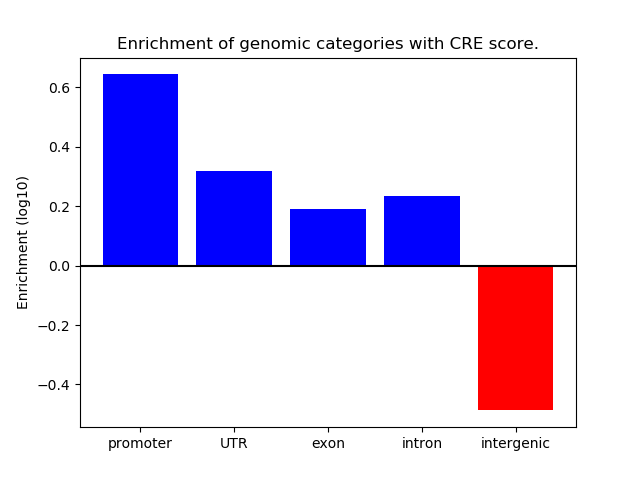
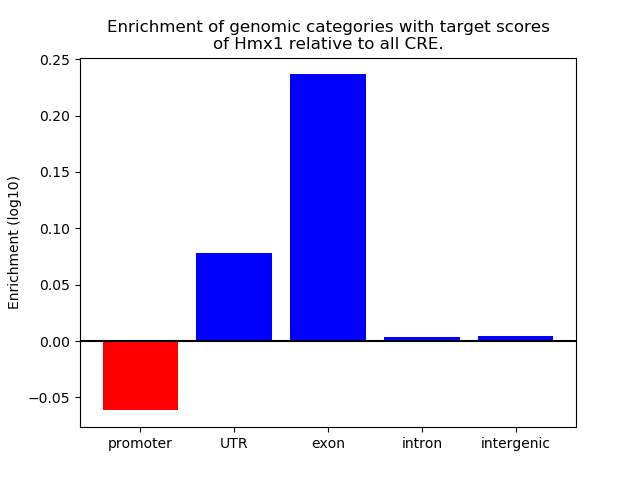
Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
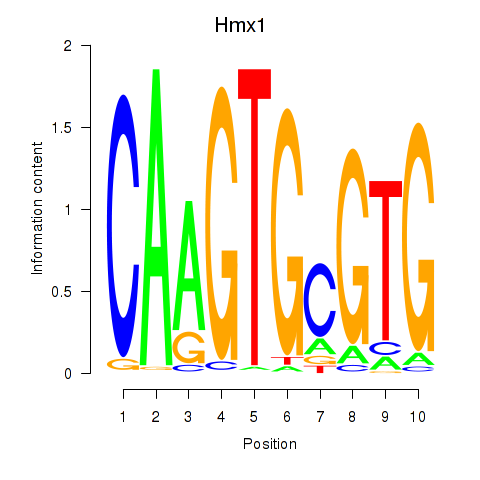
Results for Hmx1
Z-value: 0.88
Transcription factors associated with Hmx1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hmx1
|
ENSMUSG00000067438.3 | H6 homeobox 1 |
Activity of the Hmx1 motif across conditions
Conditions sorted by the z-value of the Hmx1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
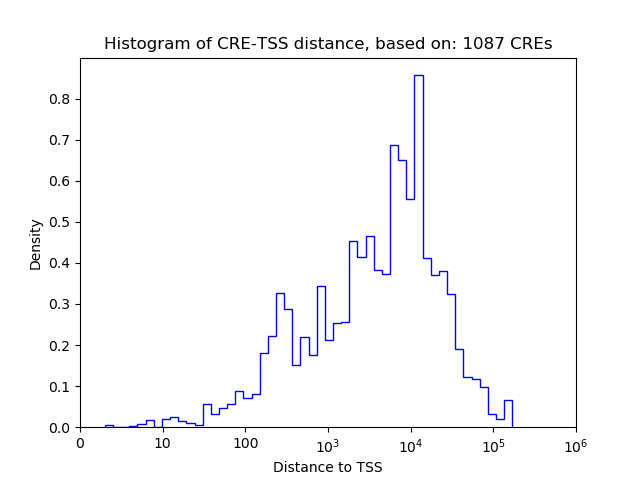
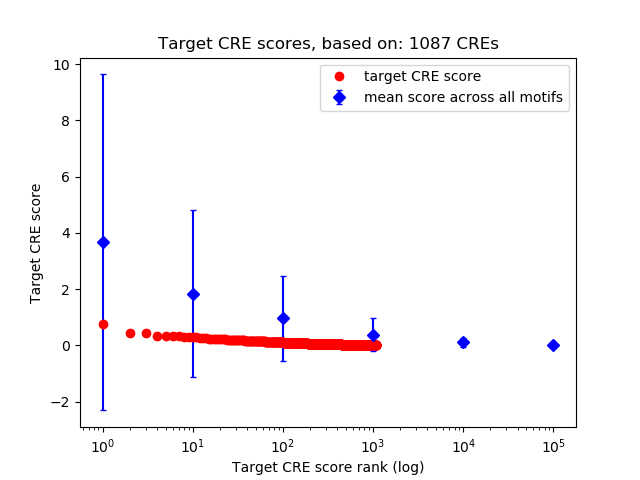
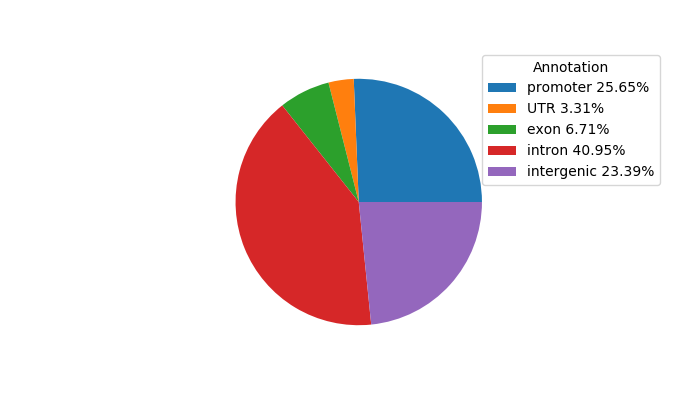
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr13_80897093_80897522 | 0.75 |
Arrdc3 |
arrestin domain containing 3 |
6789 |
0.17 |
| chr7_97414019_97414355 | 0.43 |
Thrsp |
thyroid hormone responsive |
3332 |
0.16 |
| chr17_25127135_25127453 | 0.43 |
Clcn7 |
chloride channel, voltage-sensitive 7 |
6097 |
0.1 |
| chr10_69248815_69249004 | 0.33 |
Rhobtb1 |
Rho-related BTB domain containing 1 |
16728 |
0.18 |
| chr1_132075707_132075908 | 0.32 |
Mfsd4a |
major facilitator superfamily domain containing 4A |
7745 |
0.13 |
| chr11_107317810_107317984 | 0.32 |
Gm11719 |
predicted gene 11719 |
12811 |
0.14 |
| chr8_72665230_72665413 | 0.31 |
Nwd1 |
NACHT and WD repeat domain containing 1 |
8235 |
0.15 |
| chr7_141460760_141460954 | 0.30 |
Cracr2b |
calcium release activated channel regulator 2B |
237 |
0.77 |
| chr2_25384633_25384784 | 0.30 |
Sapcd2 |
suppressor APC domain containing 2 |
8748 |
0.07 |
| chr3_107654890_107655041 | 0.29 |
Ahcyl1 |
S-adenosylhomocysteine hydrolase-like 1 |
10631 |
0.14 |
| chr9_32695602_32695787 | 0.29 |
Ets1 |
E26 avian leukemia oncogene 1, 5' domain |
328 |
0.89 |
| chr17_25981156_25981307 | 0.27 |
Capn15 |
calpain 15 |
1338 |
0.22 |
| chr13_43795452_43795640 | 0.27 |
Cd83 |
CD83 antigen |
10361 |
0.19 |
| chr3_84484936_84485087 | 0.25 |
Fhdc1 |
FH2 domain containing 1 |
4582 |
0.25 |
| chr18_76242274_76242708 | 0.23 |
Smad2 |
SMAD family member 2 |
317 |
0.89 |
| chr9_121794943_121795277 | 0.23 |
Hhatl |
hedgehog acyltransferase-like |
2603 |
0.13 |
| chr9_61596903_61597054 | 0.23 |
Gm34424 |
predicted gene, 34424 |
63563 |
0.12 |
| chr1_91437326_91437628 | 0.22 |
Per2 |
period circadian clock 2 |
12388 |
0.11 |
| chr13_21914635_21914872 | 0.22 |
Gm44456 |
predicted gene, 44456 |
10263 |
0.06 |
| chr9_57214145_57214320 | 0.22 |
Trcg1 |
taste receptor cell gene 1 |
22324 |
0.12 |
| chr15_59195501_59195712 | 0.22 |
Rpl7-ps8 |
ribosomal protein L7, pseudogene 8 |
15302 |
0.19 |
| chr11_43960310_43960461 | 0.21 |
Gm12153 |
predicted gene 12153 |
1296 |
0.56 |
| chr11_11848585_11848736 | 0.21 |
Ddc |
dopa decarboxylase |
12380 |
0.17 |
| chr9_22017521_22017672 | 0.20 |
Elavl3 |
ELAV like RNA binding protein 3 |
8778 |
0.08 |
| chr5_134626803_134626969 | 0.20 |
Lat2 |
linker for activation of T cells family, member 2 |
11861 |
0.11 |
| chr2_105252035_105252217 | 0.20 |
Them7 |
thioesterase superfamily member 7 |
27784 |
0.2 |
| chr10_89619251_89619434 | 0.19 |
Slc17a8 |
solute carrier family 17 (sodium-dependent inorganic phosphate cotransporter), member 8 |
1911 |
0.34 |
| chr10_127492352_127492555 | 0.19 |
R3hdm2 |
R3H domain containing 2 |
204 |
0.89 |
| chr15_27850966_27851117 | 0.19 |
Trio |
triple functional domain (PTPRF interacting) |
2336 |
0.28 |
| chr16_11410474_11410628 | 0.19 |
Snx29 |
sorting nexin 29 |
3894 |
0.28 |
| chr9_104260385_104260565 | 0.19 |
Gm37563 |
predicted gene, 37563 |
1630 |
0.31 |
| chr15_60823042_60823212 | 0.18 |
9930014A18Rik |
RIKEN cDNA 9930014A18 gene |
174 |
0.94 |
| chr3_18151182_18151333 | 0.18 |
Gm23686 |
predicted gene, 23686 |
26368 |
0.2 |
| chr1_55685863_55686014 | 0.18 |
Plcl1 |
phospholipase C-like 1 |
16087 |
0.27 |
| chr15_80260597_80260758 | 0.18 |
Rps19bp1 |
ribosomal protein S19 binding protein 1 |
3569 |
0.13 |
| chr11_97958258_97958415 | 0.17 |
Gm11633 |
predicted gene 11633 |
10456 |
0.1 |
| chr8_12927350_12927516 | 0.17 |
Mcf2l |
mcf.2 transforming sequence-like |
801 |
0.5 |
| chr1_151243463_151243618 | 0.17 |
Gm24402 |
predicted gene, 24402 |
17374 |
0.12 |
| chr8_44373378_44373540 | 0.17 |
Gm37972 |
predicted gene, 37972 |
144914 |
0.04 |
| chr2_170501364_170501515 | 0.17 |
Cyp24a1 |
cytochrome P450, family 24, subfamily a, polypeptide 1 |
4294 |
0.17 |
| chr10_121349498_121349682 | 0.16 |
Gm48435 |
predicted gene, 48435 |
870 |
0.44 |
| chr7_132778377_132778559 | 0.16 |
Fam53b |
family with sequence similarity 53, member B |
1552 |
0.4 |
| chr16_70354766_70354917 | 0.16 |
Gbe1 |
glucan (1,4-alpha-), branching enzyme 1 |
40707 |
0.19 |
| chr2_166155377_166155528 | 0.16 |
Sulf2 |
sulfatase 2 |
167 |
0.95 |
| chr11_102792950_102793101 | 0.16 |
Gjc1 |
gap junction protein, gamma 1 |
11876 |
0.11 |
| chr2_20909493_20909665 | 0.16 |
Arhgap21 |
Rho GTPase activating protein 21 |
5115 |
0.23 |
| chr14_122946058_122946219 | 0.16 |
4930594M22Rik |
RIKEN cDNA 4930594M22 gene |
5981 |
0.19 |
| chr2_117108153_117108332 | 0.16 |
Spred1 |
sprouty protein with EVH-1 domain 1, related sequence |
13132 |
0.21 |
| chr2_103462568_103462754 | 0.15 |
Elf5 |
E74-like factor 5 |
13923 |
0.17 |
| chr9_66887602_66887790 | 0.15 |
Rab8b |
RAB8B, member RAS oncogene family |
31991 |
0.12 |
| chr10_99201854_99202041 | 0.15 |
Poc1b |
POC1 centriolar protein B |
9055 |
0.12 |
| chr9_62340934_62341117 | 0.15 |
Anp32a |
acidic (leucine-rich) nuclear phosphoprotein 32 family, member A |
268 |
0.92 |
| chr6_113828282_113828660 | 0.15 |
Gm44167 |
predicted gene, 44167 |
28787 |
0.13 |
| chr6_87389476_87389814 | 0.14 |
Gkn3 |
gastrokine 3 |
710 |
0.57 |
| chr14_119829629_119829803 | 0.14 |
4930404K13Rik |
RIKEN cDNA 4930404K13 gene |
30738 |
0.19 |
| chr16_23061607_23061804 | 0.14 |
Kng1 |
kininogen 1 |
3312 |
0.1 |
| chr13_46929135_46929650 | 0.14 |
Kif13a |
kinesin family member 13A |
325 |
0.84 |
| chr14_8223695_8223950 | 0.14 |
Kctd6 |
potassium channel tetramerisation domain containing 6 |
9320 |
0.18 |
| chr19_21275641_21275792 | 0.14 |
Zfand5 |
zinc finger, AN1-type domain 5 |
264 |
0.91 |
| chr1_134574617_134574768 | 0.14 |
Kdm5b |
lysine (K)-specific demethylase 5B |
13285 |
0.13 |
| chr2_170054980_170055177 | 0.14 |
Zfp217 |
zinc finger protein 217 |
76142 |
0.11 |
| chr11_20396557_20396740 | 0.14 |
Gm12034 |
predicted gene 12034 |
49764 |
0.12 |
| chr3_88254407_88254577 | 0.14 |
Rhbg |
Rhesus blood group-associated B glycoprotein |
217 |
0.84 |
| chr18_65989569_65989860 | 0.14 |
Lman1 |
lectin, mannose-binding, 1 |
3946 |
0.15 |
| chr1_75285156_75285327 | 0.13 |
Resp18 |
regulated endocrine-specific protein 18 |
6826 |
0.09 |
| chr19_3684251_3684432 | 0.13 |
Lrp5 |
low density lipoprotein receptor-related protein 5 |
2215 |
0.19 |
| chr6_114922468_114922619 | 0.13 |
Vgll4 |
vestigial like family member 4 |
722 |
0.73 |
| chr6_145469244_145469410 | 0.13 |
Gm25373 |
predicted gene, 25373 |
5138 |
0.18 |
| chr2_80038899_80039095 | 0.13 |
Pde1a |
phosphodiesterase 1A, calmodulin-dependent |
32 |
0.99 |
| chr4_155676097_155676257 | 0.13 |
Mib2 |
mindbomb E3 ubiquitin protein ligase 2 |
6979 |
0.08 |
| chr18_12716045_12717219 | 0.12 |
Mir1948 |
microRNA 1948 |
1821 |
0.27 |
| chr11_78344051_78344376 | 0.12 |
Unc119 |
unc-119 lipid binding chaperone |
668 |
0.49 |
| chr13_23285684_23285835 | 0.12 |
4933404K08Rik |
RIKEN cDNA 4933404K08 gene |
7409 |
0.08 |
| chr8_109997497_109997880 | 0.12 |
Tat |
tyrosine aminotransferase |
7182 |
0.12 |
| chr1_85624553_85624756 | 0.12 |
Sp140 |
Sp140 nuclear body protein |
7824 |
0.1 |
| chr14_64139029_64139418 | 0.12 |
9630015K15Rik |
RIKEN cDNA 9630015K15 gene |
22909 |
0.13 |
| chr4_153019323_153019495 | 0.12 |
Gm25779 |
predicted gene, 25779 |
5825 |
0.32 |
| chr2_181461420_181461583 | 0.12 |
Zbtb46 |
zinc finger and BTB domain containing 46 |
2075 |
0.19 |
| chr18_30530220_30530381 | 0.11 |
Gm7936 |
predicted pseudogene 7936 |
34927 |
0.16 |
| chr15_27492250_27492429 | 0.11 |
B230362B09Rik |
RIKEN cDNA B230362B09 gene |
11573 |
0.15 |
| chr19_29042459_29042628 | 0.11 |
1700018L02Rik |
RIKEN cDNA 1700018L02 gene |
3702 |
0.14 |
| chr2_132169135_132169286 | 0.11 |
Slc23a2 |
solute carrier family 23 (nucleobase transporters), member 2 |
24102 |
0.14 |
| chr10_81415647_81415812 | 0.11 |
Mir1191b |
microRNA 1191b |
568 |
0.46 |
| chr5_148982497_148982697 | 0.11 |
Gm42791 |
predicted gene 42791 |
1504 |
0.21 |
| chr11_61374533_61374684 | 0.11 |
Slc47a1 |
solute carrier family 47, member 1 |
3456 |
0.17 |
| chr7_66801636_66801787 | 0.11 |
Cers3 |
ceramide synthase 3 |
37542 |
0.13 |
| chr6_140347096_140347262 | 0.11 |
n-R5s168 |
nuclear encoded rRNA 5S 168 |
27549 |
0.16 |
| chr9_21837693_21838006 | 0.11 |
Angptl8 |
angiopoietin-like 8 |
2339 |
0.17 |
| chr17_83898655_83898817 | 0.11 |
1810073O08Rik |
RIKEN cDNA 1810073O08 gene |
19201 |
0.13 |
| chr15_25651633_25651912 | 0.11 |
Gm48996 |
predicted gene, 48996 |
17145 |
0.17 |
| chr15_83158429_83158596 | 0.11 |
Rnu12 |
RNA U12, small nuclear |
8868 |
0.1 |
| chr12_54735073_54735224 | 0.10 |
Gm24305 |
predicted gene, 24305 |
430 |
0.6 |
| chr14_122451191_122451371 | 0.10 |
Gm5089 |
predicted gene 5089 |
166 |
0.92 |
| chr1_189908243_189908415 | 0.10 |
Gm38245 |
predicted gene, 38245 |
7630 |
0.17 |
| chr2_158122416_158122699 | 0.10 |
Gm20412 |
predicted gene 20412 |
5403 |
0.17 |
| chr12_111743398_111743567 | 0.10 |
Gm15996 |
predicted gene 15996 |
11651 |
0.1 |
| chr14_28638881_28639055 | 0.10 |
Gm35164 |
predicted gene, 35164 |
10704 |
0.26 |
| chr1_74375022_74375208 | 0.10 |
Slc11a1 |
solute carrier family 11 (proton-coupled divalent metal ion transporters), member 1 |
80 |
0.94 |
| chr2_27223223_27223409 | 0.10 |
Sardh |
sarcosine dehydrogenase |
1876 |
0.28 |
| chr11_51856076_51856454 | 0.10 |
Jade2 |
jade family PHD finger 2 |
860 |
0.59 |
| chr1_86175497_86175764 | 0.10 |
Armc9 |
armadillo repeat containing 9 |
20760 |
0.12 |
| chr4_55014920_55015072 | 0.10 |
Zfp462 |
zinc finger protein 462 |
3516 |
0.32 |
| chr8_33832056_33832231 | 0.10 |
Rbpms |
RNA binding protein gene with multiple splicing |
11528 |
0.16 |
| chr3_10239666_10239823 | 0.10 |
1700029B24Rik |
RIKEN cDNA 1700029B24 gene |
2068 |
0.18 |
| chr7_109976050_109976201 | 0.10 |
Tmem41b |
transmembrane protein 41B |
2804 |
0.15 |
| chr5_139812152_139812320 | 0.10 |
Tmem184a |
transmembrane protein 184a |
1007 |
0.41 |
| chr8_121648538_121648748 | 0.10 |
Zcchc14 |
zinc finger, CCHC domain containing 14 |
4258 |
0.14 |
| chr7_84129291_84129727 | 0.10 |
Abhd17c |
abhydrolase domain containing 17C |
18306 |
0.14 |
| chr13_89782815_89783020 | 0.10 |
Vcan |
versican |
40408 |
0.16 |
| chr13_6635720_6635884 | 0.10 |
Pfkp |
phosphofructokinase, platelet |
848 |
0.62 |
| chr17_45573199_45573482 | 0.10 |
Hsp90ab1 |
heat shock protein 90 alpha (cytosolic), class B member 1 |
69 |
0.94 |
| chr8_84704510_84705270 | 0.10 |
Nfix |
nuclear factor I/X |
2826 |
0.13 |
| chr18_46341760_46341934 | 0.09 |
4930415P13Rik |
RIKEN cDNA 4930415P13 gene |
1418 |
0.34 |
| chr12_81862549_81862733 | 0.09 |
Pcnx |
pecanex homolog |
2300 |
0.32 |
| chr18_39394059_39394221 | 0.09 |
Gm15337 |
predicted gene 15337 |
4715 |
0.24 |
| chr13_110667648_110667811 | 0.09 |
Gm33172 |
predicted gene, 33172 |
16183 |
0.23 |
| chr18_84210867_84211018 | 0.09 |
Gm50311 |
predicted gene, 50311 |
8018 |
0.25 |
| chr15_100319010_100319169 | 0.09 |
Mettl7a1 |
methyltransferase like 7A1 |
13918 |
0.1 |
| chr2_101727956_101728139 | 0.09 |
Traf6 |
TNF receptor-associated factor 6 |
43832 |
0.14 |
| chr7_44668104_44668269 | 0.09 |
2310016G11Rik |
RIKEN cDNA 2310016G11 gene |
47 |
0.93 |
| chr11_120819707_120820136 | 0.09 |
Fasn |
fatty acid synthase |
3859 |
0.11 |
| chr5_140831353_140831504 | 0.09 |
Gna12 |
guanine nucleotide binding protein, alpha 12 |
997 |
0.6 |
| chr3_107695641_107695815 | 0.09 |
Ahcyl1 |
S-adenosylhomocysteine hydrolase-like 1 |
235 |
0.92 |
| chr10_62231949_62232100 | 0.09 |
Tspan15 |
tetraspanin 15 |
773 |
0.6 |
| chr2_26445181_26445721 | 0.09 |
Sec16a |
SEC16 homolog A, endoplasmic reticulum export factor |
235 |
0.47 |
| chr11_120099084_120099272 | 0.09 |
Ndufaf8 |
NADH:ubiquinone oxidoreductase complex assembly factor 8 |
79 |
0.84 |
| chr15_98582497_98582655 | 0.09 |
Gm29331 |
predicted gene 29331 |
7414 |
0.08 |
| chr17_63488193_63488355 | 0.08 |
Fbxl17 |
F-box and leucine-rich repeat protein 17 |
4479 |
0.29 |
| chr15_64155927_64156078 | 0.08 |
Asap1 |
ArfGAP with SH3 domain, ankyrin repeat and PH domain1 |
32196 |
0.17 |
| chr15_82241552_82241727 | 0.08 |
Cenpm |
centromere protein M |
2693 |
0.12 |
| chr13_23843910_23844061 | 0.08 |
Slc17a3 |
solute carrier family 17 (sodium phosphate), member 3 |
4395 |
0.1 |
| chr3_27321509_27321660 | 0.08 |
Tnfsf10 |
tumor necrosis factor (ligand) superfamily, member 10 |
4523 |
0.21 |
| chr14_32708910_32709089 | 0.08 |
Gm28651 |
predicted gene 28651 |
22441 |
0.16 |
| chrX_169981103_169981260 | 0.08 |
Mid1 |
midline 1 |
1730 |
0.26 |
| chr6_66910538_66910915 | 0.08 |
Gng12 |
guanine nucleotide binding protein (G protein), gamma 12 |
13835 |
0.14 |
| chr17_29157409_29157621 | 0.08 |
Cpne5 |
copine V |
1990 |
0.18 |
| chr4_118425783_118425968 | 0.08 |
Elovl1 |
elongation of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 1 |
2218 |
0.17 |
| chr2_154942991_154943142 | 0.08 |
a |
nonagouti |
8114 |
0.2 |
| chr2_125792282_125792442 | 0.08 |
Secisbp2l |
SECIS binding protein 2-like |
9492 |
0.22 |
| chr2_67972706_67972961 | 0.08 |
Gm37964 |
predicted gene, 37964 |
74237 |
0.1 |
| chr9_21429799_21429981 | 0.08 |
Dnm2 |
dynamin 2 |
4646 |
0.12 |
| chr12_54713676_54713835 | 0.08 |
Gm23804 |
predicted gene, 23804 |
463 |
0.55 |
| chr1_131963604_131963963 | 0.08 |
Slc45a3 |
solute carrier family 45, member 3 |
816 |
0.35 |
| chr13_63296898_63297049 | 0.08 |
Aopep |
aminopeptidase O |
1820 |
0.15 |
| chr11_120807985_120808259 | 0.08 |
Fasn |
fatty acid synthase |
483 |
0.62 |
| chr11_59428179_59428372 | 0.08 |
Snap47 |
synaptosomal-associated protein, 47 |
19216 |
0.1 |
| chr1_40217857_40218008 | 0.08 |
Il1r1 |
interleukin 1 receptor, type I |
7148 |
0.21 |
| chr6_90571749_90571967 | 0.08 |
Aldh1l1 |
aldehyde dehydrogenase 1 family, member L1 |
4152 |
0.17 |
| chr14_24960151_24960328 | 0.08 |
Gm10398 |
predicted gene 10398 |
50825 |
0.13 |
| chr19_16495538_16495696 | 0.08 |
Gm8222 |
predicted gene 8222 |
4553 |
0.24 |
| chr2_122158054_122158205 | 0.08 |
Trim69 |
tripartite motif-containing 69 |
2571 |
0.17 |
| chr7_127773891_127774064 | 0.08 |
Setd1a |
SET domain containing 1A |
2693 |
0.1 |
| chr13_33935942_33936110 | 0.08 |
1110046J04Rik |
RIKEN cDNA 1110046J04 gene |
6 |
0.5 |
| chr17_65885492_65885896 | 0.08 |
Gm49870 |
predicted gene, 49870 |
61 |
0.53 |
| chr4_10972186_10972337 | 0.08 |
Rps11-ps3 |
ribosomal protein S11, pseudogene 3 |
25653 |
0.15 |
| chr13_111872156_111872322 | 0.07 |
Gm15326 |
predicted gene 15326 |
1099 |
0.41 |
| chr1_195013951_195014268 | 0.07 |
9630010A21Rik |
RIKEN cDNA 9630010A21 gene |
363 |
0.72 |
| chr5_96437978_96438159 | 0.07 |
Gm33050 |
predicted gene, 33050 |
19675 |
0.21 |
| chr6_115480017_115480335 | 0.07 |
Gm44079 |
predicted gene, 44079 |
15186 |
0.16 |
| chr14_72710122_72710340 | 0.07 |
Fndc3a |
fibronectin type III domain containing 3A |
228 |
0.94 |
| chr13_31546784_31547087 | 0.07 |
Foxq1 |
forkhead box Q1 |
9199 |
0.14 |
| chr12_13097085_13097304 | 0.07 |
Gm35298 |
predicted gene, 35298 |
5651 |
0.18 |
| chr10_42239037_42239279 | 0.07 |
Foxo3 |
forkhead box O3 |
19208 |
0.24 |
| chr6_17488292_17488473 | 0.07 |
Met |
met proto-oncogene |
2859 |
0.31 |
| chr2_118537338_118537525 | 0.07 |
Bmf |
BCL2 modifying factor |
7156 |
0.17 |
| chrX_10719688_10719839 | 0.07 |
Gm14493 |
predicted gene 14493 |
1053 |
0.45 |
| chr7_112212933_112213105 | 0.07 |
Mical2 |
microtubule associated monooxygenase, calponin and LIM domain containing 2 |
12837 |
0.25 |
| chr1_85164204_85164433 | 0.07 |
Gm6264 |
predicted gene 6264 |
3455 |
0.14 |
| chr15_58053454_58053645 | 0.07 |
Gm22299 |
predicted gene, 22299 |
11898 |
0.12 |
| chr1_91062463_91062650 | 0.07 |
Lrrfip1 |
leucine rich repeat (in FLII) interacting protein 1 |
8970 |
0.2 |
| chr6_86218224_86218416 | 0.07 |
Tgfa |
transforming growth factor alpha |
6726 |
0.17 |
| chr2_65222858_65223009 | 0.07 |
Cobll1 |
Cobl-like 1 |
13062 |
0.19 |
| chr6_135542716_135542892 | 0.07 |
Gm25136 |
predicted gene, 25136 |
41388 |
0.17 |
| chr5_33722581_33722796 | 0.07 |
Fgfr3 |
fibroblast growth factor receptor 3 |
132 |
0.9 |
| chr16_96082081_96082232 | 0.07 |
Brwd1 |
bromodomain and WD repeat domain containing 1 |
272 |
0.68 |
| chr18_21207409_21207563 | 0.07 |
Garem1 |
GRB2 associated regulator of MAPK1 subtype 1 |
92637 |
0.07 |
| chr11_72739594_72739780 | 0.07 |
Ankfy1 |
ankyrin repeat and FYVE domain containing 1 |
20229 |
0.16 |
| chr10_17552124_17552284 | 0.07 |
Gm47770 |
predicted gene, 47770 |
25290 |
0.17 |
| chr19_42750313_42750502 | 0.07 |
Pyroxd2 |
pyridine nucleotide-disulphide oxidoreductase domain 2 |
2065 |
0.25 |
| chr14_27367879_27368030 | 0.07 |
Arhgef3 |
Rho guanine nucleotide exchange factor (GEF) 3 |
6565 |
0.2 |
| chr5_114932334_114932489 | 0.07 |
Oasl1 |
2'-5' oligoadenylate synthetase-like 1 |
2562 |
0.13 |
| chr2_32256394_32256572 | 0.07 |
Uck1 |
uridine-cytidine kinase 1 |
186 |
0.87 |
| chr4_135787434_135787634 | 0.07 |
Myom3 |
myomesin family, member 3 |
10656 |
0.14 |
| chr2_91478238_91478389 | 0.07 |
Lrp4 |
low density lipoprotein receptor-related protein 4 |
1994 |
0.28 |
| chr2_6118230_6118416 | 0.07 |
Proser2 |
proline and serine rich 2 |
11816 |
0.15 |
| chrX_142389835_142389999 | 0.06 |
Acsl4 |
acyl-CoA synthetase long-chain family member 4 |
427 |
0.8 |
| chr13_82226171_82226354 | 0.06 |
Gm48155 |
predicted gene, 48155 |
136505 |
0.05 |
| chr5_52982111_52982447 | 0.06 |
Gm30301 |
predicted gene, 30301 |
242 |
0.91 |
| chr6_139896009_139896196 | 0.06 |
Pik3c2g |
phosphatidylinositol-4-phosphate 3-kinase catalytic subunit type 2 gamma |
21881 |
0.2 |
| chr8_109374001_109374166 | 0.06 |
Gm1943 |
predicted gene 1943 |
33219 |
0.17 |
| chr7_16844727_16845054 | 0.06 |
Prkd2 |
protein kinase D2 |
343 |
0.77 |
| chr11_100919372_100919549 | 0.06 |
Stat3 |
signal transducer and activator of transcription 3 |
11236 |
0.14 |
| chr11_101971493_101971644 | 0.06 |
Gm20659 |
predicted gene 20659 |
425 |
0.68 |
| chr17_53618913_53619202 | 0.06 |
Gm6919 |
predicted gene 6919 |
20394 |
0.13 |
| chr1_151289316_151289514 | 0.06 |
Gm24402 |
predicted gene, 24402 |
28501 |
0.12 |
| chr11_49615889_49616040 | 0.06 |
Flt4 |
FMS-like tyrosine kinase 4 |
6701 |
0.12 |
| chr14_7943834_7943992 | 0.06 |
Gm45521 |
predicted gene 45521 |
13375 |
0.17 |
| chr4_40823217_40823652 | 0.06 |
Mir5123 |
microRNA 5123 |
26704 |
0.09 |
| chr4_118217577_118217806 | 0.06 |
Ptprf |
protein tyrosine phosphatase, receptor type, F |
11 |
0.97 |
| chr11_110367467_110367657 | 0.06 |
Abca5 |
ATP-binding cassette, sub-family A (ABC1), member 5 |
29846 |
0.19 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0035860 | glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.1 | 0.3 | GO:0030578 | PML body organization(GO:0030578) |
| 0.0 | 0.1 | GO:0071879 | positive regulation of adrenergic receptor signaling pathway(GO:0071879) |
| 0.0 | 0.2 | GO:2000382 | positive regulation of mesoderm development(GO:2000382) |
| 0.0 | 0.1 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 0.0 | 0.1 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.2 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.0 | 0.1 | GO:0072386 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.0 | 0.1 | GO:1903660 | regulation of complement-dependent cytotoxicity(GO:1903659) negative regulation of complement-dependent cytotoxicity(GO:1903660) |
| 0.0 | 0.1 | GO:0002309 | T cell proliferation involved in immune response(GO:0002309) |
| 0.0 | 0.1 | GO:1900186 | negative regulation of clathrin-mediated endocytosis(GO:1900186) |
| 0.0 | 0.1 | GO:0061144 | alveolar secondary septum development(GO:0061144) |
| 0.0 | 0.0 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.0 | 0.0 | GO:0006172 | ADP biosynthetic process(GO:0006172) |
| 0.0 | 0.1 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.0 | 0.1 | GO:0043097 | pyrimidine-containing compound salvage(GO:0008655) pyrimidine nucleoside salvage(GO:0043097) |
| 0.0 | 0.0 | GO:1901492 | positive regulation of lymphangiogenesis(GO:1901492) |
| 0.0 | 0.1 | GO:0009181 | purine nucleoside diphosphate catabolic process(GO:0009137) purine ribonucleoside diphosphate catabolic process(GO:0009181) |
| 0.0 | 0.0 | GO:0061502 | early endosome to recycling endosome transport(GO:0061502) |
| 0.0 | 0.1 | GO:0032304 | negative regulation of icosanoid secretion(GO:0032304) |
| 0.0 | 0.0 | GO:0042998 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) |
| 0.0 | 0.0 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.0 | 0.0 | GO:0042427 | serotonin biosynthetic process(GO:0042427) primary amino compound biosynthetic process(GO:1901162) |
| 0.0 | 0.0 | GO:0018992 | germ-line sex determination(GO:0018992) |
| 0.0 | 0.1 | GO:0051387 | negative regulation of neurotrophin TRK receptor signaling pathway(GO:0051387) |
| 0.0 | 0.0 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.0 | 0.0 | GO:0015766 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.0 | 0.1 | GO:1900122 | positive regulation of receptor binding(GO:1900122) |
| 0.0 | 0.0 | GO:1904395 | regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904393) positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) |
| 0.0 | 0.1 | GO:0032488 | Cdc42 protein signal transduction(GO:0032488) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.0 | 0.1 | GO:0097441 | basilar dendrite(GO:0097441) |
| 0.0 | 0.1 | GO:0030313 | cell outer membrane(GO:0009279) cell envelope(GO:0030313) external encapsulating structure part(GO:0044462) |
| 0.0 | 0.1 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.0 | 0.1 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 0.1 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 0.0 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.1 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.0 | 0.1 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.2 | GO:0005072 | transforming growth factor beta receptor, cytoplasmic mediator activity(GO:0005072) |
| 0.0 | 0.1 | GO:0017098 | sulfonylurea receptor binding(GO:0017098) |
| 0.0 | 0.1 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) |
| 0.0 | 0.1 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.0 | 0.1 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 0.0 | 0.1 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.0 | 0.1 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.1 | GO:0043262 | adenosine-diphosphatase activity(GO:0043262) |
| 0.0 | 0.0 | GO:0016401 | palmitoyl-CoA oxidase activity(GO:0016401) |
| 0.0 | 0.0 | GO:0015157 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.0 | 0.0 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.0 | 0.0 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.0 | 0.1 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |