Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
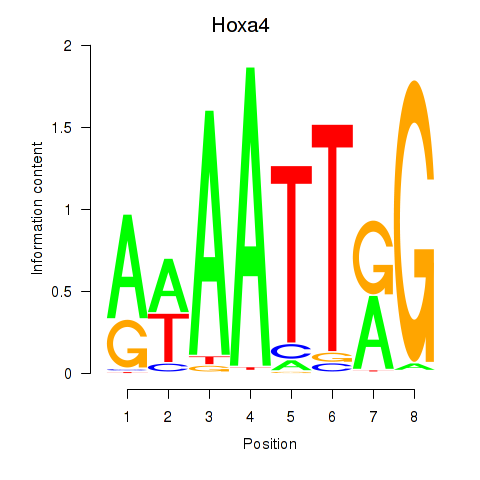
Results for Hoxa4
Z-value: 0.97
Transcription factors associated with Hoxa4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hoxa4
|
ENSMUSG00000000942.10 | homeobox A4 |
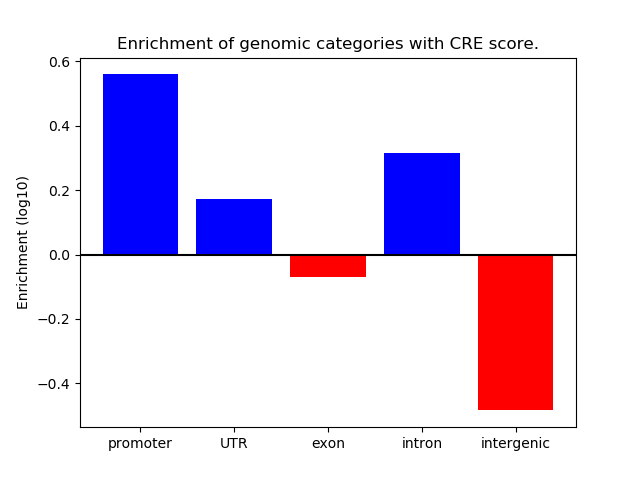
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr6_52191906_52192068 | Hoxa4 | 234 | 0.752593 | -0.18 | 7.4e-01 | Click! |
Activity of the Hoxa4 motif across conditions
Conditions sorted by the z-value of the Hoxa4 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
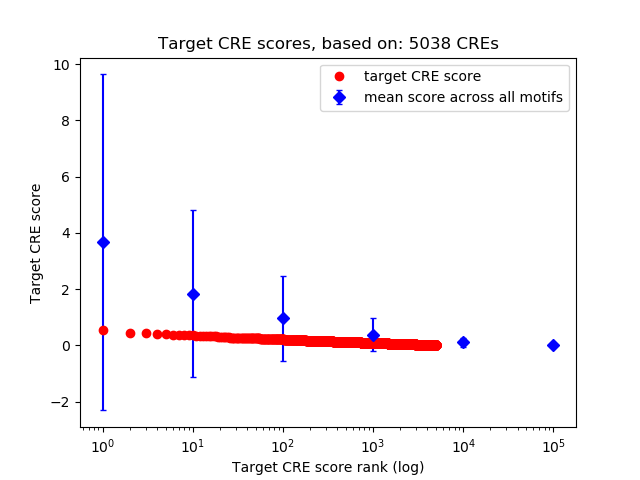
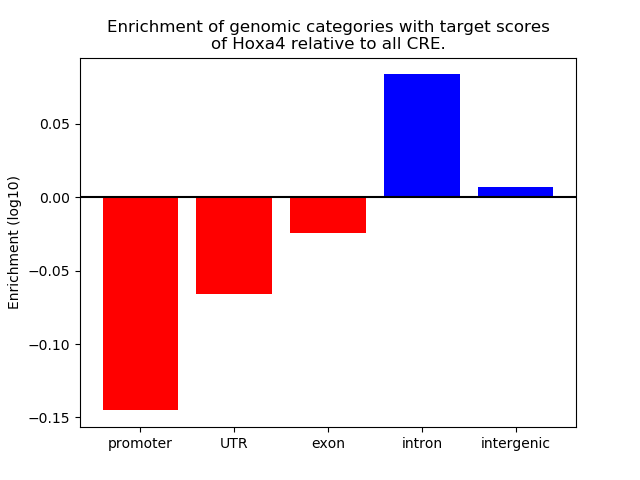
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr19_44401904_44402310 | 0.53 |
Scd1 |
stearoyl-Coenzyme A desaturase 1 |
4583 |
0.17 |
| chr19_36264512_36264835 | 0.44 |
Gm32027 |
predicted gene, 32027 |
236 |
0.93 |
| chr17_25446180_25446342 | 0.44 |
Tekt4 |
tektin 4 |
8354 |
0.11 |
| chr8_93167580_93167731 | 0.41 |
Ces1d |
carboxylesterase 1D |
2320 |
0.22 |
| chr3_118606217_118606485 | 0.40 |
Dpyd |
dihydropyrimidine dehydrogenase |
44165 |
0.15 |
| chr15_3475054_3475205 | 0.38 |
Ghr |
growth hormone receptor |
3485 |
0.35 |
| chr15_4641921_4642072 | 0.38 |
C6 |
complement component 6 |
85179 |
0.09 |
| chr8_93176743_93176912 | 0.37 |
Ces1d |
carboxylesterase 1D |
1538 |
0.3 |
| chr12_104087776_104088197 | 0.36 |
Serpina4-ps1 |
serine (or cysteine) peptidase inhibitor, clade A, member 4, pseudogene 1 |
7337 |
0.1 |
| chr9_122848915_122849207 | 0.36 |
Gm47140 |
predicted gene, 47140 |
643 |
0.55 |
| chr19_44402814_44402965 | 0.34 |
Scd1 |
stearoyl-Coenzyme A desaturase 1 |
3801 |
0.18 |
| chr3_51230467_51230865 | 0.34 |
Gm38357 |
predicted gene, 38357 |
1251 |
0.37 |
| chr10_4613331_4613709 | 0.33 |
Esr1 |
estrogen receptor 1 (alpha) |
1499 |
0.46 |
| chr3_76637152_76637321 | 0.33 |
Fstl5 |
follistatin-like 5 |
43686 |
0.19 |
| chr5_125524109_125525122 | 0.33 |
Tmem132b |
transmembrane protein 132B |
7159 |
0.16 |
| chr12_51689904_51690055 | 0.33 |
Ap4s1 |
adaptor-related protein complex AP-4, sigma 1 |
1054 |
0.46 |
| chr10_87896571_87896873 | 0.32 |
Igf1os |
insulin-like growth factor 1, opposite strand |
33341 |
0.15 |
| chr15_3463620_3463812 | 0.31 |
Ghr |
growth hormone receptor |
7928 |
0.3 |
| chr11_28690043_28690210 | 0.31 |
2810471M01Rik |
RIKEN cDNA 2810471M01 gene |
8562 |
0.19 |
| chr2_58790704_58790855 | 0.30 |
Upp2 |
uridine phosphorylase 2 |
25454 |
0.18 |
| chr7_19508824_19509003 | 0.30 |
Trappc6a |
trafficking protein particle complex 6A |
183 |
0.76 |
| chr8_93258234_93258385 | 0.30 |
Ces1f |
carboxylesterase 1F |
430 |
0.78 |
| chr1_67236092_67236243 | 0.29 |
Gm15668 |
predicted gene 15668 |
13033 |
0.22 |
| chr4_102576883_102577034 | 0.28 |
Pde4b |
phosphodiesterase 4B, cAMP specific |
6863 |
0.31 |
| chr8_23031563_23031855 | 0.28 |
Ank1 |
ankyrin 1, erythroid |
3390 |
0.23 |
| chr4_46832518_46832720 | 0.28 |
Gabbr2 |
gamma-aminobutyric acid (GABA) B receptor, 2 |
27283 |
0.22 |
| chr12_80336463_80336614 | 0.28 |
Dcaf5 |
DDB1 and CUL4 associated factor 5 |
40154 |
0.1 |
| chr2_35193405_35193556 | 0.27 |
Rab14 |
RAB14, member RAS oncogene family |
817 |
0.57 |
| chr8_93165469_93165620 | 0.27 |
Ces1d |
carboxylesterase 1D |
4431 |
0.16 |
| chr6_22001861_22002013 | 0.27 |
Cped1 |
cadherin-like and PC-esterase domain containing 1 |
15045 |
0.24 |
| chr2_50283993_50284144 | 0.27 |
Mmadhc |
methylmalonic aciduria (cobalamin deficiency) cblD type, with homocystinuria |
5131 |
0.24 |
| chr1_67227779_67227978 | 0.27 |
Gm15668 |
predicted gene 15668 |
21322 |
0.2 |
| chr19_4766055_4766206 | 0.27 |
Gm37206 |
predicted gene, 37206 |
959 |
0.35 |
| chr9_23436438_23436789 | 0.27 |
Bmper |
BMP-binding endothelial regulator |
62681 |
0.16 |
| chr5_51876698_51876972 | 0.27 |
Gm42616 |
predicted gene 42616 |
2767 |
0.24 |
| chr1_55699716_55699934 | 0.26 |
Plcl1 |
phospholipase C-like 1 |
2200 |
0.43 |
| chr4_102558090_102558267 | 0.26 |
Pde4b |
phosphodiesterase 4B, cAMP specific |
11917 |
0.3 |
| chr13_96674138_96674322 | 0.26 |
Hmgcr |
3-hydroxy-3-methylglutaryl-Coenzyme A reductase |
3294 |
0.19 |
| chr4_47366562_47366857 | 0.26 |
Tgfbr1 |
transforming growth factor, beta receptor I |
13099 |
0.22 |
| chr14_116240919_116241084 | 0.26 |
Gm20713 |
predicted gene 20713 |
369723 |
0.01 |
| chr2_58773576_58773851 | 0.26 |
Upp2 |
uridine phosphorylase 2 |
8388 |
0.21 |
| chr15_38049613_38049764 | 0.26 |
Gm22353 |
predicted gene, 22353 |
8597 |
0.16 |
| chr12_99858543_99858709 | 0.26 |
Efcab11 |
EF-hand calcium binding domain 11 |
24231 |
0.11 |
| chr3_111102424_111102592 | 0.26 |
Gm43407 |
predicted gene 43407 |
5992 |
0.34 |
| chr1_50685464_50685672 | 0.26 |
Gm28321 |
predicted gene 28321 |
129807 |
0.05 |
| chr13_95792630_95792781 | 0.25 |
Iqgap2 |
IQ motif containing GTPase activating protein 2 |
28308 |
0.16 |
| chr3_158035265_158035416 | 0.25 |
Gm43362 |
predicted gene 43362 |
1069 |
0.28 |
| chr2_34775525_34776380 | 0.25 |
Hspa5 |
heat shock protein 5 |
1105 |
0.4 |
| chr2_122251200_122251358 | 0.25 |
Sord |
sorbitol dehydrogenase |
16530 |
0.1 |
| chr11_117599771_117599931 | 0.25 |
2900041M22Rik |
RIKEN cDNA 2900041M22 gene |
11396 |
0.18 |
| chr18_34948918_34949069 | 0.25 |
Hspa9 |
heat shock protein 9 |
869 |
0.45 |
| chr13_46153184_46153335 | 0.25 |
Gm10113 |
predicted gene 10113 |
37787 |
0.19 |
| chr16_26619429_26619580 | 0.25 |
Il1rap |
interleukin 1 receptor accessory protein |
4652 |
0.32 |
| chr17_28502859_28503010 | 0.24 |
Fkbp5 |
FK506 binding protein 5 |
4472 |
0.09 |
| chr10_31517928_31518079 | 0.24 |
Gm47693 |
predicted gene, 47693 |
39437 |
0.12 |
| chr6_72098587_72098738 | 0.24 |
St3gal5 |
ST3 beta-galactoside alpha-2,3-sialyltransferase 5 |
905 |
0.44 |
| chr6_29820680_29820831 | 0.24 |
Ahcyl2 |
S-adenosylhomocysteine hydrolase-like 2 |
18896 |
0.17 |
| chr6_67268305_67268456 | 0.24 |
Serbp1 |
serpine1 mRNA binding protein 1 |
1050 |
0.44 |
| chr4_122983111_122983491 | 0.24 |
Mycl |
v-myc avian myelocytomatosis viral oncogene lung carcinoma derived |
12351 |
0.13 |
| chr3_57448360_57448527 | 0.24 |
Tm4sf4 |
transmembrane 4 superfamily member 4 |
23129 |
0.19 |
| chr14_67053186_67053337 | 0.24 |
Ppp2r2a |
protein phosphatase 2, regulatory subunit B, alpha |
9732 |
0.17 |
| chr2_64059065_64059258 | 0.24 |
Fign |
fidgetin |
38827 |
0.23 |
| chr12_104346905_104347119 | 0.24 |
Serpina3k |
serine (or cysteine) peptidase inhibitor, clade A, member 3K |
8526 |
0.12 |
| chr14_21033937_21034339 | 0.23 |
Vcl |
vinculin |
12135 |
0.18 |
| chr19_44404359_44404510 | 0.23 |
Scd1 |
stearoyl-Coenzyme A desaturase 1 |
2256 |
0.23 |
| chr1_67208135_67208300 | 0.23 |
Gm15668 |
predicted gene 15668 |
40983 |
0.16 |
| chr9_48737248_48737430 | 0.23 |
Zbtb16 |
zinc finger and BTB domain containing 16 |
98606 |
0.07 |
| chr6_138145599_138145922 | 0.23 |
Mgst1 |
microsomal glutathione S-transferase 1 |
2906 |
0.37 |
| chr2_122737415_122737579 | 0.23 |
Bloc1s6os |
biogenesis of lysosomal organelles complex-1, subunit 6, pallidin, opposite strand |
865 |
0.39 |
| chr12_99861381_99861532 | 0.23 |
Efcab11 |
EF-hand calcium binding domain 11 |
21401 |
0.12 |
| chr6_66963164_66963414 | 0.23 |
Gm36816 |
predicted gene, 36816 |
45124 |
0.08 |
| chr9_74881714_74881880 | 0.23 |
Onecut1 |
one cut domain, family member 1 |
15313 |
0.15 |
| chr5_125522928_125523080 | 0.22 |
Aacs |
acetoacetyl-CoA synthetase |
7761 |
0.16 |
| chr7_49290770_49290970 | 0.22 |
Nav2 |
neuron navigator 2 |
13268 |
0.23 |
| chr2_118306989_118307215 | 0.22 |
1700054M17Rik |
RIKEN cDNA 1700054M17 gene |
2188 |
0.23 |
| chr7_118181720_118181871 | 0.22 |
Smg1 |
SMG1 homolog, phosphatidylinositol 3-kinase-related kinase (C. elegans) |
10496 |
0.16 |
| chr6_72125643_72125794 | 0.22 |
4933431G14Rik |
RIKEN cDNA 4933431G14 gene |
2521 |
0.19 |
| chr3_121735675_121736039 | 0.22 |
F3 |
coagulation factor III |
6310 |
0.11 |
| chr7_64802263_64802423 | 0.22 |
Fam189a1 |
family with sequence similarity 189, member A1 |
28691 |
0.19 |
| chr15_10485447_10485602 | 0.22 |
Brix1 |
BRX1, biogenesis of ribosomes |
139 |
0.83 |
| chr5_51162183_51162334 | 0.22 |
Gm44377 |
predicted gene, 44377 |
61416 |
0.14 |
| chr17_28428613_28428954 | 0.22 |
Fkbp5 |
FK506 binding protein 5 |
154 |
0.91 |
| chr12_72768361_72768564 | 0.22 |
Ppm1a |
protein phosphatase 1A, magnesium dependent, alpha isoform |
7147 |
0.2 |
| chr19_17599285_17599457 | 0.22 |
Gm17819 |
predicted gene, 17819 |
93392 |
0.08 |
| chr2_122206384_122206535 | 0.21 |
Gm14050 |
predicted gene 14050 |
1461 |
0.25 |
| chr10_28123601_28123752 | 0.21 |
Ptprk |
protein tyrosine phosphatase, receptor type, K |
48666 |
0.15 |
| chr13_9145204_9145368 | 0.21 |
Gm28155 |
predicted gene 28155 |
4405 |
0.19 |
| chr11_80383442_80384000 | 0.21 |
Zfp207 |
zinc finger protein 207 |
324 |
0.87 |
| chr6_124758482_124758633 | 0.21 |
Atn1 |
atrophin 1 |
2033 |
0.1 |
| chr13_63665698_63665976 | 0.21 |
Gm47387 |
predicted gene, 47387 |
4040 |
0.18 |
| chr7_73608284_73608633 | 0.21 |
Gm44734 |
predicted gene 44734 |
490 |
0.68 |
| chr19_44416577_44416796 | 0.21 |
Gm50337 |
predicted gene, 50337 |
2924 |
0.2 |
| chr13_44437957_44438156 | 0.21 |
1700029N11Rik |
RIKEN cDNA 1700029N11 gene |
1656 |
0.27 |
| chr3_60965282_60965682 | 0.21 |
P2ry1 |
purinergic receptor P2Y, G-protein coupled 1 |
37313 |
0.16 |
| chr10_37139504_37139765 | 0.21 |
5930403N24Rik |
RIKEN cDNA 5930403N24 gene |
76 |
0.93 |
| chr7_119978745_119978903 | 0.21 |
Gm25217 |
predicted gene, 25217 |
9003 |
0.16 |
| chr10_67107041_67107217 | 0.21 |
Reep3 |
receptor accessory protein 3 |
10184 |
0.2 |
| chr10_53373661_53374052 | 0.21 |
Cep85l |
centrosomal protein 85-like |
2258 |
0.21 |
| chr3_18123292_18123443 | 0.21 |
Gm23726 |
predicted gene, 23726 |
54041 |
0.12 |
| chr5_147545326_147545477 | 0.21 |
Pan3 |
PAN3 poly(A) specific ribonuclease subunit |
6124 |
0.21 |
| chr10_87051307_87051578 | 0.21 |
1700113H08Rik |
RIKEN cDNA 1700113H08 gene |
6603 |
0.2 |
| chr2_121419496_121419659 | 0.21 |
Pdia3 |
protein disulfide isomerase associated 3 |
5053 |
0.08 |
| chr12_52464277_52464428 | 0.21 |
Gm47431 |
predicted gene, 47431 |
16227 |
0.17 |
| chr2_24892753_24892952 | 0.21 |
Gm37139 |
predicted gene, 37139 |
1014 |
0.3 |
| chr1_100656422_100656580 | 0.20 |
Gm29334 |
predicted gene 29334 |
2701 |
0.31 |
| chr11_17736440_17736720 | 0.20 |
Gm12016 |
predicted gene 12016 |
97397 |
0.08 |
| chr7_78896045_78896424 | 0.20 |
Aen |
apoptosis enhancing nuclease |
225 |
0.89 |
| chr5_134295659_134295849 | 0.20 |
Gtf2i |
general transcription factor II I |
146 |
0.93 |
| chr13_96742797_96742948 | 0.20 |
Ankrd31 |
ankyrin repeat domain 31 |
5400 |
0.2 |
| chr3_52270975_52271214 | 0.20 |
Gm20402 |
predicted gene 20402 |
1659 |
0.26 |
| chr15_3500655_3500833 | 0.20 |
Ghr |
growth hormone receptor |
29100 |
0.23 |
| chr2_121438376_121438543 | 0.20 |
Ell3 |
elongation factor RNA polymerase II-like 3 |
2692 |
0.11 |
| chr2_31485846_31486386 | 0.20 |
Ass1 |
argininosuccinate synthetase 1 |
11656 |
0.18 |
| chr2_30378803_30378994 | 0.20 |
Miga2 |
mitoguardin 2 |
663 |
0.51 |
| chr11_110431257_110431440 | 0.20 |
Map2k6 |
mitogen-activated protein kinase kinase 6 |
32089 |
0.21 |
| chr16_30110807_30110970 | 0.20 |
Gm20040 |
predicted gene, 20040 |
18006 |
0.13 |
| chr2_128378785_128378975 | 0.20 |
Morrbid |
myeloid RNA regulator of BCL2L11 induced cell death |
24085 |
0.18 |
| chr14_122878269_122878430 | 0.20 |
Pcca |
propionyl-Coenzyme A carboxylase, alpha polypeptide |
2414 |
0.29 |
| chr10_15746557_15746708 | 0.20 |
Gm32283 |
predicted gene, 32283 |
7810 |
0.18 |
| chr5_89462792_89462945 | 0.20 |
Gc |
vitamin D binding protein |
4970 |
0.25 |
| chr9_41328613_41328764 | 0.20 |
Mir100hg |
Mir100 Mirlet7a-2 Mir125b-1 cluster host gene |
78 |
0.97 |
| chr3_60576910_60577122 | 0.20 |
Mbnl1 |
muscleblind like splicing factor 1 |
18625 |
0.19 |
| chr11_28685801_28685952 | 0.20 |
2810471M01Rik |
RIKEN cDNA 2810471M01 gene |
4312 |
0.22 |
| chr3_29732375_29732526 | 0.19 |
Gm37557 |
predicted gene, 37557 |
38834 |
0.19 |
| chr14_8003568_8003795 | 0.19 |
Abhd6 |
abhydrolase domain containing 6 |
715 |
0.66 |
| chr6_85095404_85095555 | 0.19 |
Gm43955 |
predicted gene, 43955 |
1448 |
0.21 |
| chr3_97632897_97633210 | 0.19 |
Fmo5 |
flavin containing monooxygenase 5 |
4170 |
0.16 |
| chr17_46046958_46047109 | 0.19 |
Vegfa |
vascular endothelial growth factor A |
14664 |
0.14 |
| chr7_140724826_140724998 | 0.19 |
Olfr542-ps1 |
olfactory receptor 542, pseudogene 1 |
3630 |
0.12 |
| chr17_28025808_28026411 | 0.19 |
Anks1 |
ankyrin repeat and SAM domain containing 1 |
14666 |
0.11 |
| chr8_25776385_25776540 | 0.19 |
Bag4 |
BCL2-associated athanogene 4 |
1120 |
0.32 |
| chr2_156152255_156152428 | 0.19 |
Romo1 |
reactive oxygen species modulator 1 |
8101 |
0.1 |
| chr2_128377567_128377751 | 0.19 |
Morrbid |
myeloid RNA regulator of BCL2L11 induced cell death |
25306 |
0.18 |
| chr2_84637297_84637592 | 0.19 |
Ctnnd1 |
catenin (cadherin associated protein), delta 1 |
661 |
0.54 |
| chr10_28166717_28166892 | 0.19 |
Gm22370 |
predicted gene, 22370 |
47317 |
0.17 |
| chr2_31513022_31513372 | 0.19 |
Ass1 |
argininosuccinate synthetase 1 |
5293 |
0.19 |
| chr10_24486336_24486502 | 0.19 |
Gm15272 |
predicted gene 15272 |
28092 |
0.16 |
| chr3_136853089_136853240 | 0.19 |
Ppp3ca |
protein phosphatase 3, catalytic subunit, alpha isoform |
18471 |
0.2 |
| chr19_56533507_56533720 | 0.19 |
Dclre1a |
DNA cross-link repair 1A |
3769 |
0.23 |
| chr9_48733643_48733794 | 0.19 |
Zbtb16 |
zinc finger and BTB domain containing 16 |
102227 |
0.06 |
| chr5_139559359_139559510 | 0.19 |
Uncx |
UNC homeobox |
15536 |
0.17 |
| chr10_15748693_15748844 | 0.19 |
Gm32283 |
predicted gene, 32283 |
9946 |
0.18 |
| chr11_90686948_90687471 | 0.18 |
Tom1l1 |
target of myb1-like 1 (chicken) |
370 |
0.89 |
| chr4_108425790_108426145 | 0.18 |
Gpx7 |
glutathione peroxidase 7 |
19006 |
0.1 |
| chr6_29571610_29572022 | 0.18 |
Tnpo3 |
transportin 3 |
297 |
0.87 |
| chr10_89530174_89530325 | 0.18 |
Nr1h4 |
nuclear receptor subfamily 1, group H, member 4 |
3336 |
0.26 |
| chr6_37542487_37542638 | 0.18 |
Gm7463 |
predicted gene 7463 |
1148 |
0.52 |
| chr10_87532342_87532676 | 0.18 |
Pah |
phenylalanine hydroxylase |
10482 |
0.2 |
| chr12_84361536_84361697 | 0.18 |
Coq6 |
coenzyme Q6 monooxygenase |
41 |
0.67 |
| chr8_40564203_40564371 | 0.18 |
Vps37a |
vacuolar protein sorting 37A |
20863 |
0.15 |
| chr9_86486034_86486185 | 0.18 |
Dop1a |
DOP1 leucine zipper like protein A |
702 |
0.68 |
| chr11_88841479_88841856 | 0.18 |
Akap1 |
A kinase (PRKA) anchor protein 1 |
127 |
0.96 |
| chr15_3512657_3512808 | 0.18 |
Ghr |
growth hormone receptor |
41088 |
0.19 |
| chr15_35886635_35886786 | 0.18 |
Vps13b |
vacuolar protein sorting 13B |
14988 |
0.16 |
| chr10_34326839_34326990 | 0.18 |
Nt5dc1 |
5'-nucleotidase domain containing 1 |
13231 |
0.13 |
| chr19_32622727_32622878 | 0.18 |
Papss2 |
3'-phosphoadenosine 5'-phosphosulfate synthase 2 |
2797 |
0.32 |
| chr3_34483929_34484290 | 0.18 |
Gm29135 |
predicted gene 29135 |
1902 |
0.34 |
| chr7_115795674_115795828 | 0.18 |
Sox6 |
SRY (sex determining region Y)-box 6 |
28959 |
0.24 |
| chr7_136457949_136458342 | 0.18 |
Gm36849 |
predicted gene, 36849 |
104781 |
0.07 |
| chr6_145855503_145855668 | 0.18 |
Gm43909 |
predicted gene, 43909 |
7712 |
0.17 |
| chr19_48883437_48883771 | 0.18 |
Gm50436 |
predicted gene, 50436 |
72468 |
0.11 |
| chr3_118592516_118592739 | 0.18 |
Dpyd |
dihydropyrimidine dehydrogenase |
30441 |
0.17 |
| chr7_136406426_136406609 | 0.18 |
Gm36849 |
predicted gene, 36849 |
53153 |
0.14 |
| chr12_41068657_41068813 | 0.18 |
Immp2l |
IMP2 inner mitochondrial membrane peptidase-like (S. cerevisiae) |
423 |
0.86 |
| chr15_3463467_3463618 | 0.18 |
Ghr |
growth hormone receptor |
8102 |
0.29 |
| chr16_54907388_54907558 | 0.18 |
Gm22977 |
predicted gene, 22977 |
64958 |
0.13 |
| chr16_35401691_35401895 | 0.17 |
Pdia5 |
protein disulfide isomerase associated 5 |
11382 |
0.13 |
| chr4_95992355_95992506 | 0.17 |
Hook1 |
hook microtubule tethering protein 1 |
10016 |
0.18 |
| chr12_70999972_71000123 | 0.17 |
Psma3 |
proteasome subunit alpha 3 |
13155 |
0.14 |
| chr11_16780814_16780965 | 0.17 |
Egfr |
epidermal growth factor receptor |
28659 |
0.16 |
| chr3_51204305_51204466 | 0.17 |
Noct |
nocturnin |
20062 |
0.14 |
| chr8_94399242_94399642 | 0.17 |
Ap3s1-ps2 |
adaptor-related protein complex 3, sigma 1 subunit, pseudogene 2 |
5769 |
0.11 |
| chr3_107279311_107279673 | 0.17 |
Lamtor5 |
late endosomal/lysosomal adaptor, MAPK and MTOR activator 5 |
493 |
0.74 |
| chr13_119966643_119967735 | 0.17 |
Gm20784 |
predicted gene, 20784 |
4945 |
0.11 |
| chr9_122127991_122128201 | 0.17 |
4632418H02Rik |
RIKEN cDNA 4632418H02 gene |
802 |
0.5 |
| chr11_106889132_106889465 | 0.17 |
Smurf2 |
SMAD specific E3 ubiquitin protein ligase 2 |
30977 |
0.11 |
| chr4_123990670_123991070 | 0.17 |
Gm12902 |
predicted gene 12902 |
64636 |
0.08 |
| chr16_36987638_36987887 | 0.17 |
Fbxo40 |
F-box protein 40 |
2705 |
0.19 |
| chr3_18138170_18138321 | 0.17 |
Gm23686 |
predicted gene, 23686 |
39380 |
0.16 |
| chr5_87089108_87089304 | 0.17 |
Ugt2b36 |
UDP glucuronosyltransferase 2 family, polypeptide B36 |
1951 |
0.22 |
| chr11_5096479_5096889 | 0.17 |
Rhbdd3 |
rhomboid domain containing 3 |
2242 |
0.19 |
| chr19_20614762_20614977 | 0.17 |
Aldh1a1 |
aldehyde dehydrogenase family 1, subfamily A1 |
12908 |
0.21 |
| chr19_17599754_17599928 | 0.17 |
Gm17819 |
predicted gene, 17819 |
92922 |
0.09 |
| chr6_37782285_37782449 | 0.17 |
Atp6v0c-ps2 |
ATPase, H+ transporting, lysosomal V0 subunit C, pseudogene 2 |
23719 |
0.21 |
| chr9_113127097_113127258 | 0.17 |
Gm36251 |
predicted gene, 36251 |
4148 |
0.32 |
| chr3_97643032_97643186 | 0.17 |
Fmo5 |
flavin containing monooxygenase 5 |
14226 |
0.12 |
| chr13_119966017_119966201 | 0.17 |
Gm20784 |
predicted gene, 20784 |
3865 |
0.12 |
| chr3_142862185_142862381 | 0.17 |
Pkn2 |
protein kinase N2 |
6548 |
0.15 |
| chr9_43258125_43258276 | 0.17 |
D630033O11Rik |
RIKEN cDNA D630033O11 gene |
1680 |
0.32 |
| chr2_34828383_34828580 | 0.17 |
Fbxw2 |
F-box and WD-40 domain protein 2 |
2170 |
0.17 |
| chr19_40168031_40168182 | 0.17 |
Cyp2c70 |
cytochrome P450, family 2, subfamily c, polypeptide 70 |
19180 |
0.15 |
| chr2_109714999_109715174 | 0.17 |
Bdnf |
brain derived neurotrophic factor |
5703 |
0.22 |
| chr14_40032524_40032701 | 0.17 |
Gm25012 |
predicted gene, 25012 |
22710 |
0.21 |
| chr4_62102548_62102834 | 0.17 |
Gm12910 |
predicted gene 12910 |
10240 |
0.13 |
| chr3_97638864_97639015 | 0.17 |
Fmo5 |
flavin containing monooxygenase 5 |
10056 |
0.13 |
| chr1_106267814_106267970 | 0.17 |
Phlpp1 |
PH domain and leucine rich repeat protein phosphatase 1 |
13533 |
0.22 |
| chr6_72865575_72865746 | 0.17 |
Kcmf1 |
potassium channel modulatory factor 1 |
6766 |
0.18 |
| chr16_93353844_93354010 | 0.17 |
1810053B23Rik |
RIKEN cDNA 1810053B23 gene |
134 |
0.96 |
| chr7_100609367_100609521 | 0.17 |
Mrpl48 |
mitochondrial ribosomal protein L48 |
1143 |
0.29 |
| chr5_99262128_99262472 | 0.16 |
Rasgef1b |
RasGEF domain family, member 1B |
9373 |
0.22 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.1 | 0.5 | GO:0046449 | creatinine metabolic process(GO:0046449) cellular lactam metabolic process(GO:0072338) |
| 0.1 | 0.3 | GO:0060523 | prostate epithelial cord elongation(GO:0060523) |
| 0.1 | 0.3 | GO:1903964 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.1 | 0.2 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.1 | 0.2 | GO:1901078 | negative regulation of relaxation of muscle(GO:1901078) |
| 0.1 | 0.4 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.1 | 0.2 | GO:2000211 | regulation of glutamate metabolic process(GO:2000211) |
| 0.1 | 0.2 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.1 | 0.2 | GO:0071926 | cannabinoid signaling pathway(GO:0038171) endocannabinoid signaling pathway(GO:0071926) |
| 0.1 | 0.2 | GO:0060741 | prostate gland stromal morphogenesis(GO:0060741) |
| 0.0 | 0.1 | GO:1904154 | positive regulation of retrograde protein transport, ER to cytosol(GO:1904154) |
| 0.0 | 0.2 | GO:0006166 | purine ribonucleoside salvage(GO:0006166) |
| 0.0 | 0.1 | GO:0006208 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) thymine metabolic process(GO:0019859) |
| 0.0 | 0.1 | GO:0006982 | response to lipid hydroperoxide(GO:0006982) |
| 0.0 | 0.2 | GO:0045875 | negative regulation of sister chromatid cohesion(GO:0045875) |
| 0.0 | 0.1 | GO:0044337 | canonical Wnt signaling pathway involved in positive regulation of apoptotic process(GO:0044337) |
| 0.0 | 0.2 | GO:0016557 | peroxisome membrane biogenesis(GO:0016557) |
| 0.0 | 0.1 | GO:0034395 | regulation of transcription from RNA polymerase II promoter in response to iron(GO:0034395) |
| 0.0 | 0.0 | GO:0006714 | sesquiterpenoid metabolic process(GO:0006714) |
| 0.0 | 0.1 | GO:1903334 | positive regulation of protein folding(GO:1903334) |
| 0.0 | 0.1 | GO:0097680 | double-strand break repair via classical nonhomologous end joining(GO:0097680) |
| 0.0 | 0.1 | GO:0097460 | ferrous iron import into cell(GO:0097460) |
| 0.0 | 0.1 | GO:0072318 | clathrin coat disassembly(GO:0072318) |
| 0.0 | 0.1 | GO:0006987 | activation of signaling protein activity involved in unfolded protein response(GO:0006987) |
| 0.0 | 0.1 | GO:2001274 | negative regulation of glucose import in response to insulin stimulus(GO:2001274) |
| 0.0 | 0.1 | GO:0048696 | regulation of collateral sprouting in absence of injury(GO:0048696) |
| 0.0 | 0.1 | GO:0090187 | positive regulation of pancreatic juice secretion(GO:0090187) |
| 0.0 | 0.2 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.0 | 0.1 | GO:0015747 | urate transport(GO:0015747) |
| 0.0 | 0.1 | GO:1902915 | negative regulation of protein K63-linked ubiquitination(GO:1900045) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.0 | 0.1 | GO:0061043 | regulation of vascular wound healing(GO:0061043) |
| 0.0 | 0.1 | GO:0055005 | ventricular cardiac myofibril assembly(GO:0055005) |
| 0.0 | 0.1 | GO:0006393 | termination of mitochondrial transcription(GO:0006393) |
| 0.0 | 0.1 | GO:0070681 | glutaminyl-tRNAGln biosynthesis via transamidation(GO:0070681) |
| 0.0 | 0.1 | GO:1900226 | negative regulation of NLRP3 inflammasome complex assembly(GO:1900226) |
| 0.0 | 0.1 | GO:0000430 | regulation of transcription from RNA polymerase II promoter by glucose(GO:0000430) positive regulation of transcription from RNA polymerase II promoter by glucose(GO:0000432) |
| 0.0 | 0.1 | GO:0003213 | cardiac right atrium morphogenesis(GO:0003213) |
| 0.0 | 0.1 | GO:0018894 | dibenzo-p-dioxin metabolic process(GO:0018894) |
| 0.0 | 0.1 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.0 | 0.1 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.0 | 0.1 | GO:0042780 | tRNA 3'-end processing(GO:0042780) |
| 0.0 | 0.1 | GO:1900122 | positive regulation of receptor binding(GO:1900122) |
| 0.0 | 0.1 | GO:0043465 | regulation of fermentation(GO:0043465) regulation of NAD metabolic process(GO:1902688) regulation of glucose catabolic process to lactate via pyruvate(GO:1904023) |
| 0.0 | 0.1 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.0 | 0.1 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.0 | 0.2 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.0 | 0.1 | GO:0061419 | positive regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061419) |
| 0.0 | 0.1 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.0 | 0.1 | GO:0050427 | 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) |
| 0.0 | 0.1 | GO:0006499 | N-terminal protein myristoylation(GO:0006499) |
| 0.0 | 0.1 | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.0 | 0.1 | GO:0060689 | cell differentiation involved in salivary gland development(GO:0060689) |
| 0.0 | 0.1 | GO:0042524 | negative regulation of tyrosine phosphorylation of Stat5 protein(GO:0042524) |
| 0.0 | 0.2 | GO:0010944 | negative regulation of transcription by competitive promoter binding(GO:0010944) |
| 0.0 | 0.1 | GO:0072602 | interleukin-4 secretion(GO:0072602) |
| 0.0 | 0.3 | GO:0006743 | ubiquinone metabolic process(GO:0006743) |
| 0.0 | 0.1 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.0 | 0.1 | GO:0010912 | regulation of isomerase activity(GO:0010911) positive regulation of isomerase activity(GO:0010912) |
| 0.0 | 0.0 | GO:0021524 | visceral motor neuron differentiation(GO:0021524) |
| 0.0 | 0.0 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 0.0 | 0.1 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.0 | 0.1 | GO:0046881 | positive regulation of follicle-stimulating hormone secretion(GO:0046881) |
| 0.0 | 0.1 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.0 | 0.0 | GO:0031087 | nuclear-transcribed mRNA catabolic process, deadenylation-independent decay(GO:0031086) deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.0 | 0.0 | GO:1903587 | regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903587) |
| 0.0 | 0.1 | GO:0021779 | oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.0 | 0.1 | GO:0060155 | platelet dense granule organization(GO:0060155) |
| 0.0 | 0.0 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.0 | 0.1 | GO:0034310 | primary alcohol catabolic process(GO:0034310) |
| 0.0 | 0.1 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.0 | 0.0 | GO:1901187 | regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.0 | 0.1 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.0 | 0.2 | GO:0045176 | apical protein localization(GO:0045176) |
| 0.0 | 0.1 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.0 | 0.1 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.0 | 0.0 | GO:0048850 | hypophysis morphogenesis(GO:0048850) |
| 0.0 | 0.1 | GO:0072309 | mesenchymal stem cell maintenance involved in metanephric nephron morphogenesis(GO:0072309) |
| 0.0 | 0.1 | GO:2000609 | regulation of thyroid hormone generation(GO:2000609) |
| 0.0 | 0.1 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.0 | 0.1 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.0 | 0.1 | GO:0042045 | epithelial fluid transport(GO:0042045) |
| 0.0 | 0.0 | GO:0070375 | ERK5 cascade(GO:0070375) |
| 0.0 | 0.1 | GO:0032849 | positive regulation of cellular pH reduction(GO:0032849) |
| 0.0 | 0.1 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.0 | 0.5 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.0 | 0.0 | GO:0070103 | regulation of interleukin-6-mediated signaling pathway(GO:0070103) |
| 0.0 | 0.1 | GO:0045900 | negative regulation of translational elongation(GO:0045900) |
| 0.0 | 0.0 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.0 | 0.3 | GO:0061462 | protein localization to lysosome(GO:0061462) |
| 0.0 | 0.0 | GO:0006059 | hexitol metabolic process(GO:0006059) |
| 0.0 | 0.1 | GO:0097105 | presynaptic membrane assembly(GO:0097105) |
| 0.0 | 0.3 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.1 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.0 | 0.0 | GO:0034140 | negative regulation of toll-like receptor 3 signaling pathway(GO:0034140) |
| 0.0 | 0.0 | GO:1904222 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) regulation of serine C-palmitoyltransferase activity(GO:1904220) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.0 | 0.0 | GO:0021847 | ventricular zone neuroblast division(GO:0021847) |
| 0.0 | 0.0 | GO:0030397 | membrane disassembly(GO:0030397) nuclear envelope disassembly(GO:0051081) |
| 0.0 | 0.0 | GO:0051890 | regulation of cardioblast differentiation(GO:0051890) |
| 0.0 | 0.1 | GO:0034773 | histone H4-K20 trimethylation(GO:0034773) |
| 0.0 | 0.0 | GO:0071455 | cellular response to increased oxygen levels(GO:0036295) cellular response to hyperoxia(GO:0071455) |
| 0.0 | 0.0 | GO:0042360 | vitamin E metabolic process(GO:0042360) |
| 0.0 | 0.0 | GO:1902309 | negative regulation of peptidyl-serine dephosphorylation(GO:1902309) |
| 0.0 | 0.1 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.0 | GO:0045726 | positive regulation of integrin biosynthetic process(GO:0045726) |
| 0.0 | 0.1 | GO:0033299 | secretion of lysosomal enzymes(GO:0033299) |
| 0.0 | 0.1 | GO:0001880 | Mullerian duct regression(GO:0001880) |
| 0.0 | 0.1 | GO:2000370 | positive regulation of clathrin-mediated endocytosis(GO:2000370) |
| 0.0 | 0.2 | GO:0051481 | negative regulation of cytosolic calcium ion concentration(GO:0051481) |
| 0.0 | 0.0 | GO:0045112 | integrin biosynthetic process(GO:0045112) |
| 0.0 | 0.1 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.0 | 0.1 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.0 | 0.0 | GO:0044597 | polyketide metabolic process(GO:0030638) aminoglycoside antibiotic metabolic process(GO:0030647) daunorubicin metabolic process(GO:0044597) doxorubicin metabolic process(GO:0044598) |
| 0.0 | 0.1 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.0 | 0.1 | GO:0008354 | germ cell migration(GO:0008354) |
| 0.0 | 0.1 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.0 | 0.0 | GO:2000809 | positive regulation of synaptic vesicle clustering(GO:2000809) |
| 0.0 | 0.0 | GO:0001712 | ectodermal cell fate commitment(GO:0001712) |
| 0.0 | 0.1 | GO:0072592 | oxygen metabolic process(GO:0072592) |
| 0.0 | 0.0 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.0 | 0.0 | GO:0032789 | saturated monocarboxylic acid metabolic process(GO:0032788) unsaturated monocarboxylic acid metabolic process(GO:0032789) |
| 0.0 | 0.2 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.2 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.0 | 0.0 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.0 | 0.0 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.0 | 0.0 | GO:0003164 | His-Purkinje system development(GO:0003164) |
| 0.0 | 0.0 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.0 | 0.0 | GO:0003175 | tricuspid valve development(GO:0003175) tricuspid valve morphogenesis(GO:0003186) tricuspid valve formation(GO:0003195) |
| 0.0 | 0.1 | GO:0033147 | negative regulation of intracellular estrogen receptor signaling pathway(GO:0033147) |
| 0.0 | 0.2 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.0 | 0.1 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.0 | 0.0 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.0 | 0.0 | GO:1904502 | lipophagy(GO:0061724) regulation of lipophagy(GO:1904502) positive regulation of lipophagy(GO:1904504) |
| 0.0 | 0.0 | GO:0060398 | regulation of growth hormone receptor signaling pathway(GO:0060398) |
| 0.0 | 0.0 | GO:0034436 | glycoprotein transport(GO:0034436) |
| 0.0 | 0.1 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.0 | 0.0 | GO:0033092 | positive regulation of immature T cell proliferation in thymus(GO:0033092) |
| 0.0 | 0.1 | GO:0070189 | kynurenine metabolic process(GO:0070189) |
| 0.0 | 0.2 | GO:0010257 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.1 | GO:0071397 | cellular response to cholesterol(GO:0071397) |
| 0.0 | 0.0 | GO:0009629 | response to gravity(GO:0009629) |
| 0.0 | 0.0 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 0.0 | 0.1 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 0.0 | GO:0071394 | cellular response to testosterone stimulus(GO:0071394) |
| 0.0 | 0.1 | GO:0098908 | regulation of neuronal action potential(GO:0098908) |
| 0.0 | 0.0 | GO:2000659 | regulation of interleukin-1-mediated signaling pathway(GO:2000659) |
| 0.0 | 0.0 | GO:0070340 | detection of bacterial lipopeptide(GO:0070340) |
| 0.0 | 0.0 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.0 | 0.0 | GO:0036016 | response to interleukin-3(GO:0036015) cellular response to interleukin-3(GO:0036016) |
| 0.0 | 0.0 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.0 | 0.1 | GO:0006353 | DNA-templated transcription, termination(GO:0006353) |
| 0.0 | 0.1 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.0 | 0.0 | GO:1903899 | positive regulation of PERK-mediated unfolded protein response(GO:1903899) |
| 0.0 | 0.1 | GO:0097084 | vascular smooth muscle cell development(GO:0097084) |
| 0.0 | 0.1 | GO:0060020 | Bergmann glial cell differentiation(GO:0060020) |
| 0.0 | 0.0 | GO:0006573 | valine metabolic process(GO:0006573) |
| 0.0 | 0.1 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 0.1 | GO:0042256 | mature ribosome assembly(GO:0042256) |
| 0.0 | 0.0 | GO:0043174 | nucleoside salvage(GO:0043174) |
| 0.0 | 0.1 | GO:0080154 | regulation of fertilization(GO:0080154) |
| 0.0 | 0.0 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.0 | GO:0035519 | protein K29-linked ubiquitination(GO:0035519) |
| 0.0 | 0.0 | GO:1904479 | negative regulation of intestinal absorption(GO:1904479) |
| 0.0 | 0.0 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.0 | 0.1 | GO:0019184 | nonribosomal peptide biosynthetic process(GO:0019184) |
| 0.0 | 0.0 | GO:0097212 | lysosomal membrane organization(GO:0097212) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.0 | 0.1 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 0.0 | 0.1 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.0 | 0.3 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.2 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.0 | 0.1 | GO:0030956 | glutamyl-tRNA(Gln) amidotransferase complex(GO:0030956) |
| 0.0 | 0.1 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.0 | 0.1 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.0 | 0.1 | GO:0042825 | TAP complex(GO:0042825) |
| 0.0 | 0.1 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.0 | 0.2 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.0 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.1 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.0 | 0.1 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.1 | GO:0000306 | extrinsic component of vacuolar membrane(GO:0000306) |
| 0.0 | 0.2 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 0.1 | GO:0038037 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor complex(GO:0097648) |
| 0.0 | 0.0 | GO:0034679 | integrin alpha9-beta1 complex(GO:0034679) |
| 0.0 | 0.1 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.0 | 0.1 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.0 | 0.2 | GO:0097208 | alveolar lamellar body(GO:0097208) |
| 0.0 | 0.1 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 0.1 | GO:0031211 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.0 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.0 | 0.1 | GO:0032797 | SMN complex(GO:0032797) |
| 0.0 | 0.0 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 0.0 | GO:0070110 | ciliary neurotrophic factor receptor complex(GO:0070110) |
| 0.0 | 0.1 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.0 | 0.2 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 0.1 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.0 | 0.1 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 0.1 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 0.0 | GO:0031094 | platelet dense tubular network(GO:0031094) |
| 0.0 | 0.2 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.1 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.0 | 0.0 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.1 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.1 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.1 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.0 | 0.0 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.0 | 0.0 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.0 | 0.1 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.0 | GO:1990851 | Wnt-Frizzled-LRP5/6 complex(GO:1990851) |
| 0.0 | 0.0 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.0 | 0.0 | GO:0072588 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 0.0 | 0.1 | GO:0045261 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.0 | 0.2 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.0 | 0.0 | GO:0035355 | Toll-like receptor 2-Toll-like receptor 6 protein complex(GO:0035355) |
| 0.0 | 0.0 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.0 | 0.0 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.0 | 0.0 | GO:0097629 | extrinsic component of omegasome membrane(GO:0097629) |
| 0.0 | 0.1 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.0 | 0.1 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 0.1 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.1 | 0.3 | GO:0032896 | palmitoyl-CoA 9-desaturase activity(GO:0032896) |
| 0.1 | 0.3 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.1 | 0.3 | GO:0004075 | biotin carboxylase activity(GO:0004075) |
| 0.1 | 0.2 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.0 | 0.1 | GO:0016743 | carboxyl- or carbamoyltransferase activity(GO:0016743) |
| 0.0 | 0.0 | GO:0050051 | leukotriene-B4 20-monooxygenase activity(GO:0050051) |
| 0.0 | 0.1 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.1 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.0 | 0.1 | GO:0019153 | protein-disulfide reductase (glutathione) activity(GO:0019153) |
| 0.0 | 0.1 | GO:0034191 | apolipoprotein A-I receptor binding(GO:0034191) |
| 0.0 | 0.1 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 0.2 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.1 | GO:0038181 | bile acid receptor activity(GO:0038181) |
| 0.0 | 0.1 | GO:0004096 | catalase activity(GO:0004096) |
| 0.0 | 0.2 | GO:0005072 | transforming growth factor beta receptor, cytoplasmic mediator activity(GO:0005072) |
| 0.0 | 0.1 | GO:0050567 | glutaminyl-tRNA synthase (glutamine-hydrolyzing) activity(GO:0050567) |
| 0.0 | 0.1 | GO:0016899 | oxidoreductase activity, acting on the CH-OH group of donors, oxygen as acceptor(GO:0016899) |
| 0.0 | 0.1 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.0 | 0.1 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.0 | 0.1 | GO:0004083 | bisphosphoglycerate 2-phosphatase activity(GO:0004083) |
| 0.0 | 0.2 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.0 | 0.1 | GO:0003846 | 2-acylglycerol O-acyltransferase activity(GO:0003846) |
| 0.0 | 0.1 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.0 | 0.1 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.0 | 0.1 | GO:0030158 | protein xylosyltransferase activity(GO:0030158) |
| 0.0 | 0.1 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) |
| 0.0 | 0.1 | GO:0033695 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 0.0 | 0.2 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.1 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 0.1 | GO:0047023 | androsterone dehydrogenase activity(GO:0047023) |
| 0.0 | 0.1 | GO:0030620 | U2 snRNA binding(GO:0030620) |
| 0.0 | 0.3 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.1 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.0 | 0.1 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.3 | GO:1900750 | glutathione binding(GO:0043295) oligopeptide binding(GO:1900750) |
| 0.0 | 0.0 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.0 | 0.1 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 0.1 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.0 | 0.1 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.0 | 0.2 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.1 | GO:0018633 | 3-(3-hydroxyphenyl)propionate hydroxylase activity(GO:0008688) 4-chlorobenzaldehyde oxidase activity(GO:0018471) 3,5-xylenol methylhydroxylase activity(GO:0018630) phenylacetate hydroxylase activity(GO:0018631) 4-nitrophenol 4-monooxygenase activity(GO:0018632) dimethyl sulfide monooxygenase activity(GO:0018633) alpha-pinene monooxygenase [NADH] activity(GO:0018634) phenanthrene 9,10-monooxygenase activity(GO:0018636) 1-hydroxy-2-naphthoate hydroxylase activity(GO:0018637) toluene 4-monooxygenase activity(GO:0018638) xylene monooxygenase activity(GO:0018639) dibenzothiophene monooxygenase activity(GO:0018640) 6-hydroxy-3-methyl-2-oxo-1,2-dihydroquinoline 6-monooxygenase activity(GO:0018641) chlorophenol 4-monooxygenase activity(GO:0018642) carbon disulfide oxygenase activity(GO:0018643) toluene 2-monooxygenase activity(GO:0018644) 1-hydroxy-2-oxolimonene 1,2-monooxygenase activity(GO:0018646) phenanthrene 1,2-monooxygenase activity(GO:0018647) tetrahydrofuran hydroxylase activity(GO:0018649) styrene monooxygenase activity(GO:0018650) toluene-4-sulfonate monooxygenase activity(GO:0018651) toluene-sulfonate methyl-monooxygenase activity(GO:0018652) 3-methyl-2-oxo-1,2-dihydroquinoline 6-monooxygenase activity(GO:0018653) 2-hydroxy-phenylacetate hydroxylase activity(GO:0018654) 2-oxo-delta3-4,5,5-trimethylcyclopentenylacetyl-CoA 1,2-monooxygenase activity(GO:0018655) phenanthrene 3,4-monooxygenase activity(GO:0018656) toluene 3-monooxygenase activity(GO:0018657) 4-hydroxyphenylacetate,NADH:oxygen oxidoreductase (3-hydroxylating) activity(GO:0018660) limonene monooxygenase activity(GO:0019113) 2-methylnaphthalene hydroxylase activity(GO:0034526) 1-methylnaphthalene hydroxylase activity(GO:0034534) bisphenol A hydroxylase A activity(GO:0034560) salicylate 5-hydroxylase activity(GO:0034785) isobutylamine N-hydroxylase activity(GO:0034791) branched-chain dodecylbenzene sulfonate monooxygenase activity(GO:0034802) 3-HSA hydroxylase activity(GO:0034819) 4-hydroxypyridine-3-hydroxylase activity(GO:0034894) 2-octaprenyl-3-methyl-6-methoxy-1,4-benzoquinol hydroxylase activity(GO:0043719) 6-hydroxynicotinate 3-monooxygenase activity(GO:0043731) thalianol hydroxylase activity(GO:0080014) |
| 0.0 | 0.1 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.0 | 0.6 | GO:0080031 | prenylcysteine methylesterase activity(GO:0010296) 1-oxa-2-oxocycloheptane lactonase activity(GO:0018731) sulfolactone hydrolase activity(GO:0018732) butyrolactone hydrolase activity(GO:0018734) endosulfan lactone lactonase activity(GO:0034892) L-ascorbate 6-phosphate lactonase activity(GO:0035460) Ser-tRNA(Thr) hydrolase activity(GO:0043905) Ala-tRNA(Pro) hydrolase activity(GO:0043906) Cys-tRNA(Pro) hydrolase activity(GO:0043907) Ser(Gly)-tRNA(Ala) hydrolase activity(GO:0043908) all-trans-retinyl-palmitate hydrolase, all-trans-retinol forming activity(GO:0047376) mannosyl-oligosaccharide 1,6-alpha-mannosidase activity(GO:0052767) mannosyl-oligosaccharide 1,3-alpha-mannosidase activity(GO:0052768) methyl indole-3-acetate esterase activity(GO:0080030) methyl salicylate esterase activity(GO:0080031) methyl jasmonate esterase activity(GO:0080032) |
| 0.0 | 0.1 | GO:0047760 | butyrate-CoA ligase activity(GO:0047760) |
| 0.0 | 0.1 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 0.0 | 0.1 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.0 | 0.1 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 0.1 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 0.1 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.0 | 0.1 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 0.0 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.0 | 0.3 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.1 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.0 | 0.1 | GO:0034548 | N-cyclopropylmelamine deaminase activity(GO:0034547) N-cyclopropylammeline deaminase activity(GO:0034548) N-cyclopropylammelide alkylamino hydrolase activity(GO:0034549) 2,5-diamino-6-ribitylamino-4(3H)-pyrimidinone 5'-phosphate deaminase activity(GO:0043723) tRNA-specific adenosine-37 deaminase activity(GO:0043829) archaeal-specific GTP cyclohydrolase activity(GO:0044682) tRNA-specific adenosine-34 deaminase activity(GO:0052717) |
| 0.0 | 0.2 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.0 | 0.0 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.0 | 0.0 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.0 | 0.0 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.0 | 0.1 | GO:0072542 | protein phosphatase activator activity(GO:0072542) |
| 0.0 | 0.0 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.0 | 0.0 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.0 | 0.1 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.0 | 0.1 | GO:0016679 | oxidoreductase activity, acting on diphenols and related substances as donors(GO:0016679) |
| 0.0 | 0.0 | GO:0031686 | A1 adenosine receptor binding(GO:0031686) |
| 0.0 | 0.0 | GO:0032767 | copper-dependent protein binding(GO:0032767) |
| 0.0 | 0.1 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.0 | 0.1 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.0 | 0.1 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.0 | 0.1 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 0.0 | 0.1 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.0 | 0.1 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.0 | 0.0 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.0 | GO:0001602 | pancreatic polypeptide receptor activity(GO:0001602) |
| 0.0 | 0.1 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 0.1 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.0 | 0.0 | GO:0004897 | ciliary neurotrophic factor receptor activity(GO:0004897) |
| 0.0 | 0.0 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.0 | GO:0031697 | beta-1 adrenergic receptor binding(GO:0031697) |
| 0.0 | 0.0 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.0 | 0.0 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.0 | 0.0 | GO:0035663 | Toll-like receptor 2 binding(GO:0035663) |
| 0.0 | 0.3 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 0.4 | GO:0008392 | arachidonic acid epoxygenase activity(GO:0008392) |
| 0.0 | 0.1 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.0 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.1 | GO:0043426 | MRF binding(GO:0043426) |
| 0.0 | 0.1 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.0 | 0.0 | GO:1990239 | steroid hormone binding(GO:1990239) |
| 0.0 | 0.2 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 0.0 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.0 | 0.1 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.0 | 0.0 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.0 | 0.0 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.0 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.0 | GO:0003985 | acetyl-CoA C-acetyltransferase activity(GO:0003985) C-acetyltransferase activity(GO:0016453) |
| 0.0 | 0.3 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.1 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 0.4 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.5 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.1 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.1 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 0.1 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.0 | PID AVB3 INTEGRIN PATHWAY | Integrins in angiogenesis |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.3 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.3 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.2 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.1 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.0 | 0.4 | REACTOME GRB2 EVENTS IN ERBB2 SIGNALING | Genes involved in GRB2 events in ERBB2 signaling |
| 0.0 | 0.1 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.0 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.2 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.0 | 0.3 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 0.2 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.1 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.1 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.1 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.2 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.0 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 0.1 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.2 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.0 | 0.1 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.1 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.2 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |