Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
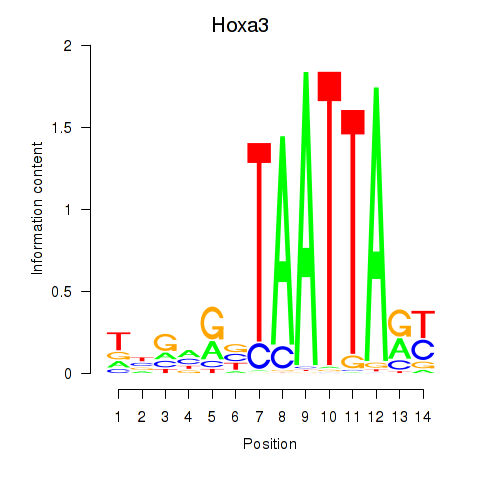
Results for Hoxa3
Z-value: 0.59
Transcription factors associated with Hoxa3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hoxa3
|
ENSMUSG00000079560.7 | homeobox A3 |
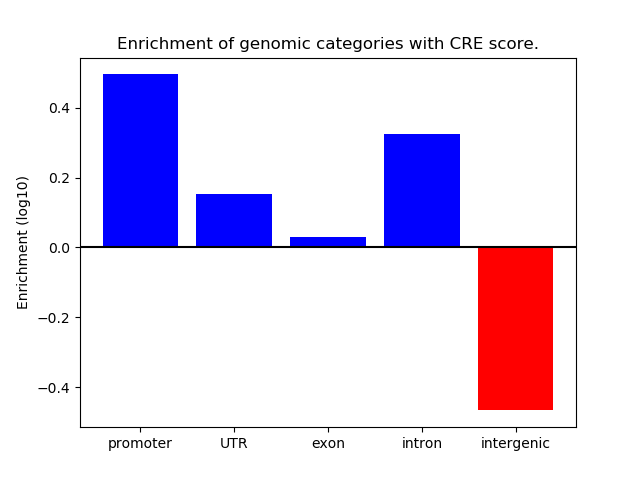
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr6_52211661_52211823 | Hoxa3 | 1373 | 0.132305 | -0.65 | 1.6e-01 | Click! |
Activity of the Hoxa3 motif across conditions
Conditions sorted by the z-value of the Hoxa3 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
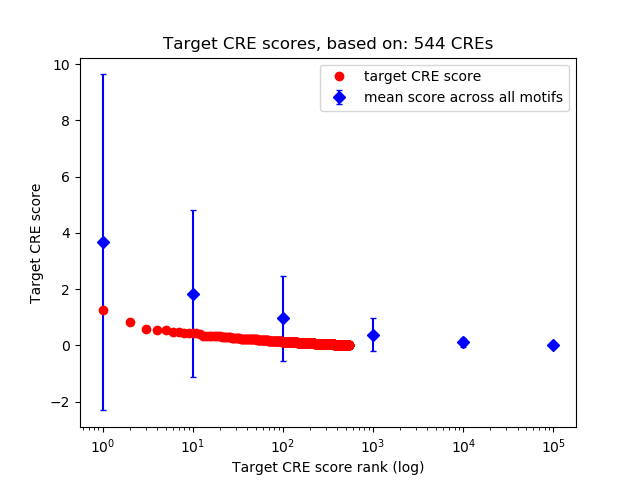
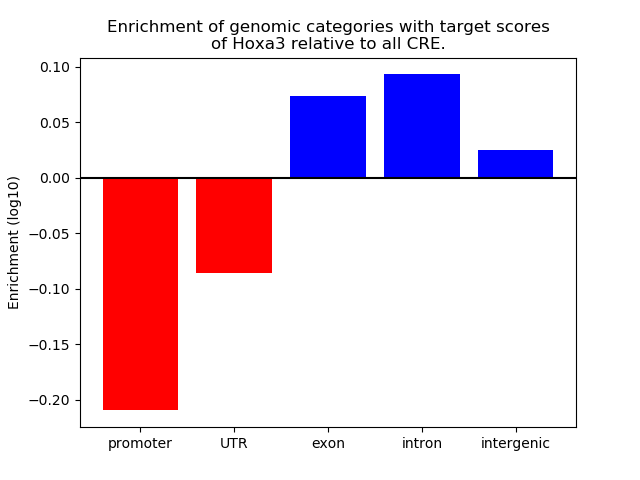
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr5_125526040_125526191 | 1.27 |
Tmem132b |
transmembrane protein 132B |
5659 |
0.17 |
| chr7_140722780_140722931 | 0.81 |
Olfr542-ps1 |
olfactory receptor 542, pseudogene 1 |
1573 |
0.22 |
| chr13_44437957_44438156 | 0.57 |
1700029N11Rik |
RIKEN cDNA 1700029N11 gene |
1656 |
0.27 |
| chr19_44401346_44401497 | 0.54 |
Scd1 |
stearoyl-Coenzyme A desaturase 1 |
5269 |
0.16 |
| chr2_38823684_38823849 | 0.53 |
Nr6a1os |
nuclear receptor subfamily 6, group A, member 1, opposite strand |
425 |
0.74 |
| chr16_42947764_42948084 | 0.49 |
Ndufs5-ps |
NADH:ubiquinone oxidoreductase core subunit S5, pseudogene |
7707 |
0.21 |
| chr2_32606655_32606830 | 0.47 |
St6galnac6 |
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 6 |
219 |
0.84 |
| chr11_16854059_16854210 | 0.44 |
Egfros |
epidermal growth factor receptor, opposite strand |
23432 |
0.17 |
| chr2_18048778_18049088 | 0.43 |
Skida1 |
SKI/DACH domain containing 1 |
98 |
0.94 |
| chr9_74791015_74791321 | 0.43 |
Gm22315 |
predicted gene, 22315 |
9098 |
0.19 |
| chr6_52610697_52610915 | 0.42 |
Gm44434 |
predicted gene, 44434 |
7561 |
0.15 |
| chr10_4605236_4605428 | 0.41 |
Esr1 |
estrogen receptor 1 (alpha) |
6261 |
0.23 |
| chr3_18250842_18251072 | 0.35 |
Cyp7b1 |
cytochrome P450, family 7, subfamily b, polypeptide 1 |
7619 |
0.24 |
| chr14_21435070_21435221 | 0.34 |
Gm25864 |
predicted gene, 25864 |
15329 |
0.19 |
| chr4_101198122_101198415 | 0.34 |
Gm24468 |
predicted gene, 24468 |
11504 |
0.14 |
| chr19_40153747_40153898 | 0.33 |
Cyp2c70 |
cytochrome P450, family 2, subfamily c, polypeptide 70 |
33464 |
0.13 |
| chr17_81386211_81386362 | 0.33 |
Gm50044 |
predicted gene, 50044 |
15453 |
0.24 |
| chr16_87627804_87628113 | 0.33 |
Gm22808 |
predicted gene, 22808 |
6838 |
0.21 |
| chr5_102009929_102010121 | 0.32 |
Wdfy3 |
WD repeat and FYVE domain containing 3 |
28467 |
0.16 |
| chr17_89646332_89646522 | 0.32 |
Gm50061 |
predicted gene, 50061 |
106099 |
0.08 |
| chr10_87932167_87932318 | 0.31 |
Tyms-ps |
thymidylate synthase, pseudogene |
34605 |
0.14 |
| chr11_121349825_121350205 | 0.30 |
Wdr45b |
WD repeat domain 45B |
4326 |
0.16 |
| chr11_35750589_35750898 | 0.30 |
Pank3 |
pantothenate kinase 3 |
18741 |
0.16 |
| chr15_59316490_59316736 | 0.30 |
Sqle |
squalene epoxidase |
1506 |
0.35 |
| chr6_58611375_58611579 | 0.30 |
Abcg2 |
ATP binding cassette subfamily G member 2 (Junior blood group) |
14806 |
0.19 |
| chr3_18163689_18163955 | 0.29 |
Gm23686 |
predicted gene, 23686 |
13803 |
0.23 |
| chr13_4270161_4270342 | 0.28 |
Akr1c12 |
aldo-keto reductase family 1, member C12 |
9182 |
0.15 |
| chr7_139119516_139119677 | 0.27 |
Dpysl4 |
dihydropyrimidinase-like 4 |
30349 |
0.14 |
| chr4_109104769_109104920 | 0.27 |
Osbpl9 |
oxysterol binding protein-like 9 |
3187 |
0.27 |
| chr15_9076174_9076325 | 0.27 |
Nadk2 |
NAD kinase 2, mitochondrial |
972 |
0.61 |
| chr1_100298746_100298897 | 0.25 |
Gm29667 |
predicted gene 29667 |
16030 |
0.18 |
| chr14_32720173_32720370 | 0.25 |
Gm28651 |
predicted gene 28651 |
33713 |
0.14 |
| chr2_107628721_107628872 | 0.24 |
Gm9864 |
predicted gene 9864 |
28257 |
0.26 |
| chr3_79563335_79563486 | 0.24 |
Fnip2 |
folliculin interacting protein 2 |
4269 |
0.14 |
| chr4_116689870_116690150 | 0.23 |
Prdx1 |
peroxiredoxin 1 |
2698 |
0.15 |
| chr17_46054574_46054727 | 0.23 |
Vegfa |
vascular endothelial growth factor A |
22281 |
0.12 |
| chr10_89563955_89564134 | 0.23 |
Gm48087 |
predicted gene, 48087 |
8325 |
0.2 |
| chrX_72953520_72953708 | 0.22 |
Nsdhl |
NAD(P) dependent steroid dehydrogenase-like |
23862 |
0.1 |
| chr2_58787332_58787483 | 0.22 |
Upp2 |
uridine phosphorylase 2 |
22082 |
0.19 |
| chr2_67977965_67978321 | 0.22 |
Gm37964 |
predicted gene, 37964 |
79547 |
0.1 |
| chr14_7834896_7835063 | 0.22 |
Flnb |
filamin, beta |
17022 |
0.15 |
| chr7_135720125_135720308 | 0.22 |
Mki67 |
antigen identified by monoclonal antibody Ki 67 |
3855 |
0.2 |
| chr2_31513022_31513372 | 0.22 |
Ass1 |
argininosuccinate synthetase 1 |
5293 |
0.19 |
| chr5_62761138_62761503 | 0.22 |
Arap2 |
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
4800 |
0.29 |
| chr19_37507552_37507703 | 0.21 |
Exoc6 |
exocyst complex component 6 |
13252 |
0.16 |
| chr2_24040580_24040761 | 0.21 |
Hnmt |
histamine N-methyltransferase |
8196 |
0.23 |
| chr2_18063532_18063683 | 0.21 |
Mllt10 |
myeloid/lymphoid or mixed-lineage leukemia; translocated to, 10 |
787 |
0.48 |
| chr16_65665507_65665824 | 0.21 |
Gm49633 |
predicted gene, 49633 |
52246 |
0.14 |
| chr4_147433621_147433808 | 0.21 |
Gm13161 |
predicted gene 13161 |
9757 |
0.14 |
| chr11_16883025_16883203 | 0.21 |
Egfr |
epidermal growth factor receptor |
4964 |
0.23 |
| chr3_69280240_69280391 | 0.21 |
Ppm1l |
protein phosphatase 1 (formerly 2C)-like |
36546 |
0.13 |
| chr4_136470355_136470864 | 0.20 |
Luzp1 |
leucine zipper protein 1 |
759 |
0.6 |
| chr2_5939653_5939804 | 0.20 |
Dhtkd1 |
dehydrogenase E1 and transketolase domain containing 1 |
3015 |
0.22 |
| chr15_54933581_54933745 | 0.20 |
Enpp2 |
ectonucleotide pyrophosphatase/phosphodiesterase 2 |
11050 |
0.17 |
| chr6_27967716_27967883 | 0.20 |
Mir592 |
microRNA 592 |
31049 |
0.23 |
| chr17_43275589_43275758 | 0.20 |
Adgrf1 |
adhesion G protein-coupled receptor F1 |
5344 |
0.29 |
| chr12_83905337_83905488 | 0.19 |
Numb |
NUMB endocytic adaptor protein |
16321 |
0.11 |
| chr12_4220651_4220920 | 0.19 |
Gm48210 |
predicted gene, 48210 |
1317 |
0.27 |
| chr14_66125086_66125237 | 0.19 |
Ephx2 |
epoxide hydrolase 2, cytoplasmic |
661 |
0.67 |
| chr4_124809297_124809476 | 0.18 |
Mtf1 |
metal response element binding transcription factor 1 |
6708 |
0.1 |
| chr12_104341895_104342090 | 0.18 |
Serpina3k |
serine (or cysteine) peptidase inhibitor, clade A, member 3K |
3506 |
0.14 |
| chr5_36135969_36136149 | 0.18 |
Psapl1 |
prosaposin-like 1 |
67962 |
0.11 |
| chr11_30792834_30792985 | 0.18 |
Psme4 |
proteasome (prosome, macropain) activator subunit 4 |
18628 |
0.14 |
| chr7_115858420_115858571 | 0.17 |
Sox6 |
SRY (sex determining region Y)-box 6 |
1357 |
0.57 |
| chr18_9833982_9834146 | 0.17 |
Gm23637 |
predicted gene, 23637 |
6945 |
0.15 |
| chr18_10707142_10707343 | 0.17 |
4930563E18Rik |
RIKEN cDNA 4930563E18 gene |
382 |
0.52 |
| chr4_105100779_105100973 | 0.17 |
Prkaa2 |
protein kinase, AMP-activated, alpha 2 catalytic subunit |
9014 |
0.27 |
| chr8_44955232_44955417 | 0.17 |
Fat1 |
FAT atypical cadherin 1 |
5110 |
0.23 |
| chr17_33667328_33667604 | 0.17 |
Hnrnpm |
heterogeneous nuclear ribonucleoprotein M |
896 |
0.43 |
| chr11_118508000_118508168 | 0.17 |
Rbfox3 |
RNA binding protein, fox-1 homolog (C. elegans) 3 |
731 |
0.62 |
| chr18_51150358_51150691 | 0.17 |
Prr16 |
proline rich 16 |
32786 |
0.23 |
| chr1_88055296_88055515 | 0.17 |
Ugt1a10 |
UDP glycosyltransferase 1 family, polypeptide A10 |
6 |
0.94 |
| chr2_103461502_103462486 | 0.17 |
Elf5 |
E74-like factor 5 |
13256 |
0.17 |
| chr1_13322152_13322303 | 0.16 |
Gm23169 |
predicted gene, 23169 |
12382 |
0.11 |
| chr7_101353319_101353516 | 0.16 |
Gm45548 |
predicted gene 45548 |
1022 |
0.35 |
| chr14_65375161_65375312 | 0.16 |
Zfp395 |
zinc finger protein 395 |
157 |
0.95 |
| chr17_64607771_64607970 | 0.16 |
Man2a1 |
mannosidase 2, alpha 1 |
7134 |
0.26 |
| chr15_31250860_31251157 | 0.16 |
Dap |
death-associated protein |
17328 |
0.15 |
| chr4_44777019_44777170 | 0.16 |
Zcchc7 |
zinc finger, CCHC domain containing 7 |
16650 |
0.16 |
| chr11_120523107_120523310 | 0.16 |
Gm11789 |
predicted gene 11789 |
7346 |
0.07 |
| chr14_7852949_7853102 | 0.16 |
Flnb |
filamin, beta |
31716 |
0.14 |
| chr12_28636385_28636550 | 0.15 |
Rps7 |
ribosomal protein S7 |
514 |
0.71 |
| chr17_49304784_49304935 | 0.15 |
Gm17830 |
predicted gene, 17830 |
7382 |
0.19 |
| chr5_108694583_108695258 | 0.15 |
Fgfrl1 |
fibroblast growth factor receptor-like 1 |
418 |
0.74 |
| chr7_118715006_118715199 | 0.15 |
Ccp110 |
centriolar coiled coil protein 110 |
272 |
0.86 |
| chr9_60982764_60983106 | 0.15 |
Gm5122 |
predicted gene 5122 |
30546 |
0.14 |
| chr14_17864277_17864546 | 0.15 |
Gm48320 |
predicted gene, 48320 |
93289 |
0.08 |
| chr14_30530847_30531018 | 0.14 |
Dcp1a |
decapping mRNA 1A |
7225 |
0.15 |
| chr10_89397980_89398148 | 0.14 |
Gas2l3 |
growth arrest-specific 2 like 3 |
35778 |
0.18 |
| chr9_119368235_119368400 | 0.14 |
Slc22a14 |
solute carrier family 22 (organic cation transporter), member 14 |
2764 |
0.16 |
| chr9_64175252_64175472 | 0.14 |
Snord16a |
small nucleolar RNA, C/D box 16A |
61 |
0.7 |
| chr9_112785054_112785229 | 0.14 |
Gm24957 |
predicted gene, 24957 |
262612 |
0.02 |
| chr1_16746119_16746270 | 0.14 |
Gm29649 |
predicted gene 29649 |
12429 |
0.16 |
| chr16_38246128_38246487 | 0.14 |
Gsk3b |
glycogen synthase kinase 3 beta |
38269 |
0.11 |
| chr12_52448577_52448767 | 0.14 |
Gm47431 |
predicted gene, 47431 |
547 |
0.79 |
| chr9_62023104_62023499 | 0.14 |
Paqr5 |
progestin and adipoQ receptor family member V |
3500 |
0.27 |
| chr7_49764294_49764503 | 0.13 |
Htatip2 |
HIV-1 Tat interactive protein 2 |
5245 |
0.23 |
| chr19_53904567_53904727 | 0.13 |
Pdcd4 |
programmed cell death 4 |
1231 |
0.39 |
| chr1_67197806_67198015 | 0.13 |
Gm15668 |
predicted gene 15668 |
51290 |
0.13 |
| chr9_46479855_46480133 | 0.13 |
Gm47144 |
predicted gene, 47144 |
25079 |
0.15 |
| chr3_119165577_119165728 | 0.13 |
Gm43410 |
predicted gene 43410 |
297408 |
0.01 |
| chr13_91208892_91209073 | 0.13 |
Gm17450 |
predicted gene, 17450 |
11470 |
0.19 |
| chr2_31505484_31505674 | 0.13 |
Ass1 |
argininosuccinate synthetase 1 |
4853 |
0.2 |
| chr5_77131739_77131917 | 0.13 |
Chaer1 |
cardiac hypertrophy associated epigenetic regulator 1 |
8424 |
0.13 |
| chr13_41197676_41197955 | 0.13 |
Gm48510 |
predicted gene, 48510 |
20927 |
0.11 |
| chr10_51722569_51722927 | 0.13 |
Rfx6 |
regulatory factor X, 6 |
28977 |
0.12 |
| chr14_119831250_119831463 | 0.13 |
4930404K13Rik |
RIKEN cDNA 4930404K13 gene |
32378 |
0.19 |
| chr5_64933539_64933713 | 0.13 |
Tlr1 |
toll-like receptor 1 |
63 |
0.96 |
| chr1_21252081_21252232 | 0.13 |
Gsta3 |
glutathione S-transferase, alpha 3 |
1365 |
0.29 |
| chr4_87899011_87899212 | 0.13 |
Mllt3 |
myeloid/lymphoid or mixed-lineage leukemia; translocated to, 3 |
24569 |
0.24 |
| chr2_31489958_31490302 | 0.12 |
Ass1 |
argininosuccinate synthetase 1 |
7642 |
0.19 |
| chr18_60606060_60606525 | 0.12 |
Synpo |
synaptopodin |
3813 |
0.19 |
| chr2_160865219_160865594 | 0.12 |
Zhx3 |
zinc fingers and homeoboxes 3 |
5775 |
0.14 |
| chr12_73341772_73342027 | 0.12 |
Slc38a6 |
solute carrier family 38, member 6 |
2468 |
0.25 |
| chr2_157131528_157131679 | 0.12 |
Samhd1 |
SAM domain and HD domain, 1 |
1546 |
0.33 |
| chr11_120819707_120820136 | 0.12 |
Fasn |
fatty acid synthase |
3859 |
0.11 |
| chr9_55221060_55221220 | 0.12 |
Fbxo22 |
F-box protein 22 |
7477 |
0.17 |
| chr4_40729981_40730352 | 0.12 |
Dnaja1 |
DnaJ heat shock protein family (Hsp40) member A1 |
6117 |
0.12 |
| chr12_87387039_87387194 | 0.12 |
Sptlc2 |
serine palmitoyltransferase, long chain base subunit 2 |
1125 |
0.29 |
| chr11_22844106_22844418 | 0.12 |
Gm23772 |
predicted gene, 23772 |
2606 |
0.17 |
| chr5_122876895_122877046 | 0.12 |
Kdm2b |
lysine (K)-specific demethylase 2B |
4115 |
0.16 |
| chr2_28618402_28618756 | 0.12 |
Gfi1b |
growth factor independent 1B |
3174 |
0.15 |
| chr1_63136268_63136443 | 0.12 |
Ndufs1 |
NADH:ubiquinone oxidoreductase core subunit S1 |
8606 |
0.09 |
| chr3_51277891_51278084 | 0.12 |
Elf2 |
E74-like factor 2 |
412 |
0.7 |
| chr17_32935365_32935709 | 0.11 |
Cyp4f13 |
cytochrome P450, family 4, subfamily f, polypeptide 13 |
4529 |
0.11 |
| chr6_67441579_67441755 | 0.11 |
Gm44083 |
predicted gene, 44083 |
23616 |
0.11 |
| chr9_119401641_119401816 | 0.11 |
Acvr2b |
activin receptor IIB |
390 |
0.77 |
| chrX_77601507_77601673 | 0.11 |
Gm23121 |
predicted gene, 23121 |
22103 |
0.2 |
| chr7_35802047_35802654 | 0.11 |
E130304I02Rik |
RIKEN cDNA E130304I02 gene |
242 |
0.77 |
| chr2_150913728_150914090 | 0.11 |
Gins1 |
GINS complex subunit 1 (Psf1 homolog) |
1090 |
0.37 |
| chr9_15301981_15302132 | 0.11 |
4931406C07Rik |
RIKEN cDNA 4931406C07 gene |
500 |
0.46 |
| chr1_145995029_145995180 | 0.11 |
Gm5263 |
predicted gene 5263 |
425332 |
0.01 |
| chr4_6239539_6239806 | 0.11 |
Gm11798 |
predicted gene 11798 |
21293 |
0.18 |
| chr8_62185245_62185446 | 0.11 |
Gm2961 |
predicted gene 2961 |
8867 |
0.27 |
| chr2_165869197_165869348 | 0.11 |
Zmynd8 |
zinc finger, MYND-type containing 8 |
6505 |
0.15 |
| chr19_31886956_31887132 | 0.11 |
A1cf |
APOBEC1 complementation factor |
18263 |
0.2 |
| chr11_32651005_32651314 | 0.11 |
Fbxw11 |
F-box and WD-40 domain protein 11 |
8255 |
0.23 |
| chr9_36726106_36726470 | 0.11 |
Chek1 |
checkpoint kinase 1 |
86 |
0.95 |
| chr4_134843941_134844233 | 0.10 |
Maco1 |
macoilin 1 |
9062 |
0.18 |
| chr19_29271349_29271500 | 0.10 |
Jak2 |
Janus kinase 2 |
18983 |
0.16 |
| chr11_103132517_103132723 | 0.10 |
Hexim2 |
hexamethylene bis-acetamide inducible 2 |
191 |
0.91 |
| chr3_138223407_138223558 | 0.10 |
Adh7 |
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
2011 |
0.23 |
| chr4_10459968_10460323 | 0.10 |
Gm11814 |
predicted gene 11814 |
20402 |
0.22 |
| chr12_73861884_73862127 | 0.10 |
Gm15283 |
predicted gene 15283 |
7607 |
0.18 |
| chr8_75450708_75450868 | 0.10 |
Iqcm |
IQ motif containing M |
1805 |
0.34 |
| chr8_22623689_22623868 | 0.10 |
Dkk4 |
dickkopf WNT signaling pathway inhibitor 4 |
265 |
0.89 |
| chr8_3276470_3276698 | 0.10 |
Insr |
insulin receptor |
2967 |
0.26 |
| chr6_119404440_119404707 | 0.10 |
Adipor2 |
adiponectin receptor 2 |
12902 |
0.2 |
| chr12_112590755_112591153 | 0.10 |
Inf2 |
inverted formin, FH2 and WH2 domain containing |
1688 |
0.29 |
| chr10_82740996_82741147 | 0.10 |
Nfyb |
nuclear transcription factor-Y beta |
10588 |
0.13 |
| chr12_33285610_33285770 | 0.10 |
Atxn7l1 |
ataxin 7-like 1 |
16825 |
0.2 |
| chr4_148590620_148590972 | 0.10 |
Srm |
spermidine synthase |
707 |
0.49 |
| chr1_13368915_13369352 | 0.10 |
Ncoa2 |
nuclear receptor coactivator 2 |
3296 |
0.16 |
| chr13_31550371_31550524 | 0.10 |
Foxq1 |
forkhead box Q1 |
5687 |
0.15 |
| chr9_89882776_89882950 | 0.09 |
Rasgrf1 |
RAS protein-specific guanine nucleotide-releasing factor 1 |
27045 |
0.16 |
| chr15_53238274_53238425 | 0.09 |
Ext1 |
exostosin glycosyltransferase 1 |
42094 |
0.21 |
| chr2_64093669_64093820 | 0.09 |
Fign |
fidgetin |
4244 |
0.37 |
| chr1_151718954_151719119 | 0.09 |
2810414N06Rik |
RIKEN cDNA 2810414N06 gene |
35982 |
0.14 |
| chr16_52374896_52375129 | 0.09 |
Alcam |
activated leukocyte cell adhesion molecule |
77453 |
0.12 |
| chr6_108135683_108135868 | 0.09 |
Rpl36-ps12 |
ribosomal protein L36, pseudogene 12 |
4734 |
0.23 |
| chr19_21654390_21654886 | 0.09 |
Abhd17b |
abhydrolase domain containing 17B |
1113 |
0.45 |
| chr10_10448945_10449124 | 0.09 |
Gm48326 |
predicted gene, 48326 |
16500 |
0.19 |
| chr17_43113705_43113864 | 0.09 |
E130008D07Rik |
RIKEN cDNA E130008D07 gene |
44412 |
0.18 |
| chr10_87872347_87872727 | 0.09 |
Igf1os |
insulin-like growth factor 1, opposite strand |
9156 |
0.2 |
| chr4_123990393_123990580 | 0.09 |
Gm12902 |
predicted gene 12902 |
64252 |
0.08 |
| chr6_122148793_122148971 | 0.09 |
Gm10319 |
predicted pseudogene 10319 |
12150 |
0.17 |
| chr1_136007677_136007912 | 0.09 |
Tmem9 |
transmembrane protein 9 |
356 |
0.78 |
| chr2_34049474_34049625 | 0.09 |
C230014O12Rik |
RIKEN cDNA C230014O12 gene |
58180 |
0.11 |
| chr8_40418187_40418345 | 0.09 |
Zdhhc2 |
zinc finger, DHHC domain containing 2 |
5549 |
0.24 |
| chr12_85291673_85291832 | 0.09 |
Zc2hc1c |
zinc finger, C2HC-type containing 1C |
3161 |
0.14 |
| chr1_162813245_162813512 | 0.09 |
Fmo4 |
flavin containing monooxygenase 4 |
294 |
0.9 |
| chr1_134567570_134567757 | 0.09 |
Kdm5b |
lysine (K)-specific demethylase 5B |
7456 |
0.14 |
| chr4_80919092_80919518 | 0.09 |
Lurap1l |
leucine rich adaptor protein 1-like |
8659 |
0.26 |
| chr3_118443694_118443845 | 0.08 |
Gm9916 |
predicted gene 9916 |
8999 |
0.15 |
| chr11_22493852_22494015 | 0.08 |
Gm12051 |
predicted gene 12051 |
10325 |
0.19 |
| chr13_75387054_75387205 | 0.08 |
Gm48234 |
predicted gene, 48234 |
92258 |
0.07 |
| chr10_24362567_24362800 | 0.08 |
Gm15271 |
predicted gene 15271 |
84807 |
0.08 |
| chr16_72808058_72808236 | 0.08 |
Robo1 |
roundabout guidance receptor 1 |
144943 |
0.05 |
| chr19_38339776_38339993 | 0.08 |
Gm50150 |
predicted gene, 50150 |
2730 |
0.22 |
| chr9_50500490_50500842 | 0.08 |
Plet1os |
placenta expressed transcript 1, opposite strand |
4139 |
0.14 |
| chr7_114420709_114420920 | 0.08 |
Pde3b |
phosphodiesterase 3B, cGMP-inhibited |
5533 |
0.23 |
| chr12_80972933_80973282 | 0.08 |
Slc10a1 |
solute carrier family 10 (sodium/bile acid cotransporter family), member 1 |
4402 |
0.17 |
| chr7_30095803_30095954 | 0.08 |
Zfp260 |
zinc finger protein 260 |
693 |
0.43 |
| chr5_134177966_134178117 | 0.08 |
Rcc1l |
reculator of chromosome condensation 1 like |
1267 |
0.3 |
| chr7_80052331_80052493 | 0.08 |
Gm44951 |
predicted gene 44951 |
15707 |
0.11 |
| chr18_39438249_39438499 | 0.08 |
Nr3c1 |
nuclear receptor subfamily 3, group C, member 1 |
48858 |
0.13 |
| chr3_97596140_97596302 | 0.08 |
Gm43075 |
predicted gene 43075 |
13711 |
0.15 |
| chr2_128966761_128966912 | 0.08 |
Zc3h6 |
zinc finger CCCH type containing 6 |
566 |
0.49 |
| chr12_77681559_77681753 | 0.08 |
4930458K08Rik |
RIKEN cDNA 4930458K08 gene |
1405 |
0.49 |
| chr2_125781835_125782214 | 0.08 |
Secisbp2l |
SECIS binding protein 2-like |
846 |
0.66 |
| chr4_130663944_130664724 | 0.08 |
Pum1 |
pumilio RNA-binding family member 1 |
928 |
0.61 |
| chr17_74255165_74255351 | 0.08 |
Memo1 |
mediator of cell motility 1 |
727 |
0.62 |
| chr4_48555827_48555978 | 0.08 |
Msantd3 |
Myb/SANT-like DNA-binding domain containing 3 |
4045 |
0.25 |
| chr15_50267936_50268109 | 0.08 |
Gm49198 |
predicted gene, 49198 |
18876 |
0.3 |
| chr13_51111229_51111529 | 0.08 |
Spin1 |
spindlin 1 |
10499 |
0.22 |
| chr5_147902213_147902380 | 0.08 |
Slc46a3 |
solute carrier family 46, member 3 |
7481 |
0.17 |
| chr3_75530671_75531016 | 0.07 |
Pdcd10 |
programmed cell death 10 |
9273 |
0.2 |
| chr6_86523276_86523978 | 0.07 |
1600020E01Rik |
RIKEN cDNA 1600020E01 gene |
2620 |
0.12 |
| chr2_35093747_35093964 | 0.07 |
AI182371 |
expressed sequence AI182371 |
2000 |
0.28 |
| chr3_52323120_52323465 | 0.07 |
Gm38034 |
predicted gene, 38034 |
41051 |
0.12 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 0.1 | GO:0070340 | detection of bacterial lipopeptide(GO:0070340) |
| 0.0 | 0.1 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.0 | 0.4 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.0 | 0.1 | GO:0046271 | phenylpropanoid catabolic process(GO:0046271) |
| 0.0 | 0.1 | GO:0010046 | response to mycotoxin(GO:0010046) |
| 0.0 | 0.1 | GO:1903966 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.0 | 0.2 | GO:0006975 | DNA damage induced protein phosphorylation(GO:0006975) |
| 0.0 | 0.1 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.0 | 0.1 | GO:0060523 | prostate epithelial cord elongation(GO:0060523) prostate gland morphogenetic growth(GO:0060737) |
| 0.0 | 0.1 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.0 | 0.1 | GO:0036016 | response to interleukin-3(GO:0036015) cellular response to interleukin-3(GO:0036016) |
| 0.0 | 0.2 | GO:0052697 | xenobiotic glucuronidation(GO:0052697) |
| 0.0 | 0.0 | GO:1904192 | cholangiocyte apoptotic process(GO:1902488) regulation of cholangiocyte apoptotic process(GO:1904192) negative regulation of cholangiocyte apoptotic process(GO:1904193) |
| 0.0 | 0.1 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.0 | 0.1 | GO:0001842 | neural fold formation(GO:0001842) |
| 0.0 | 0.0 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.0 | 0.1 | GO:0060836 | lymphatic endothelial cell differentiation(GO:0060836) |
| 0.0 | 0.0 | GO:1990705 | cholangiocyte proliferation(GO:1990705) |
| 0.0 | 0.1 | GO:0042737 | drug catabolic process(GO:0042737) |
| 0.0 | 0.1 | GO:0051799 | negative regulation of hair follicle development(GO:0051799) |
| 0.0 | 0.1 | GO:0046618 | drug export(GO:0046618) |
| 0.0 | 0.0 | GO:0006203 | dGTP catabolic process(GO:0006203) |
| 0.0 | 0.0 | GO:1904016 | response to Thyroglobulin triiodothyronine(GO:1904016) cellular response to Thyroglobulin triiodothyronine(GO:1904017) |
| 0.0 | 0.0 | GO:2000211 | regulation of glutamate metabolic process(GO:2000211) |
| 0.0 | 0.0 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.0 | 0.1 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.0 | 0.0 | GO:1904504 | regulation of lipophagy(GO:1904502) positive regulation of lipophagy(GO:1904504) |
| 0.0 | 0.0 | GO:0045896 | regulation of transcription during mitosis(GO:0045896) positive regulation of transcription during mitosis(GO:0045897) |
| 0.0 | 0.0 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0035355 | Toll-like receptor 2-Toll-like receptor 6 protein complex(GO:0035355) |
| 0.0 | 0.1 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.0 | 0.0 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.0 | 0.0 | GO:0000811 | GINS complex(GO:0000811) |
| 0.0 | 0.0 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.1 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.0 | GO:0097441 | basilar dendrite(GO:0097441) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.0 | 0.1 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.0 | 0.1 | GO:0035663 | Toll-like receptor 2 binding(GO:0035663) |
| 0.0 | 0.2 | GO:0047760 | butyrate-CoA ligase activity(GO:0047760) |
| 0.0 | 0.1 | GO:0032896 | palmitoyl-CoA 9-desaturase activity(GO:0032896) |
| 0.0 | 0.1 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.0 | 0.1 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.0 | 0.1 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.0 | 0.1 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.0 | 0.1 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.1 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 0.1 | GO:0071614 | linoleic acid epoxygenase activity(GO:0071614) |
| 0.0 | 0.1 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.0 | 0.0 | GO:0038181 | bile acid receptor activity(GO:0038181) |
| 0.0 | 0.1 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.0 | 0.1 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.0 | 0.0 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.0 | GO:0004096 | catalase activity(GO:0004096) |
| 0.0 | 0.0 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.1 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.0 | 0.0 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.0 | 0.1 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 0.1 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |