3 | GO:0030519 | snoRNP binding(GO:0030519) |
| 0.1 | 0.3 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.1 | 1.2 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.1 | 0.2 | GO:0061513 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.1 | 0.7 | GO:0034522 | pinocarveol dehydrogenase activity(GO:0018446) chloral hydrate dehydrogenase activity(GO:0018447) hydroxymethylmethylsilanediol oxidase activity(GO:0018448) 1-phenylethanol dehydrogenase activity(GO:0018449) myrtenol dehydrogenase activity(GO:0018450) cis-1,2-dihydroxy-1,2-dihydro-8-carboxynaphthalene dehydrogenase activity(GO:0034522) 3-hydroxy-4-methyloctanoyl-CoA dehydrogenase activity(GO:0034582) 2-hydroxy-4-isopropenylcyclohexane-1-carboxyl-CoA dehydrogenase activity(GO:0034778) cis-9,10-dihydroanthracene-9,10-diol dehydrogenase activity(GO:0034817) citronellol dehydrogenase activity(GO:0034821) naphthyl-2-hydroxymethyl-succinyl-CoA dehydrogenase activity(GO:0034847) 2,4,4-trimethyl-1-pentanol dehydrogenase activity(GO:0034863) 2,4,4-trimethyl-3-hydroxypentanoyl-CoA dehydrogenase activity(GO:0034868) 1-hydroxy-4,4-dimethylpentan-3-one dehydrogenase activity(GO:0034871) endosulfan diol dehydrogenase activity(GO:0034891) endosulfan hydroxyether dehydrogenase activity(GO:0034901) 3-hydroxy-2-methylhexanoyl-CoA dehydrogenase activity(GO:0034918) 3-hydroxy-2,6-dimethyl-5-methylene-heptanoyl-CoA dehydrogenase activity(GO:0034944) versicolorin reductase activity(GO:0042469) ketoreductase activity(GO:0045703) |
| 0.1 | 0.2 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.1 | 0.2 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.1 | 0.2 | GO:0019959 | interleukin-8 binding(GO:0019959) |
| 0.1 | 0.3 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.1 | 0.3 | GO:0004645 | phosphorylase activity(GO:0004645) |
| 0.1 | 0.4 | GO:0102337 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.1 | 0.3 | GO:0070728 | leucine binding(GO:0070728) |
| 0.1 | 0.2 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.1 | 0.3 | GO:0022820 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.1 | 0.3 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.1 | 0.2 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 2.0 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.3 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 0.2 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.6 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.0 | 0.4 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.3 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.0 | 0.3 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 0.4 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.0 | 0.1 | GO:0051718 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) DNA (cytosine-5-)-methyltransferase activity, acting on CpG substrates(GO:0051718) |
| 0.0 | 0.2 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.0 | 0.6 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 0.1 | GO:0004802 | transketolase activity(GO:0004802) |
| 0.0 | 0.1 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.6 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.0 | 0.2 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.0 | 0.1 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.0 | 0.5 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.4 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.1 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.0 | 0.1 | GO:0004104 | cholinesterase activity(GO:0004104) |
| 0.0 | 0.4 | GO:0016780 | phosphotransferase activity, for other substituted phosphate groups(GO:0016780) |
| 0.0 | 0.3 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 0.1 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.0 | 0.3 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 0.5 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.0 | 0.1 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.0 | 0.1 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 0.1 | GO:0003958 | NADPH-hemoprotein reductase activity(GO:0003958) |
| 0.0 | 0.1 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.1 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.0 | 0.2 | GO:0034791 | 3-(3-hydroxyphenyl)propionate hydroxylase activity(GO:0008688) 4-chlorobenzaldehyde oxidase activity(GO:0018471) 3,5-xylenol methylhydroxylase activity(GO:0018630) phenylacetate hydroxylase activity(GO:0018631) 4-nitrophenol 4-monooxygenase activity(GO:0018632) dimethyl sulfide monooxygenase activity(GO:0018633) alpha-pinene monooxygenase [NADH] activity(GO:0018634) 1-hydroxy-2-naphthoate hydroxylase activity(GO:0018637) toluene 4-monooxygenase activity(GO:0018638) xylene monooxygenase activity(GO:0018639) dibenzothiophene monooxygenase activity(GO:0018640) 6-hydroxy-3-methyl-2-oxo-1,2-dihydroquinoline 6-monooxygenase activity(GO:0018641) chlorophenol 4-monooxygenase activity(GO:0018642) carbon disulfide oxygenase activity(GO:0018643) toluene 2-monooxygenase activity(GO:0018644) 1-hydroxy-2-oxolimonene 1,2-monooxygenase activity(GO:0018646) phenanthrene 1,2-monooxygenase activity(GO:0018647) tetrahydrofuran hydroxylase activity(GO:0018649) styrene monooxygenase activity(GO:0018650) toluene-4-sulfonate monooxygenase activity(GO:0018651) toluene-sulfonate methyl-monooxygenase activity(GO:0018652) 3-methyl-2-oxo-1,2-dihydroquinoline 6-monooxygenase activity(GO:0018653) 2-hydroxy-phenylacetate hydroxylase activity(GO:0018654) 2-oxo-delta3-4,5,5-trimethylcyclopentenylacetyl-CoA 1,2-monooxygenase activity(GO:0018655) phenanthrene 3,4-monooxygenase activity(GO:0018656) toluene 3-monooxygenase activity(GO:0018657) 4-hydroxyphenylacetate,NADH:oxygen oxidoreductase (3-hydroxylating) activity(GO:0018660) limonene monooxygenase activity(GO:0019113) 2-methylnaphthalene hydroxylase activity(GO:0034526) 1-methylnaphthalene hydroxylase activity(GO:0034534) bisphenol A hydroxylase A activity(GO:0034560) salicylate 5-hydroxylase activity(GO:0034785) isobutylamine N-hydroxylase activity(GO:0034791) branched-chain dodecylbenzene sulfonate monooxygenase activity(GO:0034802) 3-HSA hydroxylase activity(GO:0034819) 4-hydroxypyridine-3-hydroxylase activity(GO:0034894) 2-octaprenyl-3-methyl-6-methoxy-1,4-benzoquinol hydroxylase activity(GO:0043719) 6-hydroxynicotinate 3-monooxygenase activity(GO:0043731) thalianol hydroxylase activity(GO:0080014) |
| 0.0 | 0.1 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 0.0 | 0.1 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.0 | 0.2 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.1 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.0 | 0.1 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.0 | 0.1 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.0 | 0.2 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.0 | 0.4 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.1 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.4 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.0 | 0.1 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.1 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 0.3 | GO:0072349 | modified amino acid transmembrane transporter activity(GO:0072349) |
| 0.0 | 0.1 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.0 | 0.1 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.0 | 0.0 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.0 | 0.1 | GO:0019145 | aminobutyraldehyde dehydrogenase activity(GO:0019145) 4-trimethylammoniobutyraldehyde dehydrogenase activity(GO:0047105) |
| 0.0 | 0.1 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.0 | 0.1 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.0 | 0.1 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.0 | 0.2 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 0.0 | 0.3 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.0 | 0.1 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.2 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.3 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.1 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.0 | 0.1 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.0 | 0.2 | GO:0004844 | uracil DNA N-glycosylase activity(GO:0004844) deaminated base DNA N-glycosylase activity(GO:0097506) |
| 0.0 | 0.1 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.3 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.1 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.0 | 0.2 | GO:0008601 | protein phosphatase type 2A regulator activity(GO:0008601) |
| 0.0 | 0.0 | GO:0050252 | retinol O-fatty-acyltransferase activity(GO:0050252) |
| 0.0 | 0.0 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 0.1 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.1 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.0 | 0.2 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.1 | GO:0048038 | quinone binding(GO:0048038) |
| 0.0 | 0.1 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.0 | 0.1 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.0 | 0.1 | GO:0090482 | vitamin transmembrane transporter activity(GO:0090482) |
| 0.0 | 1.0 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.0 | 0.0 | GO:0070137 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.0 | 0.1 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.0 | 0.2 | GO:0008198 | ferrous iron binding(GO:0008198) |
| 0.0 | 0.1 | GO:0047760 | butyrate-CoA ligase activity(GO:0047760) |
| 0.0 | 0.0 | GO:0003865 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) cholestenone 5-alpha-reductase activity(GO:0047751) |
| 0.0 | 0.1 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.1 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.0 | 0.2 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.0 | 0.2 | GO:1990939 | ATP-dependent microtubule motor activity(GO:1990939) |
| 0.0 | 0.0 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.0 | 0.1 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.0 | 0.3 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.1 | GO:0015922 | aspartate oxidase activity(GO:0015922) |
| 0.0 | 0.2 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.0 | 0.2 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.0 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.0 | 0.1 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.1 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.2 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 0.1 | GO:0052758 | 4-methyloctanoyl-CoA dehydrogenase activity(GO:0034580) naphthyl-2-methyl-succinyl-CoA dehydrogenase activity(GO:0034845) 2-methylhexanoyl-CoA dehydrogenase activity(GO:0034916) propionyl-CoA dehydrogenase activity(GO:0043820) thiol-driven fumarate reductase activity(GO:0043830) coenzyme F420-dependent 2,4,6-trinitrophenol reductase activity(GO:0052758) coenzyme F420-dependent 2,4,6-trinitrophenol hydride reductase activity(GO:0052759) coenzyme F420-dependent 2,4-dinitrophenol reductase activity(GO:0052760) |
| 0.0 | 0.1 | GO:0016624 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, disulfide as acceptor(GO:0016624) |
| 0.0 | 0.0 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.0 | 0.1 | GO:0015194 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.2 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.1 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.0 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.1 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.0 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.0 | 0.0 | GO:0003696 | satellite DNA binding(GO:0003696) |
| 0.0 | 0.1 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.1 | GO:0051183 | vitamin transporter activity(GO:0051183) |
| 0.0 | 0.0 | GO:0017098 | sulfonylurea receptor binding(GO:0017098) |
| 0.0 | 0.0 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.0 | 0.0 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.0 | 0.3 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.9 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.0 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.1 | 1.1 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.1 | 0.3 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.1 | 0.1 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 0.7 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 0.3 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 0.6 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.1 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.0 | 0.7 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.4 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.7 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 1.2 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.0 | 0.5 | ST P38 MAPK PATHWAY | p38 MAPK Pathway |
| 0.0 | 0.3 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.4 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.7 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.2 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.1 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 0.0 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 0.3 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 0.5 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 0.2 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.1 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.0 | 0.0 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 0.1 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.0 | 0.1 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.4 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.1 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.0 | 0.9 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.3 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.4 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.1 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 0.1 | REACTOME FATTY ACID TRIACYLGLYCEROL AND KETONE BODY METABOLISM | Genes involved in Fatty acid, triacylglycerol, and ketone body metabolism |
| 0.0 | 0.6 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 0.3 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.0 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.2 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.2 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.5 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 0.1 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 0.3 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.5 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.2 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.0 | 0.2 | REACTOME SIGNALING BY NOTCH4 | Genes involved in Signaling by NOTCH4 |
| 0.0 | 0.3 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.2 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.0 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.0 | 0.2 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.0 | 0.2 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.1 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.4 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 1.1 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 0.0 | 0.1 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.2 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.3 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.0 | 0.1 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.0 | REACTOME CD28 DEPENDENT PI3K AKT SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.0 | 0.1 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.1 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |