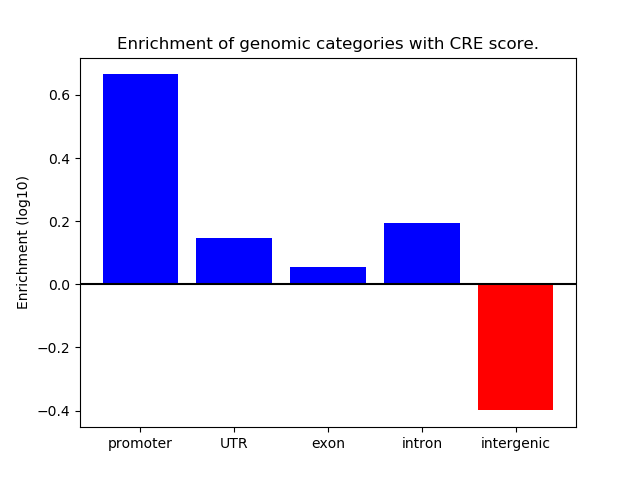
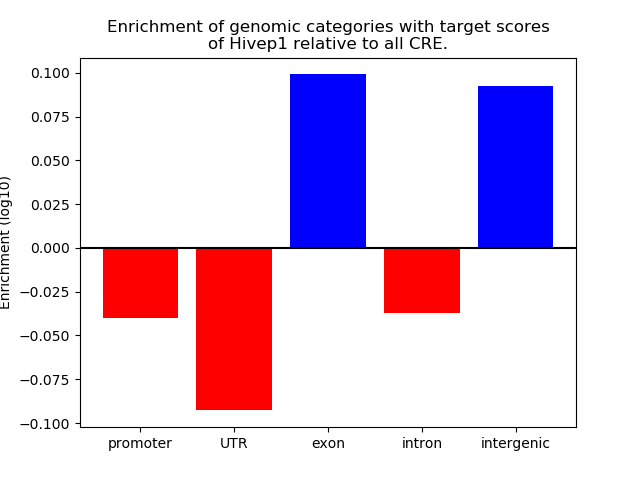
Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
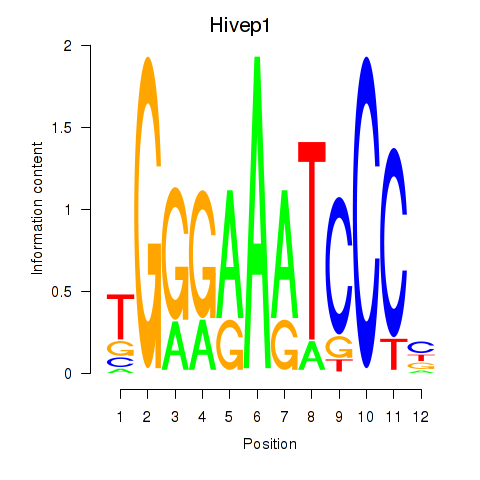
Results for Hivep1
Z-value: 1.68
Transcription factors associated with Hivep1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hivep1
|
ENSMUSG00000021366.7 | human immunodeficiency virus type I enhancer binding protein 1 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr13_42052837_42053016 | Hivep1 | 522 | 0.803363 | 0.92 | 9.0e-03 | Click! |
| chr13_42051493_42051760 | Hivep1 | 395 | 0.853693 | 0.80 | 5.5e-02 | Click! |
| chr13_42062809_42062993 | Hivep1 | 10497 | 0.219785 | 0.75 | 8.4e-02 | Click! |
| chr13_42053164_42053359 | Hivep1 | 857 | 0.639304 | -0.75 | 8.9e-02 | Click! |
| chr13_42051976_42052134 | Hivep1 | 34 | 0.978295 | 0.74 | 9.1e-02 | Click! |
Activity of the Hivep1 motif across conditions
Conditions sorted by the z-value of the Hivep1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr7_141628129_141628296 | 1.33 |
Mir7063 |
microRNA 7063 |
7511 |
0.13 |
| chr6_72606152_72606303 | 0.87 |
Retsat |
retinol saturase (all trans retinol 13,14 reductase) |
255 |
0.72 |
| chr1_127014601_127014752 | 0.85 |
Gm5261 |
predicted gene 5261 |
36020 |
0.21 |
| chr12_98901637_98901855 | 0.79 |
Eml5 |
echinoderm microtubule associated protein like 5 |
262 |
0.91 |
| chr9_50887504_50887656 | 0.79 |
Gm25558 |
predicted gene, 25558 |
370 |
0.85 |
| chr5_8964466_8964675 | 0.76 |
Abcb4 |
ATP-binding cassette, sub-family B (MDR/TAP), member 4 |
6018 |
0.13 |
| chr4_155245715_155246079 | 0.71 |
Faap20 |
Fanconi anemia core complex associated protein 20 |
3905 |
0.19 |
| chr19_5430069_5430231 | 0.68 |
AI837181 |
expressed sequence AI837181 |
4993 |
0.05 |
| chr18_65099329_65099521 | 0.67 |
Nedd4l |
neural precursor cell expressed, developmentally down-regulated gene 4-like |
16371 |
0.23 |
| chr6_120540294_120540445 | 0.64 |
Hdhd5 |
haloacid dehalogenase like hydrolase domain containing 5 |
9050 |
0.12 |
| chr2_156451427_156451737 | 0.62 |
Gm14225 |
predicted gene 14225 |
3974 |
0.13 |
| chr2_135692897_135693099 | 0.61 |
Gm14211 |
predicted gene 14211 |
156 |
0.96 |
| chr11_106528533_106528684 | 0.60 |
Gm22711 |
predicted gene, 22711 |
27363 |
0.13 |
| chr10_24103718_24103905 | 0.56 |
Taar8c |
trace amine-associated receptor 8C |
1899 |
0.18 |
| chr17_30590513_30590860 | 0.55 |
Gm50244 |
predicted gene, 50244 |
822 |
0.45 |
| chr12_21160412_21160576 | 0.55 |
Asap2 |
ArfGAP with SH3 domain, ankyrin repeat and PH domain 2 |
48540 |
0.12 |
| chr13_69583732_69583897 | 0.54 |
Srd5a1 |
steroid 5 alpha-reductase 1 |
11015 |
0.12 |
| chr2_158302944_158303106 | 0.54 |
Lbp |
lipopolysaccharide binding protein |
3468 |
0.15 |
| chr10_24074506_24074765 | 0.52 |
Taar8a |
trace amine-associated receptor 8A |
1865 |
0.16 |
| chr19_42612808_42613017 | 0.52 |
Loxl4 |
lysyl oxidase-like 4 |
99 |
0.97 |
| chr2_32607757_32607919 | 0.52 |
St6galnac6 |
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 6 |
233 |
0.82 |
| chr19_23035930_23036081 | 0.51 |
Gm50136 |
predicted gene, 50136 |
25449 |
0.19 |
| chr5_105830258_105830416 | 0.49 |
Lrrc8d |
leucine rich repeat containing 8D |
5826 |
0.14 |
| chr9_5308705_5308876 | 0.49 |
Casp4 |
caspase 4, apoptosis-related cysteine peptidase |
38 |
0.98 |
| chr11_79673804_79673986 | 0.48 |
Rab11fip4os2 |
RAB11 family interacting protein 4 (class II), opposite strand 2 |
1188 |
0.34 |
| chr19_10690326_10690666 | 0.48 |
Vps37c |
vacuolar protein sorting 37C |
1731 |
0.21 |
| chr1_93145749_93145928 | 0.47 |
Agxt |
alanine-glyoxylate aminotransferase |
5959 |
0.13 |
| chr6_5496841_5497314 | 0.47 |
Pdk4 |
pyruvate dehydrogenase kinase, isoenzyme 4 |
768 |
0.76 |
| chr10_24097357_24097545 | 0.47 |
Taar8c |
trace amine-associated receptor 8C |
4461 |
0.1 |
| chr7_68709256_68709407 | 0.47 |
Gm44692 |
predicted gene 44692 |
17136 |
0.21 |
| chr13_47150132_47150307 | 0.46 |
A930002C04Rik |
RIKEN cDNA A930002C04 gene |
1549 |
0.3 |
| chr4_137497524_137497692 | 0.46 |
Hspg2 |
perlecan (heparan sulfate proteoglycan 2) |
24851 |
0.12 |
| chr11_95350469_95350629 | 0.46 |
Fam117a |
family with sequence similarity 117, member A |
10587 |
0.12 |
| chr13_41334685_41334849 | 0.46 |
Nedd9 |
neural precursor cell expressed, developmentally down-regulated gene 9 |
24480 |
0.12 |
| chr10_94072375_94072538 | 0.45 |
Fgd6 |
FYVE, RhoGEF and PH domain containing 6 |
86 |
0.96 |
| chr8_70564788_70565043 | 0.45 |
Ell |
elongation factor RNA polymerase II |
25227 |
0.07 |
| chr11_94346567_94346751 | 0.45 |
Abcc3 |
ATP-binding cassette, sub-family C (CFTR/MRP), member 3 |
4527 |
0.17 |
| chr8_33902604_33902775 | 0.45 |
Rbpms |
RNA binding protein gene with multiple splicing |
10925 |
0.17 |
| chr10_59786230_59786381 | 0.45 |
Gm17059 |
predicted gene 17059 |
13949 |
0.14 |
| chr9_73105883_73106055 | 0.44 |
Khdc3 |
KH domain containing 3, subcortical maternal complex member |
3459 |
0.12 |
| chr5_146687074_146687253 | 0.44 |
4930573C15Rik |
RIKEN cDNA 4930573C15 gene |
19459 |
0.16 |
| chr1_36064998_36065160 | 0.44 |
Hs6st1 |
heparan sulfate 6-O-sulfotransferase 1 |
3321 |
0.19 |
| chr8_105820252_105820414 | 0.43 |
Ranbp10 |
RAN binding protein 10 |
6872 |
0.09 |
| chr12_55492611_55492778 | 0.43 |
Nfkbia |
nuclear factor of kappa light polypeptide gene enhancer in B cells inhibitor, alpha |
47 |
0.97 |
| chr4_60659551_60660018 | 0.43 |
Mup11 |
major urinary protein 11 |
46 |
0.97 |
| chr18_39693461_39693788 | 0.43 |
Gm50398 |
predicted gene, 50398 |
32747 |
0.16 |
| chr10_23842034_23842208 | 0.42 |
Vnn3 |
vanin 3 |
9341 |
0.12 |
| chr4_44990961_44991112 | 0.42 |
Grhpr |
glyoxylate reductase/hydroxypyruvate reductase |
9536 |
0.11 |
| chr2_43548371_43548543 | 0.42 |
Kynu |
kynureninase |
6872 |
0.28 |
| chr10_59403529_59403689 | 0.41 |
Pla2g12b |
phospholipase A2, group XIIB |
51 |
0.97 |
| chr8_119423599_119423760 | 0.41 |
Osgin1 |
oxidative stress induced growth inhibitor 1 |
10445 |
0.15 |
| chr8_75092030_75092181 | 0.41 |
Hmox1 |
heme oxygenase 1 |
1516 |
0.3 |
| chr9_64506948_64507234 | 0.41 |
Megf11 |
multiple EGF-like-domains 11 |
5490 |
0.28 |
| chr7_74647398_74647580 | 0.41 |
Gm7726 |
predicted gene 7726 |
52367 |
0.15 |
| chr8_106500079_106500253 | 0.41 |
6030452D12Rik |
RIKEN cDNA 6030452D12 gene |
339 |
0.87 |
| chr8_124634513_124634671 | 0.41 |
Capn9 |
calpain 9 |
21152 |
0.14 |
| chr1_180163928_180164201 | 0.40 |
Cdc42bpa |
CDC42 binding protein kinase alpha |
3057 |
0.2 |
| chr19_18627525_18627688 | 0.40 |
Ostf1 |
osteoclast stimulating factor 1 |
4112 |
0.19 |
| chr16_30163409_30163569 | 0.39 |
n-R5s32 |
nuclear encoded rRNA 5S 32 |
21544 |
0.12 |
| chr3_36551644_36551818 | 0.39 |
Exosc9 |
exosome component 9 |
875 |
0.45 |
| chr1_165486032_165486258 | 0.39 |
Adcy10 |
adenylate cyclase 10 |
934 |
0.38 |
| chr13_69264071_69264248 | 0.39 |
Gm4812 |
predicted gene 4812 |
81496 |
0.09 |
| chr11_75231643_75231820 | 0.39 |
Rtn4rl1 |
reticulon 4 receptor-like 1 |
37948 |
0.08 |
| chr6_71204222_71204852 | 0.39 |
Fabp1 |
fatty acid binding protein 1, liver |
4710 |
0.14 |
| chr18_36703302_36703470 | 0.39 |
E230025N22Rik |
Riken cDNA E230025N22 gene |
7461 |
0.08 |
| chr4_106845457_106845733 | 0.39 |
Gm12746 |
predicted gene 12746 |
2325 |
0.28 |
| chr6_48700323_48700513 | 0.38 |
Gimap6 |
GTPase, IMAP family member 6 |
5818 |
0.08 |
| chr3_105803714_105803865 | 0.38 |
Rap1a |
RAS-related protein 1a |
2453 |
0.17 |
| chr7_143792435_143792623 | 0.38 |
Nadsyn1 |
NAD synthetase 1 |
6312 |
0.11 |
| chr12_104262958_104263126 | 0.38 |
Serpina3i |
serine (or cysteine) peptidase inhibitor, clade A, member 3I |
80 |
0.94 |
| chr11_16919693_16920246 | 0.37 |
Egfr |
epidermal growth factor receptor |
14784 |
0.17 |
| chr13_98595769_98595925 | 0.37 |
Gm4815 |
predicted gene 4815 |
17654 |
0.12 |
| chr12_105046297_105046454 | 0.37 |
Glrx5 |
glutaredoxin 5 |
11159 |
0.1 |
| chr5_118294350_118294584 | 0.37 |
Gm25076 |
predicted gene, 25076 |
28018 |
0.15 |
| chr19_47330818_47330996 | 0.37 |
Sh3pxd2a |
SH3 and PX domains 2A |
16156 |
0.18 |
| chr12_29137061_29137232 | 0.37 |
Gm31333 |
predicted gene, 31333 |
2021 |
0.39 |
| chr7_44861566_44861717 | 0.36 |
Pnkp |
polynucleotide kinase 3'- phosphatase |
56 |
0.92 |
| chr14_46261947_46262118 | 0.36 |
Gm15217 |
predicted gene 15217 |
117382 |
0.05 |
| chr1_165849593_165849764 | 0.36 |
Gm37469 |
predicted gene, 37469 |
634 |
0.54 |
| chr9_20781366_20781683 | 0.35 |
Col5a3 |
collagen, type V, alpha 3 |
33543 |
0.09 |
| chr15_97184306_97184490 | 0.35 |
Gm32885 |
predicted gene, 32885 |
17154 |
0.22 |
| chr2_128738093_128738244 | 0.35 |
Gm14011 |
predicted gene 14011 |
15537 |
0.15 |
| chr15_79234881_79235032 | 0.35 |
Pick1 |
protein interacting with C kinase 1 |
5254 |
0.1 |
| chr8_123777594_123777758 | 0.35 |
Gm45865 |
predicted gene 45865 |
10125 |
0.08 |
| chr14_25710571_25710770 | 0.35 |
1700054O19Rik |
RIKEN cDNA 1700054O19 gene |
3397 |
0.17 |
| chr9_20804931_20805096 | 0.35 |
Col5a3 |
collagen, type V, alpha 3 |
10054 |
0.11 |
| chr4_41555100_41555359 | 0.35 |
Fam219a |
family with sequence similarity 219, member A |
14280 |
0.1 |
| chr17_29395434_29395615 | 0.35 |
Fgd2 |
FYVE, RhoGEF and PH domain containing 2 |
19023 |
0.11 |
| chr5_112486568_112486719 | 0.35 |
Sez6l |
seizure related 6 homolog like |
11516 |
0.16 |
| chr15_25914259_25914426 | 0.34 |
Retreg1 |
reticulophagy regulator 1 |
3918 |
0.25 |
| chr10_19942019_19942177 | 0.34 |
Map3k5 |
mitogen-activated protein kinase kinase kinase 5 |
6875 |
0.22 |
| chr15_78995837_78995995 | 0.34 |
Triobp |
TRIO and F-actin binding protein |
6066 |
0.08 |
| chr13_111809911_111810122 | 0.33 |
Gm15327 |
predicted gene 15327 |
452 |
0.67 |
| chr2_25095384_25095554 | 0.33 |
Noxa1 |
NADPH oxidase activator 1 |
320 |
0.75 |
| chr2_50964157_50964313 | 0.33 |
Gm13498 |
predicted gene 13498 |
54551 |
0.16 |
| chr11_78697500_78697668 | 0.33 |
Nlk |
nemo like kinase |
211 |
0.92 |
| chr13_43236729_43237131 | 0.32 |
Tbc1d7 |
TBC1 domain family, member 7 |
65429 |
0.1 |
| chr13_113530199_113530540 | 0.32 |
4921509O07Rik |
RIKEN cDNA 4921509O07 gene |
27525 |
0.13 |
| chr9_47254607_47254758 | 0.32 |
Gm31816 |
predicted gene, 31816 |
16061 |
0.23 |
| chr1_88976364_88976553 | 0.32 |
1700067G17Rik |
RIKEN cDNA 1700067G17 gene |
39655 |
0.14 |
| chr15_82147771_82148080 | 0.32 |
Srebf2 |
sterol regulatory element binding factor 2 |
88 |
0.94 |
| chr4_60419310_60419496 | 0.32 |
Mup9 |
major urinary protein 9 |
1181 |
0.4 |
| chr16_48767631_48767807 | 0.31 |
Trat1 |
T cell receptor associated transmembrane adaptor 1 |
4237 |
0.22 |
| chr6_88737071_88737256 | 0.31 |
Gm44005 |
predicted gene, 44005 |
1029 |
0.4 |
| chr6_125361870_125362038 | 0.31 |
Tnfrsf1a |
tumor necrosis factor receptor superfamily, member 1a |
1527 |
0.28 |
| chr5_89033271_89033463 | 0.31 |
Slc4a4 |
solute carrier family 4 (anion exchanger), member 4 |
5275 |
0.32 |
| chr11_87025481_87025638 | 0.31 |
Gm22387 |
predicted gene, 22387 |
28296 |
0.11 |
| chr8_13670213_13670364 | 0.31 |
Rasa3 |
RAS p21 protein activator 3 |
7163 |
0.18 |
| chr2_52918246_52918431 | 0.31 |
Fmnl2 |
formin-like 2 |
60470 |
0.14 |
| chr1_90909242_90909420 | 0.31 |
Mlph |
melanophilin |
5754 |
0.17 |
| chr3_65958418_65958847 | 0.31 |
Ccnl1 |
cyclin L1 |
383 |
0.66 |
| chr14_47057457_47057608 | 0.30 |
Samd4 |
sterile alpha motif domain containing 4 |
11442 |
0.16 |
| chr2_181677237_181677441 | 0.30 |
Tcea2 |
transcription elongation factor A (SII), 2 |
2971 |
0.12 |
| chr19_26733384_26733559 | 0.30 |
Smarca2 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
3472 |
0.3 |
| chr12_28898892_28899474 | 0.30 |
Gm31508 |
predicted gene, 31508 |
11046 |
0.17 |
| chr12_110219862_110220027 | 0.30 |
Gm40576 |
predicted gene, 40576 |
16418 |
0.11 |
| chr13_42052837_42053016 | 0.30 |
Hivep1 |
human immunodeficiency virus type I enhancer binding protein 1 |
522 |
0.8 |
| chr2_25534308_25534469 | 0.30 |
Traf2 |
TNF receptor-associated factor 2 |
4752 |
0.08 |
| chr14_117648617_117648937 | 0.30 |
Mir6239 |
microRNA 6239 |
305070 |
0.01 |
| chr15_82390095_82390257 | 0.30 |
Mir7684 |
microRNA 7684 |
3802 |
0.07 |
| chr8_45267218_45267382 | 0.30 |
F11 |
coagulation factor XI |
5269 |
0.18 |
| chr16_4029500_4029663 | 0.30 |
Dnase1 |
deoxyribonuclease I |
7361 |
0.1 |
| chr3_97620588_97620739 | 0.30 |
Fmo5 |
flavin containing monooxygenase 5 |
8141 |
0.14 |
| chr9_35318763_35318962 | 0.29 |
Gm33838 |
predicted gene, 33838 |
11263 |
0.13 |
| chr6_116654223_116654391 | 0.29 |
Depp1 |
DEPP1 autophagy regulator |
3611 |
0.13 |
| chr8_84660024_84660186 | 0.29 |
Ier2 |
immediate early response 2 |
2749 |
0.14 |
| chr11_7198962_7199113 | 0.29 |
Igfbp1 |
insulin-like growth factor binding protein 1 |
1255 |
0.43 |
| chr16_35363303_35363500 | 0.29 |
Sec22a |
SEC22 homolog A, vesicle trafficking protein |
414 |
0.79 |
| chr10_81430090_81430281 | 0.29 |
Nfic |
nuclear factor I/C |
820 |
0.33 |
| chr8_117368850_117369012 | 0.29 |
Cmip |
c-Maf inducing protein |
19761 |
0.22 |
| chr11_82016294_82016449 | 0.29 |
Gm31522 |
predicted gene, 31522 |
3596 |
0.16 |
| chr10_80764726_80764877 | 0.29 |
Gm17151 |
predicted gene 17151 |
4186 |
0.1 |
| chr6_119402004_119402155 | 0.29 |
Adipor2 |
adiponectin receptor 2 |
11831 |
0.2 |
| chr13_93989307_93989458 | 0.29 |
Gm47216 |
predicted gene, 47216 |
2386 |
0.28 |
| chr6_119902151_119902479 | 0.29 |
4930540M05Rik |
RIKEN cDNA 4930540M05 gene |
338 |
0.51 |
| chr3_83029428_83029640 | 0.29 |
Fga |
fibrinogen alpha chain |
3319 |
0.18 |
| chr19_46133855_46134398 | 0.28 |
Elovl3 |
elongation of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 3 |
2229 |
0.19 |
| chr19_57553071_57553238 | 0.28 |
Gm50285 |
predicted gene, 50285 |
8931 |
0.15 |
| chr11_49088297_49088883 | 0.28 |
Gm12188 |
predicted gene 12188 |
47 |
0.79 |
| chr19_46137654_46137874 | 0.28 |
Pitx3 |
paired-like homeodomain transcription factor 3 |
475 |
0.71 |
| chr4_61779591_61780024 | 0.28 |
Mup19 |
major urinary protein 19 |
2417 |
0.19 |
| chr10_127067027_127067252 | 0.28 |
Cdk4 |
cyclin-dependent kinase 4 |
1063 |
0.24 |
| chr5_53042406_53042593 | 0.28 |
Slc34a2 |
solute carrier family 34 (sodium phosphate), member 2 |
4418 |
0.19 |
| chr2_85107051_85107210 | 0.28 |
4930443O20Rik |
RIKEN cDNA 4930443O20 gene |
20669 |
0.11 |
| chr13_112192610_112192944 | 0.27 |
Gm48802 |
predicted gene, 48802 |
20709 |
0.16 |
| chr17_46674852_46675062 | 0.27 |
Rrp36 |
ribosomal RNA processing 36 homolog (S. cerevisiae) |
605 |
0.51 |
| chr14_76179509_76179660 | 0.27 |
Rps2-ps6 |
ribosomal protein S2, pseudogene 6 |
57961 |
0.11 |
| chr11_4974491_4974717 | 0.27 |
Ap1b1 |
adaptor protein complex AP-1, beta 1 subunit |
12220 |
0.13 |
| chr4_136174341_136174511 | 0.27 |
E2f2 |
E2F transcription factor 2 |
2032 |
0.24 |
| chr7_115564788_115564958 | 0.27 |
Sox6 |
SRY (sex determining region Y)-box 6 |
33023 |
0.24 |
| chr9_108083627_108083864 | 0.26 |
Rnf123 |
ring finger protein 123 |
399 |
0.46 |
| chr19_42780063_42780262 | 0.26 |
Hps1 |
HPS1, biogenesis of lysosomal organelles complex 3 subunit 1 |
184 |
0.9 |
| chr5_89355824_89355979 | 0.26 |
Gc |
vitamin D binding protein |
79727 |
0.11 |
| chr4_61301111_61301509 | 0.26 |
Mup14 |
major urinary protein 14 |
2055 |
0.32 |
| chr2_52930153_52930305 | 0.26 |
Fmnl2 |
formin-like 2 |
72361 |
0.12 |
| chr6_125575019_125575372 | 0.26 |
Vwf |
Von Willebrand factor |
8944 |
0.21 |
| chr5_151345483_151345662 | 0.26 |
1700028E10Rik |
RIKEN cDNA 1700028E10 gene |
23103 |
0.16 |
| chr10_84886268_84886436 | 0.26 |
Ric8b |
RIC8 guanine nucleotide exchange factor B |
31264 |
0.17 |
| chr14_60414955_60415106 | 0.26 |
Gm6066 |
predicted gene 6066 |
26949 |
0.17 |
| chr16_91355282_91355468 | 0.26 |
Gm24695 |
predicted gene, 24695 |
5591 |
0.13 |
| chr1_40680680_40680831 | 0.26 |
Slc9a2 |
solute carrier family 9 (sodium/hydrogen exchanger), member 2 |
181 |
0.95 |
| chr7_19817545_19817767 | 0.26 |
Gm16174 |
predicted gene 16174 |
1103 |
0.24 |
| chr2_32875662_32875824 | 0.25 |
Gm13523 |
predicted gene 13523 |
258 |
0.58 |
| chr1_72832378_72832635 | 0.25 |
Igfbp2 |
insulin-like growth factor binding protein 2 |
7184 |
0.23 |
| chr4_120283294_120283461 | 0.25 |
Foxo6 |
forkhead box O6 |
3972 |
0.25 |
| chr11_114867640_114867800 | 0.25 |
Gprc5c |
G protein-coupled receptor, family C, group 5, member C |
3228 |
0.17 |
| chr12_85698292_85698454 | 0.25 |
Batf |
basic leucine zipper transcription factor, ATF-like |
9081 |
0.13 |
| chr8_106138653_106138804 | 0.25 |
1810019D21Rik |
RIKEN cDNA 1810019D21 gene |
1663 |
0.21 |
| chr4_20017927_20018324 | 0.25 |
Ttpa |
tocopherol (alpha) transfer protein |
9697 |
0.19 |
| chr7_101302097_101302264 | 0.25 |
Atg16l2 |
autophagy related 16-like 2 (S. cerevisiae) |
71 |
0.95 |
| chr9_66620785_66620957 | 0.25 |
Usp3 |
ubiquitin specific peptidase 3 |
27729 |
0.17 |
| chr4_43492855_43493028 | 0.25 |
Ccdc107 |
coiled-coil domain containing 107 |
41 |
0.58 |
| chr12_102453853_102454036 | 0.25 |
Gm30198 |
predicted gene, 30198 |
6616 |
0.17 |
| chr2_116990270_116990421 | 0.25 |
Gm29340 |
predicted gene 29340 |
13917 |
0.19 |
| chr2_167853451_167853633 | 0.25 |
Gm14319 |
predicted gene 14319 |
5043 |
0.19 |
| chr2_133427180_133427333 | 0.25 |
A430048G15Rik |
RIKEN cDNA A430048G15 gene |
4839 |
0.26 |
| chr2_38378853_38379004 | 0.24 |
Gm13584 |
predicted gene 13584 |
14948 |
0.14 |
| chr18_16213890_16214071 | 0.24 |
Gm7665 |
predicted pseudogene 7665 |
60964 |
0.14 |
| chr5_35755279_35755445 | 0.24 |
Ablim2 |
actin-binding LIM protein 2 |
2518 |
0.26 |
| chr4_61436878_61437320 | 0.24 |
Mup15 |
major urinary protein 15 |
2644 |
0.27 |
| chr10_60280489_60280673 | 0.24 |
Psap |
prosaposin |
2953 |
0.25 |
| chr11_102409825_102409994 | 0.24 |
Slc25a39 |
solute carrier family 25, member 39 |
1963 |
0.17 |
| chr11_97920869_97921025 | 0.24 |
Gm22461 |
predicted gene, 22461 |
20793 |
0.1 |
| chr7_141335382_141335533 | 0.24 |
Tmem80 |
transmembrane protein 80 |
1799 |
0.16 |
| chr19_36627002_36627176 | 0.24 |
Hectd2os |
Hectd2, opposite strand |
1065 |
0.57 |
| chr2_119546518_119546691 | 0.24 |
Exd1 |
exonuclease 3'-5' domain containing 1 |
590 |
0.55 |
| chr6_94169886_94170336 | 0.24 |
Magi1 |
membrane associated guanylate kinase, WW and PDZ domain containing 1 |
112914 |
0.06 |
| chr12_26356269_26356452 | 0.23 |
4930549C15Rik |
RIKEN cDNA 4930549C15 gene |
9643 |
0.19 |
| chr8_75385238_75385564 | 0.23 |
Umpk-ps |
uridine monophosphate kinase, pseudogene |
1599 |
0.31 |
| chr10_93493384_93493577 | 0.23 |
Hal |
histidine ammonia lyase |
2925 |
0.2 |
| chr1_131473746_131474085 | 0.23 |
Gm24273 |
predicted gene, 24273 |
24887 |
0.12 |
| chr16_35742175_35742539 | 0.23 |
Gm25967 |
predicted gene, 25967 |
20124 |
0.13 |
| chr18_46718542_46718894 | 0.23 |
Cdo1 |
cysteine dioxygenase 1, cytosolic |
9311 |
0.13 |
| chr4_133084344_133084978 | 0.23 |
Ahdc1 |
AT hook, DNA binding motif, containing 1 |
18412 |
0.17 |
| chr19_5810333_5810765 | 0.23 |
Malat1 |
metastasis associated lung adenocarcinoma transcript 1 (non-coding RNA) |
7877 |
0.08 |
| chr4_40239674_40239847 | 0.23 |
Ddx58 |
DEAD (Asp-Glu-Ala-Asp) box polypeptide 58 |
5 |
0.97 |
| chr17_24204987_24205163 | 0.23 |
Tbc1d24 |
TBC1 domain family, member 24 |
407 |
0.66 |
| chr10_67288327_67288543 | 0.23 |
Nrbf2 |
nuclear receptor binding factor 2 |
3130 |
0.22 |
| chrX_20862192_20862360 | 0.23 |
Mir5617 |
microRNA 5617 |
906 |
0.41 |
| chr18_4964609_4964760 | 0.23 |
Svil |
supervillin |
29129 |
0.24 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.1 | 0.4 | GO:0070427 | nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070427) |
| 0.1 | 0.3 | GO:0072367 | regulation of lipid transport by regulation of transcription from RNA polymerase II promoter(GO:0072367) |
| 0.1 | 0.3 | GO:0052173 | response to defenses of other organism involved in symbiotic interaction(GO:0052173) response to host defenses(GO:0052200) response to host(GO:0075136) |
| 0.1 | 0.3 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.1 | 0.1 | GO:0032079 | positive regulation of endodeoxyribonuclease activity(GO:0032079) |
| 0.1 | 0.2 | GO:0019448 | cysteine catabolic process(GO:0009093) L-cysteine catabolic process(GO:0019448) L-cysteine metabolic process(GO:0046439) |
| 0.1 | 0.3 | GO:0060486 | Clara cell differentiation(GO:0060486) |
| 0.1 | 0.1 | GO:0032911 | negative regulation of transforming growth factor beta1 production(GO:0032911) |
| 0.1 | 0.2 | GO:0045472 | response to ether(GO:0045472) |
| 0.1 | 0.2 | GO:1904742 | regulation of telomeric DNA binding(GO:1904742) |
| 0.1 | 0.3 | GO:0052805 | histidine catabolic process(GO:0006548) imidazole-containing compound catabolic process(GO:0052805) |
| 0.1 | 0.2 | GO:0034473 | U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) |
| 0.1 | 0.5 | GO:0035234 | ectopic germ cell programmed cell death(GO:0035234) |
| 0.1 | 0.3 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.1 | 0.2 | GO:0034395 | regulation of transcription from RNA polymerase II promoter in response to iron(GO:0034395) |
| 0.1 | 0.2 | GO:0009597 | detection of virus(GO:0009597) |
| 0.1 | 0.1 | GO:1904179 | positive regulation of adipose tissue development(GO:1904179) |
| 0.0 | 0.1 | GO:2000313 | fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060825) regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000313) |
| 0.0 | 0.5 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.0 | 0.2 | GO:0060264 | regulation of respiratory burst involved in inflammatory response(GO:0060264) |
| 0.0 | 0.1 | GO:0003289 | atrial septum primum morphogenesis(GO:0003289) |
| 0.0 | 0.2 | GO:0044034 | negative stranded viral RNA replication(GO:0039689) multi-organism biosynthetic process(GO:0044034) |
| 0.0 | 0.1 | GO:0010916 | regulation of very-low-density lipoprotein particle clearance(GO:0010915) negative regulation of very-low-density lipoprotein particle clearance(GO:0010916) |
| 0.0 | 0.2 | GO:0070842 | aggresome assembly(GO:0070842) |
| 0.0 | 0.2 | GO:0048069 | eye pigmentation(GO:0048069) |
| 0.0 | 0.1 | GO:0006701 | progesterone biosynthetic process(GO:0006701) |
| 0.0 | 0.3 | GO:0033136 | serine phosphorylation of STAT3 protein(GO:0033136) |
| 0.0 | 0.1 | GO:2000823 | regulation of androgen receptor activity(GO:2000823) |
| 0.0 | 0.1 | GO:0006768 | biotin metabolic process(GO:0006768) |
| 0.0 | 0.2 | GO:1901896 | positive regulation of calcium-transporting ATPase activity(GO:1901896) |
| 0.0 | 0.1 | GO:0060375 | regulation of mast cell differentiation(GO:0060375) |
| 0.0 | 0.1 | GO:0031508 | pericentric heterochromatin assembly(GO:0031508) |
| 0.0 | 0.1 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.0 | 0.2 | GO:1902237 | positive regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway(GO:1902237) |
| 0.0 | 0.2 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.0 | 0.2 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.0 | 0.6 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.0 | 0.1 | GO:2001171 | positive regulation of ATP biosynthetic process(GO:2001171) |
| 0.0 | 0.1 | GO:0035771 | interleukin-4-mediated signaling pathway(GO:0035771) |
| 0.0 | 0.0 | GO:0045002 | double-strand break repair via single-strand annealing(GO:0045002) |
| 0.0 | 0.1 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 0.0 | 0.1 | GO:2000564 | regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000564) |
| 0.0 | 0.1 | GO:0035880 | embryonic nail plate morphogenesis(GO:0035880) |
| 0.0 | 0.2 | GO:0009115 | xanthine catabolic process(GO:0009115) |
| 0.0 | 0.1 | GO:0009414 | response to water deprivation(GO:0009414) |
| 0.0 | 0.1 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.0 | 0.1 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.0 | 0.1 | GO:1904479 | negative regulation of intestinal absorption(GO:1904479) |
| 0.0 | 0.2 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.0 | 0.1 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.0 | 0.1 | GO:2000553 | positive regulation of T-helper 2 cell cytokine production(GO:2000553) |
| 0.0 | 0.1 | GO:0072531 | pyrimidine-containing compound transmembrane transport(GO:0072531) |
| 0.0 | 0.1 | GO:0035425 | autocrine signaling(GO:0035425) |
| 0.0 | 0.0 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.0 | 0.3 | GO:0042511 | positive regulation of tyrosine phosphorylation of Stat1 protein(GO:0042511) |
| 0.0 | 0.1 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 | 0.2 | GO:0099515 | actin filament-based transport(GO:0099515) |
| 0.0 | 0.1 | GO:1990314 | cellular response to insulin-like growth factor stimulus(GO:1990314) |
| 0.0 | 0.2 | GO:1901409 | regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.0 | 0.1 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.2 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.0 | 0.1 | GO:0035790 | platelet-derived growth factor receptor-alpha signaling pathway(GO:0035790) |
| 0.0 | 0.1 | GO:0045950 | negative regulation of mitotic recombination(GO:0045950) |
| 0.0 | 0.1 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.0 | 0.4 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.0 | 0.1 | GO:0045347 | negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 0.0 | 0.1 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.0 | 0.1 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 0.0 | 0.2 | GO:0060263 | regulation of respiratory burst(GO:0060263) |
| 0.0 | 0.1 | GO:0010626 | negative regulation of Schwann cell proliferation(GO:0010626) |
| 0.0 | 0.1 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.0 | 0.1 | GO:1902809 | regulation of skeletal muscle fiber differentiation(GO:1902809) |
| 0.0 | 0.2 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.0 | 0.1 | GO:0006566 | threonine metabolic process(GO:0006566) |
| 0.0 | 0.1 | GO:0043133 | hindgut contraction(GO:0043133) regulation of hindgut contraction(GO:0043134) |
| 0.0 | 0.0 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 0.1 | GO:0036394 | amylase secretion(GO:0036394) |
| 0.0 | 0.1 | GO:0055005 | ventricular cardiac myofibril assembly(GO:0055005) |
| 0.0 | 0.2 | GO:0060391 | positive regulation of SMAD protein import into nucleus(GO:0060391) |
| 0.0 | 0.0 | GO:0048320 | axial mesoderm formation(GO:0048320) |
| 0.0 | 0.1 | GO:0065001 | specification of axis polarity(GO:0065001) |
| 0.0 | 0.2 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 | 0.0 | GO:0003162 | atrioventricular node development(GO:0003162) |
| 0.0 | 0.1 | GO:1990086 | lens fiber cell apoptotic process(GO:1990086) |
| 0.0 | 0.1 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.0 | 0.0 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.0 | 0.1 | GO:0006982 | response to lipid hydroperoxide(GO:0006982) |
| 0.0 | 0.1 | GO:0051791 | medium-chain fatty acid metabolic process(GO:0051791) |
| 0.0 | 0.2 | GO:2001223 | negative regulation of neuron migration(GO:2001223) |
| 0.0 | 0.0 | GO:0071725 | response to triacyl bacterial lipopeptide(GO:0071725) cellular response to triacyl bacterial lipopeptide(GO:0071727) |
| 0.0 | 0.1 | GO:2001286 | regulation of caveolin-mediated endocytosis(GO:2001286) |
| 0.0 | 0.2 | GO:0098719 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.1 | GO:0097252 | oligodendrocyte apoptotic process(GO:0097252) |
| 0.0 | 0.1 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.0 | 0.1 | GO:0038171 | cannabinoid signaling pathway(GO:0038171) endocannabinoid signaling pathway(GO:0071926) |
| 0.0 | 0.1 | GO:0031547 | brain-derived neurotrophic factor receptor signaling pathway(GO:0031547) |
| 0.0 | 0.1 | GO:0070948 | regulation of neutrophil mediated cytotoxicity(GO:0070948) regulation of neutrophil mediated killing of symbiont cell(GO:0070949) |
| 0.0 | 0.1 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.0 | 0.1 | GO:0032417 | positive regulation of sodium:proton antiporter activity(GO:0032417) |
| 0.0 | 0.1 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.0 | 0.1 | GO:0071830 | chylomicron remnant clearance(GO:0034382) triglyceride-rich lipoprotein particle clearance(GO:0071830) |
| 0.0 | 0.1 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.1 | GO:0033504 | floor plate development(GO:0033504) |
| 0.0 | 0.0 | GO:0045054 | constitutive secretory pathway(GO:0045054) |
| 0.0 | 0.1 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.0 | 0.2 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.0 | 0.0 | GO:1901679 | nucleotide transmembrane transport(GO:1901679) |
| 0.0 | 0.1 | GO:0061052 | negative regulation of cell growth involved in cardiac muscle cell development(GO:0061052) |
| 0.0 | 0.1 | GO:2000483 | negative regulation of interleukin-8 secretion(GO:2000483) |
| 0.0 | 0.4 | GO:0055090 | acylglycerol homeostasis(GO:0055090) triglyceride homeostasis(GO:0070328) |
| 0.0 | 0.1 | GO:0003072 | regulation of blood vessel size by renin-angiotensin(GO:0002034) renal control of peripheral vascular resistance involved in regulation of systemic arterial blood pressure(GO:0003072) |
| 0.0 | 0.1 | GO:0000820 | regulation of glutamine family amino acid metabolic process(GO:0000820) |
| 0.0 | 0.1 | GO:0002726 | positive regulation of T cell cytokine production(GO:0002726) |
| 0.0 | 0.1 | GO:0008611 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) cellular lipid biosynthetic process(GO:0097384) ether biosynthetic process(GO:1901503) |
| 0.0 | 0.0 | GO:0001828 | inner cell mass cellular morphogenesis(GO:0001828) |
| 0.0 | 0.3 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.0 | 0.1 | GO:0030576 | Cajal body organization(GO:0030576) |
| 0.0 | 0.1 | GO:0060737 | prostate gland morphogenetic growth(GO:0060737) |
| 0.0 | 0.1 | GO:0018199 | peptidyl-glutamine modification(GO:0018199) |
| 0.0 | 0.1 | GO:0034162 | toll-like receptor 9 signaling pathway(GO:0034162) |
| 0.0 | 0.0 | GO:0015879 | carnitine transport(GO:0015879) |
| 0.0 | 0.1 | GO:0010288 | response to lead ion(GO:0010288) |
| 0.0 | 0.2 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.0 | 0.2 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 0.0 | GO:1902263 | apoptotic process involved in embryonic digit morphogenesis(GO:1902263) |
| 0.0 | 0.0 | GO:0000189 | MAPK import into nucleus(GO:0000189) |
| 0.0 | 0.0 | GO:0003130 | BMP signaling pathway involved in heart induction(GO:0003130) endodermal-mesodermal cell signaling(GO:0003133) endodermal-mesodermal cell signaling involved in heart induction(GO:0003134) |
| 0.0 | 0.1 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 0.0 | 0.1 | GO:1900025 | negative regulation of substrate adhesion-dependent cell spreading(GO:1900025) |
| 0.0 | 0.1 | GO:0034116 | positive regulation of heterotypic cell-cell adhesion(GO:0034116) |
| 0.0 | 0.1 | GO:0032075 | positive regulation of nuclease activity(GO:0032075) |
| 0.0 | 0.2 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 0.0 | GO:0090210 | regulation of establishment of blood-brain barrier(GO:0090210) |
| 0.0 | 0.0 | GO:1903689 | regulation of wound healing, spreading of epidermal cells(GO:1903689) |
| 0.0 | 0.0 | GO:0035963 | response to interleukin-13(GO:0035962) cellular response to interleukin-13(GO:0035963) |
| 0.0 | 0.0 | GO:1902564 | negative regulation of neutrophil activation(GO:1902564) |
| 0.0 | 0.0 | GO:0006114 | glycerol biosynthetic process(GO:0006114) alditol biosynthetic process(GO:0019401) |
| 0.0 | 0.1 | GO:1904415 | regulation of xenophagy(GO:1904415) positive regulation of xenophagy(GO:1904417) |
| 0.0 | 0.1 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.0 | 0.1 | GO:1904707 | positive regulation of vascular smooth muscle cell proliferation(GO:1904707) |
| 0.0 | 0.0 | GO:0021775 | smoothened signaling pathway involved in ventral spinal cord interneuron specification(GO:0021775) smoothened signaling pathway involved in spinal cord motor neuron cell fate specification(GO:0021776) |
| 0.0 | 0.1 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.0 | 0.1 | GO:2001214 | positive regulation of vasculogenesis(GO:2001214) |
| 0.0 | 0.0 | GO:0033091 | positive regulation of immature T cell proliferation(GO:0033091) |
| 0.0 | 0.2 | GO:0019985 | translesion synthesis(GO:0019985) |
| 0.0 | 0.0 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.0 | 0.3 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.0 | 0.1 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.0 | 0.1 | GO:0048318 | axial mesoderm development(GO:0048318) |
| 0.0 | 0.1 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.0 | GO:2000726 | negative regulation of cardiac muscle cell differentiation(GO:2000726) |
| 0.0 | 0.1 | GO:0021914 | negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021914) |
| 0.0 | 0.0 | GO:0001712 | ectodermal cell fate commitment(GO:0001712) |
| 0.0 | 0.0 | GO:0032202 | telomere assembly(GO:0032202) |
| 0.0 | 0.2 | GO:0003351 | epithelial cilium movement(GO:0003351) |
| 0.0 | 0.0 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.0 | 0.1 | GO:1902033 | regulation of hematopoietic stem cell proliferation(GO:1902033) |
| 0.0 | 0.1 | GO:0044068 | modulation by symbiont of host cellular process(GO:0044068) |
| 0.0 | 0.1 | GO:0032286 | central nervous system myelin maintenance(GO:0032286) |
| 0.0 | 0.0 | GO:2000418 | positive regulation of eosinophil migration(GO:2000418) |
| 0.0 | 0.0 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.0 | 0.1 | GO:0072675 | osteoclast fusion(GO:0072675) |
| 0.0 | 0.0 | GO:0060051 | negative regulation of protein glycosylation(GO:0060051) |
| 0.0 | 0.0 | GO:0060956 | endocardial cell differentiation(GO:0060956) |
| 0.0 | 0.0 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.0 | 0.0 | GO:0014045 | establishment of endothelial blood-brain barrier(GO:0014045) |
| 0.0 | 0.0 | GO:0009756 | carbohydrate mediated signaling(GO:0009756) hexose mediated signaling(GO:0009757) sugar mediated signaling pathway(GO:0010182) glucose mediated signaling pathway(GO:0010255) |
| 0.0 | 0.0 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.0 | 0.0 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 0.0 | 0.1 | GO:2000169 | regulation of peptidyl-cysteine S-nitrosylation(GO:2000169) |
| 0.0 | 0.0 | GO:0015846 | polyamine transport(GO:0015846) |
| 0.0 | 0.2 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.0 | 0.1 | GO:0031584 | activation of phospholipase D activity(GO:0031584) |
| 0.0 | 0.1 | GO:0042985 | negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 0.0 | 0.0 | GO:0051387 | negative regulation of neurotrophin TRK receptor signaling pathway(GO:0051387) |
| 0.0 | 0.0 | GO:0097155 | fasciculation of sensory neuron axon(GO:0097155) |
| 0.0 | 0.0 | GO:0043379 | memory T cell differentiation(GO:0043379) |
| 0.0 | 0.0 | GO:0048254 | snoRNA localization(GO:0048254) |
| 0.0 | 0.0 | GO:2000304 | positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 0.0 | 0.0 | GO:0033030 | negative regulation of neutrophil apoptotic process(GO:0033030) |
| 0.0 | 0.0 | GO:0070343 | white fat cell proliferation(GO:0070343) regulation of white fat cell proliferation(GO:0070350) |
| 0.0 | 0.0 | GO:0034729 | histone H3-K79 methylation(GO:0034729) |
| 0.0 | 0.1 | GO:0045717 | negative regulation of fatty acid biosynthetic process(GO:0045717) |
| 0.0 | 0.0 | GO:0008228 | opsonization(GO:0008228) |
| 0.0 | 0.0 | GO:0071816 | protein insertion into ER membrane(GO:0045048) tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 0.0 | 0.1 | GO:1902036 | regulation of hematopoietic stem cell differentiation(GO:1902036) |
| 0.0 | 0.0 | GO:0035739 | CD4-positive, alpha-beta T cell proliferation(GO:0035739) |
| 0.0 | 0.0 | GO:0023021 | termination of signal transduction(GO:0023021) |
| 0.0 | 0.0 | GO:0010886 | positive regulation of cholesterol storage(GO:0010886) |
| 0.0 | 0.0 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.1 | GO:0045064 | T-helper 2 cell differentiation(GO:0045064) |
| 0.0 | 0.0 | GO:0050904 | diapedesis(GO:0050904) |
| 0.0 | 0.0 | GO:2000394 | regulation of lamellipodium morphogenesis(GO:2000392) positive regulation of lamellipodium morphogenesis(GO:2000394) |
| 0.0 | 0.0 | GO:0070102 | interleukin-6-mediated signaling pathway(GO:0070102) |
| 0.0 | 0.0 | GO:0045583 | regulation of cytotoxic T cell differentiation(GO:0045583) positive regulation of cytotoxic T cell differentiation(GO:0045585) |
| 0.0 | 0.1 | GO:0072673 | lamellipodium morphogenesis(GO:0072673) |
| 0.0 | 0.0 | GO:0060596 | mammary placode formation(GO:0060596) |
| 0.0 | 0.0 | GO:0006069 | ethanol metabolic process(GO:0006067) ethanol oxidation(GO:0006069) |
| 0.0 | 0.0 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.0 | 0.0 | GO:1902445 | regulation of mitochondrial membrane permeability involved in programmed necrotic cell death(GO:1902445) |
| 0.0 | 0.0 | GO:0086045 | membrane depolarization during AV node cell action potential(GO:0086045) |
| 0.0 | 0.1 | GO:0015871 | choline transport(GO:0015871) |
| 0.0 | 0.1 | GO:0009437 | carnitine metabolic process(GO:0009437) |
| 0.0 | 0.1 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.0 | 0.0 | GO:0032802 | low-density lipoprotein particle receptor catabolic process(GO:0032802) regulation of low-density lipoprotein particle receptor catabolic process(GO:0032803) |
| 0.0 | 0.1 | GO:0046322 | negative regulation of fatty acid oxidation(GO:0046322) |
| 0.0 | 0.3 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.0 | 0.1 | GO:0019363 | pyridine nucleotide biosynthetic process(GO:0019363) |
| 0.0 | 0.0 | GO:0046499 | S-adenosylmethioninamine metabolic process(GO:0046499) |
| 0.0 | 0.0 | GO:0002434 | immune complex clearance(GO:0002434) |
| 0.0 | 0.0 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.0 | 0.0 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.0 | 0.0 | GO:0032792 | negative regulation of CREB transcription factor activity(GO:0032792) |
| 0.0 | 0.1 | GO:0033601 | positive regulation of mammary gland epithelial cell proliferation(GO:0033601) |
| 0.0 | 0.1 | GO:0060484 | lung-associated mesenchyme development(GO:0060484) |
| 0.0 | 0.2 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.0 | 0.0 | GO:0010898 | positive regulation of triglyceride catabolic process(GO:0010898) |
| 0.0 | 0.0 | GO:0051562 | negative regulation of mitochondrial calcium ion concentration(GO:0051562) |
| 0.0 | 0.1 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.0 | 0.0 | GO:0072429 | response to intra-S DNA damage checkpoint signaling(GO:0072429) |
| 0.0 | 0.0 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 0.1 | 0.3 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.1 | 0.2 | GO:0035355 | Toll-like receptor 2-Toll-like receptor 6 protein complex(GO:0035355) |
| 0.1 | 0.3 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.1 | 0.2 | GO:0097451 | glial limiting end-foot(GO:0097451) |
| 0.1 | 0.8 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.1 | GO:1990923 | PET complex(GO:1990923) |
| 0.0 | 0.1 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.0 | 0.2 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.0 | 0.2 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.0 | 0.3 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.2 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.1 | GO:0032127 | dense core granule membrane(GO:0032127) |
| 0.0 | 0.3 | GO:0045179 | apical cortex(GO:0045179) |
| 0.0 | 0.1 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 0.2 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.1 | GO:0098642 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.0 | 0.0 | GO:0032010 | phagolysosome(GO:0032010) |
| 0.0 | 0.2 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.0 | 0.1 | GO:0030891 | VCB complex(GO:0030891) |
| 0.0 | 0.1 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.0 | 0.2 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.1 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.1 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.0 | 0.3 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.1 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.0 | 0.3 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 0.3 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.3 | GO:0031082 | BLOC complex(GO:0031082) |
| 0.0 | 0.0 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.0 | 0.2 | GO:0005766 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.0 | 0.0 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.0 | 0.1 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 0.1 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.0 | 0.1 | GO:0000778 | condensed nuclear chromosome kinetochore(GO:0000778) |
| 0.0 | 0.2 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.2 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 0.1 | GO:0031010 | ISWI-type complex(GO:0031010) |
| 0.0 | 0.1 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.1 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.2 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.1 | GO:0005818 | aster(GO:0005818) |
| 0.0 | 0.0 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.0 | 0.1 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.0 | 0.0 | GO:0072379 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.0 | 0.0 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.1 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.1 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.0 | 0.2 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.1 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.0 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.0 | 0.0 | GO:0044194 | cytolytic granule(GO:0044194) |
| 0.0 | 0.0 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.1 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.0 | 0.1 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.3 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.1 | 0.3 | GO:0046404 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) |
| 0.1 | 0.4 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.1 | 0.3 | GO:0008525 | phosphatidylcholine transporter activity(GO:0008525) |
| 0.1 | 0.3 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.1 | 0.4 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.1 | 1.0 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.1 | 0.2 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.1 | 0.2 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 0.1 | 0.2 | GO:0004920 | interleukin-10 receptor activity(GO:0004920) |
| 0.0 | 0.2 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.0 | 0.3 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.1 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 0.1 | GO:0000171 | ribonuclease MRP activity(GO:0000171) |
| 0.0 | 0.1 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.0 | 0.1 | GO:0043120 | tumor necrosis factor binding(GO:0043120) |
| 0.0 | 0.2 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.0 | 0.1 | GO:0016401 | palmitoyl-CoA oxidase activity(GO:0016401) |
| 0.0 | 0.3 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.1 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.0 | 0.2 | GO:0004854 | xanthine dehydrogenase activity(GO:0004854) oxidoreductase activity, acting on CH or CH2 groups, NAD or NADP as acceptor(GO:0016726) |
| 0.0 | 0.2 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.1 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.0 | 0.0 | GO:0043546 | molybdopterin cofactor binding(GO:0043546) |
| 0.0 | 0.1 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.0 | 0.1 | GO:0043199 | sulfate binding(GO:0043199) |
| 0.0 | 0.1 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.0 | 0.1 | GO:0004528 | phosphodiesterase I activity(GO:0004528) |
| 0.0 | 0.1 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.0 | 0.1 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.0 | 0.2 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.2 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.0 | 0.1 | GO:0000403 | Y-form DNA binding(GO:0000403) |
| 0.0 | 0.1 | GO:0016822 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.0 | 0.1 | GO:0047751 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) cholestenone 5-alpha-reductase activity(GO:0047751) |
| 0.0 | 0.0 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.0 | 0.2 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.1 | GO:0000099 | sulfur amino acid transmembrane transporter activity(GO:0000099) |
| 0.0 | 0.1 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.0 | 0.1 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 0.0 | 0.1 | GO:0015119 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.0 | 0.1 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.0 | 0.1 | GO:0071535 | RING-like zinc finger domain binding(GO:0071535) |
| 0.0 | 0.2 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.0 | 0.1 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.0 | 0.1 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.0 | 0.0 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.0 | 0.1 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 0.1 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.0 | 0.1 | GO:0004611 | phosphoenolpyruvate carboxykinase activity(GO:0004611) |
| 0.0 | 0.1 | GO:0051880 | G-quadruplex DNA binding(GO:0051880) |
| 0.0 | 0.2 | GO:0015386 | sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.2 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.0 | 0.1 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.1 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.0 | 0.2 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.1 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.0 | 0.1 | GO:0070905 | serine binding(GO:0070905) |
| 0.0 | 0.1 | GO:0015205 | purine nucleobase transmembrane transporter activity(GO:0005345) pyrimidine nucleobase transmembrane transporter activity(GO:0005350) nucleobase transmembrane transporter activity(GO:0015205) |
| 0.0 | 0.1 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 0.1 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.0 | 0.2 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 0.1 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.1 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.0 | 0.1 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 0.0 | 0.1 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.1 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.0 | 0.1 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 0.0 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.0 | 0.1 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.0 | 0.3 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.0 | GO:0070546 | L-phenylalanine aminotransferase activity(GO:0070546) L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.0 | 0.1 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.1 | GO:0018644 | 3-(3-hydroxyphenyl)propionate hydroxylase activity(GO:0008688) 4-chlorobenzaldehyde oxidase activity(GO:0018471) 3,5-xylenol methylhydroxylase activity(GO:0018630) phenylacetate hydroxylase activity(GO:0018631) 4-nitrophenol 4-monooxygenase activity(GO:0018632) dimethyl sulfide monooxygenase activity(GO:0018633) alpha-pinene monooxygenase [NADH] activity(GO:0018634) 1-hydroxy-2-naphthoate hydroxylase activity(GO:0018637) toluene 4-monooxygenase activity(GO:0018638) xylene monooxygenase activity(GO:0018639) dibenzothiophene monooxygenase activity(GO:0018640) 6-hydroxy-3-methyl-2-oxo-1,2-dihydroquinoline 6-monooxygenase activity(GO:0018641) chlorophenol 4-monooxygenase activity(GO:0018642) carbon disulfide oxygenase activity(GO:0018643) toluene 2-monooxygenase activity(GO:0018644) 1-hydroxy-2-oxolimonene 1,2-monooxygenase activity(GO:0018646) phenanthrene 1,2-monooxygenase activity(GO:0018647) tetrahydrofuran hydroxylase activity(GO:0018649) styrene monooxygenase activity(GO:0018650) toluene-4-sulfonate monooxygenase activity(GO:0018651) toluene-sulfonate methyl-monooxygenase activity(GO:0018652) 3-methyl-2-oxo-1,2-dihydroquinoline 6-monooxygenase activity(GO:0018653) 2-hydroxy-phenylacetate hydroxylase activity(GO:0018654) 2-oxo-delta3-4,5,5-trimethylcyclopentenylacetyl-CoA 1,2-monooxygenase activity(GO:0018655) phenanthrene 3,4-monooxygenase activity(GO:0018656) toluene 3-monooxygenase activity(GO:0018657) 4-hydroxyphenylacetate,NADH:oxygen oxidoreductase (3-hydroxylating) activity(GO:0018660) limonene monooxygenase activity(GO:0019113) 2-methylnaphthalene hydroxylase activity(GO:0034526) 1-methylnaphthalene hydroxylase activity(GO:0034534) bisphenol A hydroxylase A activity(GO:0034560) salicylate 5-hydroxylase activity(GO:0034785) isobutylamine N-hydroxylase activity(GO:0034791) branched-chain dodecylbenzene sulfonate monooxygenase activity(GO:0034802) 3-HSA hydroxylase activity(GO:0034819) 4-hydroxypyridine-3-hydroxylase activity(GO:0034894) 2-octaprenyl-3-methyl-6-methoxy-1,4-benzoquinol hydroxylase activity(GO:0043719) 6-hydroxynicotinate 3-monooxygenase activity(GO:0043731) thalianol hydroxylase activity(GO:0080014) |
| 0.0 | 0.4 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.0 | 0.3 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 0.0 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.0 | 0.1 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) |
| 0.0 | 0.1 | GO:0015321 | sodium-dependent phosphate transmembrane transporter activity(GO:0015321) |
| 0.0 | 0.1 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.0 | 0.2 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.1 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.0 | 0.0 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) DNA (cytosine-5-)-methyltransferase activity, acting on CpG substrates(GO:0051718) |
| 0.0 | 0.0 | GO:0019961 | interferon binding(GO:0019961) |
| 0.0 | 0.0 | GO:0016635 | oxidoreductase activity, acting on the CH-CH group of donors, quinone or related compound as acceptor(GO:0016635) |
| 0.0 | 0.1 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.0 | 0.1 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.1 | GO:0048273 | mitogen-activated protein kinase p38 binding(GO:0048273) |
| 0.0 | 0.0 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.0 | 0.1 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.0 | 0.0 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 0.0 | 0.3 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.0 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.0 | 0.0 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.0 | 0.0 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.0 | 0.1 | GO:0034902 | alkyl sulfatase activity(GO:0018741) endosulfan hemisulfate sulfatase activity(GO:0034889) endosulfan sulfate hydrolase activity(GO:0034902) |
| 0.0 | 0.0 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.0 | 0.1 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.0 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 0.4 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.0 | GO:0010858 | calcium-dependent protein kinase regulator activity(GO:0010858) |
| 0.0 | 0.0 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.0 | 0.0 | GO:0000702 | oxidized base lesion DNA N-glycosylase activity(GO:0000702) |
| 0.0 | 0.0 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.0 | 0.1 | GO:0017127 | cholesterol transporter activity(GO:0017127) |
| 0.0 | 0.0 | GO:0042979 | ornithine decarboxylase regulator activity(GO:0042979) |
| 0.0 | 0.3 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 0.0 | GO:0004346 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.0 | 0.1 | GO:0015106 | bicarbonate transmembrane transporter activity(GO:0015106) |
| 0.0 | 0.0 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) |
| 0.0 | 0.2 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 0.4 | GO:0031593 | polyubiquitin binding(GO:0031593) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.9 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 0.0 | PID TCPTP PATHWAY | Signaling events mediated by TCPTP |
| 0.0 | 0.5 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.6 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 0.2 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.0 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 0.3 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.2 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 0.5 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.2 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 0.2 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.3 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 0.1 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
| 0.0 | 0.0 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.0 | 0.4 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.1 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.0 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 0.4 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.0 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 0.2 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 0.3 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.1 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.2 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.0 | 0.7 | REACTOME TRAF6 MEDIATED NFKB ACTIVATION | Genes involved in TRAF6 mediated NF-kB activation |
| 0.0 | 0.2 | REACTOME REGULATED PROTEOLYSIS OF P75NTR | Genes involved in Regulated proteolysis of p75NTR |
| 0.0 | 0.3 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.1 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 0.3 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.2 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 0.3 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.4 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.0 | 0.3 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.0 | 0.0 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.3 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.0 | 0.2 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.0 | REACTOME GPCR DOWNSTREAM SIGNALING | Genes involved in GPCR downstream signaling |
| 0.0 | 0.1 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.1 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.1 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.0 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 0.0 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.0 | 0.1 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.1 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 0.1 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.0 | 0.1 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.0 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.0 | 0.2 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.1 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.0 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.2 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.2 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.1 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.0 | 0.1 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.1 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.1 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.0 | 0.1 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.1 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.0 | REACTOME P75NTR RECRUITS SIGNALLING COMPLEXES | Genes involved in p75NTR recruits signalling complexes |