Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
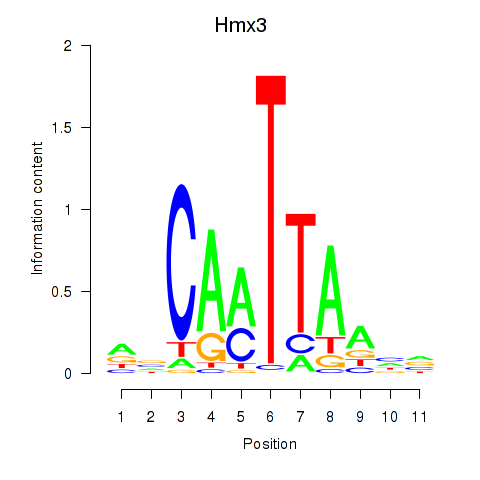
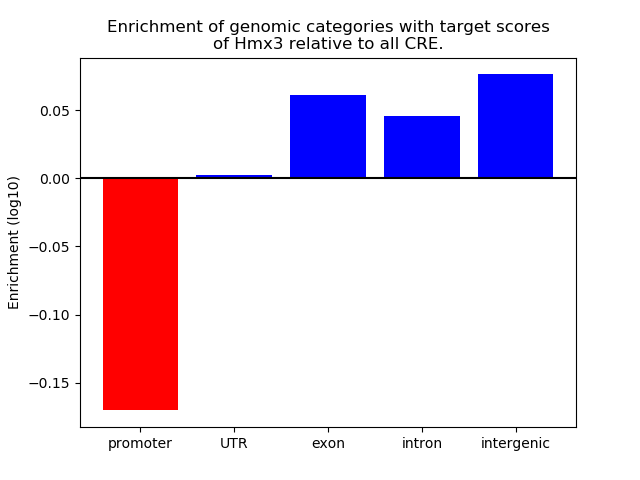
Results for Hmx3
Z-value: 1.24
Transcription factors associated with Hmx3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hmx3
|
ENSMUSG00000040148.5 | H6 homeobox 3 |
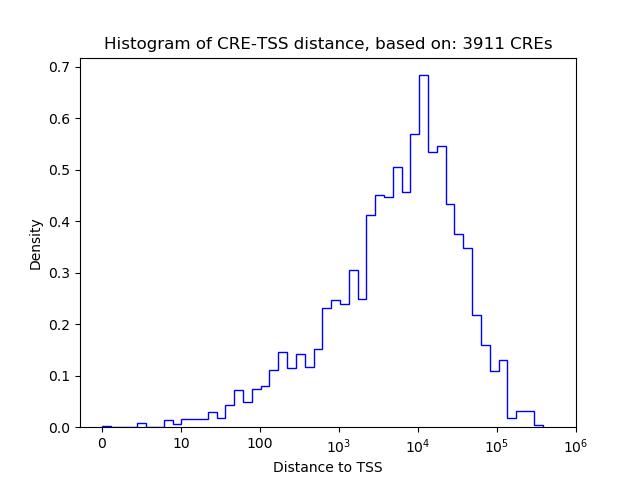
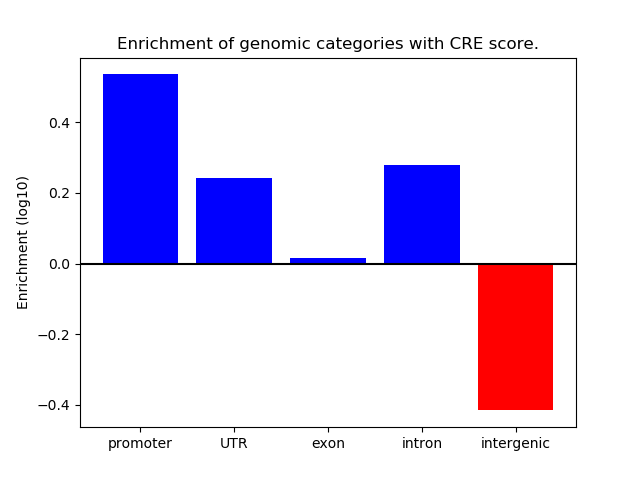
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr7_131540075_131540234 | Hmx3 | 2713 | 0.187430 | -0.50 | 3.1e-01 | Click! |
Activity of the Hmx3 motif across conditions
Conditions sorted by the z-value of the Hmx3 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
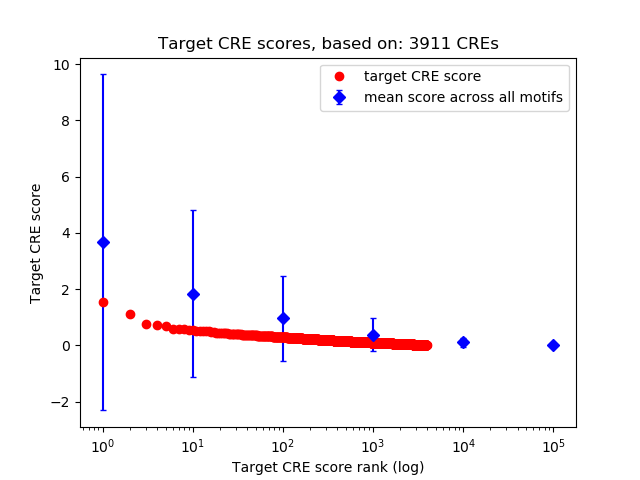
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr7_145056434_145056606 | 1.54 |
Gm45181 |
predicted gene 45181 |
106476 |
0.05 |
| chr13_80895235_80895839 | 1.12 |
Arrdc3 |
arrestin domain containing 3 |
5019 |
0.18 |
| chr15_27993720_27993871 | 0.76 |
Trio |
triple functional domain (PTPRF interacting) |
1568 |
0.45 |
| chr8_13160592_13160981 | 0.74 |
Lamp1 |
lysosomal-associated membrane protein 1 |
1625 |
0.22 |
| chr2_75202592_75202875 | 0.68 |
Gm13653 |
predicted gene 13653 |
10456 |
0.15 |
| chr1_24612849_24613004 | 0.59 |
Gm28438 |
predicted gene 28438 |
193 |
0.45 |
| chr7_140112202_140112371 | 0.57 |
Paox |
polyamine oxidase (exo-N4-amino) |
3513 |
0.11 |
| chr17_81006523_81006701 | 0.57 |
Thumpd2 |
THUMP domain containing 2 |
58455 |
0.12 |
| chr9_105878487_105878719 | 0.56 |
Col6a5 |
collagen, type VI, alpha 5 |
3203 |
0.27 |
| chr9_102516246_102516552 | 0.56 |
Ky |
kyphoscoliosis peptidase |
4937 |
0.14 |
| chr1_67124558_67125159 | 0.51 |
Cps1 |
carbamoyl-phosphate synthetase 1 |
1832 |
0.42 |
| chr5_74649245_74649406 | 0.50 |
Lnx1 |
ligand of numb-protein X 1 |
25804 |
0.15 |
| chr1_154143789_154144011 | 0.50 |
A830008E24Rik |
RIKEN cDNA A830008E24 gene |
25949 |
0.16 |
| chr11_113725344_113725520 | 0.49 |
Cdc42ep4 |
CDC42 effector protein (Rho GTPase binding) 4 |
13890 |
0.12 |
| chr11_16908621_16908873 | 0.49 |
Egfr |
epidermal growth factor receptor |
3562 |
0.23 |
| chr7_140772355_140773139 | 0.48 |
Cyp2e1 |
cytochrome P450, family 2, subfamily e, polypeptide 1 |
8266 |
0.09 |
| chr13_85126873_85127061 | 0.47 |
Gm4076 |
predicted gene 4076 |
547 |
0.76 |
| chr6_85823511_85823680 | 0.45 |
Nat8f5 |
N-acetyltransferase 8 (GCN5-related) family member 5 |
2623 |
0.13 |
| chr18_8939044_8939213 | 0.45 |
Gm37148 |
predicted gene, 37148 |
10893 |
0.24 |
| chr8_91333225_91333390 | 0.45 |
Fto |
fat mass and obesity associated |
19693 |
0.13 |
| chr11_75750659_75750835 | 0.45 |
Ywhae |
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, epsilon polypeptide |
857 |
0.56 |
| chr6_28681979_28682177 | 0.44 |
Snd1 |
staphylococcal nuclease and tudor domain containing 1 |
22831 |
0.21 |
| chr17_4004285_4004445 | 0.44 |
4930470H14Rik |
RIKEN cDNA 4930470H14 gene |
78630 |
0.11 |
| chr9_116136676_116136901 | 0.44 |
Tgfbr2 |
transforming growth factor, beta receptor II |
38477 |
0.15 |
| chr19_28520868_28521034 | 0.42 |
Glis3 |
GLIS family zinc finger 3 |
19469 |
0.24 |
| chr10_108350464_108350766 | 0.42 |
Gm23105 |
predicted gene, 23105 |
13050 |
0.19 |
| chrX_84312323_84312721 | 0.40 |
Dmd |
dystrophin, muscular dystrophy |
235873 |
0.02 |
| chr19_16450910_16451085 | 0.39 |
Gna14 |
guanine nucleotide binding protein, alpha 14 |
11697 |
0.17 |
| chr11_16704048_16704199 | 0.39 |
Gm25698 |
predicted gene, 25698 |
28588 |
0.15 |
| chr1_9906956_9907361 | 0.39 |
Mcmdc2 |
minichromosome maintenance domain containing 2 |
1480 |
0.29 |
| chr13_102858710_102858861 | 0.39 |
Mast4 |
microtubule associated serine/threonine kinase family member 4 |
47001 |
0.17 |
| chr5_91200336_91200632 | 0.39 |
Gm23092 |
predicted gene, 23092 |
17520 |
0.2 |
| chr4_59292456_59292607 | 0.39 |
Susd1 |
sushi domain containing 1 |
23435 |
0.17 |
| chr15_100349593_100349772 | 0.38 |
Methig1 |
methyltransferase hypoxia inducible domain containing 1 |
3467 |
0.13 |
| chr10_68137101_68137496 | 0.38 |
Arid5b |
AT rich interactive domain 5B (MRF1-like) |
672 |
0.78 |
| chr4_80003665_80003823 | 0.38 |
Gm11408 |
predicted gene 11408 |
170 |
0.59 |
| chr13_20205112_20205287 | 0.38 |
Elmo1 |
engulfment and cell motility 1 |
19992 |
0.25 |
| chr9_56260139_56260312 | 0.38 |
Peak1os |
pseudopodium-enriched atypical kinase 1, opposite strand |
2335 |
0.21 |
| chr14_22269831_22270192 | 0.38 |
Lrmda |
leucine rich melanocyte differentiation associated |
40154 |
0.2 |
| chr18_9632745_9632896 | 0.38 |
Gm4834 |
predicted gene 4834 |
3958 |
0.16 |
| chr3_69778568_69778734 | 0.38 |
Nmd3 |
NMD3 ribosome export adaptor |
32219 |
0.16 |
| chr1_20481729_20481880 | 0.38 |
Gm24162 |
predicted gene, 24162 |
36892 |
0.15 |
| chr4_131939568_131939730 | 0.38 |
Epb41 |
erythrocyte membrane protein band 4.1 |
2262 |
0.18 |
| chr2_146577293_146577459 | 0.37 |
4933406D12Rik |
RIKEN cDNA 4933406D12 gene |
34445 |
0.2 |
| chr1_168502271_168502436 | 0.37 |
Mir6348 |
microRNA 6348 |
11710 |
0.27 |
| chr9_55233053_55233204 | 0.37 |
Nrg4 |
neuregulin 4 |
9333 |
0.17 |
| chr9_25460856_25461181 | 0.36 |
Gm8049 |
predicted gene 8049 |
10556 |
0.2 |
| chr9_112743581_112744166 | 0.36 |
Gm24957 |
predicted gene, 24957 |
221344 |
0.02 |
| chr15_54870838_54871021 | 0.36 |
Enpp2 |
ectonucleotide pyrophosphatase/phosphodiesterase 2 |
8243 |
0.24 |
| chr15_59391938_59392317 | 0.35 |
Nsmce2 |
NSE2/MMS21 homolog, SMC5-SMC6 complex SUMO ligase |
17840 |
0.18 |
| chr6_35253418_35253775 | 0.35 |
1810058I24Rik |
RIKEN cDNA 1810058I24 gene |
864 |
0.54 |
| chr12_17601719_17602041 | 0.35 |
Gm48313 |
predicted gene, 48313 |
3426 |
0.21 |
| chr19_29968799_29968964 | 0.35 |
Il33 |
interleukin 33 |
17065 |
0.16 |
| chr9_59942649_59942840 | 0.34 |
Gm20275 |
predicted gene, 20275 |
1667 |
0.26 |
| chrX_136171222_136171400 | 0.34 |
Tceal8 |
transcription elongation factor A (SII)-like 8 |
890 |
0.43 |
| chr6_140187005_140187211 | 0.34 |
Gm24174 |
predicted gene, 24174 |
20463 |
0.18 |
| chr8_53780941_53781092 | 0.34 |
Gm19921 |
predicted gene, 19921 |
130723 |
0.05 |
| chr19_39463095_39463278 | 0.34 |
Cyp2c38 |
cytochrome P450, family 2, subfamily c, polypeptide 38 |
111 |
0.98 |
| chr17_62171907_62172058 | 0.34 |
Gm49860 |
predicted gene, 49860 |
264075 |
0.02 |
| chr13_34313351_34313741 | 0.34 |
Gm47086 |
predicted gene, 47086 |
5262 |
0.21 |
| chr11_116206084_116206272 | 0.34 |
Ten1 |
TEN1 telomerase capping complex subunit |
4756 |
0.1 |
| chr2_159755776_159755955 | 0.33 |
Gm11445 |
predicted gene 11445 |
22342 |
0.25 |
| chr19_40191993_40192245 | 0.33 |
Gm5827 |
predicted gene 5827 |
1081 |
0.44 |
| chr6_59032854_59033027 | 0.33 |
Gm19165 |
predicted gene, 19165 |
3949 |
0.2 |
| chr3_116409014_116409188 | 0.33 |
Cdc14a |
CDC14 cell division cycle 14A |
3613 |
0.2 |
| chr14_69628830_69628981 | 0.33 |
Gm27177 |
predicted gene 27177 |
11369 |
0.11 |
| chr2_125826215_125826366 | 0.33 |
Cops2 |
COP9 signalosome subunit 2 |
32706 |
0.15 |
| chr5_96222210_96222544 | 0.32 |
Mrpl1 |
mitochondrial ribosomal protein L1 |
12233 |
0.22 |
| chr12_17770698_17770853 | 0.32 |
Hpcal1 |
hippocalcin-like 1 |
42549 |
0.14 |
| chr9_74881023_74881569 | 0.32 |
Onecut1 |
one cut domain, family member 1 |
14812 |
0.15 |
| chr5_22255477_22255648 | 0.32 |
Gm16113 |
predicted gene 16113 |
26020 |
0.16 |
| chr1_89667541_89667692 | 0.32 |
4933400F21Rik |
RIKEN cDNA 4933400F21 gene |
16272 |
0.23 |
| chr1_60716026_60716187 | 0.32 |
Cd28 |
CD28 antigen |
694 |
0.59 |
| chr18_34868711_34868905 | 0.32 |
Egr1 |
early growth response 1 |
7601 |
0.14 |
| chr13_37468607_37468985 | 0.32 |
Gm47731 |
predicted gene, 47731 |
3242 |
0.13 |
| chr18_69663255_69663406 | 0.31 |
Tcf4 |
transcription factor 4 |
6663 |
0.31 |
| chr1_86700604_86700771 | 0.31 |
Dis3l2 |
DIS3 like 3'-5' exoribonuclease 2 |
3121 |
0.19 |
| chr1_52597389_52597548 | 0.31 |
Gm5527 |
predicted gene 5527 |
12950 |
0.13 |
| chr11_107452533_107452722 | 0.31 |
Pitpnc1 |
phosphatidylinositol transfer protein, cytoplasmic 1 |
18072 |
0.11 |
| chr10_67048205_67048592 | 0.31 |
Reep3 |
receptor accessory protein 3 |
11076 |
0.19 |
| chr4_81591457_81591882 | 0.31 |
Gm11411 |
predicted gene 11411 |
104772 |
0.07 |
| chr12_80956924_80957266 | 0.31 |
Srsf5 |
serine and arginine-rich splicing factor 5 |
10719 |
0.13 |
| chr9_69204954_69205120 | 0.31 |
Rora |
RAR-related orphan receptor alpha |
9496 |
0.29 |
| chr5_150912926_150913090 | 0.31 |
Gm43298 |
predicted gene 43298 |
30366 |
0.16 |
| chr5_75961858_75962064 | 0.31 |
Kdr |
kinase insert domain protein receptor |
16497 |
0.17 |
| chrX_140387482_140387633 | 0.30 |
Frmpd3 |
FERM and PDZ domain containing 3 |
5586 |
0.23 |
| chr5_89416345_89416742 | 0.30 |
Gc |
vitamin D binding protein |
19085 |
0.24 |
| chr2_173449296_173449460 | 0.30 |
Gm14642 |
predicted gene 14642 |
54812 |
0.12 |
| chr2_163661430_163661594 | 0.30 |
Pkig |
protein kinase inhibitor, gamma |
13 |
0.97 |
| chr4_141955631_141955810 | 0.29 |
Fhad1 |
forkhead-associated (FHA) phosphopeptide binding domain 1 |
1526 |
0.3 |
| chr19_26754124_26754675 | 0.29 |
Smarca2 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
799 |
0.69 |
| chr7_80741087_80741271 | 0.29 |
Iqgap1 |
IQ motif containing GTPase activating protein 1 |
2701 |
0.24 |
| chr13_24945328_24945479 | 0.29 |
Gpld1 |
glycosylphosphatidylinositol specific phospholipase D1 |
2251 |
0.21 |
| chrX_74270256_74270420 | 0.29 |
Rpl10 |
ribosomal protein L10 |
474 |
0.52 |
| chr18_16753631_16753792 | 0.29 |
Gm15485 |
predicted gene 15485 |
24978 |
0.2 |
| chr5_90424369_90424630 | 0.29 |
Gm43363 |
predicted gene 43363 |
27226 |
0.13 |
| chr11_16908151_16908431 | 0.29 |
Egfr |
epidermal growth factor receptor |
3106 |
0.25 |
| chr13_58999542_58999699 | 0.29 |
Gm34245 |
predicted gene, 34245 |
78676 |
0.08 |
| chr15_91275437_91275588 | 0.29 |
CN725425 |
cDNA sequence CN725425 |
43934 |
0.16 |
| chr5_72763116_72763280 | 0.29 |
Txk |
TXK tyrosine kinase |
10421 |
0.15 |
| chr3_66594684_66595084 | 0.29 |
Gm17952 |
predicted gene, 17952 |
31333 |
0.23 |
| chr7_4532478_4532639 | 0.28 |
Dnaaf3 |
dynein, axonemal assembly factor 3 |
105 |
0.91 |
| chr2_158859600_158859775 | 0.28 |
Dhx35 |
DEAH (Asp-Glu-Ala-His) box polypeptide 35 |
9058 |
0.25 |
| chrX_42148063_42148502 | 0.28 |
Stag2 |
stromal antigen 2 |
1035 |
0.46 |
| chr3_65357881_65358061 | 0.28 |
Kcnab1 |
potassium voltage-gated channel, shaker-related subfamily, beta member 1 |
4204 |
0.18 |
| chr2_61138777_61138928 | 0.28 |
Gm13581 |
predicted gene 13581 |
85940 |
0.09 |
| chr2_122266063_122266214 | 0.28 |
Duox2 |
dual oxidase 2 |
19431 |
0.1 |
| chr16_24019943_24020095 | 0.28 |
Gm49518 |
predicted gene, 49518 |
19887 |
0.15 |
| chr2_137422722_137422873 | 0.28 |
Gm14055 |
predicted gene 14055 |
134294 |
0.05 |
| chr11_109733399_109733703 | 0.28 |
Fam20a |
family with sequence similarity 20, member A |
11272 |
0.17 |
| chr1_85199249_85199400 | 0.28 |
AC123856.1 |
nuclear antigen Sp100 (Sp100) pseudogene |
289 |
0.46 |
| chr4_138065715_138066098 | 0.28 |
Eif4g3 |
eukaryotic translation initiation factor 4 gamma, 3 |
61154 |
0.1 |
| chr9_13681735_13681905 | 0.27 |
Maml2 |
mastermind like transcriptional coactivator 2 |
19360 |
0.18 |
| chr2_156585343_156585515 | 0.27 |
Gm14168 |
predicted gene 14168 |
13637 |
0.11 |
| chr14_25385763_25386034 | 0.27 |
Gm26660 |
predicted gene, 26660 |
6289 |
0.21 |
| chr7_30957359_30958052 | 0.27 |
Gm4673 |
predicted gene 4673 |
296 |
0.67 |
| chr18_7847563_7847725 | 0.27 |
Gm50027 |
predicted gene, 50027 |
2709 |
0.23 |
| chr17_64643456_64643864 | 0.27 |
Man2a1 |
mannosidase 2, alpha 1 |
42924 |
0.17 |
| chr7_126636776_126636927 | 0.27 |
Nupr1 |
nuclear protein transcription regulator 1 |
5990 |
0.08 |
| chr3_101509926_101510102 | 0.27 |
Atp1a1 |
ATPase, Na+/K+ transporting, alpha 1 polypeptide |
67546 |
0.08 |
| chr1_24613582_24613754 | 0.27 |
Gm28437 |
predicted gene 28437 |
303 |
0.51 |
| chr9_107597647_107597798 | 0.26 |
Lsmem2 |
leucine-rich single-pass membrane protein 2 |
647 |
0.36 |
| chr6_54199490_54199996 | 0.26 |
Gm15526 |
predicted gene 15526 |
22060 |
0.17 |
| chr6_59052402_59052674 | 0.26 |
Gm19165 |
predicted gene, 19165 |
15649 |
0.17 |
| chr1_51501426_51501585 | 0.26 |
Gm17767 |
predicted gene, 17767 |
10824 |
0.17 |
| chr10_33945506_33945695 | 0.26 |
Zup1 |
zinc finger containing ubiquitin peptidase 1 |
4260 |
0.13 |
| chr15_98789109_98789266 | 0.26 |
Wnt1 |
wingless-type MMTV integration site family, member 1 |
670 |
0.48 |
| chr12_58323949_58324220 | 0.26 |
Clec14a |
C-type lectin domain family 14, member a |
54794 |
0.16 |
| chr13_118387400_118387593 | 0.26 |
Mrps30 |
mitochondrial ribosomal protein S30 |
244 |
0.86 |
| chr3_61217316_61217479 | 0.26 |
Gm37719 |
predicted gene, 37719 |
74049 |
0.1 |
| chr8_36457312_36457463 | 0.26 |
Trmt9b |
tRNA methyltransferase 9B |
161 |
0.96 |
| chr19_44064101_44064296 | 0.26 |
Erlin1 |
ER lipid raft associated 1 |
3485 |
0.17 |
| chr7_28411283_28411456 | 0.26 |
Samd4b |
sterile alpha motif domain containing 4B |
2538 |
0.12 |
| chr4_149453846_149453997 | 0.26 |
Rbp7 |
retinol binding protein 7, cellular |
497 |
0.65 |
| chr1_24042012_24042182 | 0.26 |
Fam135a |
family with sequence similarity 135, member A |
13011 |
0.18 |
| chr18_35681549_35681863 | 0.26 |
Dnajc18 |
DnaJ heat shock protein family (Hsp40) member C18 |
5037 |
0.1 |
| chr6_5517592_5517751 | 0.26 |
Pdk4 |
pyruvate dehydrogenase kinase, isoenzyme 4 |
21362 |
0.26 |
| chr14_45452880_45453061 | 0.26 |
Gm34250 |
predicted gene, 34250 |
5897 |
0.13 |
| chr1_66895610_66896410 | 0.25 |
Gm25832 |
predicted gene, 25832 |
29213 |
0.09 |
| chr13_12146494_12146649 | 0.25 |
Gm47323 |
predicted gene, 47323 |
14183 |
0.15 |
| chr10_68269443_68270351 | 0.25 |
Arid5b |
AT rich interactive domain 5B (MRF1-like) |
8824 |
0.23 |
| chr16_87128308_87128459 | 0.25 |
Gm32865 |
predicted gene, 32865 |
56 |
0.99 |
| chr5_90471470_90471733 | 0.25 |
Alb |
albumin |
8901 |
0.15 |
| chr15_75994277_75994580 | 0.25 |
Mapk15 |
mitogen-activated protein kinase 15 |
659 |
0.46 |
| chr3_106457659_106457818 | 0.25 |
Gm38244 |
predicted gene, 38244 |
20545 |
0.13 |
| chr14_35502504_35502655 | 0.25 |
Gm19016 |
predicted gene, 19016 |
46547 |
0.17 |
| chr7_45264516_45264678 | 0.25 |
Slc6a16 |
solute carrier family 6, member 16 |
3553 |
0.09 |
| chr6_88820995_88821174 | 0.25 |
Mgll |
monoglyceride lipase |
2046 |
0.19 |
| chr10_123588527_123588968 | 0.25 |
Gm19169 |
predicted gene, 19169 |
5159 |
0.33 |
| chr5_125491109_125491333 | 0.25 |
Gm27551 |
predicted gene, 27551 |
11844 |
0.13 |
| chr8_85088891_85089042 | 0.25 |
Man2b1 |
mannosidase 2, alpha B1 |
2508 |
0.09 |
| chr4_8361870_8362071 | 0.25 |
Gm37386 |
predicted gene, 37386 |
92277 |
0.08 |
| chr19_21152410_21152561 | 0.25 |
AC106834.1 |
novel transcript |
18356 |
0.2 |
| chr12_12509285_12509462 | 0.25 |
4921511I17Rik |
RIKEN cDNA 4921511I17 gene |
116758 |
0.06 |
| chr7_65338015_65338188 | 0.25 |
Tjp1 |
tight junction protein 1 |
4696 |
0.24 |
| chr15_10273020_10273171 | 0.25 |
Prlr |
prolactin receptor |
23535 |
0.21 |
| chr6_149344317_149344633 | 0.24 |
Gm15782 |
predicted gene 15782 |
9351 |
0.14 |
| chr16_72734333_72734499 | 0.24 |
Robo1 |
roundabout guidance receptor 1 |
71212 |
0.14 |
| chr2_39002726_39002905 | 0.24 |
Rpl35 |
ribosomal protein L35 |
2228 |
0.15 |
| chr9_70245178_70245559 | 0.24 |
Myo1e |
myosin IE |
38000 |
0.16 |
| chr17_83938813_83938978 | 0.24 |
Gm23905 |
predicted gene, 23905 |
8693 |
0.12 |
| chr12_25281511_25281662 | 0.24 |
Gm19340 |
predicted gene, 19340 |
1009 |
0.54 |
| chr4_150284112_150284294 | 0.24 |
Rere |
arginine glutamic acid dipeptide (RE) repeats |
2391 |
0.24 |
| chr6_29833568_29833845 | 0.24 |
Ahcyl2 |
S-adenosylhomocysteine hydrolase-like 2 |
20054 |
0.17 |
| chr12_110488367_110488518 | 0.24 |
Gm19605 |
predicted gene, 19605 |
2234 |
0.25 |
| chr10_86793057_86793348 | 0.24 |
Nt5dc3 |
5'-nucleotidase domain containing 3 |
14197 |
0.11 |
| chr11_87449212_87449363 | 0.24 |
Rnu3b3 |
U3B small nuclear RNA 3 |
660 |
0.48 |
| chr19_59908738_59908900 | 0.24 |
Gm17203 |
predicted gene 17203 |
7757 |
0.21 |
| chr1_182280533_182280736 | 0.24 |
Degs1 |
delta(4)-desaturase, sphingolipid 1 |
1590 |
0.35 |
| chr17_63343186_63343351 | 0.24 |
Gm17228 |
predicted gene 17228 |
4142 |
0.22 |
| chr1_24611857_24612019 | 0.24 |
Gm28439 |
predicted gene 28439 |
472 |
0.45 |
| chrX_142456153_142456325 | 0.24 |
Gm25915 |
predicted gene, 25915 |
29208 |
0.13 |
| chr8_66326515_66326676 | 0.24 |
Marchf1 |
membrane associated ring-CH-type finger 1 |
59699 |
0.12 |
| chr13_16269739_16270144 | 0.24 |
Gm48491 |
predicted gene, 48491 |
7897 |
0.23 |
| chr10_7832157_7832704 | 0.24 |
Zc3h12d |
zinc finger CCCH type containing 12D |
40 |
0.96 |
| chr12_25092894_25093391 | 0.24 |
Id2 |
inhibitor of DNA binding 2 |
2945 |
0.22 |
| chr10_9495311_9495462 | 0.24 |
Gm48751 |
predicted gene, 48751 |
8184 |
0.16 |
| chr13_100543031_100543362 | 0.23 |
Ocln |
occludin |
9264 |
0.12 |
| chr2_165999944_166000123 | 0.23 |
Ncoa3 |
nuclear receptor coactivator 3 |
7396 |
0.14 |
| chr8_117677298_117677515 | 0.23 |
Gm31717 |
predicted gene, 31717 |
197 |
0.91 |
| chr7_81243481_81243646 | 0.23 |
Gm7180 |
predicted pseudogene 7180 |
20689 |
0.14 |
| chr2_48816834_48817005 | 0.23 |
Acvr2a |
activin receptor IIA |
2810 |
0.3 |
| chr18_59082260_59082587 | 0.23 |
Minar2 |
membrane integral NOTCH2 associated receptor 2 |
19912 |
0.26 |
| chr14_20319722_20320198 | 0.23 |
Nudt13 |
nudix (nucleoside diphosphate linked moiety X)-type motif 13 |
8531 |
0.12 |
| chr12_112094362_112094513 | 0.23 |
Aspg |
asparaginase |
12242 |
0.12 |
| chr11_16890762_16890930 | 0.23 |
Egfr |
epidermal growth factor receptor |
12696 |
0.19 |
| chr10_53348308_53348525 | 0.23 |
Cep85l |
centrosomal protein 85-like |
1407 |
0.3 |
| chr13_81328304_81328472 | 0.23 |
Adgrv1 |
adhesion G protein-coupled receptor V1 |
14448 |
0.26 |
| chr9_64397221_64397372 | 0.23 |
Megf11 |
multiple EGF-like-domains 11 |
11610 |
0.2 |
| chr3_104220440_104221564 | 0.22 |
Magi3 |
membrane associated guanylate kinase, WW and PDZ domain containing 3 |
628 |
0.6 |
| chr12_80951376_80951574 | 0.22 |
Srsf5 |
serine and arginine-rich splicing factor 5 |
5099 |
0.14 |
| chr2_64462379_64462613 | 0.22 |
Gm13577 |
predicted gene 13577 |
12453 |
0.27 |
| chr4_98960279_98960453 | 0.22 |
Dock7 |
dedicator of cytokinesis 7 |
52 |
0.98 |
| chr12_41044805_41045069 | 0.22 |
Gm47375 |
predicted gene, 47375 |
2 |
0.98 |
| chr12_36155196_36155406 | 0.22 |
Bzw2 |
basic leucine zipper and W2 domains 2 |
1326 |
0.29 |
| chr12_118299403_118299566 | 0.22 |
Sp4 |
trans-acting transcription factor 4 |
1884 |
0.44 |
| chr10_116310601_116310804 | 0.22 |
Ptprb |
protein tyrosine phosphatase, receptor type, B |
9330 |
0.18 |
| chr18_14503181_14503343 | 0.22 |
Gm50098 |
predicted gene, 50098 |
11257 |
0.23 |
| chr11_19941290_19941474 | 0.22 |
Spred2 |
sprouty-related EVH1 domain containing 2 |
13188 |
0.26 |
| chr5_66081550_66081734 | 0.22 |
Rbm47 |
RNA binding motif protein 47 |
348 |
0.81 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.1 | 0.6 | GO:0060613 | fat pad development(GO:0060613) |
| 0.1 | 0.3 | GO:1902309 | negative regulation of peptidyl-serine dephosphorylation(GO:1902309) |
| 0.1 | 0.2 | GO:0045196 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.1 | 0.2 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.1 | 0.2 | GO:0000087 | mitotic M phase(GO:0000087) |
| 0.1 | 0.2 | GO:0034197 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.1 | 0.2 | GO:0060300 | regulation of cytokine activity(GO:0060300) |
| 0.1 | 0.2 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.1 | 0.1 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.1 | 0.2 | GO:0090365 | regulation of mRNA modification(GO:0090365) |
| 0.1 | 0.2 | GO:0071288 | cellular response to mercury ion(GO:0071288) |
| 0.0 | 0.1 | GO:0043321 | regulation of natural killer cell degranulation(GO:0043321) |
| 0.0 | 0.1 | GO:0061625 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) glycolytic process through fructose-1-phosphate(GO:0061625) |
| 0.0 | 0.1 | GO:0060061 | Spemann organizer formation(GO:0060061) |
| 0.0 | 0.1 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.0 | 0.3 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.1 | GO:0030070 | insulin processing(GO:0030070) |
| 0.0 | 0.3 | GO:0070269 | pyroptosis(GO:0070269) |
| 0.0 | 0.2 | GO:0016479 | negative regulation of transcription from RNA polymerase I promoter(GO:0016479) |
| 0.0 | 0.1 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.0 | 0.1 | GO:0060448 | dichotomous subdivision of terminal units involved in lung branching(GO:0060448) |
| 0.0 | 0.2 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.0 | 0.1 | GO:0010911 | regulation of isomerase activity(GO:0010911) positive regulation of isomerase activity(GO:0010912) |
| 0.0 | 0.2 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 0.1 | GO:0043134 | hindgut contraction(GO:0043133) regulation of hindgut contraction(GO:0043134) |
| 0.0 | 0.1 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.0 | 0.2 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.0 | 0.1 | GO:0006393 | termination of mitochondrial transcription(GO:0006393) |
| 0.0 | 0.9 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.0 | 0.1 | GO:0086046 | membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.0 | 0.1 | GO:0003219 | cardiac right ventricle formation(GO:0003219) |
| 0.0 | 0.1 | GO:0046499 | S-adenosylmethioninamine metabolic process(GO:0046499) |
| 0.0 | 0.1 | GO:0060178 | regulation of exocyst localization(GO:0060178) |
| 0.0 | 0.1 | GO:0071799 | response to prostaglandin D(GO:0071798) cellular response to prostaglandin D stimulus(GO:0071799) |
| 0.0 | 0.1 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.0 | 0.1 | GO:0015760 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.0 | 0.3 | GO:0051292 | nuclear pore complex assembly(GO:0051292) |
| 0.0 | 0.1 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) |
| 0.0 | 0.0 | GO:1903660 | regulation of complement-dependent cytotoxicity(GO:1903659) negative regulation of complement-dependent cytotoxicity(GO:1903660) |
| 0.0 | 0.1 | GO:1902109 | negative regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902109) |
| 0.0 | 0.0 | GO:1990705 | cholangiocyte proliferation(GO:1990705) |
| 0.0 | 0.1 | GO:0001907 | killing by symbiont of host cells(GO:0001907) disruption by symbiont of host cell(GO:0044004) |
| 0.0 | 0.1 | GO:0070671 | response to interleukin-12(GO:0070671) |
| 0.0 | 0.0 | GO:0006481 | C-terminal protein methylation(GO:0006481) |
| 0.0 | 0.0 | GO:0030210 | heparin biosynthetic process(GO:0030210) |
| 0.0 | 0.1 | GO:0046208 | spermine catabolic process(GO:0046208) |
| 0.0 | 0.1 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.0 | 0.1 | GO:0046881 | positive regulation of follicle-stimulating hormone secretion(GO:0046881) |
| 0.0 | 0.2 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.0 | 0.1 | GO:0032875 | regulation of DNA endoreduplication(GO:0032875) |
| 0.0 | 0.1 | GO:0007182 | common-partner SMAD protein phosphorylation(GO:0007182) |
| 0.0 | 0.1 | GO:0006933 | negative regulation of cell adhesion involved in substrate-bound cell migration(GO:0006933) |
| 0.0 | 0.1 | GO:0060830 | ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) |
| 0.0 | 0.0 | GO:0046100 | hypoxanthine metabolic process(GO:0046100) hypoxanthine biosynthetic process(GO:0046101) |
| 0.0 | 0.1 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.0 | 0.1 | GO:1900095 | regulation of dosage compensation by inactivation of X chromosome(GO:1900095) |
| 0.0 | 0.1 | GO:0061158 | 3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.0 | 0.1 | GO:0010255 | carbohydrate mediated signaling(GO:0009756) hexose mediated signaling(GO:0009757) sugar mediated signaling pathway(GO:0010182) glucose mediated signaling pathway(GO:0010255) |
| 0.0 | 0.1 | GO:0015808 | L-alanine transport(GO:0015808) |
| 0.0 | 0.2 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 | 0.1 | GO:0034653 | diterpenoid catabolic process(GO:0016103) retinoic acid catabolic process(GO:0034653) |
| 0.0 | 0.0 | GO:1903984 | positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) |
| 0.0 | 0.1 | GO:0034214 | protein hexamerization(GO:0034214) |
| 0.0 | 0.1 | GO:0061470 | T follicular helper cell differentiation(GO:0061470) |
| 0.0 | 0.1 | GO:0098908 | regulation of neuronal action potential(GO:0098908) |
| 0.0 | 0.1 | GO:0072675 | osteoclast fusion(GO:0072675) |
| 0.0 | 0.1 | GO:0010626 | negative regulation of Schwann cell proliferation(GO:0010626) |
| 0.0 | 0.1 | GO:0060903 | positive regulation of meiosis I(GO:0060903) |
| 0.0 | 0.1 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.0 | 0.2 | GO:0048304 | positive regulation of isotype switching to IgG isotypes(GO:0048304) |
| 0.0 | 0.1 | GO:1903416 | response to glycoside(GO:1903416) |
| 0.0 | 0.1 | GO:1904754 | positive regulation of vascular associated smooth muscle cell migration(GO:1904754) |
| 0.0 | 0.1 | GO:0019086 | late viral transcription(GO:0019086) |
| 0.0 | 0.0 | GO:0097296 | activation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097296) |
| 0.0 | 0.0 | GO:0015755 | fructose transport(GO:0015755) |
| 0.0 | 0.0 | GO:1902075 | cellular response to salt(GO:1902075) |
| 0.0 | 0.1 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.0 | 0.1 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.1 | GO:0010529 | regulation of transposition(GO:0010528) negative regulation of transposition(GO:0010529) |
| 0.0 | 0.1 | GO:0051025 | negative regulation of immunoglobulin secretion(GO:0051025) |
| 0.0 | 0.1 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.0 | 0.1 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.0 | 0.3 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.0 | 0.0 | GO:2000836 | androgen secretion(GO:0035935) regulation of androgen secretion(GO:2000834) positive regulation of androgen secretion(GO:2000836) |
| 0.0 | 0.0 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.2 | GO:0072673 | lamellipodium morphogenesis(GO:0072673) |
| 0.0 | 0.1 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.0 | 0.0 | GO:0006059 | hexitol metabolic process(GO:0006059) |
| 0.0 | 0.0 | GO:2000721 | positive regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000721) |
| 0.0 | 0.0 | GO:0042726 | flavin-containing compound metabolic process(GO:0042726) |
| 0.0 | 0.0 | GO:0006046 | N-acetylglucosamine catabolic process(GO:0006046) |
| 0.0 | 0.1 | GO:0003149 | membranous septum morphogenesis(GO:0003149) |
| 0.0 | 0.0 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.0 | 0.1 | GO:0019852 | L-ascorbic acid metabolic process(GO:0019852) |
| 0.0 | 0.0 | GO:0071926 | cannabinoid signaling pathway(GO:0038171) endocannabinoid signaling pathway(GO:0071926) |
| 0.0 | 0.1 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.0 | 0.1 | GO:0042136 | neurotransmitter biosynthetic process(GO:0042136) |
| 0.0 | 0.0 | GO:0021827 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) postnatal olfactory bulb interneuron migration(GO:0021827) |
| 0.0 | 0.0 | GO:0034093 | positive regulation of maintenance of sister chromatid cohesion(GO:0034093) positive regulation of maintenance of mitotic sister chromatid cohesion(GO:0034184) |
| 0.0 | 0.0 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.0 | 0.1 | GO:0097341 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) inhibition of cysteine-type endopeptidase activity involved in apoptotic process(GO:1990001) |
| 0.0 | 0.1 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.0 | 0.0 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.0 | 0.0 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.0 | 0.1 | GO:1904417 | regulation of xenophagy(GO:1904415) positive regulation of xenophagy(GO:1904417) |
| 0.0 | 0.1 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.0 | 0.0 | GO:0014016 | neuroblast differentiation(GO:0014016) |
| 0.0 | 0.0 | GO:0051365 | cellular response to potassium ion starvation(GO:0051365) |
| 0.0 | 0.0 | GO:0071504 | response to heparin(GO:0071503) cellular response to heparin(GO:0071504) |
| 0.0 | 0.0 | GO:0033092 | positive regulation of immature T cell proliferation(GO:0033091) positive regulation of immature T cell proliferation in thymus(GO:0033092) |
| 0.0 | 0.0 | GO:2000054 | negative regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000054) |
| 0.0 | 0.0 | GO:0036493 | positive regulation of translation in response to endoplasmic reticulum stress(GO:0036493) |
| 0.0 | 0.1 | GO:0010572 | positive regulation of platelet activation(GO:0010572) |
| 0.0 | 0.0 | GO:0021699 | cerebellar cortex maturation(GO:0021699) |
| 0.0 | 0.0 | GO:1903279 | regulation of calcium:sodium antiporter activity(GO:1903279) |
| 0.0 | 0.1 | GO:0046909 | intermembrane transport(GO:0046909) |
| 0.0 | 0.1 | GO:0034310 | primary alcohol catabolic process(GO:0034310) |
| 0.0 | 0.0 | GO:2001137 | positive regulation of endocytic recycling(GO:2001137) |
| 0.0 | 0.0 | GO:0030167 | proteoglycan catabolic process(GO:0030167) |
| 0.0 | 0.0 | GO:0045726 | positive regulation of integrin biosynthetic process(GO:0045726) |
| 0.0 | 0.1 | GO:0032570 | response to progesterone(GO:0032570) |
| 0.0 | 0.1 | GO:0060020 | Bergmann glial cell differentiation(GO:0060020) |
| 0.0 | 0.1 | GO:0097012 | response to granulocyte macrophage colony-stimulating factor(GO:0097012) |
| 0.0 | 0.1 | GO:0050916 | sensory perception of sweet taste(GO:0050916) |
| 0.0 | 0.1 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.0 | 0.0 | GO:0010025 | wax biosynthetic process(GO:0010025) wax metabolic process(GO:0010166) |
| 0.0 | 0.1 | GO:0051895 | negative regulation of focal adhesion assembly(GO:0051895) |
| 0.0 | 0.1 | GO:2001214 | positive regulation of vasculogenesis(GO:2001214) |
| 0.0 | 0.1 | GO:0051657 | maintenance of organelle location(GO:0051657) |
| 0.0 | 0.0 | GO:0035166 | post-embryonic hemopoiesis(GO:0035166) |
| 0.0 | 0.0 | GO:0060335 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.0 | 0.2 | GO:1901798 | positive regulation of signal transduction by p53 class mediator(GO:1901798) |
| 0.0 | 0.0 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.0 | 0.1 | GO:0071286 | cellular response to magnesium ion(GO:0071286) |
| 0.0 | 0.1 | GO:0070779 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.0 | 0.1 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.0 | 0.1 | GO:0014063 | negative regulation of serotonin secretion(GO:0014063) |
| 0.0 | 0.0 | GO:0048294 | negative regulation of isotype switching to IgE isotypes(GO:0048294) |
| 0.0 | 0.2 | GO:0072600 | establishment of protein localization to Golgi(GO:0072600) |
| 0.0 | 0.1 | GO:0070141 | response to UV-A(GO:0070141) |
| 0.0 | 0.1 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.3 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.1 | GO:0010992 | ubiquitin homeostasis(GO:0010992) |
| 0.0 | 0.0 | GO:0086073 | bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 0.0 | 0.1 | GO:2000786 | positive regulation of autophagosome assembly(GO:2000786) |
| 0.0 | 0.0 | GO:1904738 | vascular associated smooth muscle cell migration(GO:1904738) regulation of vascular associated smooth muscle cell migration(GO:1904752) |
| 0.0 | 0.1 | GO:2000609 | regulation of thyroid hormone generation(GO:2000609) |
| 0.0 | 0.0 | GO:0002752 | cell surface pattern recognition receptor signaling pathway(GO:0002752) |
| 0.0 | 0.1 | GO:0046984 | regulation of hemoglobin biosynthetic process(GO:0046984) |
| 0.0 | 0.0 | GO:1900739 | regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900739) positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900740) |
| 0.0 | 0.0 | GO:0043096 | purine nucleobase salvage(GO:0043096) |
| 0.0 | 0.0 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.0 | GO:0017187 | peptidyl-glutamic acid carboxylation(GO:0017187) |
| 0.0 | 0.0 | GO:0035507 | regulation of myosin-light-chain-phosphatase activity(GO:0035507) |
| 0.0 | 0.0 | GO:0090230 | regulation of centromere complex assembly(GO:0090230) |
| 0.0 | 0.0 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.0 | 0.0 | GO:0045658 | regulation of neutrophil differentiation(GO:0045658) |
| 0.0 | 0.0 | GO:0009080 | alanine catabolic process(GO:0006524) pyruvate family amino acid catabolic process(GO:0009080) |
| 0.0 | 0.1 | GO:0009115 | xanthine catabolic process(GO:0009115) xanthine metabolic process(GO:0046110) |
| 0.0 | 0.1 | GO:0000055 | ribosomal large subunit export from nucleus(GO:0000055) |
| 0.0 | 0.1 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.0 | 0.0 | GO:0006598 | polyamine catabolic process(GO:0006598) |
| 0.0 | 0.0 | GO:0061209 | cell proliferation involved in mesonephros development(GO:0061209) |
| 0.0 | 0.2 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.0 | 0.1 | GO:1901299 | negative regulation of hydrogen peroxide-mediated programmed cell death(GO:1901299) |
| 0.0 | 0.1 | GO:0045176 | apical protein localization(GO:0045176) |
| 0.0 | 0.1 | GO:0033160 | positive regulation of protein import into nucleus, translocation(GO:0033160) |
| 0.0 | 0.0 | GO:2000645 | negative regulation of receptor catabolic process(GO:2000645) |
| 0.0 | 0.1 | GO:0044387 | negative regulation of protein kinase activity by regulation of protein phosphorylation(GO:0044387) |
| 0.0 | 0.1 | GO:0030575 | nuclear body organization(GO:0030575) |
| 0.0 | 0.0 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.0 | 0.0 | GO:1902956 | regulation of mitochondrial electron transport, NADH to ubiquinone(GO:1902956) |
| 0.0 | 0.1 | GO:0043116 | negative regulation of vascular permeability(GO:0043116) |
| 0.0 | 0.0 | GO:2001168 | regulation of histone H2B ubiquitination(GO:2001166) positive regulation of histone H2B ubiquitination(GO:2001168) |
| 0.0 | 0.0 | GO:0046504 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) cellular lipid biosynthetic process(GO:0097384) ether biosynthetic process(GO:1901503) |
| 0.0 | 0.0 | GO:1904707 | positive regulation of vascular smooth muscle cell proliferation(GO:1904707) |
| 0.0 | 0.0 | GO:2000418 | positive regulation of eosinophil migration(GO:2000418) |
| 0.0 | 0.1 | GO:0000022 | mitotic spindle elongation(GO:0000022) mitotic spindle midzone assembly(GO:0051256) |
| 0.0 | 0.1 | GO:0018126 | protein hydroxylation(GO:0018126) |
| 0.0 | 0.1 | GO:0045717 | negative regulation of fatty acid biosynthetic process(GO:0045717) |
| 0.0 | 0.2 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 0.0 | GO:0035441 | cell migration involved in vasculogenesis(GO:0035441) |
| 0.0 | 0.0 | GO:0032196 | transposition(GO:0032196) |
| 0.0 | 0.0 | GO:0033030 | negative regulation of neutrophil apoptotic process(GO:0033030) |
| 0.0 | 0.1 | GO:1901407 | regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.0 | 0.0 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.1 | GO:0001553 | luteinization(GO:0001553) |
| 0.0 | 0.0 | GO:0045901 | positive regulation of translational elongation(GO:0045901) |
| 0.0 | 0.0 | GO:1904431 | positive regulation of t-circle formation(GO:1904431) |
| 0.0 | 0.0 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.0 | 0.0 | GO:1904995 | negative regulation of leukocyte tethering or rolling(GO:1903237) negative regulation of leukocyte adhesion to vascular endothelial cell(GO:1904995) |
| 0.0 | 0.0 | GO:0006216 | cytidine catabolic process(GO:0006216) cytidine deamination(GO:0009972) cytidine metabolic process(GO:0046087) |
| 0.0 | 0.0 | GO:0045626 | negative regulation of T-helper 1 cell differentiation(GO:0045626) |
| 0.0 | 0.0 | GO:1901475 | pyruvate transmembrane transport(GO:1901475) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.1 | 0.1 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 0.1 | 0.2 | GO:0032010 | phagolysosome(GO:0032010) |
| 0.0 | 0.1 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.0 | 0.2 | GO:0072557 | IPAF inflammasome complex(GO:0072557) |
| 0.0 | 0.2 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.0 | 0.1 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.0 | 0.3 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.1 | GO:0002142 | stereocilia ankle link complex(GO:0002142) |
| 0.0 | 0.1 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.0 | 0.1 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 0.1 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.0 | 0.1 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.0 | 0.2 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.3 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.1 | GO:0070820 | tertiary granule(GO:0070820) |
| 0.0 | 0.1 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 0.1 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.0 | 0.1 | GO:0097433 | dense body(GO:0097433) |
| 0.0 | 0.1 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.0 | 0.1 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.0 | 0.1 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.0 | 0.2 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.0 | 0.1 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 0.1 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.0 | 0.0 | GO:0033503 | HULC complex(GO:0033503) |
| 0.0 | 0.0 | GO:0097451 | glial limiting end-foot(GO:0097451) |
| 0.0 | 0.0 | GO:0044194 | cytolytic granule(GO:0044194) |
| 0.0 | 0.0 | GO:0035838 | growing cell tip(GO:0035838) |
| 0.0 | 0.1 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.0 | 0.0 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.0 | 0.2 | GO:0001741 | XY body(GO:0001741) |
| 0.0 | 0.1 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.0 | 0.1 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.0 | 0.1 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.0 | 0.1 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.0 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.0 | 0.1 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.0 | 0.1 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.0 | 0.0 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.0 | 0.1 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.0 | 0.0 | GO:0020016 | ciliary pocket(GO:0020016) ciliary pocket membrane(GO:0020018) |
| 0.0 | 0.0 | GO:0036488 | CHOP-C/EBP complex(GO:0036488) |
| 0.0 | 0.0 | GO:0033647 | host intracellular organelle(GO:0033647) host intracellular membrane-bounded organelle(GO:0033648) |
| 0.0 | 0.0 | GO:0046691 | intracellular canaliculus(GO:0046691) |
| 0.0 | 0.0 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.0 | 0.3 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.0 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.0 | 0.1 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.0 | 0.1 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.1 | GO:0031209 | SCAR complex(GO:0031209) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.1 | 0.5 | GO:0033695 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 0.1 | 0.2 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 0.1 | 0.2 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.1 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.0 | 0.4 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.2 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.0 | 0.1 | GO:0015526 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.0 | 0.2 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.0 | 0.1 | GO:0051718 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) DNA (cytosine-5-)-methyltransferase activity, acting on CpG substrates(GO:0051718) |
| 0.0 | 0.1 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.0 | 0.1 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.0 | 0.1 | GO:0030984 | kininogen binding(GO:0030984) |
| 0.0 | 0.2 | GO:0047760 | butyrate-CoA ligase activity(GO:0047760) |
| 0.0 | 0.1 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.1 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.1 | GO:0004528 | phosphodiesterase I activity(GO:0004528) |
| 0.0 | 0.1 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 0.1 | GO:0016899 | oxidoreductase activity, acting on the CH-OH group of donors, oxygen as acceptor(GO:0016899) |
| 0.0 | 0.2 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 0.1 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.0 | 0.1 | GO:0003696 | satellite DNA binding(GO:0003696) |
| 0.0 | 0.1 | GO:0042284 | sphingolipid delta-4 desaturase activity(GO:0042284) |
| 0.0 | 0.1 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.0 | 0.1 | GO:0032557 | pyrimidine ribonucleotide binding(GO:0032557) |
| 0.0 | 0.1 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.0 | 0.1 | GO:0050897 | cobalt ion binding(GO:0050897) |
| 0.0 | 0.1 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.0 | 0.1 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 0.1 | GO:0086007 | voltage-gated calcium channel activity involved in cardiac muscle cell action potential(GO:0086007) |
| 0.0 | 0.1 | GO:1990715 | mRNA CDS binding(GO:1990715) |
| 0.0 | 0.1 | GO:0050252 | retinol O-fatty-acyltransferase activity(GO:0050252) |
| 0.0 | 0.1 | GO:0016743 | carboxyl- or carbamoyltransferase activity(GO:0016743) |
| 0.0 | 0.1 | GO:0045340 | mercury ion binding(GO:0045340) |
| 0.0 | 0.1 | GO:1990239 | steroid hormone binding(GO:1990239) |
| 0.0 | 0.2 | GO:0001091 | RNA polymerase II basal transcription factor binding(GO:0001091) |
| 0.0 | 0.1 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.0 | 0.1 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 0.0 | 0.1 | GO:0030519 | snoRNP binding(GO:0030519) |
| 0.0 | 0.1 | GO:0070137 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.0 | 0.1 | GO:0019158 | fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.0 | 0.2 | GO:0036122 | BMP binding(GO:0036122) |
| 0.0 | 0.1 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.0 | 0.1 | GO:0043120 | tumor necrosis factor binding(GO:0043120) |
| 0.0 | 0.1 | GO:0015198 | oligopeptide transporter activity(GO:0015198) |
| 0.0 | 0.1 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.1 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.0 | 0.1 | GO:2001070 | starch binding(GO:2001070) |
| 0.0 | 0.1 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.0 | 0.1 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.0 | 0.1 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 0.0 | 0.0 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.0 | 0.0 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.2 | GO:0016717 | oxidoreductase activity, acting on paired donors, with oxidation of a pair of donors resulting in the reduction of molecular oxygen to two molecules of water(GO:0016717) |
| 0.0 | 0.0 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 0.1 | GO:0015180 | L-alanine transmembrane transporter activity(GO:0015180) alanine transmembrane transporter activity(GO:0022858) |
| 0.0 | 0.1 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.1 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) |
| 0.0 | 0.0 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.0 | 0.1 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.1 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.0 | 0.0 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.0 | 0.1 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.0 | 0.1 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.0 | 0.1 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.0 | 0.0 | GO:0004920 | interleukin-10 receptor activity(GO:0004920) |
| 0.0 | 0.1 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.0 | 0.1 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.0 | 0.0 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.0 | 0.2 | GO:0016661 | oxidoreductase activity, acting on other nitrogenous compounds as donors(GO:0016661) |
| 0.0 | 0.1 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 0.0 | 0.1 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.0 | 0.1 | GO:0060229 | lipase activator activity(GO:0060229) |
| 0.0 | 0.1 | GO:0004645 | phosphorylase activity(GO:0004645) |
| 0.0 | 0.0 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.1 | GO:0097642 | calcitonin family receptor activity(GO:0097642) |
| 0.0 | 0.3 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.0 | GO:0016778 | diphosphotransferase activity(GO:0016778) |
| 0.0 | 0.0 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.0 | 0.1 | GO:0034894 | 3-(3-hydroxyphenyl)propionate hydroxylase activity(GO:0008688) 4-chlorobenzaldehyde oxidase activity(GO:0018471) 3,5-xylenol methylhydroxylase activity(GO:0018630) phenylacetate hydroxylase activity(GO:0018631) 4-nitrophenol 4-monooxygenase activity(GO:0018632) dimethyl sulfide monooxygenase activity(GO:0018633) alpha-pinene monooxygenase [NADH] activity(GO:0018634) 1-hydroxy-2-naphthoate hydroxylase activity(GO:0018637) toluene 4-monooxygenase activity(GO:0018638) xylene monooxygenase activity(GO:0018639) dibenzothiophene monooxygenase activity(GO:0018640) 6-hydroxy-3-methyl-2-oxo-1,2-dihydroquinoline 6-monooxygenase activity(GO:0018641) chlorophenol 4-monooxygenase activity(GO:0018642) carbon disulfide oxygenase activity(GO:0018643) toluene 2-monooxygenase activity(GO:0018644) 1-hydroxy-2-oxolimonene 1,2-monooxygenase activity(GO:0018646) phenanthrene 1,2-monooxygenase activity(GO:0018647) tetrahydrofuran hydroxylase activity(GO:0018649) styrene monooxygenase activity(GO:0018650) toluene-4-sulfonate monooxygenase activity(GO:0018651) toluene-sulfonate methyl-monooxygenase activity(GO:0018652) 3-methyl-2-oxo-1,2-dihydroquinoline 6-monooxygenase activity(GO:0018653) 2-hydroxy-phenylacetate hydroxylase activity(GO:0018654) 2-oxo-delta3-4,5,5-trimethylcyclopentenylacetyl-CoA 1,2-monooxygenase activity(GO:0018655) phenanthrene 3,4-monooxygenase activity(GO:0018656) toluene 3-monooxygenase activity(GO:0018657) 4-hydroxyphenylacetate,NADH:oxygen oxidoreductase (3-hydroxylating) activity(GO:0018660) limonene monooxygenase activity(GO:0019113) 2-methylnaphthalene hydroxylase activity(GO:0034526) 1-methylnaphthalene hydroxylase activity(GO:0034534) bisphenol A hydroxylase A activity(GO:0034560) salicylate 5-hydroxylase activity(GO:0034785) isobutylamine N-hydroxylase activity(GO:0034791) branched-chain dodecylbenzene sulfonate monooxygenase activity(GO:0034802) 3-HSA hydroxylase activity(GO:0034819) 4-hydroxypyridine-3-hydroxylase activity(GO:0034894) 2-octaprenyl-3-methyl-6-methoxy-1,4-benzoquinol hydroxylase activity(GO:0043719) 6-hydroxynicotinate 3-monooxygenase activity(GO:0043731) thalianol hydroxylase activity(GO:0080014) |
| 0.0 | 0.1 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.0 | 0.2 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.0 | 0.2 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.1 | GO:0016623 | aldehyde oxidase activity(GO:0004031) oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) |
| 0.0 | 0.1 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.0 | 0.0 | GO:0016453 | acetyl-CoA C-acetyltransferase activity(GO:0003985) C-acetyltransferase activity(GO:0016453) |
| 0.0 | 0.0 | GO:0030350 | iron-responsive element binding(GO:0030350) |
| 0.0 | 0.0 | GO:0047105 | aminobutyraldehyde dehydrogenase activity(GO:0019145) 4-trimethylammoniobutyraldehyde dehydrogenase activity(GO:0047105) |
| 0.0 | 0.0 | GO:0016149 | translation release factor activity, codon specific(GO:0016149) |
| 0.0 | 0.0 | GO:0032767 | copper-dependent protein binding(GO:0032767) |
| 0.0 | 0.1 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 0.0 | 0.1 | GO:0070061 | fructose binding(GO:0070061) |
| 0.0 | 0.0 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.0 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.0 | 0.1 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 0.1 | GO:0016208 | AMP binding(GO:0016208) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.5 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.8 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.1 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 0.0 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.3 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.0 | 0.3 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 0.3 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.1 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.5 | REACTOME SHC1 EVENTS IN EGFR SIGNALING | Genes involved in SHC1 events in EGFR signaling |
| 0.0 | 0.2 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 0.2 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.3 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.0 | 0.3 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.3 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.2 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.0 | 0.1 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.2 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.2 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.0 | 0.1 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.0 | 0.1 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 0.1 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.1 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |