Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
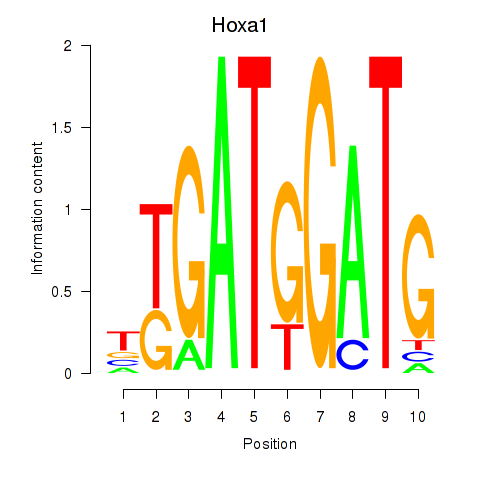
Results for Hoxa1
Z-value: 1.32
Transcription factors associated with Hoxa1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hoxa1
|
ENSMUSG00000029844.9 | homeobox A1 |
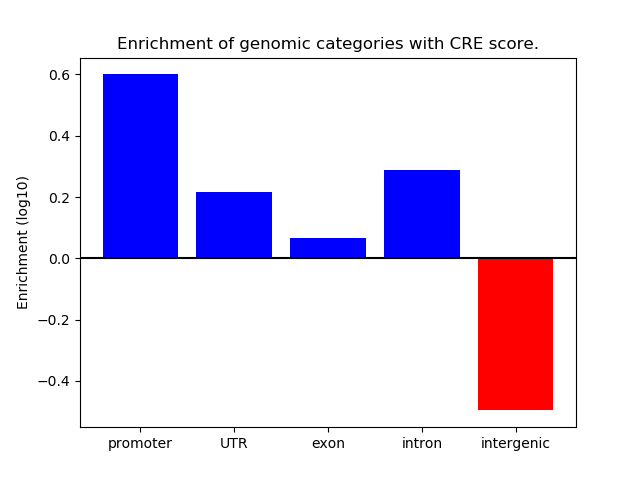
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr6_52158247_52158413 | Hoxa1 | 13 | 0.564831 | -0.02 | 9.8e-01 | Click! |
Activity of the Hoxa1 motif across conditions
Conditions sorted by the z-value of the Hoxa1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
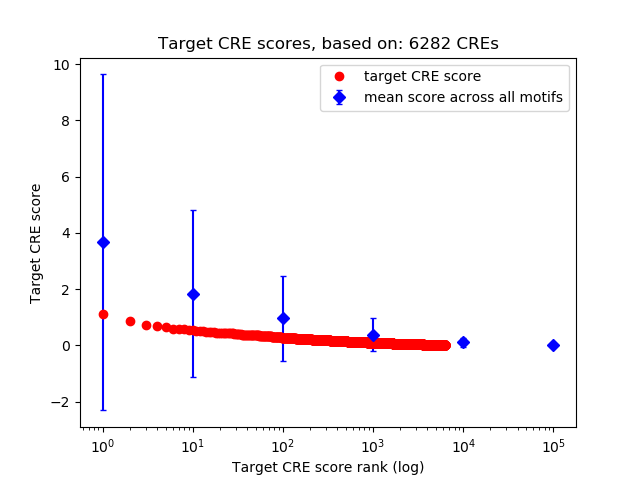
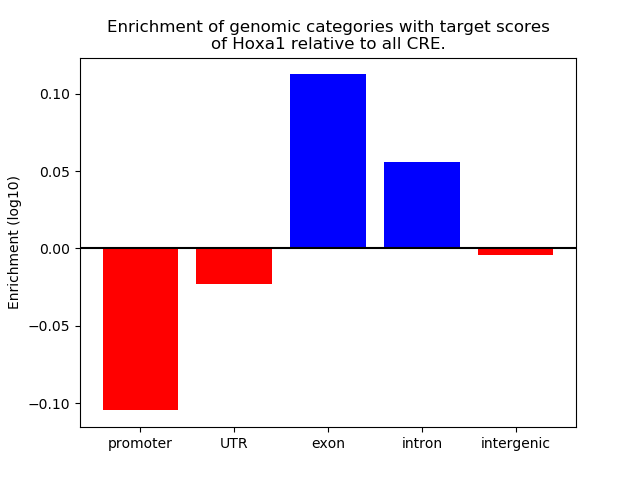
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr15_34500771_34501210 | 1.12 |
Pop1 |
processing of precursor 1, ribonuclease P/MRP family, (S. cerevisiae) |
5667 |
0.12 |
| chr11_75495112_75495263 | 0.85 |
Prpf8 |
pre-mRNA processing factor 8 |
2941 |
0.12 |
| chr6_90509396_90509549 | 0.72 |
Gm44236 |
predicted gene, 44236 |
3830 |
0.15 |
| chr11_43783562_43783743 | 0.70 |
Ttc1 |
tetratricopeptide repeat domain 1 |
35644 |
0.16 |
| chr18_54249896_54250047 | 0.65 |
Redrum |
Redrum, erythroid developmental long intergenic non-protein coding transcript |
172324 |
0.03 |
| chr7_28244671_28244868 | 0.60 |
Gm18672 |
predicted gene, 18672 |
8849 |
0.09 |
| chr7_28243862_28244153 | 0.59 |
Gm18672 |
predicted gene, 18672 |
8087 |
0.09 |
| chr13_93627456_93627944 | 0.57 |
Gm15622 |
predicted gene 15622 |
2318 |
0.25 |
| chr1_79823871_79824030 | 0.54 |
Serpine2 |
serine (or cysteine) peptidase inhibitor, clade E, member 2 |
2628 |
0.28 |
| chr19_40184875_40185171 | 0.53 |
Cyp2c70 |
cytochrome P450, family 2, subfamily c, polypeptide 70 |
2263 |
0.24 |
| chr14_65097091_65097390 | 0.53 |
Extl3 |
exostosin-like glycosyltransferase 3 |
865 |
0.63 |
| chr10_69208384_69208535 | 0.52 |
Rhobtb1 |
Rho-related BTB domain containing 1 |
93 |
0.97 |
| chr5_74907535_74907686 | 0.52 |
Gm17906 |
predicted gene, 17906 |
24275 |
0.16 |
| chr8_5106135_5106286 | 0.49 |
Slc10a2 |
solute carrier family 10, member 2 |
859 |
0.59 |
| chr6_24610042_24610446 | 0.47 |
Lmod2 |
leiomodin 2 (cardiac) |
12482 |
0.14 |
| chr10_68107778_68107929 | 0.47 |
Arid5b |
AT rich interactive domain 5B (MRF1-like) |
28773 |
0.19 |
| chr5_122455814_122455965 | 0.46 |
Anapc7 |
anaphase promoting complex subunit 7 |
14514 |
0.09 |
| chr9_48738986_48739137 | 0.45 |
Zbtb16 |
zinc finger and BTB domain containing 16 |
96884 |
0.07 |
| chr8_127515465_127515616 | 0.45 |
Gm6921 |
predicted pseudogene 6921 |
45895 |
0.17 |
| chr4_107117393_107117544 | 0.44 |
Gm12802 |
predicted gene 12802 |
9145 |
0.13 |
| chr5_66032675_66032852 | 0.44 |
Rbm47 |
RNA binding motif protein 47 |
21789 |
0.11 |
| chr14_7870473_7870795 | 0.44 |
Flnb |
filamin, beta |
14107 |
0.18 |
| chr3_52257597_52258053 | 0.44 |
Foxo1 |
forkhead box O1 |
10511 |
0.12 |
| chr3_130038452_130038603 | 0.43 |
Sec24b |
Sec24 related gene family, member B (S. cerevisiae) |
2306 |
0.27 |
| chr14_20185275_20185426 | 0.43 |
Kcnk5 |
potassium channel, subfamily K, member 5 |
3541 |
0.16 |
| chr5_92469894_92470049 | 0.43 |
Mir6415 |
microRNA 6415 |
11414 |
0.14 |
| chr10_77629279_77629435 | 0.43 |
Ube2g2 |
ubiquitin-conjugating enzyme E2G 2 |
6979 |
0.08 |
| chr8_35387028_35387898 | 0.42 |
Ppp1r3b |
protein phosphatase 1, regulatory subunit 3B |
10803 |
0.16 |
| chr7_139684055_139684211 | 0.42 |
Cfap46 |
cilia and flagella associated protein 46 |
316 |
0.88 |
| chr6_111373388_111373549 | 0.41 |
Grm7 |
glutamate receptor, metabotropic 7 |
122247 |
0.06 |
| chr10_68134623_68135230 | 0.39 |
Arid5b |
AT rich interactive domain 5B (MRF1-like) |
1700 |
0.47 |
| chr18_44661527_44661720 | 0.39 |
A930012L18Rik |
RIKEN cDNA A930012L18 gene |
42 |
0.93 |
| chr5_72948057_72948229 | 0.39 |
Slain2 |
SLAIN motif family, member 2 |
356 |
0.81 |
| chr11_59443474_59443666 | 0.39 |
Snap47 |
synaptosomal-associated protein, 47 |
3921 |
0.13 |
| chr3_65966244_65966401 | 0.38 |
Gm35106 |
predicted gene, 35106 |
2829 |
0.15 |
| chr10_69585730_69585898 | 0.38 |
Gm46231 |
predicted gene, 46231 |
19453 |
0.24 |
| chr5_112153903_112154070 | 0.38 |
1700016B01Rik |
RIKEN cDNA 1700016B01 gene |
2251 |
0.29 |
| chr18_4974876_4975041 | 0.38 |
Svil |
supervillin |
18855 |
0.27 |
| chr8_95802283_95802434 | 0.38 |
4930513N10Rik |
RIKEN cDNA 4930513N10 gene |
4386 |
0.1 |
| chr2_65212598_65212873 | 0.38 |
Cobll1 |
Cobl-like 1 |
23260 |
0.17 |
| chr1_59233451_59233602 | 0.37 |
Als2 |
alsin Rho guanine nucleotide exchange factor |
3567 |
0.21 |
| chr2_102941580_102941744 | 0.37 |
Cd44 |
CD44 antigen |
39997 |
0.15 |
| chr2_84637297_84637592 | 0.37 |
Ctnnd1 |
catenin (cadherin associated protein), delta 1 |
661 |
0.54 |
| chr12_81949344_81949529 | 0.36 |
Pcnx |
pecanex homolog |
14211 |
0.22 |
| chr3_84258598_84259173 | 0.36 |
Trim2 |
tripartite motif-containing 2 |
484 |
0.86 |
| chr10_128822735_128822886 | 0.36 |
Sarnp |
SAP domain containing ribonucleoprotein |
878 |
0.31 |
| chr2_3739821_3739975 | 0.36 |
Gm13185 |
predicted gene 13185 |
15279 |
0.17 |
| chr6_17497862_17498049 | 0.36 |
Met |
met proto-oncogene |
6714 |
0.24 |
| chr6_149146419_149146608 | 0.36 |
Etfbkmt |
electron transfer flavoprotein beta subunit lysine methyltransferase |
4867 |
0.14 |
| chr12_17471269_17471420 | 0.36 |
Gm36752 |
predicted gene, 36752 |
34726 |
0.14 |
| chr2_160868493_160868691 | 0.36 |
Zhx3 |
zinc fingers and homeoboxes 3 |
3980 |
0.16 |
| chr17_71249896_71250070 | 0.36 |
Lpin2 |
lipin 2 |
3952 |
0.19 |
| chr14_55459247_55459638 | 0.35 |
Dhrs4 |
dehydrogenase/reductase (SDR family) member 4 |
19316 |
0.09 |
| chr7_46742642_46742793 | 0.35 |
Saa1 |
serum amyloid A 1 |
225 |
0.84 |
| chr12_79673779_79674034 | 0.35 |
9430078K24Rik |
RIKEN cDNA 9430078K24 gene |
250827 |
0.02 |
| chr17_46053127_46053311 | 0.35 |
Vegfa |
vascular endothelial growth factor A |
20850 |
0.13 |
| chr7_28244175_28244405 | 0.35 |
Gm18672 |
predicted gene, 18672 |
8370 |
0.09 |
| chr3_52648763_52649088 | 0.35 |
Gm10293 |
predicted pseudogene 10293 |
36090 |
0.17 |
| chr4_95881579_95881746 | 0.35 |
Fggy |
FGGY carbohydrate kinase domain containing |
32037 |
0.19 |
| chr8_124481322_124481478 | 0.34 |
Cog2 |
component of oligomeric golgi complex 2 |
39367 |
0.13 |
| chr19_3562443_3562594 | 0.34 |
Ppp6r3 |
protein phosphatase 6, regulatory subunit 3 |
13178 |
0.15 |
| chr16_45987509_45987734 | 0.34 |
Plcxd2 |
phosphatidylinositol-specific phospholipase C, X domain containing 2 |
22597 |
0.14 |
| chr15_36578415_36578633 | 0.34 |
Gm44310 |
predicted gene, 44310 |
5020 |
0.15 |
| chr1_188125975_188126247 | 0.33 |
Gm38315 |
predicted gene, 38315 |
34596 |
0.21 |
| chr15_34501595_34501764 | 0.33 |
Pop1 |
processing of precursor 1, ribonuclease P/MRP family, (S. cerevisiae) |
6356 |
0.12 |
| chr18_65140941_65141092 | 0.33 |
Nedd4l |
neural precursor cell expressed, developmentally down-regulated gene 4-like |
2542 |
0.34 |
| chr14_70763422_70763573 | 0.33 |
Dok2 |
docking protein 2 |
2539 |
0.24 |
| chr13_36459154_36459328 | 0.33 |
Gm48763 |
predicted gene, 48763 |
39 |
0.98 |
| chr16_79059250_79059443 | 0.33 |
Tmprss15 |
transmembrane protease, serine 15 |
31746 |
0.22 |
| chr11_94370687_94371288 | 0.32 |
Abcc3 |
ATP-binding cassette, sub-family C (CFTR/MRP), member 3 |
3064 |
0.21 |
| chr5_99284011_99284176 | 0.32 |
Gm35394 |
predicted gene, 35394 |
9998 |
0.24 |
| chr8_120414524_120414709 | 0.32 |
Gm22715 |
predicted gene, 22715 |
28933 |
0.17 |
| chr6_51681649_51681815 | 0.32 |
Gm38811 |
predicted gene, 38811 |
29349 |
0.19 |
| chr15_59659799_59660177 | 0.31 |
Trib1 |
tribbles pseudokinase 1 |
11335 |
0.2 |
| chr9_66625835_66625995 | 0.31 |
Usp3 |
ubiquitin specific peptidase 3 |
32773 |
0.16 |
| chr15_3432918_3433085 | 0.31 |
Ghr |
growth hormone receptor |
38643 |
0.2 |
| chr2_147999397_147999548 | 0.31 |
9030622O22Rik |
RIKEN cDNA 9030622O22 gene |
23287 |
0.18 |
| chr10_8951296_8951475 | 0.31 |
Gm48728 |
predicted gene, 48728 |
564 |
0.78 |
| chr17_46056098_46056295 | 0.31 |
Vegfa |
vascular endothelial growth factor A |
23827 |
0.12 |
| chr4_97885703_97886090 | 0.30 |
Nfia |
nuclear factor I/A |
5777 |
0.31 |
| chr16_10648544_10649119 | 0.30 |
Clec16a |
C-type lectin domain family 16, member A |
18771 |
0.16 |
| chr16_81331237_81331468 | 0.30 |
Gm49555 |
predicted gene, 49555 |
41190 |
0.19 |
| chr15_97032998_97033149 | 0.30 |
Slc38a4 |
solute carrier family 38, member 4 |
1862 |
0.46 |
| chr19_23069533_23069684 | 0.30 |
C330002G04Rik |
RIKEN cDNA C330002G04 gene |
6245 |
0.2 |
| chr3_129331898_129332432 | 0.29 |
Enpep |
glutamyl aminopeptidase |
99 |
0.96 |
| chr13_34044303_34044743 | 0.29 |
Bphl |
biphenyl hydrolase-like (serine hydrolase, breast epithelial mucin-associated antigen) |
4682 |
0.12 |
| chr10_88201239_88201853 | 0.28 |
Washc3 |
WASH complex subunit 3 |
283 |
0.88 |
| chr17_34804558_34804741 | 0.28 |
Cyp21a1 |
cytochrome P450, family 21, subfamily a, polypeptide 1 |
88 |
0.9 |
| chr15_34501830_34501985 | 0.28 |
Pop1 |
processing of precursor 1, ribonuclease P/MRP family, (S. cerevisiae) |
6584 |
0.12 |
| chr15_59121364_59121551 | 0.28 |
Mtss1 |
MTSS I-BAR domain containing 1 |
39452 |
0.16 |
| chr9_106269146_106269548 | 0.28 |
Poc1a |
POC1 centriolar protein A |
11714 |
0.11 |
| chr14_20985530_20985681 | 0.28 |
Vcl |
vinculin |
36398 |
0.15 |
| chr13_82200174_82200343 | 0.28 |
Gm48155 |
predicted gene, 48155 |
110501 |
0.07 |
| chr6_52191906_52192068 | 0.28 |
Hoxa4 |
homeobox A4 |
234 |
0.75 |
| chr5_96304478_96304629 | 0.28 |
Fras1 |
Fraser extracellular matrix complex subunit 1 |
69402 |
0.11 |
| chr14_73776184_73776350 | 0.28 |
Gm24540 |
predicted gene, 24540 |
8299 |
0.18 |
| chr19_39675949_39676100 | 0.28 |
Cyp2c67 |
cytochrome P450, family 2, subfamily c, polypeptide 67 |
26973 |
0.21 |
| chr14_66091491_66091642 | 0.28 |
Adam2 |
a disintegrin and metallopeptidase domain 2 |
13833 |
0.15 |
| chr1_59426719_59426879 | 0.28 |
Gm37737 |
predicted gene, 37737 |
22703 |
0.15 |
| chr2_155057003_155057377 | 0.27 |
a |
nonagouti |
9575 |
0.14 |
| chr19_34525323_34525487 | 0.27 |
Lipa |
lysosomal acid lipase A |
2006 |
0.23 |
| chr6_135444931_135445089 | 0.27 |
Emp1 |
epithelial membrane protein 1 |
77517 |
0.08 |
| chr1_21245441_21245592 | 0.27 |
Gsta3 |
glutathione S-transferase, alpha 3 |
4887 |
0.13 |
| chr19_45016995_45017146 | 0.27 |
Lzts2 |
leucine zipper, putative tumor suppressor 2 |
573 |
0.58 |
| chr4_145037713_145037984 | 0.27 |
Vps13d |
vacuolar protein sorting 13D |
13186 |
0.24 |
| chr17_10253710_10253861 | 0.27 |
Qk |
quaking |
29148 |
0.22 |
| chr4_130721108_130721312 | 0.27 |
Snord85 |
small nucleolar RNA, C/D box 85 |
28424 |
0.12 |
| chr12_104087776_104088197 | 0.27 |
Serpina4-ps1 |
serine (or cysteine) peptidase inhibitor, clade A, member 4, pseudogene 1 |
7337 |
0.1 |
| chr3_51187254_51187408 | 0.27 |
Noct |
nocturnin |
37116 |
0.12 |
| chr11_28696545_28697113 | 0.27 |
2810471M01Rik |
RIKEN cDNA 2810471M01 gene |
15265 |
0.17 |
| chr2_31492850_31493284 | 0.27 |
Ass1 |
argininosuccinate synthetase 1 |
4705 |
0.21 |
| chr7_141273390_141273689 | 0.26 |
Cdhr5 |
cadherin-related family member 5 |
196 |
0.85 |
| chr11_60045283_60045434 | 0.26 |
Pemt |
phosphatidylethanolamine N-methyltransferase |
1119 |
0.47 |
| chr17_43361159_43361498 | 0.26 |
Adgrf5 |
adhesion G protein-coupled receptor F5 |
877 |
0.69 |
| chr13_36459438_36459830 | 0.26 |
Gm48763 |
predicted gene, 48763 |
354 |
0.9 |
| chr13_96981889_96982402 | 0.26 |
Gm48597 |
predicted gene, 48597 |
31231 |
0.12 |
| chr2_163301297_163301733 | 0.26 |
Tox2 |
TOX high mobility group box family member 2 |
18863 |
0.19 |
| chr13_95850293_95850468 | 0.26 |
Iqgap2 |
IQ motif containing GTPase activating protein 2 |
41377 |
0.14 |
| chr15_89315569_89316024 | 0.25 |
Sbf1 |
SET binding factor 1 |
485 |
0.62 |
| chr18_16667346_16667528 | 0.25 |
Cdh2 |
cadherin 2 |
2632 |
0.38 |
| chr6_50037597_50037914 | 0.25 |
Gm3455 |
predicted gene 3455 |
13431 |
0.26 |
| chr15_87045730_87045890 | 0.25 |
Gm23416 |
predicted gene, 23416 |
23183 |
0.18 |
| chr19_12771696_12771894 | 0.25 |
Cntf |
ciliary neurotrophic factor |
6163 |
0.12 |
| chr9_106261489_106261640 | 0.25 |
Alas1 |
aminolevulinic acid synthase 1 |
12910 |
0.1 |
| chr4_48909780_48909970 | 0.25 |
Gm12436 |
predicted gene 12436 |
11282 |
0.2 |
| chr7_120187961_120188112 | 0.25 |
Crym |
crystallin, mu |
14075 |
0.13 |
| chr4_115575098_115575249 | 0.25 |
Cyp4b1-ps2 |
cytochrome P450, family 4, subfamily b, polypeptide 1, pseudogene 2 |
7971 |
0.12 |
| chr10_53373661_53374052 | 0.25 |
Cep85l |
centrosomal protein 85-like |
2258 |
0.21 |
| chr1_40254078_40254270 | 0.25 |
Il1r1 |
interleukin 1 receptor, type I |
12412 |
0.19 |
| chr10_54058863_54059129 | 0.25 |
Gm47917 |
predicted gene, 47917 |
4815 |
0.22 |
| chr4_82453748_82453899 | 0.25 |
n-R5s188 |
nuclear encoded rRNA 5S 188 |
14413 |
0.22 |
| chr3_138285092_138285352 | 0.25 |
Adh1 |
alcohol dehydrogenase 1 (class I) |
7571 |
0.12 |
| chr8_3457973_3458135 | 0.25 |
Gm44775 |
predicted gene 44775 |
15 |
0.96 |
| chr16_57160316_57160467 | 0.25 |
Nit2 |
nitrilase family, member 2 |
250 |
0.9 |
| chr4_45492047_45492219 | 0.24 |
Shb |
src homology 2 domain-containing transforming protein B |
708 |
0.64 |
| chr9_103229092_103229718 | 0.24 |
Trf |
transferrin |
861 |
0.57 |
| chr11_121193349_121193500 | 0.24 |
Hexdc |
hexosaminidase (glycosyl hydrolase family 20, catalytic domain) containing |
11009 |
0.1 |
| chr18_4944915_4945098 | 0.24 |
Svil |
supervillin |
23280 |
0.25 |
| chr16_38433421_38433613 | 0.24 |
Pla1a |
phospholipase A1 member A |
372 |
0.8 |
| chr1_133371278_133371447 | 0.24 |
Etnk2 |
ethanolamine kinase 2 |
4075 |
0.16 |
| chr8_75090012_75090291 | 0.24 |
Hmox1 |
heme oxygenase 1 |
3470 |
0.17 |
| chr17_31446942_31447093 | 0.24 |
Pde9a |
phosphodiesterase 9A |
1350 |
0.29 |
| chr8_93257060_93257211 | 0.24 |
Ces1f |
carboxylesterase 1F |
1604 |
0.3 |
| chr3_95655803_95656050 | 0.24 |
Mcl1 |
myeloid cell leukemia sequence 1 |
2862 |
0.14 |
| chr5_118679397_118679768 | 0.24 |
Gm43052 |
predicted gene 43052 |
8036 |
0.17 |
| chr10_24229091_24229242 | 0.24 |
Moxd1 |
monooxygenase, DBH-like 1 |
5621 |
0.22 |
| chr15_34501373_34501524 | 0.24 |
Pop1 |
processing of precursor 1, ribonuclease P/MRP family, (S. cerevisiae) |
6125 |
0.12 |
| chr12_28701154_28701305 | 0.24 |
Trappc12 |
trafficking protein particle complex 12 |
380 |
0.81 |
| chr3_97903526_97903906 | 0.23 |
Sec22b |
SEC22 homolog B, vesicle trafficking protein |
2489 |
0.21 |
| chr13_34724917_34725536 | 0.23 |
Gm47151 |
predicted gene, 47151 |
4475 |
0.13 |
| chr6_100398724_100398900 | 0.23 |
Gm23234 |
predicted gene, 23234 |
96002 |
0.06 |
| chr4_49149568_49149719 | 0.23 |
Plppr1 |
phospholipid phosphatase related 1 |
90187 |
0.08 |
| chr2_134486534_134486685 | 0.23 |
Hao1 |
hydroxyacid oxidase 1, liver |
67698 |
0.14 |
| chr8_90870725_90870903 | 0.23 |
Gm45640 |
predicted gene 45640 |
5767 |
0.14 |
| chr9_57644001_57644162 | 0.23 |
Csk |
c-src tyrosine kinase |
1028 |
0.39 |
| chr8_45719244_45719451 | 0.23 |
Sorbs2 |
sorbin and SH3 domain containing 2 |
23834 |
0.18 |
| chr7_80057574_80057769 | 0.23 |
Gm21057 |
predicted gene, 21057 |
17576 |
0.1 |
| chr6_94690168_94690545 | 0.23 |
Lrig1 |
leucine-rich repeats and immunoglobulin-like domains 1 |
9726 |
0.23 |
| chr1_172082204_172082395 | 0.23 |
Copa |
coatomer protein complex subunit alpha |
230 |
0.7 |
| chr19_47317044_47317343 | 0.23 |
Sh3pxd2a |
SH3 and PX domains 2A |
2442 |
0.27 |
| chr17_28031684_28031846 | 0.23 |
Anks1 |
ankyrin repeat and SAM domain containing 1 |
9010 |
0.12 |
| chr3_144113744_144114175 | 0.23 |
Gm34078 |
predicted gene, 34078 |
21795 |
0.21 |
| chr6_86848512_86848886 | 0.23 |
2610306M01Rik |
RIKEN cDNA 2610306M01 gene |
741 |
0.4 |
| chr2_128786890_128787052 | 0.23 |
Gm23172 |
predicted gene, 23172 |
6595 |
0.15 |
| chr2_169948040_169948351 | 0.23 |
AY702102 |
cDNA sequence AY702102 |
14792 |
0.26 |
| chr6_17503345_17503773 | 0.23 |
Met |
met proto-oncogene |
12318 |
0.23 |
| chr8_36283417_36283571 | 0.23 |
Lonrf1 |
LON peptidase N-terminal domain and ring finger 1 |
33978 |
0.16 |
| chr9_35135640_35136007 | 0.23 |
Gm24262 |
predicted gene, 24262 |
8813 |
0.12 |
| chr19_37436586_37437426 | 0.23 |
Hhex |
hematopoietically expressed homeobox |
267 |
0.85 |
| chr10_81422614_81422782 | 0.23 |
Nfic |
nuclear factor I/C |
4416 |
0.07 |
| chr2_60118945_60119257 | 0.23 |
Gm13620 |
predicted gene 13620 |
5995 |
0.16 |
| chr10_115511796_115512121 | 0.23 |
Lgr5 |
leucine rich repeat containing G protein coupled receptor 5 |
48879 |
0.12 |
| chr2_18677773_18678539 | 0.23 |
Bmi1 |
Bmi1 polycomb ring finger oncogene |
349 |
0.82 |
| chr7_16463346_16463509 | 0.23 |
n-R5s151 |
nuclear encoded rRNA 5S 151 |
8386 |
0.1 |
| chr17_49455547_49455883 | 0.22 |
Mocs1 |
molybdenum cofactor synthesis 1 |
3431 |
0.28 |
| chr9_49055894_49056116 | 0.22 |
Zw10 |
zw10 kinetochore protein |
223 |
0.75 |
| chr6_15828886_15829037 | 0.22 |
Gm43990 |
predicted gene, 43990 |
7128 |
0.25 |
| chr9_107770327_107770857 | 0.22 |
Rbm5 |
RNA binding motif protein 5 |
97 |
0.94 |
| chr8_79294026_79294186 | 0.22 |
Mmaa |
methylmalonic aciduria (cobalamin deficiency) type A |
574 |
0.59 |
| chr2_23070585_23070870 | 0.22 |
Acbd5 |
acyl-Coenzyme A binding domain containing 5 |
1276 |
0.44 |
| chr3_88440726_88440965 | 0.22 |
Mir7011 |
microRNA 7011 |
106 |
0.71 |
| chr12_39373052_39373364 | 0.22 |
Gm47855 |
predicted gene, 47855 |
25348 |
0.22 |
| chr1_67185796_67186060 | 0.22 |
Cps1 |
carbamoyl-phosphate synthetase 1 |
62902 |
0.11 |
| chr5_135713964_135714115 | 0.22 |
Por |
P450 (cytochrome) oxidoreductase |
1966 |
0.2 |
| chr12_101953826_101954166 | 0.22 |
Atxn3 |
ataxin 3 |
4152 |
0.17 |
| chr10_39434414_39434685 | 0.22 |
Gm6963 |
predicted gene 6963 |
9553 |
0.2 |
| chr11_107125936_107126087 | 0.22 |
Bptf |
bromodomain PHD finger transcription factor |
5911 |
0.17 |
| chrX_49624123_49624288 | 0.22 |
Gm14719 |
predicted gene 14719 |
36755 |
0.15 |
| chr8_35383605_35384049 | 0.22 |
Ppp1r3b |
protein phosphatase 1, regulatory subunit 3B |
7167 |
0.17 |
| chr6_144895556_144895729 | 0.22 |
Gm22792 |
predicted gene, 22792 |
92010 |
0.07 |
| chr7_134742344_134742654 | 0.21 |
Dock1 |
dedicator of cytokinesis 1 |
34956 |
0.2 |
| chr11_48806124_48806302 | 0.21 |
Rack1 |
receptor for activated C kinase 1 |
765 |
0.41 |
| chr16_49862441_49862599 | 0.21 |
Cd47 |
CD47 antigen (Rh-related antigen, integrin-associated signal transducer) |
4313 |
0.32 |
| chr4_35050067_35050634 | 0.21 |
Gm12364 |
predicted gene 12364 |
88130 |
0.07 |
| chr19_37441259_37441628 | 0.21 |
Hhex |
hematopoietically expressed homeobox |
4704 |
0.13 |
| chr5_72210270_72210469 | 0.21 |
Atp10d |
ATPase, class V, type 10D |
7016 |
0.2 |
| chr6_59453276_59453536 | 0.21 |
Gprin3 |
GPRIN family member 3 |
27112 |
0.26 |
| chr7_100011175_100011513 | 0.21 |
Chrdl2 |
chordin-like 2 |
1430 |
0.34 |
| chr2_27421651_27421802 | 0.21 |
Vav2 |
vav 2 oncogene |
5186 |
0.15 |
| chr1_74230928_74231199 | 0.21 |
Arpc2 |
actin related protein 2/3 complex, subunit 2 |
5021 |
0.11 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.1 | 0.2 | GO:1904339 | negative regulation of dopaminergic neuron differentiation(GO:1904339) |
| 0.1 | 0.4 | GO:0060283 | negative regulation of oocyte development(GO:0060283) |
| 0.1 | 0.2 | GO:0030397 | membrane disassembly(GO:0030397) nuclear envelope disassembly(GO:0051081) |
| 0.1 | 0.2 | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 0.1 | 0.3 | GO:0006549 | isoleucine metabolic process(GO:0006549) |
| 0.1 | 0.2 | GO:0010046 | response to mycotoxin(GO:0010046) |
| 0.1 | 0.3 | GO:0006067 | ethanol metabolic process(GO:0006067) ethanol oxidation(GO:0006069) |
| 0.1 | 0.2 | GO:0033092 | positive regulation of immature T cell proliferation in thymus(GO:0033092) |
| 0.1 | 0.2 | GO:0060978 | angiogenesis involved in coronary vascular morphogenesis(GO:0060978) |
| 0.1 | 0.3 | GO:1900086 | positive regulation of peptidyl-tyrosine autophosphorylation(GO:1900086) |
| 0.1 | 0.2 | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.1 | 0.2 | GO:0009698 | phenylpropanoid metabolic process(GO:0009698) |
| 0.1 | 0.1 | GO:0006714 | sesquiterpenoid metabolic process(GO:0006714) |
| 0.1 | 0.2 | GO:0045297 | mating plug formation(GO:0042628) post-mating behavior(GO:0045297) |
| 0.1 | 0.2 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.2 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.0 | 0.2 | GO:0050747 | positive regulation of lipoprotein metabolic process(GO:0050747) |
| 0.0 | 0.1 | GO:0070447 | positive regulation of oligodendrocyte progenitor proliferation(GO:0070447) |
| 0.0 | 0.2 | GO:1904970 | brush border assembly(GO:1904970) |
| 0.0 | 0.2 | GO:0060431 | primary lung bud formation(GO:0060431) |
| 0.0 | 0.1 | GO:0042998 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) |
| 0.0 | 0.0 | GO:0048319 | axial mesoderm morphogenesis(GO:0048319) |
| 0.0 | 0.1 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.0 | 0.2 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.0 | 0.2 | GO:0060368 | regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060368) |
| 0.0 | 0.1 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.0 | 0.3 | GO:0016078 | tRNA catabolic process(GO:0016078) |
| 0.0 | 0.1 | GO:0048696 | regulation of collateral sprouting in absence of injury(GO:0048696) |
| 0.0 | 0.0 | GO:0021847 | ventricular zone neuroblast division(GO:0021847) |
| 0.0 | 0.1 | GO:2000097 | regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.0 | 0.2 | GO:0001996 | positive regulation of heart rate by epinephrine-norepinephrine(GO:0001996) |
| 0.0 | 0.1 | GO:1904430 | negative regulation of t-circle formation(GO:1904430) |
| 0.0 | 0.1 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.0 | 0.1 | GO:0090071 | negative regulation of ribosome biogenesis(GO:0090071) |
| 0.0 | 0.1 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.0 | 0.1 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.0 | 0.1 | GO:0003195 | tricuspid valve morphogenesis(GO:0003186) tricuspid valve formation(GO:0003195) |
| 0.0 | 0.1 | GO:0006701 | progesterone biosynthetic process(GO:0006701) |
| 0.0 | 0.1 | GO:0008228 | opsonization(GO:0008228) |
| 0.0 | 0.1 | GO:0046098 | guanine metabolic process(GO:0046098) |
| 0.0 | 0.1 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.0 | 0.1 | GO:0051025 | negative regulation of immunoglobulin secretion(GO:0051025) |
| 0.0 | 0.1 | GO:0034310 | primary alcohol catabolic process(GO:0034310) |
| 0.0 | 0.2 | GO:0060613 | fat pad development(GO:0060613) |
| 0.0 | 0.1 | GO:0031087 | nuclear-transcribed mRNA catabolic process, deadenylation-independent decay(GO:0031086) deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.0 | 0.1 | GO:0010986 | positive regulation of lipoprotein particle clearance(GO:0010986) |
| 0.0 | 0.0 | GO:0009157 | deoxyribonucleoside monophosphate biosynthetic process(GO:0009157) pyrimidine deoxyribonucleoside monophosphate biosynthetic process(GO:0009177) |
| 0.0 | 0.1 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.0 | 0.2 | GO:0042448 | progesterone metabolic process(GO:0042448) |
| 0.0 | 0.1 | GO:0045079 | negative regulation of chemokine biosynthetic process(GO:0045079) |
| 0.0 | 0.1 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.0 | 0.2 | GO:0071267 | amino acid salvage(GO:0043102) L-methionine biosynthetic process(GO:0071265) L-methionine salvage(GO:0071267) |
| 0.0 | 0.2 | GO:0009052 | pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 0.0 | 0.1 | GO:1901856 | negative regulation of cellular respiration(GO:1901856) |
| 0.0 | 0.1 | GO:1903546 | protein localization to photoreceptor outer segment(GO:1903546) |
| 0.0 | 0.1 | GO:0003431 | growth plate cartilage chondrocyte development(GO:0003431) |
| 0.0 | 0.4 | GO:0060294 | cilium movement involved in cell motility(GO:0060294) |
| 0.0 | 0.1 | GO:1900060 | negative regulation of ceramide biosynthetic process(GO:1900060) |
| 0.0 | 0.1 | GO:0033159 | negative regulation of protein import into nucleus, translocation(GO:0033159) |
| 0.0 | 0.2 | GO:0043031 | negative regulation of macrophage activation(GO:0043031) |
| 0.0 | 0.1 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.1 | GO:0021775 | smoothened signaling pathway involved in ventral spinal cord interneuron specification(GO:0021775) smoothened signaling pathway involved in spinal cord motor neuron cell fate specification(GO:0021776) |
| 0.0 | 0.2 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.1 | GO:0048320 | axial mesoderm formation(GO:0048320) |
| 0.0 | 0.1 | GO:1990036 | calcium ion import into sarcoplasmic reticulum(GO:1990036) |
| 0.0 | 0.1 | GO:0019853 | L-ascorbic acid biosynthetic process(GO:0019853) |
| 0.0 | 0.2 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.0 | 0.1 | GO:0019852 | L-ascorbic acid metabolic process(GO:0019852) |
| 0.0 | 0.1 | GO:0046084 | adenine metabolic process(GO:0046083) adenine biosynthetic process(GO:0046084) |
| 0.0 | 0.1 | GO:2000659 | regulation of interleukin-1-mediated signaling pathway(GO:2000659) |
| 0.0 | 0.1 | GO:0003383 | apical constriction(GO:0003383) |
| 0.0 | 0.1 | GO:0046618 | drug export(GO:0046618) |
| 0.0 | 0.1 | GO:0010796 | regulation of multivesicular body size(GO:0010796) |
| 0.0 | 0.1 | GO:0018364 | peptidyl-glutamine methylation(GO:0018364) |
| 0.0 | 0.0 | GO:0061668 | mitochondrial ribosome assembly(GO:0061668) |
| 0.0 | 0.1 | GO:0032898 | neurotrophin production(GO:0032898) |
| 0.0 | 0.4 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.0 | 0.1 | GO:0006528 | asparagine metabolic process(GO:0006528) |
| 0.0 | 0.2 | GO:0006207 | 'de novo' pyrimidine nucleobase biosynthetic process(GO:0006207) pyrimidine nucleobase biosynthetic process(GO:0019856) |
| 0.0 | 0.1 | GO:0090158 | endoplasmic reticulum membrane organization(GO:0090158) |
| 0.0 | 0.1 | GO:0009256 | 10-formyltetrahydrofolate metabolic process(GO:0009256) |
| 0.0 | 0.1 | GO:2001170 | negative regulation of ATP biosynthetic process(GO:2001170) |
| 0.0 | 0.1 | GO:0006553 | lysine metabolic process(GO:0006553) |
| 0.0 | 0.3 | GO:0007096 | regulation of exit from mitosis(GO:0007096) |
| 0.0 | 0.1 | GO:0019482 | beta-alanine metabolic process(GO:0019482) |
| 0.0 | 0.4 | GO:0007020 | microtubule nucleation(GO:0007020) |
| 0.0 | 0.0 | GO:0044034 | negative stranded viral RNA replication(GO:0039689) multi-organism biosynthetic process(GO:0044034) |
| 0.0 | 0.1 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.1 | GO:0045415 | negative regulation of interleukin-8 biosynthetic process(GO:0045415) |
| 0.0 | 0.1 | GO:0048597 | post-embryonic camera-type eye morphogenesis(GO:0048597) |
| 0.0 | 0.0 | GO:1904261 | regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904259) positive regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904261) basement membrane assembly involved in embryonic body morphogenesis(GO:2001197) |
| 0.0 | 0.1 | GO:0015780 | nucleotide-sugar transport(GO:0015780) pyrimidine nucleotide-sugar transport(GO:0015781) |
| 0.0 | 0.1 | GO:0070120 | ciliary neurotrophic factor-mediated signaling pathway(GO:0070120) |
| 0.0 | 0.1 | GO:1903525 | regulation of membrane tubulation(GO:1903525) |
| 0.0 | 0.1 | GO:1900623 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.0 | 0.0 | GO:0070460 | thyroid-stimulating hormone secretion(GO:0070460) |
| 0.0 | 0.0 | GO:0045658 | regulation of neutrophil differentiation(GO:0045658) |
| 0.0 | 0.1 | GO:0036265 | RNA (guanine-N7)-methylation(GO:0036265) |
| 0.0 | 0.1 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.0 | 0.1 | GO:1903223 | positive regulation of oxidative stress-induced neuron death(GO:1903223) |
| 0.0 | 0.1 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.0 | 0.1 | GO:0009957 | epidermal cell fate specification(GO:0009957) |
| 0.0 | 0.0 | GO:0010535 | positive regulation of activation of JAK2 kinase activity(GO:0010535) |
| 0.0 | 0.1 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.1 | GO:0002457 | T cell antigen processing and presentation(GO:0002457) |
| 0.0 | 0.1 | GO:1902047 | polyamine transmembrane transport(GO:1902047) regulation of polyamine transmembrane transport(GO:1902267) |
| 0.0 | 0.1 | GO:2000598 | regulation of cyclin catabolic process(GO:2000598) negative regulation of cyclin catabolic process(GO:2000599) |
| 0.0 | 0.2 | GO:0070129 | regulation of mitochondrial translation(GO:0070129) |
| 0.0 | 0.1 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.0 | 0.1 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.0 | 0.1 | GO:0032485 | regulation of Ral protein signal transduction(GO:0032485) |
| 0.0 | 0.1 | GO:0014012 | peripheral nervous system axon regeneration(GO:0014012) |
| 0.0 | 0.1 | GO:0051138 | regulation of NK T cell differentiation(GO:0051136) positive regulation of NK T cell differentiation(GO:0051138) |
| 0.0 | 0.1 | GO:0006562 | proline catabolic process(GO:0006562) |
| 0.0 | 0.1 | GO:0046218 | tryptophan catabolic process(GO:0006569) tryptophan catabolic process to kynurenine(GO:0019441) indole-containing compound catabolic process(GO:0042436) indolalkylamine catabolic process(GO:0046218) |
| 0.0 | 0.1 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.0 | 0.1 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.0 | 0.1 | GO:0019896 | axonal transport of mitochondrion(GO:0019896) |
| 0.0 | 0.1 | GO:0071679 | commissural neuron axon guidance(GO:0071679) |
| 0.0 | 0.1 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.0 | 0.1 | GO:2000574 | regulation of microtubule motor activity(GO:2000574) |
| 0.0 | 0.1 | GO:0051315 | attachment of mitotic spindle microtubules to kinetochore(GO:0051315) |
| 0.0 | 0.1 | GO:0032819 | positive regulation of natural killer cell proliferation(GO:0032819) |
| 0.0 | 0.1 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 0.1 | GO:0009448 | gamma-aminobutyric acid metabolic process(GO:0009448) |
| 0.0 | 0.0 | GO:0018894 | dibenzo-p-dioxin metabolic process(GO:0018894) |
| 0.0 | 0.0 | GO:0052422 | modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) |
| 0.0 | 0.1 | GO:0000820 | regulation of glutamine family amino acid metabolic process(GO:0000820) |
| 0.0 | 0.1 | GO:0032741 | positive regulation of interleukin-18 production(GO:0032741) |
| 0.0 | 0.1 | GO:0086046 | membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.0 | 0.0 | GO:0003419 | growth plate cartilage chondrocyte proliferation(GO:0003419) |
| 0.0 | 0.1 | GO:0009092 | homoserine metabolic process(GO:0009092) transsulfuration(GO:0019346) |
| 0.0 | 0.1 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.1 | GO:0060750 | epithelial cell proliferation involved in mammary gland duct elongation(GO:0060750) branch elongation involved in mammary gland duct branching(GO:0060751) |
| 0.0 | 0.1 | GO:1904706 | negative regulation of vascular smooth muscle cell proliferation(GO:1904706) |
| 0.0 | 0.1 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) |
| 0.0 | 0.1 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.0 | 0.0 | GO:2000686 | regulation of rubidium ion transmembrane transporter activity(GO:2000686) |
| 0.0 | 0.1 | GO:0042420 | dopamine catabolic process(GO:0042420) |
| 0.0 | 0.1 | GO:0071286 | cellular response to magnesium ion(GO:0071286) |
| 0.0 | 0.0 | GO:0060137 | maternal process involved in parturition(GO:0060137) |
| 0.0 | 0.0 | GO:0006059 | hexitol metabolic process(GO:0006059) |
| 0.0 | 0.0 | GO:0035441 | cell migration involved in vasculogenesis(GO:0035441) |
| 0.0 | 0.0 | GO:0002019 | regulation of renal output by angiotensin(GO:0002019) |
| 0.0 | 0.1 | GO:2001286 | regulation of caveolin-mediated endocytosis(GO:2001286) |
| 0.0 | 0.0 | GO:0046104 | thymidine metabolic process(GO:0046104) |
| 0.0 | 0.0 | GO:0048003 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.0 | 0.0 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.0 | 0.1 | GO:2000483 | negative regulation of interleukin-8 secretion(GO:2000483) |
| 0.0 | 0.1 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.0 | 0.0 | GO:0045869 | negative regulation of single stranded viral RNA replication via double stranded DNA intermediate(GO:0045869) |
| 0.0 | 0.1 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.0 | 0.0 | GO:0046985 | positive regulation of hemoglobin biosynthetic process(GO:0046985) |
| 0.0 | 0.2 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.0 | 0.1 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.0 | 0.2 | GO:0015012 | heparan sulfate proteoglycan biosynthetic process(GO:0015012) |
| 0.0 | 0.0 | GO:0010815 | bradykinin catabolic process(GO:0010815) |
| 0.0 | 0.0 | GO:0061535 | glutamate secretion, neurotransmission(GO:0061535) |
| 0.0 | 0.1 | GO:0090427 | activation of meiosis(GO:0090427) |
| 0.0 | 0.1 | GO:1900122 | positive regulation of receptor binding(GO:1900122) |
| 0.0 | 0.1 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.0 | 0.1 | GO:0060028 | convergent extension involved in axis elongation(GO:0060028) |
| 0.0 | 0.0 | GO:0014734 | skeletal muscle hypertrophy(GO:0014734) |
| 0.0 | 0.1 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.0 | 0.1 | GO:0030953 | astral microtubule organization(GO:0030953) |
| 0.0 | 0.0 | GO:0072497 | mesenchymal stem cell differentiation(GO:0072497) |
| 0.0 | 0.0 | GO:0010387 | COP9 signalosome assembly(GO:0010387) |
| 0.0 | 0.1 | GO:0071455 | cellular response to increased oxygen levels(GO:0036295) cellular response to hyperoxia(GO:0071455) |
| 0.0 | 0.0 | GO:0048807 | female genitalia morphogenesis(GO:0048807) |
| 0.0 | 0.1 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.0 | 0.0 | GO:0032532 | regulation of microvillus length(GO:0032532) |
| 0.0 | 0.1 | GO:2000675 | negative regulation of type B pancreatic cell apoptotic process(GO:2000675) |
| 0.0 | 0.0 | GO:0090666 | scaRNA localization to Cajal body(GO:0090666) |
| 0.0 | 0.0 | GO:2000384 | regulation of ectoderm development(GO:2000383) negative regulation of ectoderm development(GO:2000384) |
| 0.0 | 0.1 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.0 | 0.0 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.0 | 0.0 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.1 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.1 | GO:0072602 | interleukin-4 secretion(GO:0072602) |
| 0.0 | 0.0 | GO:0006393 | termination of mitochondrial transcription(GO:0006393) |
| 0.0 | 0.0 | GO:1904177 | regulation of adipose tissue development(GO:1904177) |
| 0.0 | 0.2 | GO:0000154 | rRNA modification(GO:0000154) |
| 0.0 | 0.1 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 0.1 | GO:0060510 | Type II pneumocyte differentiation(GO:0060510) |
| 0.0 | 0.0 | GO:0051547 | regulation of keratinocyte migration(GO:0051547) positive regulation of keratinocyte migration(GO:0051549) |
| 0.0 | 0.1 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.0 | 0.0 | GO:0034197 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.0 | 0.2 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.0 | 0.0 | GO:0055099 | response to high density lipoprotein particle(GO:0055099) |
| 0.0 | 0.1 | GO:0032239 | regulation of nucleobase-containing compound transport(GO:0032239) |
| 0.0 | 0.2 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.0 | 0.1 | GO:0031133 | regulation of axon diameter(GO:0031133) |
| 0.0 | 0.1 | GO:0009435 | NAD biosynthetic process(GO:0009435) |
| 0.0 | 0.0 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.0 | 0.0 | GO:0061084 | negative regulation of protein refolding(GO:0061084) |
| 0.0 | 0.1 | GO:0042447 | hormone catabolic process(GO:0042447) |
| 0.0 | 0.0 | GO:1990928 | response to amino acid starvation(GO:1990928) |
| 0.0 | 0.0 | GO:0035928 | rRNA import into mitochondrion(GO:0035928) |
| 0.0 | 0.1 | GO:0060484 | lung-associated mesenchyme development(GO:0060484) |
| 0.0 | 0.2 | GO:0030157 | pancreatic juice secretion(GO:0030157) |
| 0.0 | 0.0 | GO:0043633 | polyadenylation-dependent RNA catabolic process(GO:0043633) |
| 0.0 | 0.1 | GO:0006678 | glucosylceramide metabolic process(GO:0006678) |
| 0.0 | 0.1 | GO:0009404 | toxin metabolic process(GO:0009404) |
| 0.0 | 0.0 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.0 | 0.1 | GO:0046784 | viral mRNA export from host cell nucleus(GO:0046784) |
| 0.0 | 0.1 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.0 | 0.1 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 0.3 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.0 | 0.0 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.0 | 0.0 | GO:0070574 | cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.0 | 0.0 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.0 | 0.0 | GO:0075509 | receptor-mediated endocytosis of virus by host cell(GO:0019065) endocytosis involved in viral entry into host cell(GO:0075509) |
| 0.0 | 0.1 | GO:0045607 | regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.0 | 0.0 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.0 | 0.0 | GO:0060051 | negative regulation of protein glycosylation(GO:0060051) |
| 0.0 | 0.1 | GO:0006855 | drug transmembrane transport(GO:0006855) |
| 0.0 | 0.0 | GO:1902990 | mitotic telomere maintenance via semi-conservative replication(GO:1902990) |
| 0.0 | 0.0 | GO:0032202 | telomere assembly(GO:0032202) |
| 0.0 | 0.0 | GO:0048840 | otolith development(GO:0048840) |
| 0.0 | 0.1 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.0 | 0.0 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.0 | 0.1 | GO:0009226 | nucleotide-sugar biosynthetic process(GO:0009226) |
| 0.0 | 0.0 | GO:0000966 | RNA 5'-end processing(GO:0000966) |
| 0.0 | 0.1 | GO:0031665 | negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.0 | 0.0 | GO:0018094 | protein polyglycylation(GO:0018094) |
| 0.0 | 0.0 | GO:0071677 | positive regulation of mononuclear cell migration(GO:0071677) |
| 0.0 | 0.0 | GO:1903964 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.0 | 0.1 | GO:0070862 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.0 | 0.0 | GO:0010519 | negative regulation of phospholipase activity(GO:0010519) |
| 0.0 | 0.0 | GO:0022009 | central nervous system vasculogenesis(GO:0022009) |
| 0.0 | 0.1 | GO:0006563 | L-serine metabolic process(GO:0006563) |
| 0.0 | 0.0 | GO:2000338 | chemokine (C-X-C motif) ligand 1 production(GO:0072566) regulation of chemokine (C-X-C motif) ligand 1 production(GO:2000338) |
| 0.0 | 0.0 | GO:0035625 | receptor transactivation(GO:0035624) epidermal growth factor-activated receptor transactivation by G-protein coupled receptor signaling pathway(GO:0035625) |
| 0.0 | 0.1 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.1 | 0.3 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.1 | 0.2 | GO:0070939 | Dsl1p complex(GO:0070939) |
| 0.1 | 0.2 | GO:0042567 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) insulin-like growth factor ternary complex(GO:0042567) |
| 0.0 | 0.2 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.3 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.0 | 0.0 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.0 | 0.1 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.0 | 0.1 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.0 | 0.2 | GO:0089717 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.0 | 0.1 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.3 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.0 | 0.1 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.1 | GO:0045293 | mRNA editing complex(GO:0045293) |
| 0.0 | 0.1 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.1 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.0 | 0.2 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.0 | 0.2 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 0.1 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.1 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 0.1 | GO:0043527 | tRNA methyltransferase complex(GO:0043527) |
| 0.0 | 0.2 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.1 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.0 | 0.1 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.1 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.0 | 0.1 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.1 | GO:0070419 | nonhomologous end joining complex(GO:0070419) |
| 0.0 | 0.3 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.4 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 0.1 | GO:0036194 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.0 | 0.2 | GO:0030128 | clathrin coat of endocytic vesicle(GO:0030128) |
| 0.0 | 0.1 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.0 | 0.0 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.0 | 0.2 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.1 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.1 | GO:0001652 | granular component(GO:0001652) |
| 0.0 | 0.1 | GO:0035749 | myelin sheath adaxonal region(GO:0035749) |
| 0.0 | 0.1 | GO:0033503 | HULC complex(GO:0033503) |
| 0.0 | 0.1 | GO:0097342 | ripoptosome(GO:0097342) |
| 0.0 | 0.1 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.1 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.0 | 0.1 | GO:0000346 | transcription export complex(GO:0000346) |
| 0.0 | 0.0 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.0 | 0.0 | GO:0097629 | extrinsic component of omegasome membrane(GO:0097629) |
| 0.0 | 0.0 | GO:0097413 | Lewy body(GO:0097413) |
| 0.0 | 0.1 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.0 | 0.3 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.0 | 0.1 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 0.0 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.0 | 0.1 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 0.0 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.0 | 0.0 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.0 | 0.1 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.1 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.0 | 0.1 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.0 | 0.0 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.0 | 0.0 | GO:1990752 | microtubule end(GO:1990752) |
| 0.0 | 0.1 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.1 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.1 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.0 | 0.1 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.0 | GO:0000444 | MIS12/MIND type complex(GO:0000444) |
| 0.0 | 0.1 | GO:0000782 | telomere cap complex(GO:0000782) nuclear telomere cap complex(GO:0000783) |
| 0.0 | 0.0 | GO:0034448 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.0 | 0.1 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.0 | 0.0 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 0.0 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.1 | 0.2 | GO:0000171 | ribonuclease MRP activity(GO:0000171) |
| 0.1 | 0.3 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.1 | 0.2 | GO:0008898 | S-adenosylmethionine-homocysteine S-methyltransferase activity(GO:0008898) |
| 0.1 | 0.2 | GO:0008525 | phosphatidylcholine transporter activity(GO:0008525) |
| 0.1 | 0.2 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.1 | 0.3 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.1 | 0.2 | GO:0016743 | carboxyl- or carbamoyltransferase activity(GO:0016743) |
| 0.1 | 0.2 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 0.1 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.0 | 0.2 | GO:0016899 | oxidoreductase activity, acting on the CH-OH group of donors, oxygen as acceptor(GO:0016899) |
| 0.0 | 0.2 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.2 | GO:0016744 | transferase activity, transferring aldehyde or ketonic groups(GO:0016744) |
| 0.0 | 0.1 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.0 | 0.2 | GO:0050051 | leukotriene-B4 20-monooxygenase activity(GO:0050051) |
| 0.0 | 0.1 | GO:0003941 | L-serine ammonia-lyase activity(GO:0003941) |
| 0.0 | 0.2 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.1 | GO:0047105 | aminobutyraldehyde dehydrogenase activity(GO:0019145) 4-trimethylammoniobutyraldehyde dehydrogenase activity(GO:0047105) |
| 0.0 | 0.1 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.0 | 0.2 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.0 | 0.2 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.3 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.0 | 0.1 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.0 | 0.1 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.0 | 0.2 | GO:0047760 | butyrate-CoA ligase activity(GO:0047760) |
| 0.0 | 0.2 | GO:0034875 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 0.0 | 0.1 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.0 | 0.1 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.1 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.0 | 0.2 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.2 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.2 | GO:0016892 | endoribonuclease activity, producing 3'-phosphomonoesters(GO:0016892) |
| 0.0 | 0.1 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.0 | 0.1 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.0 | 0.3 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.0 | GO:0038181 | bile acid receptor activity(GO:0038181) |
| 0.0 | 0.1 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.0 | 0.1 | GO:0032453 | histone demethylase activity (H3-K4 specific)(GO:0032453) |
| 0.0 | 0.1 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.0 | 0.1 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.0 | 0.1 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.0 | 0.0 | GO:0051765 | inositol tetrakisphosphate kinase activity(GO:0051765) |
| 0.0 | 0.1 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.0 | 0.1 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.0 | 0.1 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.0 | 0.6 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.0 | 0.1 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.0 | 0.1 | GO:0019959 | interleukin-8 binding(GO:0019959) |
| 0.0 | 0.1 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.0 | 0.2 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.1 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 0.0 | 0.1 | GO:0043426 | MRF binding(GO:0043426) |
| 0.0 | 0.1 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.0 | 0.1 | GO:0097108 | hedgehog family protein binding(GO:0097108) |
| 0.0 | 0.0 | GO:0000701 | purine-specific mismatch base pair DNA N-glycosylase activity(GO:0000701) |
| 0.0 | 0.1 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.0 | 0.1 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 0.1 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.0 | 0.2 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.1 | GO:0035877 | death effector domain binding(GO:0035877) |
| 0.0 | 0.2 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.0 | 0.1 | GO:0003846 | 2-acylglycerol O-acyltransferase activity(GO:0003846) |
| 0.0 | 0.1 | GO:0016679 | oxidoreductase activity, acting on diphenols and related substances as donors(GO:0016679) |
| 0.0 | 0.3 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 0.1 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.0 | 0.1 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 0.1 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.1 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.0 | 0.1 | GO:0002054 | nucleobase binding(GO:0002054) |
| 0.0 | 0.1 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.1 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.0 | 0.1 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.0 | 0.1 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.0 | 0.0 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 0.0 | 0.1 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.0 | 0.2 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.1 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.0 | 0.0 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 0.0 | 0.0 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 0.0 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.0 | 0.0 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.0 | 0.0 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.0 | 0.1 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.0 | 0.2 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.0 | 0.1 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.0 | 0.0 | GO:0030620 | U2 snRNA binding(GO:0030620) |
| 0.0 | 0.0 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.0 | 0.0 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) |
| 0.0 | 0.1 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 0.0 | GO:0046538 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.0 | 0.0 | GO:0004611 | phosphoenolpyruvate carboxykinase activity(GO:0004611) |
| 0.0 | 0.1 | GO:0070878 | primary miRNA binding(GO:0070878) |
| 0.0 | 0.1 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.0 | 0.1 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.1 | GO:0003847 | 1-alkyl-2-acetylglycerophosphocholine esterase activity(GO:0003847) |
| 0.0 | 0.0 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.0 | 0.1 | GO:0003958 | NADPH-hemoprotein reductase activity(GO:0003958) |
| 0.0 | 0.1 | GO:0004792 | thiosulfate sulfurtransferase activity(GO:0004792) |
| 0.0 | 0.1 | GO:0005338 | nucleotide-sugar transmembrane transporter activity(GO:0005338) |
| 0.0 | 0.1 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.0 | GO:0016662 | oxidoreductase activity, acting on other nitrogenous compounds as donors, cytochrome as acceptor(GO:0016662) |
| 0.0 | 0.0 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.1 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.0 | GO:0034040 | lipid-transporting ATPase activity(GO:0034040) |
| 0.0 | 0.1 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.0 | GO:0070905 | serine binding(GO:0070905) |
| 0.0 | 0.0 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.0 | 0.1 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.1 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.0 | 0.2 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.0 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.0 | 0.0 | GO:0000253 | 3-keto sterol reductase activity(GO:0000253) |
| 0.0 | 0.0 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 0.0 | 0.0 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.0 | 0.0 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.0 | GO:0015375 | glycine:sodium symporter activity(GO:0015375) |
| 0.0 | 0.0 | GO:0017089 | glycolipid transporter activity(GO:0017089) |
| 0.0 | 0.2 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.1 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.0 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.0 | 0.0 | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0003829) |
| 0.0 | 0.0 | GO:0043398 | HLH domain binding(GO:0043398) |
| 0.0 | 0.2 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.0 | GO:0032896 | palmitoyl-CoA 9-desaturase activity(GO:0032896) |
| 0.0 | 0.0 | GO:0036435 | K48-linked polyubiquitin binding(GO:0036435) |
| 0.0 | 0.1 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 0.0 | 0.1 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.5 | GO:0002161 | aminoacyl-tRNA editing activity(GO:0002161) |
| 0.0 | 0.1 | GO:0008656 | cysteine-type endopeptidase activator activity involved in apoptotic process(GO:0008656) |
| 0.0 | 0.3 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 0.1 | GO:0016653 | oxidoreductase activity, acting on NAD(P)H, heme protein as acceptor(GO:0016653) |
| 0.0 | 0.1 | GO:0050072 | m7G(5')pppN diphosphatase activity(GO:0050072) |
| 0.0 | 0.1 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.0 | 0.0 | GO:0004936 | alpha-adrenergic receptor activity(GO:0004936) alpha2-adrenergic receptor activity(GO:0004938) |
| 0.0 | 0.0 | GO:0070548 | L-glutamine aminotransferase activity(GO:0070548) |
| 0.0 | 0.0 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.0 | 0.1 | GO:0043733 | alkylbase DNA N-glycosylase activity(GO:0003905) DNA-3-methylbase glycosylase activity(GO:0043733) |
| 0.0 | 0.0 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.1 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 0.3 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.1 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.0 | 0.0 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 0.5 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.1 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 0.1 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 0.4 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.1 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.0 | 0.1 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 0.5 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.2 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.0 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 0.4 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.0 | 0.2 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.2 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.3 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.4 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.0 | 0.1 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.0 | 0.2 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.1 | REACTOME NGF SIGNALLING VIA TRKA FROM THE PLASMA MEMBRANE | Genes involved in NGF signalling via TRKA from the plasma membrane |
| 0.0 | 0.2 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.2 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.1 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.0 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 0.0 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.0 | 0.1 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.0 | 0.1 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.2 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.1 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.1 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.2 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 0.2 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.0 | REACTOME CD28 CO STIMULATION | Genes involved in CD28 co-stimulation |
| 0.0 | 0.1 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.1 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.0 | 0.2 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.2 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.2 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.1 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.0 | REACTOME INTERACTIONS OF VPR WITH HOST CELLULAR PROTEINS | Genes involved in Interactions of Vpr with host cellular proteins |
| 0.0 | 0.1 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |