Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
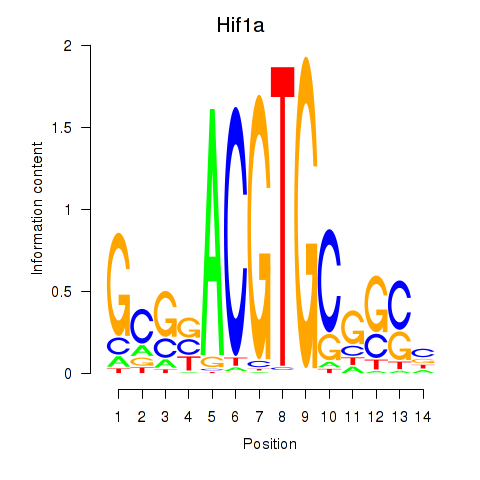
Results for Hif1a
Z-value: 1.04
Transcription factors associated with Hif1a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hif1a
|
ENSMUSG00000021109.7 | hypoxia inducible factor 1, alpha subunit |
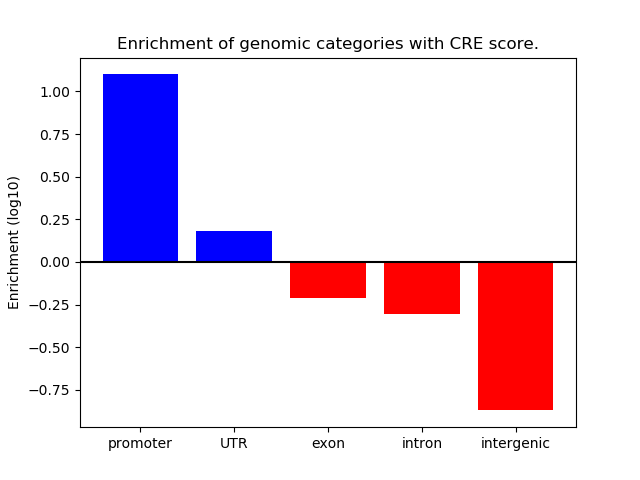
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
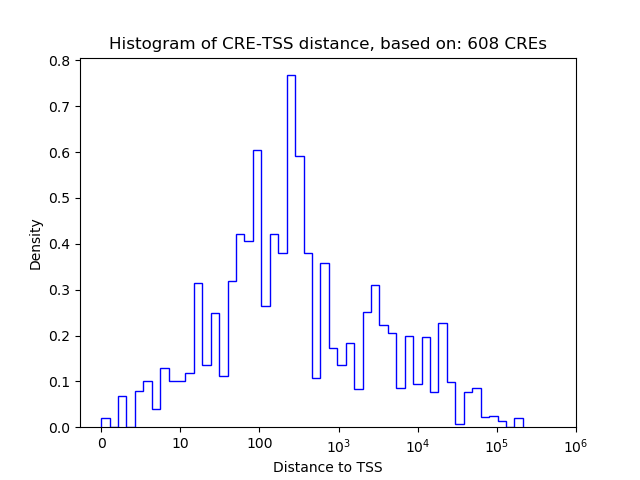
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr12_73923859_73924010 | Hif1a | 2668 | 0.215132 | -0.83 | 4.2e-02 | Click! |
| chr12_73907952_73908112 | Hif1a | 128 | 0.956916 | 0.82 | 4.6e-02 | Click! |
| chr12_73907445_73907608 | Hif1a | 378 | 0.843174 | 0.51 | 3.0e-01 | Click! |
| chr12_73909283_73909641 | Hif1a | 1558 | 0.349825 | -0.50 | 3.1e-01 | Click! |
| chr12_73907704_73907868 | Hif1a | 118 | 0.959539 | 0.46 | 3.6e-01 | Click! |
Activity of the Hif1a motif across conditions
Conditions sorted by the z-value of the Hif1a motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
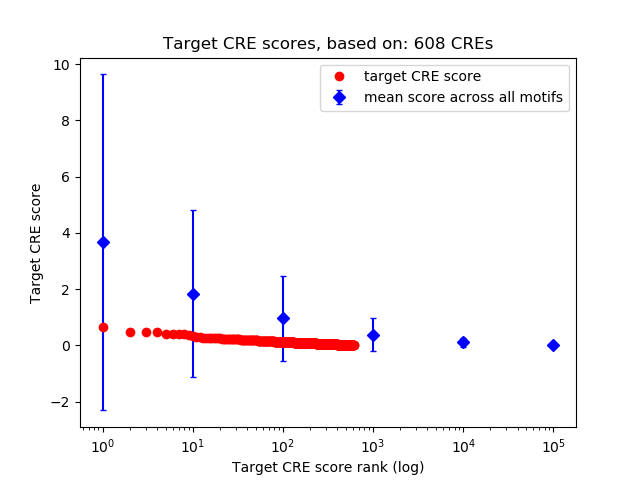
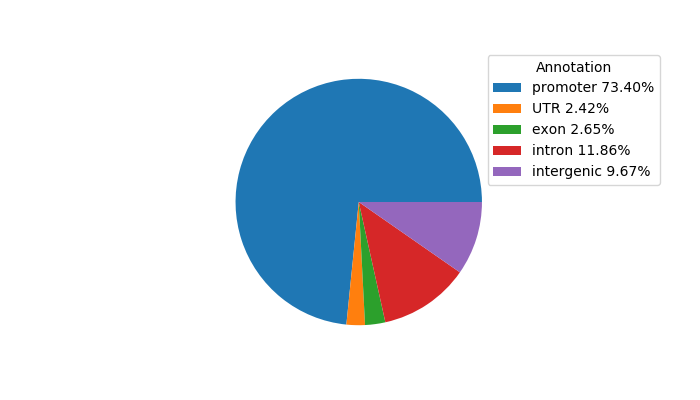
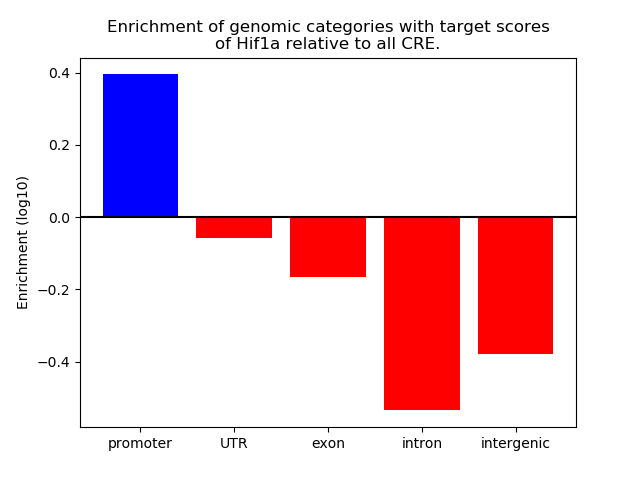
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr1_127355690_127355901 | 0.65 |
Gm23734 |
predicted gene, 23734 |
19336 |
0.15 |
| chr3_139205794_139206161 | 0.48 |
Stpg2 |
sperm tail PG rich repeat containing 2 |
84 |
0.98 |
| chr16_43979812_43979963 | 0.48 |
Zdhhc23 |
zinc finger, DHHC domain containing 23 |
96 |
0.96 |
| chr10_83337805_83337991 | 0.46 |
Slc41a2 |
solute carrier family 41, member 2 |
16 |
0.97 |
| chr8_44938067_44938242 | 0.42 |
Fat1 |
FAT atypical cadherin 1 |
2707 |
0.27 |
| chr11_53463790_53463971 | 0.41 |
Gm12212 |
predicted gene 12212 |
267 |
0.77 |
| chr11_16752814_16752965 | 0.41 |
Egfr |
epidermal growth factor receptor |
659 |
0.72 |
| chr14_65262428_65262601 | 0.39 |
Fzd3 |
frizzled class receptor 3 |
51 |
0.97 |
| chr15_83367361_83367678 | 0.35 |
1700001L05Rik |
RIKEN cDNA 1700001L05 gene |
237 |
0.9 |
| chr14_33319851_33320021 | 0.33 |
Arhgap22 |
Rho GTPase activating protein 22 |
233 |
0.93 |
| chr11_88718358_88718518 | 0.29 |
Msi2 |
musashi RNA-binding protein 2 |
75 |
0.69 |
| chr13_93499654_93499810 | 0.28 |
Jmy |
junction-mediating and regulatory protein |
5 |
0.98 |
| chr18_76378806_76378977 | 0.28 |
Gm50360 |
predicted gene, 50360 |
57067 |
0.14 |
| chr7_45466827_45467004 | 0.27 |
Bax |
BCL2-associated X protein |
17 |
0.91 |
| chr4_154946732_154946894 | 0.27 |
Hes5 |
hes family bHLH transcription factor 5 |
14110 |
0.11 |
| chr7_12780664_12780815 | 0.26 |
Zscan18 |
zinc finger and SCAN domain containing 18 |
85 |
0.53 |
| chr10_7976561_7976927 | 0.25 |
Tab2 |
TGF-beta activated kinase 1/MAP3K7 binding protein 2 |
20514 |
0.19 |
| chr11_6416246_6416529 | 0.25 |
Ppia |
peptidylprolyl isomerase A |
75 |
0.94 |
| chr13_32338323_32338641 | 0.25 |
Gmds |
GDP-mannose 4, 6-dehydratase |
103 |
0.79 |
| chr5_129020390_129020558 | 0.25 |
Ran |
RAN, member RAS oncogene family |
202 |
0.94 |
| chr6_83768773_83768930 | 0.24 |
Tex261 |
testis expressed gene 261 |
3624 |
0.1 |
| chr7_46845708_46845871 | 0.23 |
Ldha |
lactate dehydrogenase A |
50 |
0.95 |
| chr6_134566862_134567026 | 0.23 |
Lrp6 |
low density lipoprotein receptor-related protein 6 |
21 |
0.98 |
| chr10_44005716_44005885 | 0.23 |
Crybg1 |
crystallin beta-gamma domain containing 1 |
1431 |
0.34 |
| chr19_45749067_45749428 | 0.23 |
Gm15491 |
predicted gene 15491 |
261 |
0.52 |
| chrX_75155692_75155860 | 0.22 |
Gm8503 |
predicted gene 8503 |
43 |
0.95 |
| chr11_109500636_109500796 | 0.22 |
Gm22378 |
predicted gene, 22378 |
892 |
0.5 |
| chr3_86223972_86224138 | 0.22 |
Lrba |
LPS-responsive beige-like anchor |
625 |
0.66 |
| chr11_51852080_51852269 | 0.22 |
Jade2 |
jade family PHD finger 2 |
4951 |
0.2 |
| chr12_16895014_16895177 | 0.22 |
Rock2 |
Rho-associated coiled-coil containing protein kinase 2 |
72 |
0.97 |
| chr2_69897607_69897758 | 0.21 |
Ubr3 |
ubiquitin protein ligase E3 component n-recognin 3 |
379 |
0.79 |
| chr2_164785787_164785954 | 0.21 |
Snx21 |
sorting nexin family member 21 |
47 |
0.93 |
| chr4_20008188_20008393 | 0.21 |
Ttpa |
tocopherol (alpha) transfer protein |
67 |
0.97 |
| chr17_50509012_50509163 | 0.21 |
Plcl2 |
phospholipase C-like 2 |
316 |
0.94 |
| chr9_86464855_86465024 | 0.20 |
Ube2cbp |
ubiquitin-conjugating enzyme E2C binding protein |
11 |
0.97 |
| chr7_116039868_116040228 | 0.20 |
1110004F10Rik |
RIKEN cDNA 1110004F10 gene |
336 |
0.79 |
| chr5_110652988_110653519 | 0.20 |
Noc4l |
NOC4 like |
6 |
0.71 |
| chr8_106893571_106893752 | 0.20 |
Utp4 |
UTP4 small subunit processome component |
10 |
0.37 |
| chr5_139460535_139460827 | 0.20 |
3110082I17Rik |
RIKEN cDNA 3110082I17 gene |
154 |
0.93 |
| chr13_10360761_10361247 | 0.19 |
Chrm3 |
cholinergic receptor, muscarinic 3, cardiac |
157 |
0.96 |
| chr8_105852398_105852549 | 0.19 |
Cenpt |
centromere protein T |
419 |
0.64 |
| chr4_41740767_41740935 | 0.19 |
Sigmar1 |
sigma non-opioid intracellular receptor 1 |
197 |
0.87 |
| chr19_5829011_5829567 | 0.19 |
Neat1 |
nuclear paraspeckle assembly transcript 1 (non-protein coding) |
1277 |
0.22 |
| chr8_111094396_111094682 | 0.19 |
Pdpr |
pyruvate dehydrogenase phosphatase regulatory subunit |
91 |
0.94 |
| chr11_98863504_98863667 | 0.19 |
Wipf2 |
WAS/WASL interacting protein family, member 2 |
53 |
0.95 |
| chr12_110446883_110447049 | 0.18 |
Ppp2r5c |
protein phosphatase 2, regulatory subunit B', gamma |
154 |
0.95 |
| chr7_5125880_5126049 | 0.18 |
Rasl2-9 |
RAS-like, family 2, locus 9 |
14 |
0.93 |
| chr9_57940036_57940192 | 0.18 |
Sema7a |
sema domain, immunoglobulin domain (Ig), and GPI membrane anchor, (semaphorin) 7A |
1 |
0.97 |
| chr16_17210099_17210269 | 0.18 |
Rimbp3 |
RIMS binding protein 3 |
1581 |
0.21 |
| chr6_54816485_54816684 | 0.18 |
Znrf2 |
zinc and ring finger 2 |
332 |
0.91 |
| chr10_80754904_80755061 | 0.18 |
Dot1l |
DOT1-like, histone H3 methyltransferase (S. cerevisiae) |
224 |
0.85 |
| chr11_79145978_79146129 | 0.17 |
Ksr1 |
kinase suppressor of ras 1 |
354 |
0.89 |
| chr6_38551988_38552146 | 0.17 |
Luc7l2 |
LUC7-like 2 (S. cerevisiae) |
224 |
0.91 |
| chr2_180119332_180119483 | 0.16 |
Osbpl2 |
oxysterol binding protein-like 2 |
3 |
0.96 |
| chr14_24486976_24487144 | 0.16 |
Polr3a |
polymerase (RNA) III (DNA directed) polypeptide A |
2 |
0.54 |
| chr7_112225527_112225875 | 0.16 |
Mical2 |
microtubule associated monooxygenase, calponin and LIM domain containing 2 |
155 |
0.97 |
| chr5_129031100_129031253 | 0.16 |
Ran |
RAN, member RAS oncogene family |
10904 |
0.2 |
| chr2_181680135_181680294 | 0.16 |
Tcea2 |
transcription elongation factor A (SII), 2 |
96 |
0.93 |
| chr7_138538061_138538232 | 0.16 |
4930543N07Rik |
RIKEN cDNA 4930543N07 gene |
16084 |
0.26 |
| chr17_27133587_27133748 | 0.16 |
Uqcc2 |
ubiquinol-cytochrome c reductase complex assembly factor 2 |
38 |
0.95 |
| chr17_74338623_74338796 | 0.16 |
Spast |
spastin |
278 |
0.85 |
| chr1_191388747_191388913 | 0.16 |
Ppp2r5a |
protein phosphatase 2, regulatory subunit B', alpha |
3504 |
0.2 |
| chr6_29211843_29212020 | 0.16 |
Impdh1 |
inosine monophosphate dehydrogenase 1 |
318 |
0.84 |
| chr19_4196660_4196821 | 0.15 |
Ppp1ca |
protein phosphatase 1 catalytic subunit alpha |
2091 |
0.09 |
| chr15_31453304_31453493 | 0.15 |
Ropn1l |
ropporin 1-like |
339 |
0.45 |
| chr8_95107420_95107577 | 0.15 |
Kifc3 |
kinesin family member C3 |
2926 |
0.14 |
| chr11_72489944_72490114 | 0.15 |
Spns2 |
spinster homolog 2 |
125 |
0.95 |
| chr7_51861626_51861818 | 0.15 |
Gas2 |
growth arrest specific 2 |
293 |
0.68 |
| chr19_7398029_7398222 | 0.15 |
Rab11b-ps2 |
RAB11B, member RAS oncogene family, pseudogene 2 |
952 |
0.37 |
| chr11_114826796_114826950 | 0.14 |
Gprc5c |
G protein-coupled receptor, family C, group 5, member C |
24279 |
0.11 |
| chr5_139252311_139252489 | 0.14 |
Get4 |
golgi to ER traffic protein 4 |
30 |
0.97 |
| chr6_48860509_48860678 | 0.14 |
Gm7932 |
predicted gene 7932 |
169 |
0.88 |
| chr17_33747281_33747468 | 0.14 |
Rab11b |
RAB11B, member RAS oncogene family |
2471 |
0.14 |
| chr16_16213091_16213252 | 0.14 |
Pkp2 |
plakophilin 2 |
147 |
0.96 |
| chr7_67951713_67951868 | 0.14 |
Igf1r |
insulin-like growth factor I receptor |
1037 |
0.59 |
| chr10_82630042_82630204 | 0.14 |
Tdg |
thymine DNA glycosylase |
247 |
0.89 |
| chr1_135631615_135631792 | 0.14 |
Nav1 |
neuron navigator 1 |
45998 |
0.11 |
| chr17_56325722_56325896 | 0.14 |
Kdm4b |
lysine (K)-specific demethylase 4B |
253 |
0.86 |
| chr3_19310803_19310964 | 0.14 |
Pde7a |
phosphodiesterase 7A |
20 |
0.99 |
| chr6_118479189_118479349 | 0.13 |
Zfp9 |
zinc finger protein 9 |
51 |
0.97 |
| chr7_28179311_28179471 | 0.13 |
Dyrk1b |
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1b |
78 |
0.94 |
| chr4_129924204_129924361 | 0.13 |
Spocd1 |
SPOC domain containing 1 |
4967 |
0.13 |
| chr17_25837125_25837580 | 0.13 |
Rhbdl1 |
rhomboid like 1 |
57 |
0.89 |
| chr6_100287706_100287857 | 0.13 |
Rybp |
RING1 and YY1 binding protein |
296 |
0.89 |
| chr8_105279146_105279313 | 0.13 |
Nol3 |
nucleolar protein 3 (apoptosis repressor with CARD domain) |
3089 |
0.08 |
| chr11_61683998_61684190 | 0.13 |
Fam83g |
family with sequence similarity 83, member G |
3 |
0.97 |
| chr7_142097951_142098107 | 0.13 |
Dusp8 |
dual specificity phosphatase 8 |
2186 |
0.13 |
| chr13_66932977_66933151 | 0.13 |
Mterf3 |
mitochondrial transcription termination factor 3 |
3 |
0.65 |
| chr16_20621460_20621649 | 0.13 |
Camk2n2 |
calcium/calmodulin-dependent protein kinase II inhibitor 2 |
267 |
0.75 |
| chr9_94537390_94537600 | 0.12 |
Dipk2a |
divergent protein kinase domain 2A |
586 |
0.8 |
| chr5_134639466_134639692 | 0.12 |
Eif4h |
eukaryotic translation initiation factor 4H |
89 |
0.95 |
| chr11_59518331_59518501 | 0.12 |
Zkscan17 |
zinc finger with KRAB and SCAN domains 17 |
8215 |
0.1 |
| chr16_35770249_35770414 | 0.12 |
Hspbap1 |
Hspb associated protein 1 |
44 |
0.94 |
| chr9_89066820_89066977 | 0.12 |
Gm24463 |
predicted gene, 24463 |
8438 |
0.14 |
| chr4_132763837_132763988 | 0.12 |
Rpa2 |
replication protein A2 |
4420 |
0.14 |
| chr14_73325714_73325873 | 0.12 |
Rb1 |
RB transcriptional corepressor 1 |
29 |
0.98 |
| chr13_51849179_51849522 | 0.12 |
Gadd45g |
growth arrest and DNA-damage-inducible 45 gamma |
2606 |
0.33 |
| chr10_77418300_77418496 | 0.12 |
Adarb1 |
adenosine deaminase, RNA-specific, B1 |
128 |
0.96 |
| chr6_112939448_112939605 | 0.12 |
Srgap3 |
SLIT-ROBO Rho GTPase activating protein 3 |
7228 |
0.15 |
| chr1_40221808_40222119 | 0.12 |
Il1r1 |
interleukin 1 receptor, type I |
3117 |
0.26 |
| chr7_25396338_25396501 | 0.12 |
Lipe |
lipase, hormone sensitive |
40 |
0.93 |
| chr11_120542125_120542287 | 0.12 |
Mcrip1 |
MAPK regulated corepressor interacting protein 1 |
7495 |
0.06 |
| chr7_132287097_132287256 | 0.12 |
Chst15 |
carbohydrate sulfotransferase 15 |
8551 |
0.16 |
| chr4_144901641_144901842 | 0.12 |
Dhrs3 |
dehydrogenase/reductase (SDR family) member 3 |
8522 |
0.21 |
| chr15_100401091_100401280 | 0.11 |
Slc11a2 |
solute carrier family 11 (proton-coupled divalent metal ion transporters), member 2 |
2058 |
0.2 |
| chr13_113618547_113618702 | 0.11 |
Snx18 |
sorting nexin 18 |
60 |
0.97 |
| chr8_84705414_84705592 | 0.11 |
Nfix |
nuclear factor I/X |
2213 |
0.16 |
| chr17_47924756_47925097 | 0.11 |
Foxp4 |
forkhead box P4 |
281 |
0.86 |
| chr3_88835111_88835270 | 0.11 |
Gon4l |
gon-4-like (C.elegans) |
41 |
0.95 |
| chr16_55814810_55814983 | 0.11 |
Nfkbiz |
nuclear factor of kappa light polypeptide gene enhancer in B cells inhibitor, zeta |
3619 |
0.22 |
| chr7_80027017_80027182 | 0.11 |
Zfp710 |
zinc finger protein 710 |
712 |
0.56 |
| chr8_114773512_114773798 | 0.11 |
Wwox |
WW domain-containing oxidoreductase |
61488 |
0.13 |
| chr6_29272524_29272714 | 0.11 |
Hilpda |
hypoxia inducible lipid droplet associated |
7 |
0.96 |
| chr17_24753315_24753466 | 0.11 |
Hs3st6 |
heparan sulfate (glucosamine) 3-O-sulfotransferase 6 |
374 |
0.66 |
| chr6_146888080_146888231 | 0.11 |
Ppfibp1 |
PTPRF interacting protein, binding protein 1 (liprin beta 1) |
332 |
0.86 |
| chr2_114654754_114655226 | 0.11 |
Dph6 |
diphthamine biosynthesis 6 |
26 |
0.92 |
| chr4_142070841_142071028 | 0.11 |
Tmem51os1 |
Tmem51 opposite strand 1 |
13038 |
0.13 |
| chr14_67933014_67933176 | 0.11 |
Dock5 |
dedicator of cytokinesis 5 |
337 |
0.92 |
| chr6_97487494_97487819 | 0.11 |
Frmd4b |
FERM domain containing 4B |
147 |
0.96 |
| chr5_27997662_27998174 | 0.11 |
Gm4865 |
predicted gene 4865 |
3388 |
0.22 |
| chr3_89276379_89276658 | 0.11 |
Efna1 |
ephrin A1 |
3123 |
0.09 |
| chr2_181459964_181460183 | 0.11 |
Zbtb46 |
zinc finger and BTB domain containing 46 |
647 |
0.57 |
| chr16_11253479_11253674 | 0.10 |
Gspt1 |
G1 to S phase transition 1 |
633 |
0.47 |
| chr4_62524756_62524940 | 0.10 |
Pole3 |
polymerase (DNA directed), epsilon 3 (p17 subunit) |
190 |
0.77 |
| chr4_141697170_141697329 | 0.10 |
Rsc1a1 |
regulatory solute carrier protein, family 1, member 1 |
11650 |
0.12 |
| chr9_108808514_108808701 | 0.10 |
Nckipsd |
NCK interacting protein with SH3 domain |
186 |
0.88 |
| chr15_85337324_85337483 | 0.10 |
Atxn10 |
ataxin 10 |
1158 |
0.51 |
| chr7_143600068_143600237 | 0.10 |
Gm15579 |
predicted gene 15579 |
18 |
0.52 |
| chr10_95940931_95941100 | 0.10 |
Eea1 |
early endosome antigen 1 |
258 |
0.91 |
| chr11_116852385_116852543 | 0.10 |
Mfsd11 |
major facilitator superfamily domain containing 11 |
24 |
0.76 |
| chr5_124446062_124446234 | 0.10 |
Kmt5a |
lysine methyltransferase 5A |
364 |
0.75 |
| chr15_94543180_94543352 | 0.10 |
Pus7l |
pseudouridylate synthase 7-like |
220 |
0.67 |
| chr11_3963860_3964038 | 0.10 |
Pes1 |
pescadillo ribosomal biogenesis factor 1 |
26 |
0.95 |
| chr11_60811168_60811358 | 0.10 |
Shmt1 |
serine hydroxymethyltransferase 1 (soluble) |
2 |
0.95 |
| chr3_88775094_88775256 | 0.10 |
Syt11 |
synaptotagmin XI |
11 |
0.51 |
| chr7_30359743_30359928 | 0.10 |
Gm25259 |
predicted gene, 25259 |
2911 |
0.09 |
| chr11_60731090_60731277 | 0.10 |
Mir5100 |
microRNA 5100 |
2520 |
0.1 |
| chr12_105784205_105784562 | 0.10 |
Papola |
poly (A) polymerase alpha |
311 |
0.9 |
| chr17_5176692_5176856 | 0.10 |
Gm15599 |
predicted gene 15599 |
64664 |
0.13 |
| chr2_6118432_6118598 | 0.10 |
Proser2 |
proline and serine rich 2 |
11624 |
0.15 |
| chr5_5559196_5559372 | 0.10 |
Gtpbp10 |
GTP-binding protein 10 (putative) |
177 |
0.93 |
| chr16_4908036_4908194 | 0.10 |
Mgrn1 |
mahogunin, ring finger 1 |
5096 |
0.12 |
| chr4_46452273_46452437 | 0.10 |
Anp32b |
acidic (leucine-rich) nuclear phosphoprotein 32 family, member B |
1453 |
0.32 |
| chr15_73724592_73724779 | 0.10 |
Ptp4a3 |
protein tyrosine phosphatase 4a3 |
113 |
0.96 |
| chr15_88821550_88821729 | 0.09 |
Creld2 |
cysteine-rich with EGF-like domains 2 |
1993 |
0.22 |
| chr2_118972791_118972987 | 0.09 |
Gm14089 |
predicted gene 14089 |
24179 |
0.11 |
| chr3_110250186_110250444 | 0.09 |
C130013H08Rik |
RIKEN cDNA C130013H08 gene |
300 |
0.74 |
| chr2_180042367_180042531 | 0.09 |
Psma7 |
proteasome subunit alpha 7 |
16 |
0.52 |
| chrX_134276921_134277269 | 0.09 |
Trmt2b |
TRM2 tRNA methyltransferase 2B |
111 |
0.96 |
| chrX_101640210_101640705 | 0.09 |
Ogt |
O-linked N-acetylglucosamine (GlcNAc) transferase (UDP-N-acetylglucosamine:polypeptide-N-acetylglucosaminyl transferase) |
317 |
0.87 |
| chr9_65032284_65032455 | 0.09 |
Dpp8 |
dipeptidylpeptidase 8 |
45 |
0.97 |
| chr14_18238484_18238667 | 0.09 |
Nr1d2 |
nuclear receptor subfamily 1, group D, member 2 |
436 |
0.77 |
| chr5_75377548_75377800 | 0.09 |
Gm22084 |
predicted gene, 22084 |
5529 |
0.2 |
| chr17_74528690_74528846 | 0.09 |
Birc6 |
baculoviral IAP repeat-containing 6 |
216 |
0.82 |
| chr9_20607564_20607721 | 0.09 |
Gm48069 |
predicted gene, 48069 |
26 |
0.49 |
| chr6_113469767_113469928 | 0.09 |
Il17rc |
interleukin 17 receptor C |
1580 |
0.17 |
| chr4_43046114_43046311 | 0.09 |
Fam214b |
family with sequence similarity 214, member B |
8 |
0.67 |
| chr14_65358460_65358659 | 0.09 |
Zfp395 |
zinc finger protein 395 |
25 |
0.95 |
| chr13_30973965_30974242 | 0.09 |
Exoc2 |
exocyst complex component 2 |
10 |
0.53 |
| chr9_64041799_64041950 | 0.09 |
1700055C04Rik |
RIKEN cDNA 1700055C04 gene |
409 |
0.78 |
| chr11_98709546_98709720 | 0.09 |
Med24 |
mediator complex subunit 24 |
1433 |
0.2 |
| chr7_128010040_128010379 | 0.09 |
Gm19366 |
predicted gene, 19366 |
2975 |
0.09 |
| chr14_20083085_20083240 | 0.09 |
Saysd1 |
SAYSVFN motif domain containing 1 |
21 |
0.98 |
| chr10_125725411_125725590 | 0.09 |
Gm9102 |
predicted gene 9102 |
212969 |
0.02 |
| chr10_121626467_121626639 | 0.09 |
Xpot |
exportin, tRNA (nuclear export receptor for tRNAs) |
221 |
0.91 |
| chr11_102138520_102138679 | 0.09 |
Nags |
N-acetylglutamate synthase |
6914 |
0.08 |
| chr15_99126067_99126238 | 0.09 |
Spats2 |
spermatogenesis associated, serine-rich 2 |
231 |
0.86 |
| chr5_35583017_35583168 | 0.09 |
Acox3 |
acyl-Coenzyme A oxidase 3, pristanoyl |
0 |
0.97 |
| chr18_34954283_34954445 | 0.08 |
Hspa9 |
heat shock protein 9 |
7 |
0.96 |
| chr11_45008209_45008377 | 0.08 |
Ebf1 |
early B cell factor 1 |
96155 |
0.08 |
| chr7_99182977_99183283 | 0.08 |
Dgat2 |
diacylglycerol O-acyltransferase 2 |
411 |
0.78 |
| chr11_50025101_50025299 | 0.08 |
Rnf130 |
ring finger protein 130 |
146 |
0.96 |
| chr13_95525061_95525230 | 0.08 |
F2rl1 |
coagulation factor II (thrombin) receptor-like 1 |
82 |
0.96 |
| chr12_102705049_102705200 | 0.08 |
Itpk1 |
inositol 1,3,4-triphosphate 5/6 kinase |
194 |
0.89 |
| chr4_119108410_119108841 | 0.08 |
Slc2a1 |
solute carrier family 2 (facilitated glucose transporter), member 1 |
86 |
0.94 |
| chr3_127124044_127124345 | 0.08 |
Ank2 |
ankyrin 2, brain |
668 |
0.65 |
| chr11_121259763_121259955 | 0.08 |
Foxk2 |
forkhead box K2 |
131 |
0.94 |
| chr1_180179778_180180226 | 0.08 |
Coq8a |
coenzyme Q8A |
1080 |
0.44 |
| chr11_82908100_82908264 | 0.08 |
Nle1 |
notchless homolog 1 |
160 |
0.89 |
| chr11_101507204_101507377 | 0.08 |
Brca1 |
breast cancer 1, early onset |
888 |
0.38 |
| chr17_47864605_47864762 | 0.08 |
Foxp4 |
forkhead box P4 |
14270 |
0.12 |
| chr5_64092821_64092990 | 0.08 |
Pgm2 |
phosphoglucomutase 2 |
45 |
0.96 |
| chr7_105736434_105736596 | 0.08 |
Ilk |
integrin linked kinase |
77 |
0.58 |
| chr5_129725307_129725503 | 0.08 |
Nipsnap2 |
nipsnap homolog 2 |
304 |
0.8 |
| chr10_82868804_82868994 | 0.08 |
Eid3 |
EP300 interacting inhibitor of differentiation 3 |
2273 |
0.21 |
| chr17_80728037_80728199 | 0.08 |
Gm9959 |
predicted gene 9959 |
13 |
0.5 |
| chr19_8880846_8881009 | 0.08 |
Uqcc3 |
ubiquinol-cytochrome c reductase complex assembly factor 3 |
4 |
0.9 |
| chr13_48968536_48968687 | 0.08 |
Fam120a |
family with sequence similarity 120, member A |
594 |
0.8 |
| chr11_120549471_120549635 | 0.08 |
Mcrip1 |
MAPK regulated corepressor interacting protein 1 |
148 |
0.85 |
| chr19_6057770_6057938 | 0.08 |
Fau |
Finkel-Biskis-Reilly murine sarcoma virus (FBR-MuSV) ubiquitously expressed (fox derived) |
10 |
0.52 |
| chr17_66123664_66123841 | 0.08 |
Ddx11 |
DEAD/H (Asp-Glu-Ala-Asp/His) box helicase 11 |
139 |
0.93 |
| chr9_70503778_70503941 | 0.08 |
Rnf111 |
ring finger 111 |
134 |
0.94 |
| chr6_88841586_88841956 | 0.08 |
Abtb1 |
ankyrin repeat and BTB (POZ) domain containing 1 |
59 |
0.93 |
| chr7_45000808_45000995 | 0.08 |
Irf3 |
interferon regulatory factor 3 |
153 |
0.84 |
| chr8_124344380_124344599 | 0.07 |
Gm15775 |
predicted gene 15775 |
12694 |
0.14 |
| chr8_123948951_123949132 | 0.07 |
Nup133 |
nucleoporin 133 |
214 |
0.52 |
| chr3_49756473_49756624 | 0.07 |
Pcdh18 |
protocadherin 18 |
735 |
0.7 |
| chr3_27710416_27710593 | 0.07 |
Fndc3b |
fibronectin type III domain containing 3B |
65 |
0.98 |
| chr11_57801415_57801582 | 0.07 |
2010001A14Rik |
RIKEN cDNA 2010001A14 gene |
81 |
0.58 |
| chr8_10899998_10900419 | 0.07 |
4833411C07Rik |
RIKEN cDNA 4833411C07 gene |
271 |
0.54 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.2 | GO:0061341 | non-canonical Wnt signaling pathway involved in heart development(GO:0061341) planar cell polarity pathway involved in heart morphogenesis(GO:0061346) |
| 0.1 | 0.3 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.1 | 0.3 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.1 | 0.4 | GO:0036515 | serotonergic neuron axon guidance(GO:0036515) |
| 0.1 | 0.2 | GO:1902445 | regulation of mitochondrial membrane permeability involved in programmed necrotic cell death(GO:1902445) |
| 0.1 | 0.2 | GO:0002337 | B-1a B cell differentiation(GO:0002337) |
| 0.1 | 0.2 | GO:0042360 | vitamin E metabolic process(GO:0042360) |
| 0.1 | 0.2 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.1 | 0.2 | GO:0019249 | lactate biosynthetic process from pyruvate(GO:0019244) lactate biosynthetic process(GO:0019249) |
| 0.1 | 0.2 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.1 | 0.2 | GO:0071394 | cellular response to testosterone stimulus(GO:0071394) |
| 0.0 | 0.1 | GO:0034729 | histone H3-K79 methylation(GO:0034729) |
| 0.0 | 0.1 | GO:0006285 | base-excision repair, AP site formation(GO:0006285) |
| 0.0 | 0.1 | GO:0045048 | protein insertion into ER membrane(GO:0045048) tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 0.0 | 0.1 | GO:0071930 | negative regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071930) |
| 0.0 | 0.1 | GO:0042758 | long-chain fatty acid catabolic process(GO:0042758) |
| 0.0 | 0.1 | GO:0035425 | autocrine signaling(GO:0035425) |
| 0.0 | 0.3 | GO:0003056 | regulation of vascular smooth muscle contraction(GO:0003056) |
| 0.0 | 0.1 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.0 | 0.1 | GO:0034214 | protein hexamerization(GO:0034214) |
| 0.0 | 0.1 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.0 | 0.2 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.0 | 0.1 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 0.0 | 0.1 | GO:0009157 | deoxyribonucleoside monophosphate biosynthetic process(GO:0009157) pyrimidine deoxyribonucleoside monophosphate biosynthetic process(GO:0009177) |
| 0.0 | 0.1 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.0 | 0.1 | GO:1904192 | cholangiocyte apoptotic process(GO:1902488) regulation of cholangiocyte apoptotic process(GO:1904192) negative regulation of cholangiocyte apoptotic process(GO:1904193) |
| 0.0 | 0.2 | GO:0019673 | GDP-mannose metabolic process(GO:0019673) |
| 0.0 | 0.2 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.0 | 0.1 | GO:0043309 | regulation of eosinophil degranulation(GO:0043309) |
| 0.0 | 0.1 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.0 | 0.2 | GO:0060907 | positive regulation of macrophage cytokine production(GO:0060907) |
| 0.0 | 0.1 | GO:0070544 | histone H3-K36 demethylation(GO:0070544) |
| 0.0 | 0.1 | GO:0036089 | cleavage furrow formation(GO:0036089) |
| 0.0 | 0.1 | GO:0048069 | eye pigmentation(GO:0048069) |
| 0.0 | 0.0 | GO:0043308 | eosinophil activation involved in immune response(GO:0002278) eosinophil mediated immunity(GO:0002447) eosinophil degranulation(GO:0043308) |
| 0.0 | 0.1 | GO:0019064 | fusion of virus membrane with host plasma membrane(GO:0019064) membrane fusion involved in viral entry into host cell(GO:0039663) multi-organism membrane fusion(GO:0044800) |
| 0.0 | 0.1 | GO:0033750 | ribosomal subunit export from nucleus(GO:0000054) ribosome localization(GO:0033750) establishment of ribosome localization(GO:0033753) |
| 0.0 | 0.1 | GO:0051031 | tRNA transport(GO:0051031) |
| 0.0 | 0.4 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.1 | GO:2001199 | negative regulation of dendritic cell differentiation(GO:2001199) |
| 0.0 | 0.1 | GO:0051697 | protein delipidation(GO:0051697) |
| 0.0 | 0.1 | GO:1905154 | negative regulation of membrane invagination(GO:1905154) |
| 0.0 | 0.0 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 0.1 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 | 0.1 | GO:0050689 | negative regulation of defense response to virus by host(GO:0050689) |
| 0.0 | 0.0 | GO:0006166 | purine ribonucleoside salvage(GO:0006166) |
| 0.0 | 0.1 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.0 | 0.1 | GO:0043622 | cortical microtubule organization(GO:0043622) |
| 0.0 | 0.1 | GO:0060480 | lung goblet cell differentiation(GO:0060480) |
| 0.0 | 0.1 | GO:0015760 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.0 | 0.1 | GO:2001268 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001268) |
| 0.0 | 0.2 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.0 | 0.0 | GO:0042891 | antibiotic transport(GO:0042891) |
| 0.0 | 0.0 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.0 | 0.1 | GO:0098910 | regulation of atrial cardiac muscle cell action potential(GO:0098910) |
| 0.0 | 0.1 | GO:0035356 | cellular triglyceride homeostasis(GO:0035356) |
| 0.0 | 0.1 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.0 | 0.0 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.0 | 0.0 | GO:0071336 | regulation of hair follicle cell proliferation(GO:0071336) |
| 0.0 | 0.1 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.0 | 0.0 | GO:0001927 | exocyst assembly(GO:0001927) |
| 0.0 | 0.1 | GO:0000972 | transcription-dependent tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000972) |
| 0.0 | 0.1 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.0 | 0.1 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.0 | 0.0 | GO:1900738 | positive regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900738) |
| 0.0 | 0.1 | GO:0060510 | Type II pneumocyte differentiation(GO:0060510) |
| 0.0 | 0.0 | GO:0001983 | baroreceptor response to increased systemic arterial blood pressure(GO:0001983) |
| 0.0 | 0.1 | GO:0035518 | histone H2A monoubiquitination(GO:0035518) |
| 0.0 | 0.1 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.0 | 0.0 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.0 | 0.0 | GO:0051572 | negative regulation of histone H3-K4 methylation(GO:0051572) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.1 | 0.2 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.1 | 0.2 | GO:1990851 | Wnt-Frizzled-LRP5/6 complex(GO:1990851) |
| 0.1 | 0.2 | GO:0031074 | nucleocytoplasmic shuttling complex(GO:0031074) |
| 0.0 | 0.1 | GO:0071818 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.0 | 0.1 | GO:0030689 | Noc complex(GO:0030689) |
| 0.0 | 0.1 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.1 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.0 | 0.4 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 0.1 | GO:0044308 | axonal spine(GO:0044308) |
| 0.0 | 0.1 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 0.1 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.0 | 0.1 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.0 | 0.2 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.1 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 0.2 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.0 | 0.2 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.0 | 0.0 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.0 | 0.2 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.0 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.0 | 0.1 | GO:0070688 | MLL5-L complex(GO:0070688) |
| 0.0 | 0.0 | GO:0000811 | GINS complex(GO:0000811) |
| 0.0 | 0.1 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.6 | GO:0005811 | lipid particle(GO:0005811) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.1 | 0.2 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.1 | 0.2 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.1 | 0.2 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.1 | 0.2 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.0 | 0.1 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.1 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.0 | 0.2 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.0 | 0.1 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) |
| 0.0 | 0.1 | GO:0070905 | serine binding(GO:0070905) |
| 0.0 | 0.6 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.1 | GO:0033592 | RNA strand annealing activity(GO:0033592) |
| 0.0 | 0.2 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.0 | 0.1 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) |
| 0.0 | 0.1 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.0 | 0.2 | GO:0018643 | 3-(3-hydroxyphenyl)propionate hydroxylase activity(GO:0008688) 4-chlorobenzaldehyde oxidase activity(GO:0018471) 3,5-xylenol methylhydroxylase activity(GO:0018630) phenylacetate hydroxylase activity(GO:0018631) 4-nitrophenol 4-monooxygenase activity(GO:0018632) dimethyl sulfide monooxygenase activity(GO:0018633) alpha-pinene monooxygenase [NADH] activity(GO:0018634) 1-hydroxy-2-naphthoate hydroxylase activity(GO:0018637) toluene 4-monooxygenase activity(GO:0018638) xylene monooxygenase activity(GO:0018639) dibenzothiophene monooxygenase activity(GO:0018640) 6-hydroxy-3-methyl-2-oxo-1,2-dihydroquinoline 6-monooxygenase activity(GO:0018641) chlorophenol 4-monooxygenase activity(GO:0018642) carbon disulfide oxygenase activity(GO:0018643) toluene 2-monooxygenase activity(GO:0018644) 1-hydroxy-2-oxolimonene 1,2-monooxygenase activity(GO:0018646) phenanthrene 1,2-monooxygenase activity(GO:0018647) tetrahydrofuran hydroxylase activity(GO:0018649) styrene monooxygenase activity(GO:0018650) toluene-4-sulfonate monooxygenase activity(GO:0018651) toluene-sulfonate methyl-monooxygenase activity(GO:0018652) 3-methyl-2-oxo-1,2-dihydroquinoline 6-monooxygenase activity(GO:0018653) 2-hydroxy-phenylacetate hydroxylase activity(GO:0018654) 2-oxo-delta3-4,5,5-trimethylcyclopentenylacetyl-CoA 1,2-monooxygenase activity(GO:0018655) phenanthrene 3,4-monooxygenase activity(GO:0018656) toluene 3-monooxygenase activity(GO:0018657) 4-hydroxyphenylacetate,NADH:oxygen oxidoreductase (3-hydroxylating) activity(GO:0018660) limonene monooxygenase activity(GO:0019113) 2-methylnaphthalene hydroxylase activity(GO:0034526) 1-methylnaphthalene hydroxylase activity(GO:0034534) bisphenol A hydroxylase A activity(GO:0034560) salicylate 5-hydroxylase activity(GO:0034785) isobutylamine N-hydroxylase activity(GO:0034791) branched-chain dodecylbenzene sulfonate monooxygenase activity(GO:0034802) 3-HSA hydroxylase activity(GO:0034819) 4-hydroxypyridine-3-hydroxylase activity(GO:0034894) 2-octaprenyl-3-methyl-6-methoxy-1,4-benzoquinol hydroxylase activity(GO:0043719) 6-hydroxynicotinate 3-monooxygenase activity(GO:0043731) thalianol hydroxylase activity(GO:0080014) |
| 0.0 | 0.1 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.0 | 0.1 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.0 | 0.1 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.0 | 0.2 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.1 | GO:0050252 | retinol O-fatty-acyltransferase activity(GO:0050252) |
| 0.0 | 0.1 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.0 | 0.2 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.1 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.0 | 0.1 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.0 | 0.1 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.1 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.1 | GO:0015526 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.0 | 0.4 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) |
| 0.0 | 0.1 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.1 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.0 | 0.1 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.2 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.0 | GO:0000253 | 3-keto sterol reductase activity(GO:0000253) |
| 0.0 | 0.2 | GO:0008601 | protein phosphatase type 2A regulator activity(GO:0008601) |
| 0.0 | 0.1 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.2 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.0 | 0.1 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.0 | 0.1 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.1 | GO:0043559 | insulin binding(GO:0043559) |
| 0.0 | 0.0 | GO:0042895 | antibiotic transporter activity(GO:0042895) |
| 0.0 | 0.0 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.0 | GO:0070548 | L-glutamine aminotransferase activity(GO:0070548) |
| 0.0 | 0.1 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.0 | 0.0 | GO:0016649 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.0 | 0.2 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.2 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.0 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 0.0 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.0 | 0.0 | GO:0052833 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.3 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.2 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 0.2 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.2 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 0.4 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 0.3 | REACTOME SHC1 EVENTS IN EGFR SIGNALING | Genes involved in SHC1 events in EGFR signaling |
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.1 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.0 | 0.0 | REACTOME SOS MEDIATED SIGNALLING | Genes involved in SOS-mediated signalling |
| 0.0 | 0.2 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.1 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 0.2 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.1 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.3 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.0 | 0.1 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |