Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
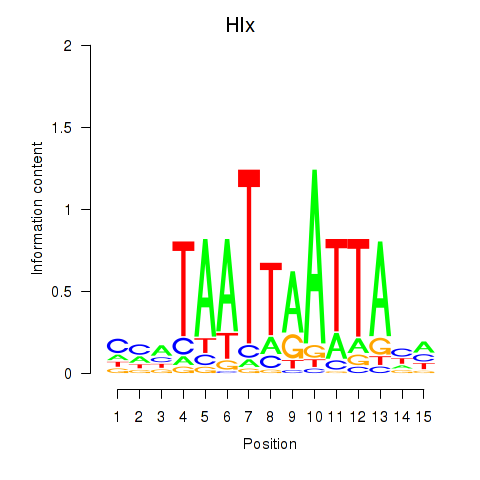
Results for Hlx
Z-value: 0.65
Transcription factors associated with Hlx
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hlx
|
ENSMUSG00000039377.6 | H2.0-like homeobox |
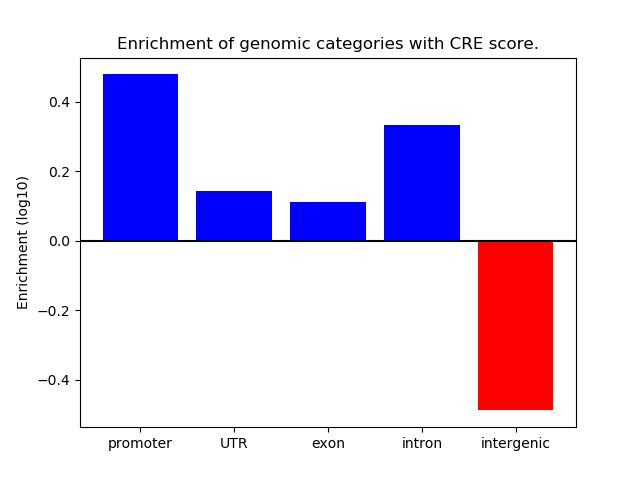
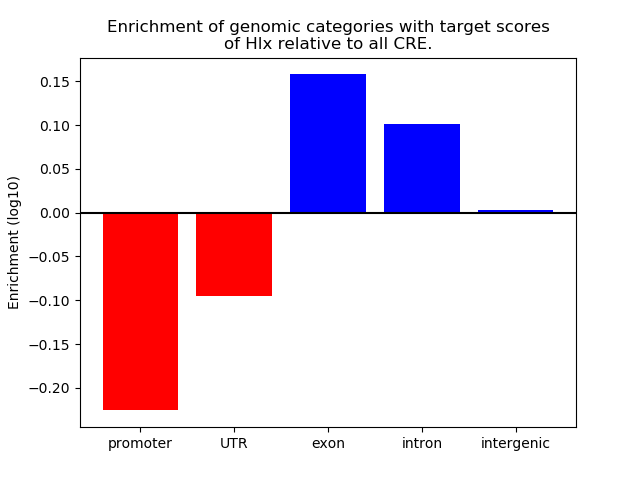
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr1_184732600_184732772 | Hlx | 67 | 0.962378 | -0.78 | 6.7e-02 | Click! |
| chr1_184729358_184729760 | Hlx | 2039 | 0.244262 | -0.78 | 6.8e-02 | Click! |
| chr1_184729963_184730114 | Hlx | 1560 | 0.305726 | 0.74 | 9.3e-02 | Click! |
| chr1_184733359_184733533 | Hlx | 827 | 0.525337 | -0.61 | 2.0e-01 | Click! |
| chr1_184734331_184734518 | Hlx | 1805 | 0.253962 | 0.56 | 2.5e-01 | Click! |
Activity of the Hlx motif across conditions
Conditions sorted by the z-value of the Hlx motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
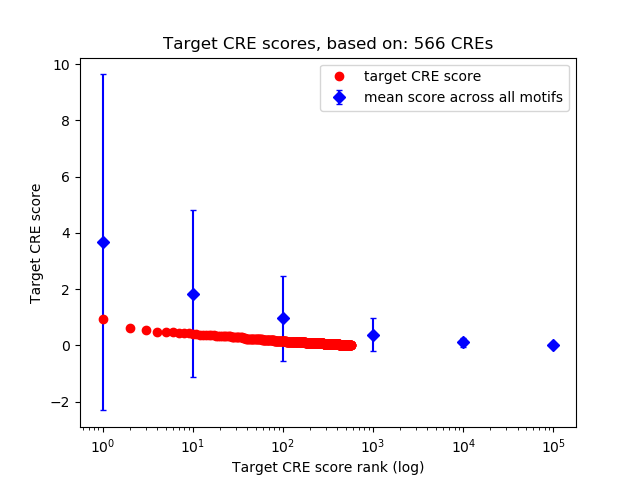
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr5_90338896_90339063 | 0.93 |
Ankrd17 |
ankyrin repeat domain 17 |
764 |
0.67 |
| chr9_106264996_106265377 | 0.60 |
Poc1a |
POC1 centriolar protein A |
15875 |
0.1 |
| chr10_89573725_89573906 | 0.54 |
Gm48087 |
predicted gene, 48087 |
18096 |
0.17 |
| chr1_195223205_195223404 | 0.48 |
Gm37887 |
predicted gene, 37887 |
538 |
0.73 |
| chr17_64607771_64607970 | 0.48 |
Man2a1 |
mannosidase 2, alpha 1 |
7134 |
0.26 |
| chr9_15301981_15302132 | 0.46 |
4931406C07Rik |
RIKEN cDNA 4931406C07 gene |
500 |
0.46 |
| chr16_24449611_24449762 | 0.45 |
Lpp |
LIM domain containing preferred translocation partner in lipoma |
1595 |
0.42 |
| chr5_86919547_86919811 | 0.44 |
Gm25211 |
predicted gene, 25211 |
3357 |
0.13 |
| chr2_34049474_34049625 | 0.43 |
C230014O12Rik |
RIKEN cDNA C230014O12 gene |
58180 |
0.11 |
| chr8_61563294_61563445 | 0.40 |
Palld |
palladin, cytoskeletal associated protein |
27770 |
0.22 |
| chrX_38400123_38400274 | 0.39 |
Gm4853 |
predicted pseudogene 4853 |
3263 |
0.22 |
| chr11_28696253_28696446 | 0.38 |
2810471M01Rik |
RIKEN cDNA 2810471M01 gene |
14785 |
0.17 |
| chr6_72120521_72121047 | 0.38 |
4933431G14Rik |
RIKEN cDNA 4933431G14 gene |
2156 |
0.2 |
| chr1_119646750_119646988 | 0.37 |
Epb41l5 |
erythrocyte membrane protein band 4.1 like 5 |
1748 |
0.32 |
| chr1_127787130_127787281 | 0.37 |
Ccnt2 |
cyclin T2 |
9366 |
0.16 |
| chr16_94606252_94606403 | 0.36 |
Gm15971 |
predicted gene 15971 |
14050 |
0.19 |
| chr5_150650487_150650670 | 0.36 |
N4bp2l2 |
NEDD4 binding protein 2-like 2 |
2317 |
0.19 |
| chr6_37510078_37510463 | 0.35 |
Akr1d1 |
aldo-keto reductase family 1, member D1 |
19903 |
0.21 |
| chr4_123990393_123990580 | 0.35 |
Gm12902 |
predicted gene 12902 |
64252 |
0.08 |
| chr2_157367239_157367418 | 0.34 |
Manbal |
mannosidase, beta A, lysosomal-like |
266 |
0.9 |
| chr6_119380906_119381057 | 0.34 |
Adipor2 |
adiponectin receptor 2 |
7705 |
0.21 |
| chr12_70985566_70985717 | 0.33 |
Psma3 |
proteasome subunit alpha 3 |
1251 |
0.39 |
| chr14_119831250_119831463 | 0.33 |
4930404K13Rik |
RIKEN cDNA 4930404K13 gene |
32378 |
0.19 |
| chrX_49489499_49489695 | 0.33 |
Arhgap36 |
Rho GTPase activating protein 36 |
3922 |
0.27 |
| chr19_29285240_29285391 | 0.32 |
Jak2 |
Janus kinase 2 |
32874 |
0.12 |
| chr7_83634897_83635261 | 0.32 |
Gm45838 |
predicted gene 45838 |
1708 |
0.22 |
| chr7_70356055_70356242 | 0.31 |
Nr2f2 |
nuclear receptor subfamily 2, group F, member 2 |
911 |
0.45 |
| chr14_79629212_79629374 | 0.30 |
Sugt1 |
SGT1, suppressor of G2 allele of SKP1 (S. cerevisiae) |
4654 |
0.19 |
| chr4_32300145_32300312 | 0.29 |
Bach2it1 |
BTB and CNC homology 2, intronic transcript 1 |
48467 |
0.14 |
| chr14_45487800_45488006 | 0.29 |
Fermt2 |
fermitin family member 2 |
3934 |
0.14 |
| chr6_17059434_17059627 | 0.29 |
Gm15473 |
predicted gene 15473 |
5466 |
0.23 |
| chr2_12925309_12925626 | 0.29 |
Pter |
phosphotriesterase related |
1366 |
0.49 |
| chr12_75472570_75472746 | 0.28 |
Gm47690 |
predicted gene, 47690 |
36087 |
0.17 |
| chr7_89401915_89402333 | 0.28 |
Tmem135 |
transmembrane protein 135 |
2098 |
0.22 |
| chr4_137313822_137313984 | 0.28 |
Gm25772 |
predicted gene, 25772 |
11656 |
0.14 |
| chr19_55111923_55112097 | 0.27 |
Gpam |
glycerol-3-phosphate acyltransferase, mitochondrial |
8919 |
0.2 |
| chr10_125785978_125786129 | 0.27 |
Lrig3 |
leucine-rich repeats and immunoglobulin-like domains 3 |
180115 |
0.03 |
| chr13_95326893_95327258 | 0.26 |
Zbed3 |
zinc finger, BED type containing 3 |
308 |
0.86 |
| chr6_38794603_38794986 | 0.24 |
Hipk2 |
homeodomain interacting protein kinase 2 |
23552 |
0.21 |
| chr11_85131557_85131734 | 0.24 |
Usp32 |
ubiquitin specific peptidase 32 |
8284 |
0.14 |
| chr3_138278438_138278607 | 0.24 |
Adh1 |
alcohol dehydrogenase 1 (class I) |
871 |
0.47 |
| chr2_127608341_127608498 | 0.24 |
Mrps5 |
mitochondrial ribosomal protein S5 |
8386 |
0.13 |
| chr11_4095549_4095867 | 0.23 |
Mtfp1 |
mitochondrial fission process 1 |
263 |
0.83 |
| chr2_166153854_166154005 | 0.23 |
Sulf2 |
sulfatase 2 |
494 |
0.79 |
| chr1_21315523_21315674 | 0.23 |
Gm21909 |
predicted gene, 21909 |
722 |
0.5 |
| chr12_51274477_51274668 | 0.23 |
Rps11-ps4 |
ribosomal protein S11, pseudogene 4 |
22915 |
0.22 |
| chr11_72010467_72010628 | 0.23 |
Gm23226 |
predicted gene, 23226 |
12208 |
0.15 |
| chr16_21826703_21826857 | 0.22 |
Map3k13 |
mitogen-activated protein kinase kinase kinase 13 |
838 |
0.48 |
| chr10_10426013_10426174 | 0.22 |
Adgb |
androglobin |
3746 |
0.26 |
| chr10_87040962_87041138 | 0.22 |
1700113H08Rik |
RIKEN cDNA 1700113H08 gene |
16995 |
0.16 |
| chr2_154875946_154876097 | 0.22 |
Eif2s2 |
eukaryotic translation initiation factor 2, subunit 2 (beta) |
16761 |
0.19 |
| chr8_109803384_109803547 | 0.22 |
Ap1g1 |
adaptor protein complex AP-1, gamma 1 subunit |
24559 |
0.12 |
| chr4_124809297_124809476 | 0.21 |
Mtf1 |
metal response element binding transcription factor 1 |
6708 |
0.1 |
| chr11_32651005_32651314 | 0.21 |
Fbxw11 |
F-box and WD-40 domain protein 11 |
8255 |
0.23 |
| chr2_144369139_144369688 | 0.21 |
Kat14 |
lysine acetyltransferase 14 |
414 |
0.43 |
| chr8_71699244_71699423 | 0.21 |
B3gnt3 |
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 3 |
2456 |
0.13 |
| chr4_109104769_109104920 | 0.21 |
Osbpl9 |
oxysterol binding protein-like 9 |
3187 |
0.27 |
| chr19_32778255_32778413 | 0.21 |
Pten |
phosphatase and tensin homolog |
20739 |
0.22 |
| chr11_35732740_35732891 | 0.21 |
Pank3 |
pantothenate kinase 3 |
36669 |
0.14 |
| chr2_103177461_103177622 | 0.20 |
Gm13874 |
predicted gene 13874 |
246 |
0.92 |
| chr16_43375815_43375981 | 0.20 |
Zbtb20 |
zinc finger and BTB domain containing 20 |
11694 |
0.17 |
| chr2_64026273_64026476 | 0.20 |
Fign |
fidgetin |
71614 |
0.14 |
| chr2_103790393_103790544 | 0.20 |
Caprin1 |
cell cycle associated protein 1 |
6076 |
0.11 |
| chr14_21211171_21211322 | 0.20 |
Adk |
adenosine kinase |
106879 |
0.06 |
| chr19_20608706_20609243 | 0.19 |
Aldh1a1 |
aldehyde dehydrogenase family 1, subfamily A1 |
7013 |
0.22 |
| chr12_33285610_33285770 | 0.19 |
Atxn7l1 |
ataxin 7-like 1 |
16825 |
0.2 |
| chr1_88055296_88055515 | 0.19 |
Ugt1a10 |
UDP glycosyltransferase 1 family, polypeptide A10 |
6 |
0.94 |
| chr10_8928866_8929030 | 0.19 |
Gm48728 |
predicted gene, 48728 |
23001 |
0.15 |
| chr16_37566906_37567176 | 0.19 |
Rabl3 |
RAB, member RAS oncogene family-like 3 |
3307 |
0.18 |
| chr18_4929619_4929788 | 0.18 |
Svil |
supervillin |
7977 |
0.3 |
| chr9_7858246_7858397 | 0.18 |
Birc3 |
baculoviral IAP repeat-containing 3 |
2005 |
0.3 |
| chr9_74885383_74885534 | 0.18 |
Onecut1 |
one cut domain, family member 1 |
18974 |
0.14 |
| chr2_155136281_155136621 | 0.18 |
Itch |
itchy, E3 ubiquitin protein ligase |
2890 |
0.22 |
| chr13_48543258_48543458 | 0.18 |
Gm37238 |
predicted gene, 37238 |
1675 |
0.18 |
| chr18_64619314_64619465 | 0.18 |
Gm6978 |
predicted gene 6978 |
6290 |
0.17 |
| chr17_45570529_45570680 | 0.18 |
Hsp90ab1 |
heat shock protein 90 alpha (cytosolic), class B member 1 |
868 |
0.38 |
| chr1_13715965_13716146 | 0.18 |
Xkr9 |
X-linked Kx blood group related 9 |
47284 |
0.12 |
| chr6_141994065_141994238 | 0.18 |
Gm6614 |
predicted gene 6614 |
14594 |
0.21 |
| chr7_3664020_3664377 | 0.18 |
Leng1 |
leukocyte receptor cluster (LRC) member 1 |
1642 |
0.13 |
| chr18_33210537_33210688 | 0.17 |
Stard4 |
StAR-related lipid transfer (START) domain containing 4 |
3154 |
0.37 |
| chr12_57515437_57515691 | 0.17 |
Gm2568 |
predicted gene 2568 |
5060 |
0.18 |
| chr3_63961910_63962265 | 0.17 |
Gm26850 |
predicted gene, 26850 |
1546 |
0.27 |
| chr14_25538345_25538496 | 0.17 |
Mir3075 |
microRNA 3075 |
3981 |
0.21 |
| chr8_111533821_111533983 | 0.17 |
Znrf1 |
zinc and ring finger 1 |
2195 |
0.29 |
| chr2_5939653_5939804 | 0.16 |
Dhtkd1 |
dehydrogenase E1 and transketolase domain containing 1 |
3015 |
0.22 |
| chr2_18587299_18587467 | 0.16 |
Gm13353 |
predicted gene 13353 |
19257 |
0.18 |
| chr16_17069481_17069751 | 0.16 |
Ypel1 |
yippee like 1 |
80 |
0.93 |
| chr3_138223407_138223558 | 0.16 |
Adh7 |
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
2011 |
0.23 |
| chr5_91204972_91205145 | 0.16 |
Gm23092 |
predicted gene, 23092 |
12946 |
0.21 |
| chr5_88581999_88582150 | 0.16 |
Rufy3 |
RUN and FYVE domain containing 3 |
1439 |
0.36 |
| chr16_24477436_24477695 | 0.16 |
Lpp |
LIM domain containing preferred translocation partner in lipoma |
29474 |
0.18 |
| chr1_67150788_67150939 | 0.16 |
Cps1 |
carbamoyl-phosphate synthetase 1 |
27837 |
0.2 |
| chr12_16586341_16586492 | 0.15 |
Lpin1 |
lipin 1 |
3304 |
0.3 |
| chr6_51269674_51270293 | 0.15 |
Mir148a |
microRNA 148a |
73 |
0.98 |
| chr7_100610040_100610203 | 0.15 |
Mrpl48 |
mitochondrial ribosomal protein L48 |
1820 |
0.19 |
| chr2_173741924_173742368 | 0.15 |
Vapb |
vesicle-associated membrane protein, associated protein B and C |
4635 |
0.17 |
| chr14_47295088_47295256 | 0.15 |
Mapk1ip1l |
mitogen-activated protein kinase 1 interacting protein 1-like |
3094 |
0.11 |
| chr7_123642615_123642780 | 0.15 |
Zkscan2 |
zinc finger with KRAB and SCAN domains 2 |
142248 |
0.04 |
| chr5_123975512_123975863 | 0.15 |
Hip1r |
huntingtin interacting protein 1 related |
2059 |
0.19 |
| chr2_170091915_170092085 | 0.15 |
Zfp217 |
zinc finger protein 217 |
39220 |
0.2 |
| chr14_67900348_67900512 | 0.15 |
Dock5 |
dedicator of cytokinesis 5 |
33002 |
0.17 |
| chr13_31779124_31779297 | 0.15 |
Gm11379 |
predicted gene 11379 |
18329 |
0.16 |
| chr19_36835799_36836029 | 0.14 |
Tnks2 |
tankyrase, TRF1-interacting ankyrin-related ADP-ribose polymerase 2 |
1510 |
0.42 |
| chr3_60141187_60141605 | 0.14 |
Gm24382 |
predicted gene, 24382 |
13649 |
0.22 |
| chr4_76350085_76350300 | 0.14 |
Ptprd |
protein tyrosine phosphatase, receptor type, D |
5949 |
0.28 |
| chr7_132941355_132941506 | 0.14 |
Zranb1 |
zinc finger, RAN-binding domain containing 1 |
6738 |
0.15 |
| chr16_57130407_57130558 | 0.14 |
Tomm70a |
translocase of outer mitochondrial membrane 70A |
7016 |
0.17 |
| chr4_11614681_11614832 | 0.14 |
Gm11832 |
predicted gene 11832 |
251 |
0.9 |
| chr1_119661376_119661532 | 0.14 |
Epb41l5 |
erythrocyte membrane protein band 4.1 like 5 |
12454 |
0.16 |
| chr2_114788136_114788290 | 0.13 |
Gm13974 |
predicted gene 13974 |
65880 |
0.12 |
| chr9_74328635_74328791 | 0.13 |
Gm24141 |
predicted gene, 24141 |
33897 |
0.17 |
| chr18_3465417_3465568 | 0.13 |
Gm50088 |
predicted gene, 50088 |
1010 |
0.43 |
| chr6_99189402_99189564 | 0.13 |
Foxp1 |
forkhead box P1 |
26465 |
0.25 |
| chr7_140723570_140723813 | 0.13 |
Olfr542-ps1 |
olfactory receptor 542, pseudogene 1 |
2409 |
0.15 |
| chr6_50041341_50041531 | 0.13 |
Gm3455 |
predicted gene 3455 |
9750 |
0.26 |
| chr7_96957836_96958018 | 0.13 |
C230038L03Rik |
RIKEN cDNA C230038L03 gene |
6041 |
0.19 |
| chr11_30792834_30792985 | 0.13 |
Psme4 |
proteasome (prosome, macropain) activator subunit 4 |
18628 |
0.14 |
| chr4_53134451_53134701 | 0.13 |
Abca1 |
ATP-binding cassette, sub-family A (ABC1), member 1 |
25319 |
0.19 |
| chr3_51202400_51202602 | 0.13 |
Noct |
nocturnin |
21946 |
0.14 |
| chr16_25221904_25222082 | 0.12 |
Tprg |
transformation related protein 63 regulated |
64824 |
0.14 |
| chr14_20112304_20112464 | 0.12 |
Saysd1 |
SAYSVFN motif domain containing 1 |
26150 |
0.14 |
| chr3_138281002_138281176 | 0.12 |
Adh1 |
alcohol dehydrogenase 1 (class I) |
3438 |
0.15 |
| chr4_109655723_109655874 | 0.12 |
9630013D21Rik |
RIKEN cDNA 9630013D21 gene |
1667 |
0.31 |
| chr1_161244723_161244874 | 0.12 |
Prdx6 |
peroxiredoxin 6 |
5756 |
0.18 |
| chr17_10187170_10187336 | 0.12 |
Qk |
quaking |
26321 |
0.25 |
| chr9_69044250_69044433 | 0.12 |
Rora |
RAR-related orphan receptor alpha |
151200 |
0.04 |
| chr18_85490238_85490398 | 0.12 |
Gm7612 |
predicted gene 7612 |
226407 |
0.02 |
| chr11_115919240_115919538 | 0.12 |
Recql5os1 |
RecQ-like 5, opposite strand 1 |
5638 |
0.09 |
| chr11_109771773_109771962 | 0.12 |
Gm11684 |
predicted gene 11684 |
3381 |
0.21 |
| chr8_95449738_95450026 | 0.12 |
Csnk2a2 |
casein kinase 2, alpha prime polypeptide |
10832 |
0.12 |
| chr9_103625706_103625857 | 0.12 |
Gm38032 |
predicted gene, 38032 |
25617 |
0.16 |
| chr9_63327486_63327998 | 0.12 |
Map2k5 |
mitogen-activated protein kinase kinase 5 |
24599 |
0.19 |
| chr1_180179778_180180226 | 0.12 |
Coq8a |
coenzyme Q8A |
1080 |
0.44 |
| chr2_43592382_43592533 | 0.12 |
Kynu |
kynureninase |
7158 |
0.3 |
| chr1_162867938_162868165 | 0.12 |
Fmo1 |
flavin containing monooxygenase 1 |
1441 |
0.39 |
| chr9_122355767_122356085 | 0.12 |
Abhd5 |
abhydrolase domain containing 5 |
4137 |
0.17 |
| chr4_131876493_131876847 | 0.12 |
A930004J17Rik |
RIKEN cDNA A930004J17 gene |
1958 |
0.19 |
| chr8_93235851_93236023 | 0.12 |
Ces1e |
carboxylesterase 1E |
6318 |
0.14 |
| chr17_88462554_88462727 | 0.12 |
Foxn2 |
forkhead box N2 |
21865 |
0.17 |
| chr2_165869197_165869348 | 0.12 |
Zmynd8 |
zinc finger, MYND-type containing 8 |
6505 |
0.15 |
| chr15_53179541_53179708 | 0.12 |
Ext1 |
exostosin glycosyltransferase 1 |
16631 |
0.3 |
| chr10_127158373_127158600 | 0.12 |
B4galnt1 |
beta-1,4-N-acetyl-galactosaminyl transferase 1 |
6739 |
0.08 |
| chr11_21121403_21121623 | 0.11 |
Peli1 |
pellino 1 |
13956 |
0.18 |
| chr10_59149959_59150116 | 0.11 |
Septin10 |
septin 10 |
28820 |
0.16 |
| chr4_118031195_118031493 | 0.11 |
St3gal3 |
ST3 beta-galactoside alpha-2,3-sialyltransferase 3 |
538 |
0.78 |
| chr18_11496730_11496892 | 0.11 |
Gm50067 |
predicted gene, 50067 |
128104 |
0.05 |
| chr6_86523276_86523978 | 0.11 |
1600020E01Rik |
RIKEN cDNA 1600020E01 gene |
2620 |
0.12 |
| chr9_122874871_122875092 | 0.11 |
Zfp445 |
zinc finger protein 445 |
8975 |
0.1 |
| chr10_67104058_67104209 | 0.11 |
Reep3 |
receptor accessory protein 3 |
7188 |
0.2 |
| chr1_179108579_179108982 | 0.11 |
Smyd3 |
SET and MYND domain containing 3 |
119435 |
0.06 |
| chr2_75718851_75719260 | 0.11 |
Gm13656 |
predicted gene 13656 |
11117 |
0.12 |
| chr16_37635855_37636049 | 0.11 |
Ndufb4 |
NADH:ubiquinone oxidoreductase subunit B4 |
13407 |
0.15 |
| chr10_59277548_59277712 | 0.11 |
P4ha1 |
procollagen-proline, 2-oxoglutarate 4-dioxygenase (proline 4-hydroxylase), alpha 1 polypeptide |
45666 |
0.11 |
| chr3_149240098_149240249 | 0.11 |
Gm10287 |
predicted gene 10287 |
14428 |
0.2 |
| chr16_84807428_84807749 | 0.11 |
Jam2 |
junction adhesion molecule 2 |
1819 |
0.26 |
| chr16_65665507_65665824 | 0.11 |
Gm49633 |
predicted gene, 49633 |
52246 |
0.14 |
| chr17_28428613_28428954 | 0.11 |
Fkbp5 |
FK506 binding protein 5 |
154 |
0.91 |
| chr12_57537784_57538085 | 0.11 |
Foxa1 |
forkhead box A1 |
8187 |
0.15 |
| chr3_19961027_19961202 | 0.11 |
Cp |
ceruloplasmin |
3830 |
0.21 |
| chr15_3463467_3463618 | 0.11 |
Ghr |
growth hormone receptor |
8102 |
0.29 |
| chr13_101801879_101802177 | 0.11 |
Pik3r1 |
phosphoinositide-3-kinase regulatory subunit 1 |
33811 |
0.15 |
| chr3_84860618_84860912 | 0.11 |
Fbxw7 |
F-box and WD-40 domain protein 7 |
3380 |
0.29 |
| chr8_114706794_114706945 | 0.11 |
Gm16118 |
predicted gene 16118 |
1137 |
0.54 |
| chr15_3560210_3560361 | 0.11 |
Ghr |
growth hormone receptor |
21557 |
0.23 |
| chr17_12941646_12941826 | 0.11 |
Acat3 |
acetyl-Coenzyme A acetyltransferase 3 |
1136 |
0.27 |
| chr17_88605968_88606119 | 0.11 |
Gm9406 |
predicted gene 9406 |
2501 |
0.24 |
| chr19_12638609_12638960 | 0.10 |
Glyat |
glycine-N-acyltransferase |
708 |
0.5 |
| chr8_3276470_3276698 | 0.10 |
Insr |
insulin receptor |
2967 |
0.26 |
| chr4_105076264_105076471 | 0.10 |
Prkaa2 |
protein kinase, AMP-activated, alpha 2 catalytic subunit |
33523 |
0.2 |
| chr9_50500490_50500842 | 0.10 |
Plet1os |
placenta expressed transcript 1, opposite strand |
4139 |
0.14 |
| chr10_10843070_10843258 | 0.10 |
4930567K20Rik |
RIKEN cDNA 4930567K20 gene |
92084 |
0.07 |
| chr19_55560363_55560546 | 0.10 |
Vti1a |
vesicle transport through interaction with t-SNAREs 1A |
179440 |
0.03 |
| chr18_51164059_51164479 | 0.10 |
Prr16 |
proline rich 16 |
46531 |
0.18 |
| chr13_117679367_117679569 | 0.10 |
4933413L06Rik |
RIKEN cDNA 4933413L06 gene |
40519 |
0.2 |
| chr11_74924478_74924629 | 0.10 |
Srr |
serine racemase |
1105 |
0.27 |
| chr3_145762528_145762679 | 0.10 |
Ddah1 |
dimethylarginine dimethylaminohydrolase 1 |
1855 |
0.37 |
| chr7_99974308_99974843 | 0.10 |
Rnf169 |
ring finger protein 169 |
5873 |
0.14 |
| chr2_174774160_174774316 | 0.10 |
Edn3 |
endothelin 3 |
4316 |
0.25 |
| chr5_96206945_96207101 | 0.10 |
Mrpl1 |
mitochondrial ribosomal protein L1 |
2470 |
0.32 |
| chr19_47436846_47437016 | 0.10 |
Sh3pxd2a |
SH3 and PX domains 2A |
26572 |
0.17 |
| chr8_69545222_69545373 | 0.10 |
Rps7-ps1 |
ribosomal protein S7, pseudogene 1 |
4340 |
0.17 |
| chr9_74963277_74963428 | 0.10 |
Fam214a |
family with sequence similarity 214, member A |
10280 |
0.2 |
| chr2_3760026_3760210 | 0.10 |
Gm13185 |
predicted gene 13185 |
4941 |
0.2 |
| chr6_94358809_94358972 | 0.10 |
Gm19908 |
predicted gene, 19908 |
9356 |
0.21 |
| chr6_58611375_58611579 | 0.10 |
Abcg2 |
ATP binding cassette subfamily G member 2 (Junior blood group) |
14806 |
0.19 |
| chr5_106573383_106573688 | 0.10 |
Gm28050 |
predicted gene, 28050 |
1180 |
0.32 |
| chr2_135799055_135799206 | 0.09 |
Plcb4 |
phospholipase C, beta 4 |
6418 |
0.23 |
| chr2_119662546_119662712 | 0.09 |
Ndufaf1 |
NADH:ubiquinone oxidoreductase complex assembly factor 1 |
169 |
0.9 |
| chr3_87907622_87908270 | 0.09 |
Hdgf |
heparin binding growth factor |
412 |
0.71 |
| chr11_4832480_4832698 | 0.09 |
Nf2 |
neurofibromin 2 |
544 |
0.74 |
| chr1_181842789_181842980 | 0.09 |
Lbr |
lamin B receptor |
25 |
0.96 |
| chr16_26669241_26669605 | 0.09 |
Il1rap |
interleukin 1 receptor accessory protein |
45267 |
0.18 |
| chr9_108687622_108687990 | 0.09 |
Prkar2a |
protein kinase, cAMP dependent regulatory, type II alpha |
1508 |
0.24 |
| chr7_30967090_30967447 | 0.09 |
Lsr |
lipolysis stimulated lipoprotein receptor |
6054 |
0.07 |
| chr15_51865415_51865710 | 0.09 |
Eif3h |
eukaryotic translation initiation factor 3, subunit H |
39 |
0.97 |
| chr3_79518695_79518990 | 0.09 |
Fnip2 |
folliculin interacting protein 2 |
3590 |
0.21 |
| chr7_19119331_19119555 | 0.09 |
Fbxo46 |
F-box protein 46 |
416 |
0.59 |
| chr6_129228000_129228171 | 0.09 |
2310001H17Rik |
RIKEN cDNA 2310001H17 gene |
5889 |
0.13 |
| chr18_5013827_5013978 | 0.09 |
Svil |
supervillin |
18966 |
0.27 |
| chr10_24362567_24362800 | 0.09 |
Gm15271 |
predicted gene 15271 |
84807 |
0.08 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.1 | 0.2 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.1 | 0.2 | GO:0035860 | glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.0 | 0.1 | GO:0009826 | unidimensional cell growth(GO:0009826) |
| 0.0 | 0.1 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 0.0 | 0.1 | GO:0060839 | endothelial cell fate commitment(GO:0060839) |
| 0.0 | 0.1 | GO:0033031 | positive regulation of neutrophil apoptotic process(GO:0033031) |
| 0.0 | 0.1 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.0 | 0.1 | GO:1903659 | regulation of complement-dependent cytotoxicity(GO:1903659) negative regulation of complement-dependent cytotoxicity(GO:1903660) |
| 0.0 | 0.1 | GO:1902036 | regulation of hematopoietic stem cell differentiation(GO:1902036) |
| 0.0 | 0.1 | GO:0030638 | polyketide metabolic process(GO:0030638) aminoglycoside antibiotic metabolic process(GO:0030647) daunorubicin metabolic process(GO:0044597) doxorubicin metabolic process(GO:0044598) |
| 0.0 | 0.1 | GO:0072361 | regulation of glycolytic process by regulation of transcription from RNA polymerase II promoter(GO:0072361) |
| 0.0 | 0.2 | GO:0052697 | xenobiotic glucuronidation(GO:0052697) |
| 0.0 | 0.0 | GO:0018879 | biphenyl metabolic process(GO:0018879) |
| 0.0 | 0.1 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.0 | 0.1 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.0 | 0.1 | GO:0042524 | negative regulation of tyrosine phosphorylation of Stat5 protein(GO:0042524) |
| 0.0 | 0.1 | GO:0097105 | presynaptic membrane assembly(GO:0097105) |
| 0.0 | 0.1 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.0 | 0.0 | GO:2000211 | regulation of glutamate metabolic process(GO:2000211) |
| 0.0 | 0.0 | GO:0052422 | modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) |
| 0.0 | 0.1 | GO:0007223 | Wnt signaling pathway, calcium modulating pathway(GO:0007223) |
| 0.0 | 0.1 | GO:0042977 | activation of JAK2 kinase activity(GO:0042977) |
| 0.0 | 0.0 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.0 | GO:0032741 | positive regulation of interleukin-18 production(GO:0032741) |
| 0.0 | 0.0 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.1 | GO:0061042 | vascular wound healing(GO:0061042) |
| 0.0 | 0.0 | GO:0060741 | prostate gland stromal morphogenesis(GO:0060741) |
| 0.0 | 0.0 | GO:0002913 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.0 | 0.0 | GO:1903223 | positive regulation of oxidative stress-induced neuron death(GO:1903223) |
| 0.0 | 0.0 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0097451 | glial limiting end-foot(GO:0097451) |
| 0.0 | 0.1 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.0 | 0.1 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.0 | 0.1 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.1 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.0 | 0.0 | GO:0005642 | annulate lamellae(GO:0005642) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.1 | 0.2 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.1 | GO:0047045 | testosterone 17-beta-dehydrogenase (NADP+) activity(GO:0047045) |
| 0.0 | 0.1 | GO:0055100 | adiponectin binding(GO:0055100) |
| 0.0 | 0.1 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.0 | 0.1 | GO:0017098 | sulfonylurea receptor binding(GO:0017098) |
| 0.0 | 0.1 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.1 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.0 | 0.1 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.0 | 0.0 | GO:0038181 | bile acid receptor activity(GO:0038181) |
| 0.0 | 0.0 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.1 | GO:0035671 | enone reductase activity(GO:0035671) |
| 0.0 | 0.0 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.0 | 0.0 | GO:0016822 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.0 | 0.0 | GO:0016743 | carboxyl- or carbamoyltransferase activity(GO:0016743) |
| 0.0 | 0.0 | GO:0046573 | lactonohydrolase activity(GO:0046573) acyl-L-homoserine-lactone lactonohydrolase activity(GO:0102007) |
| 0.0 | 0.0 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.1 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.0 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.0 | 0.1 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 0.1 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.0 | 0.1 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.0 | 0.1 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |