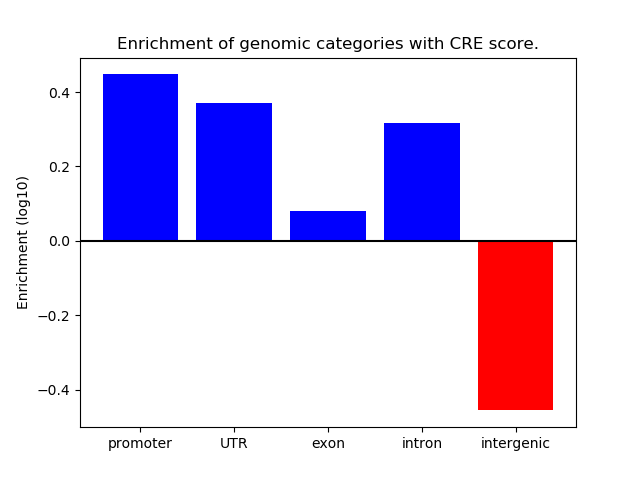
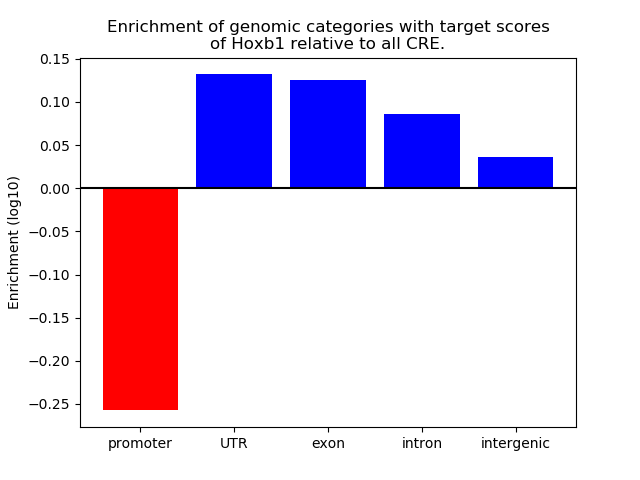
Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
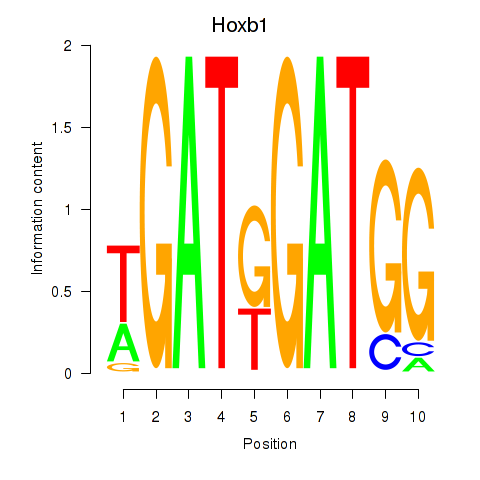
Results for Hoxb1
Z-value: 0.77
Transcription factors associated with Hoxb1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hoxb1
|
ENSMUSG00000018973.2 | homeobox B1 |
Activity of the Hoxb1 motif across conditions
Conditions sorted by the z-value of the Hoxb1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
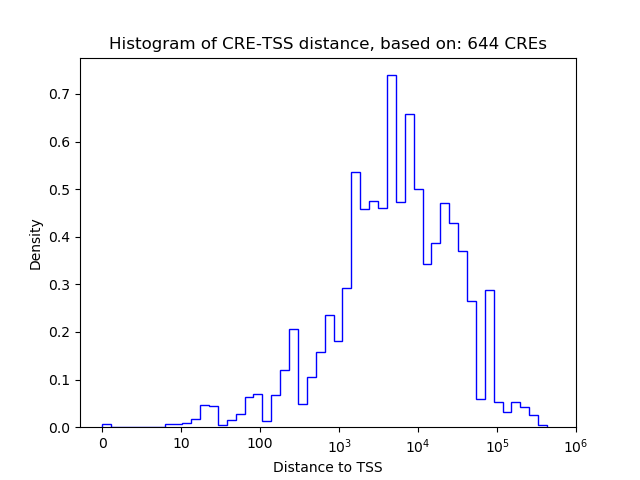
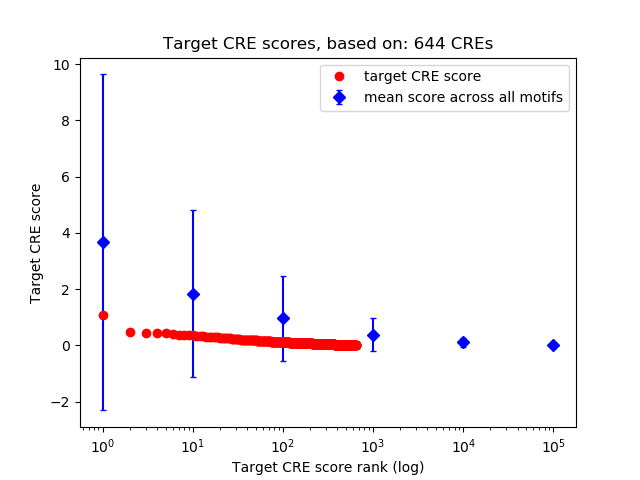
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr19_46138909_46139075 | 1.09 |
Pitx3 |
paired-like homeodomain transcription factor 3 |
1703 |
0.25 |
| chr10_66631116_66631267 | 0.48 |
Gm28139 |
predicted gene 28139 |
9727 |
0.22 |
| chr11_106580231_106580444 | 0.44 |
Tex2 |
testis expressed gene 2 |
263 |
0.92 |
| chr13_60267432_60267612 | 0.44 |
Gm24999 |
predicted gene, 24999 |
26150 |
0.16 |
| chr12_10818187_10818338 | 0.43 |
Pgk1-rs7 |
phosphoglycerate kinase-1, related sequence-7 |
81978 |
0.09 |
| chr13_30235450_30235601 | 0.40 |
Mboat1 |
membrane bound O-acyltransferase domain containing 1 |
3672 |
0.27 |
| chr7_112743142_112743293 | 0.38 |
Tead1 |
TEA domain family member 1 |
1172 |
0.52 |
| chr18_35679363_35679538 | 0.37 |
Dnajc18 |
DnaJ heat shock protein family (Hsp40) member C18 |
7293 |
0.09 |
| chr12_28905624_28905792 | 0.36 |
Gm31508 |
predicted gene, 31508 |
4521 |
0.21 |
| chr16_24400480_24400643 | 0.35 |
Gm24440 |
predicted gene, 24440 |
5856 |
0.18 |
| chr11_101372590_101372741 | 0.33 |
G6pc |
glucose-6-phosphatase, catalytic |
5104 |
0.07 |
| chr9_65327502_65327653 | 0.33 |
Gm39363 |
predicted gene, 39363 |
4943 |
0.1 |
| chr14_45491500_45491651 | 0.33 |
9630050E16Rik |
RIKEN cDNA 9630050E16 gene |
510 |
0.68 |
| chr8_33684436_33684594 | 0.31 |
Gm24727 |
predicted gene, 24727 |
7286 |
0.15 |
| chr9_114922455_114922659 | 0.31 |
Gpd1l |
glycerol-3-phosphate dehydrogenase 1-like |
3203 |
0.26 |
| chr19_20646594_20646745 | 0.30 |
Aldh1a1 |
aldehyde dehydrogenase family 1, subfamily A1 |
44708 |
0.15 |
| chr2_24474146_24474302 | 0.30 |
Pax8 |
paired box 8 |
873 |
0.5 |
| chr18_44624210_44624361 | 0.29 |
A930012L18Rik |
RIKEN cDNA A930012L18 gene |
37380 |
0.17 |
| chr4_43876248_43876406 | 0.28 |
Reck |
reversion-inducing-cysteine-rich protein with kazal motifs |
797 |
0.53 |
| chr5_33725606_33725768 | 0.27 |
Fgfr3 |
fibroblast growth factor receptor 3 |
1915 |
0.19 |
| chr12_72593347_72593534 | 0.26 |
Gm4756 |
predicted gene 4756 |
15652 |
0.17 |
| chr14_30929372_30929624 | 0.25 |
Itih3 |
inter-alpha trypsin inhibitor, heavy chain 3 |
5738 |
0.11 |
| chr11_100640165_100640335 | 0.25 |
Gm44544 |
predicted gene 44544 |
2867 |
0.14 |
| chr10_95942046_95942222 | 0.25 |
Eea1 |
early endosome antigen 1 |
1377 |
0.41 |
| chr14_31166341_31166492 | 0.25 |
Stab1 |
stabilin 1 |
2179 |
0.18 |
| chr10_8228115_8228580 | 0.24 |
Gm30906 |
predicted gene, 30906 |
52216 |
0.15 |
| chr18_37827931_37828082 | 0.23 |
Gm29994 |
predicted gene, 29994 |
2194 |
0.11 |
| chr8_35387028_35387898 | 0.23 |
Ppp1r3b |
protein phosphatase 1, regulatory subunit 3B |
10803 |
0.16 |
| chr1_165651668_165651967 | 0.22 |
Gm18407 |
predicted gene, 18407 |
7129 |
0.12 |
| chr6_93984322_93984485 | 0.22 |
Magi1 |
membrane associated guanylate kinase, WW and PDZ domain containing 1 |
70713 |
0.12 |
| chr3_96102665_96102823 | 0.21 |
Otud7b |
OTU domain containing 7B |
1783 |
0.2 |
| chr13_45627279_45627485 | 0.21 |
Gmpr |
guanosine monophosphate reductase |
81229 |
0.1 |
| chr11_97403801_97404023 | 0.21 |
Arhgap23 |
Rho GTPase activating protein 23 |
11621 |
0.14 |
| chr1_21067933_21068103 | 0.20 |
Tram2 |
translocating chain-associating membrane protein 2 |
11211 |
0.17 |
| chr19_4712495_4712727 | 0.20 |
Sptbn2 |
spectrin beta, non-erythrocytic 2 |
227 |
0.88 |
| chr8_105056108_105056869 | 0.19 |
Ces3a |
carboxylesterase 3A |
2835 |
0.14 |
| chr7_61752057_61752229 | 0.19 |
Gm23873 |
predicted gene, 23873 |
1606 |
0.2 |
| chr14_25476997_25477210 | 0.19 |
Gm47921 |
predicted gene, 47921 |
2951 |
0.19 |
| chr19_47448450_47448823 | 0.19 |
Sh3pxd2a |
SH3 and PX domains 2A |
15611 |
0.19 |
| chr4_34966679_34966979 | 0.19 |
Gm12364 |
predicted gene 12364 |
4609 |
0.19 |
| chr4_40183031_40183196 | 0.18 |
Aco1 |
aconitase 1 |
7417 |
0.19 |
| chr10_127815941_127816356 | 0.18 |
Rdh18-ps |
retinol dehydrogenase 18, pseudogene |
8136 |
0.1 |
| chr9_70898264_70898447 | 0.18 |
Gm32017 |
predicted gene, 32017 |
32133 |
0.15 |
| chr17_71250166_71250322 | 0.18 |
Lpin2 |
lipin 2 |
4213 |
0.18 |
| chr8_35406149_35406663 | 0.18 |
Gm45301 |
predicted gene 45301 |
3100 |
0.22 |
| chr9_106157796_106157960 | 0.18 |
Glyctk |
glycerate kinase |
232 |
0.82 |
| chr13_45907083_45907460 | 0.18 |
4930453C13Rik |
RIKEN cDNA 4930453C13 gene |
28769 |
0.18 |
| chr15_76113834_76113991 | 0.17 |
Eppk1 |
epiplakin 1 |
6283 |
0.08 |
| chr1_72167520_72167699 | 0.17 |
Mreg |
melanoregulin |
44698 |
0.1 |
| chr2_57148295_57148575 | 0.17 |
Nr4a2 |
nuclear receptor subfamily 4, group A, member 2 |
24432 |
0.14 |
| chr16_97751753_97751904 | 0.17 |
Ripk4 |
receptor-interacting serine-threonine kinase 4 |
11959 |
0.2 |
| chr2_59884229_59884405 | 0.17 |
Wdsub1 |
WD repeat, SAM and U-box domain containing 1 |
1726 |
0.39 |
| chr4_35000639_35000790 | 0.17 |
Gm12364 |
predicted gene 12364 |
38494 |
0.14 |
| chr10_77093982_77094133 | 0.17 |
Col18a1 |
collagen, type XVIII, alpha 1 |
4629 |
0.2 |
| chr8_115089778_115090403 | 0.17 |
Gm22556 |
predicted gene, 22556 |
37177 |
0.23 |
| chr7_19478065_19478228 | 0.17 |
Gm45167 |
predicted gene 45167 |
9918 |
0.07 |
| chr10_93525200_93525441 | 0.16 |
Amdhd1 |
amidohydrolase domain containing 1 |
14713 |
0.12 |
| chr11_7152261_7152427 | 0.16 |
Adcy1 |
adenylate cyclase 1 |
7538 |
0.21 |
| chr3_138359570_138359721 | 0.16 |
Adh6b |
alcohol dehydrogenase 6B (class V) |
7267 |
0.12 |
| chr11_101669101_101669273 | 0.16 |
Arl4d |
ADP-ribosylation factor-like 4D |
3646 |
0.12 |
| chr10_17365243_17365430 | 0.16 |
Gm47760 |
predicted gene, 47760 |
32459 |
0.19 |
| chr16_84832993_84833282 | 0.16 |
Atp5j |
ATP synthase, H+ transporting, mitochondrial F0 complex, subunit F |
739 |
0.53 |
| chr8_91374465_91374676 | 0.16 |
Fto |
fat mass and obesity associated |
15985 |
0.15 |
| chr5_137115334_137115587 | 0.16 |
Trim56 |
tripartite motif-containing 56 |
747 |
0.51 |
| chr1_184127436_184127603 | 0.15 |
Dusp10 |
dual specificity phosphatase 10 |
93138 |
0.08 |
| chr15_82400108_82400364 | 0.15 |
Cyp2d10 |
cytochrome P450, family 2, subfamily d, polypeptide 10 |
5731 |
0.06 |
| chr13_98595452_98595618 | 0.15 |
Gm4815 |
predicted gene 4815 |
17966 |
0.12 |
| chr1_136874874_136875025 | 0.14 |
Nr5a2 |
nuclear receptor subfamily 5, group A, member 2 |
65634 |
0.09 |
| chr8_72183623_72183778 | 0.14 |
Hsh2d |
hematopoietic SH2 domain containing |
5938 |
0.09 |
| chr10_28524334_28524485 | 0.14 |
Ptprk |
protein tyrosine phosphatase, receptor type, K |
35742 |
0.22 |
| chr11_109244649_109244828 | 0.14 |
Rgs9 |
regulator of G-protein signaling 9 |
2227 |
0.34 |
| chr3_98275125_98275300 | 0.14 |
Hmgcs2 |
3-hydroxy-3-methylglutaryl-Coenzyme A synthase 2 |
5223 |
0.16 |
| chr4_10988698_10988868 | 0.14 |
Plekhf2 |
pleckstrin homology domain containing, family F (with FYVE domain) member 2 |
18893 |
0.16 |
| chr8_123242718_123242892 | 0.14 |
Vps9d1 |
VPS9 domain containing 1 |
4125 |
0.08 |
| chr5_72412080_72412249 | 0.13 |
Gm19560 |
predicted gene, 19560 |
2720 |
0.22 |
| chr17_28931020_28931190 | 0.13 |
Gm16191 |
predicted gene 16191 |
3316 |
0.11 |
| chr1_39015594_39015753 | 0.13 |
Pdcl3 |
phosducin-like 3 |
20513 |
0.15 |
| chr6_17532117_17532312 | 0.13 |
Met |
met proto-oncogene |
2787 |
0.33 |
| chrX_11759036_11759212 | 0.13 |
Gm14513 |
predicted gene 14513 |
75000 |
0.1 |
| chr5_45534505_45534782 | 0.13 |
Fam184b |
family with sequence similarity 184, member B |
1899 |
0.22 |
| chr11_107384103_107384285 | 0.13 |
Gm11712 |
predicted gene 11712 |
2789 |
0.2 |
| chr12_79561056_79561219 | 0.13 |
Rad51b |
RAD51 paralog B |
233784 |
0.02 |
| chr3_89390102_89390518 | 0.13 |
Gm15417 |
predicted gene 15417 |
1540 |
0.15 |
| chr10_84381402_84381782 | 0.13 |
Nuak1 |
NUAK family, SNF1-like kinase, 1 |
10796 |
0.2 |
| chr10_108455293_108455501 | 0.12 |
Gm36283 |
predicted gene, 36283 |
8795 |
0.2 |
| chr3_10082799_10082970 | 0.12 |
Gm9833 |
predicted gene 9833 |
5226 |
0.17 |
| chr9_108305002_108305197 | 0.12 |
Tcta |
T cell leukemia translocation altered gene |
849 |
0.3 |
| chr10_84811233_84811541 | 0.12 |
Gm24226 |
predicted gene, 24226 |
204 |
0.95 |
| chr18_61653441_61653641 | 0.12 |
Mir143 |
microRNA 143 |
4283 |
0.12 |
| chr7_127819457_127819621 | 0.12 |
Stx4a |
syntaxin 4A (placental) |
4755 |
0.08 |
| chr2_59451788_59451954 | 0.12 |
Dapl1 |
death associated protein-like 1 |
32782 |
0.15 |
| chr9_59353566_59353745 | 0.12 |
Bbs4 |
Bardet-Biedl syndrome 4 (human) |
147 |
0.96 |
| chr4_47239767_47240064 | 0.12 |
Col15a1 |
collagen, type XV, alpha 1 |
6648 |
0.21 |
| chr18_62224841_62225012 | 0.12 |
Gm9949 |
predicted gene 9949 |
44800 |
0.14 |
| chr9_108783276_108783439 | 0.12 |
Ip6k2 |
inositol hexaphosphate kinase 2 |
439 |
0.68 |
| chr4_132210751_132210902 | 0.11 |
Ythdf2 |
YTH N6-methyladenosine RNA binding protein 2 |
17 |
0.95 |
| chr3_96559546_96560080 | 0.11 |
Txnip |
thioredoxin interacting protein |
90 |
0.91 |
| chr6_116348986_116349287 | 0.11 |
Marchf8 |
membrane associated ring-CH-type finger 8 |
1249 |
0.34 |
| chr19_32190045_32190211 | 0.11 |
Sgms1 |
sphingomyelin synthase 1 |
6300 |
0.24 |
| chr18_24054985_24055136 | 0.11 |
Gm18085 |
predicted gene, 18085 |
26864 |
0.14 |
| chr11_101706893_101707078 | 0.11 |
Dhx8 |
DEAH (Asp-Glu-Ala-His) box polypeptide 8 |
25934 |
0.1 |
| chr15_3232203_3232399 | 0.11 |
Gm7962 |
predicted gene 7962 |
12603 |
0.2 |
| chr8_82655212_82655370 | 0.11 |
Gm8167 |
predicted gene 8167 |
77524 |
0.1 |
| chr5_134984626_134984777 | 0.11 |
Cldn3 |
claudin 3 |
1513 |
0.19 |
| chr6_128145877_128146047 | 0.11 |
Tspan9 |
tetraspanin 9 |
2368 |
0.18 |
| chr18_54249896_54250047 | 0.11 |
Redrum |
Redrum, erythroid developmental long intergenic non-protein coding transcript |
172324 |
0.03 |
| chr10_127880036_127880349 | 0.11 |
Rdh7 |
retinol dehydrogenase 7 |
8138 |
0.1 |
| chr16_3934083_3934394 | 0.11 |
Gm15537 |
predicted gene 15537 |
4881 |
0.08 |
| chr16_76577323_76577485 | 0.11 |
Gm30726 |
predicted gene, 30726 |
28609 |
0.2 |
| chr12_79642030_79642203 | 0.11 |
9430078K24Rik |
RIKEN cDNA 9430078K24 gene |
282617 |
0.01 |
| chr5_92586271_92586466 | 0.11 |
Fam47e |
family with sequence similarity 47, member E |
14812 |
0.15 |
| chr9_110343564_110343739 | 0.11 |
Scap |
SREBF chaperone |
548 |
0.65 |
| chr6_22012994_22013221 | 0.10 |
Cped1 |
cadherin-like and PC-esterase domain containing 1 |
3959 |
0.31 |
| chr11_108306943_108307101 | 0.10 |
Apoh |
apolipoprotein H |
36332 |
0.15 |
| chr11_120531768_120531936 | 0.10 |
Gm11789 |
predicted gene 11789 |
68 |
0.9 |
| chr16_43367135_43367313 | 0.10 |
Zbtb20 |
zinc finger and BTB domain containing 20 |
3020 |
0.23 |
| chr18_80258103_80258267 | 0.10 |
Slc66a2 |
solute carrier family 66 member 2 |
1867 |
0.22 |
| chr10_80963261_80963522 | 0.10 |
Gm3828 |
predicted gene 3828 |
8681 |
0.1 |
| chr2_26435553_26435742 | 0.10 |
Sec16a |
SEC16 homolog A, endoplasmic reticulum export factor |
4108 |
0.1 |
| chr3_130038452_130038603 | 0.10 |
Sec24b |
Sec24 related gene family, member B (S. cerevisiae) |
2306 |
0.27 |
| chr4_101333343_101333527 | 0.10 |
Gm12793 |
predicted gene 12793 |
7922 |
0.11 |
| chr15_31583100_31583308 | 0.10 |
Cct5 |
chaperonin containing Tcp1, subunit 5 (epsilon) |
9857 |
0.13 |
| chr10_68168093_68168385 | 0.10 |
Arid5b |
AT rich interactive domain 5B (MRF1-like) |
31613 |
0.21 |
| chr15_83800905_83801056 | 0.10 |
Mpped1 |
metallophosphoesterase domain containing 1 |
950 |
0.64 |
| chr16_8541145_8541296 | 0.10 |
Abat |
4-aminobutyrate aminotransferase |
27735 |
0.13 |
| chr10_99150947_99151159 | 0.10 |
Poc1b |
POC1 centriolar protein B |
20231 |
0.12 |
| chr7_141638201_141638394 | 0.10 |
Muc6 |
mucin 6, gastric |
17011 |
0.12 |
| chr10_80071687_80071929 | 0.09 |
Sbno2 |
strawberry notch 2 |
3592 |
0.11 |
| chr8_33904205_33904688 | 0.09 |
Rbpms |
RNA binding protein gene with multiple splicing |
12682 |
0.17 |
| chr1_21027602_21027756 | 0.09 |
Tram2 |
translocating chain-associating membrane protein 2 |
15977 |
0.17 |
| chr1_179279335_179279501 | 0.09 |
Smyd3 |
SET and MYND domain containing 3 |
30578 |
0.24 |
| chr13_23729800_23729980 | 0.09 |
Gm11338 |
predicted gene 11338 |
1342 |
0.13 |
| chr10_95238706_95239119 | 0.09 |
Gm48880 |
predicted gene, 48880 |
75941 |
0.06 |
| chr8_84236114_84236304 | 0.09 |
Zswim4 |
zinc finger SWIM-type containing 4 |
846 |
0.33 |
| chr16_97962560_97962728 | 0.09 |
Zbtb21 |
zinc finger and BTB domain containing 21 |
22 |
0.51 |
| chr11_32150361_32150527 | 0.09 |
Gm12109 |
predicted gene 12109 |
34561 |
0.11 |
| chr5_106466342_106466661 | 0.09 |
Gm26872 |
predicted gene, 26872 |
4506 |
0.19 |
| chr11_109691833_109692167 | 0.09 |
Fam20a |
family with sequence similarity 20, member A |
9021 |
0.18 |
| chr19_47317044_47317343 | 0.09 |
Sh3pxd2a |
SH3 and PX domains 2A |
2442 |
0.27 |
| chr2_72306157_72306335 | 0.09 |
Map3k20 |
mitogen-activated protein kinase kinase kinase 20 |
8345 |
0.2 |
| chr1_72220096_72220263 | 0.09 |
Gm25360 |
predicted gene, 25360 |
6061 |
0.13 |
| chr8_76985045_76985216 | 0.09 |
Nr3c2 |
nuclear receptor subfamily 3, group C, member 2 |
76858 |
0.1 |
| chr11_94575112_94575273 | 0.09 |
Acsf2 |
acyl-CoA synthetase family member 2 |
4414 |
0.13 |
| chr10_128409887_128410052 | 0.09 |
Nabp2 |
nucleic acid binding protein 2 |
95 |
0.89 |
| chr18_65104255_65104458 | 0.08 |
Nedd4l |
neural precursor cell expressed, developmentally down-regulated gene 4-like |
21302 |
0.21 |
| chr5_130074851_130075046 | 0.08 |
Tpst1 |
protein-tyrosine sulfotransferase 1 |
1622 |
0.29 |
| chr18_60814075_60814226 | 0.08 |
Mir5107 |
microRNA 5107 |
2074 |
0.23 |
| chr13_109881168_109881343 | 0.08 |
Pde4d |
phosphodiesterase 4D, cAMP specific |
21847 |
0.23 |
| chr18_74231507_74231667 | 0.08 |
Gm5690 |
predicted gene 5690 |
9303 |
0.15 |
| chr2_26477777_26477967 | 0.08 |
Notch1 |
notch 1 |
7374 |
0.1 |
| chr4_34981947_34982131 | 0.08 |
Gm12364 |
predicted gene 12364 |
19819 |
0.16 |
| chr13_70611184_70611345 | 0.08 |
Ice1 |
interactor of little elongation complex ELL subunit 1 |
2445 |
0.27 |
| chr16_43335899_43336088 | 0.08 |
Gm15711 |
predicted gene 15711 |
23399 |
0.14 |
| chr6_115884148_115884359 | 0.08 |
Ift122 |
intraflagellar transport 122 |
1655 |
0.26 |
| chr10_93112384_93112535 | 0.08 |
Gm32468 |
predicted gene, 32468 |
27208 |
0.15 |
| chrX_12358352_12358503 | 0.08 |
Gm14635 |
predicted gene 14635 |
3431 |
0.34 |
| chr1_85199424_85199611 | 0.08 |
AC123856.1 |
nuclear antigen Sp100 (Sp100) pseudogene |
96 |
0.49 |
| chr16_94369838_94370349 | 0.08 |
Pigp |
phosphatidylinositol glycan anchor biosynthesis, class P |
196 |
0.78 |
| chr1_193625744_193625936 | 0.08 |
Mir205hg |
Mir205 host gene |
115666 |
0.05 |
| chr9_119160366_119160634 | 0.08 |
Gm47289 |
predicted gene, 47289 |
1339 |
0.26 |
| chr1_58149355_58149553 | 0.08 |
Gm24548 |
predicted gene, 24548 |
5634 |
0.2 |
| chr6_99085209_99085370 | 0.08 |
Gm49348 |
predicted gene, 49348 |
9232 |
0.24 |
| chr7_27473957_27474137 | 0.08 |
Sertad3 |
SERTA domain containing 3 |
279 |
0.8 |
| chr11_21402382_21402856 | 0.08 |
Ugp2 |
UDP-glucose pyrophosphorylase 2 |
31418 |
0.11 |
| chr3_95643059_95643376 | 0.08 |
E330034L11Rik |
RIKEN cDNA E330034L11 gene |
3913 |
0.12 |
| chr14_75869894_75870089 | 0.08 |
Snora31 |
small nucleolar RNA, H/ACA box 31 |
22086 |
0.13 |
| chr18_65059600_65059753 | 0.08 |
Nedd4l |
neural precursor cell expressed, developmentally down-regulated gene 4-like |
8686 |
0.24 |
| chr19_42611869_42612025 | 0.08 |
Loxl4 |
lysyl oxidase-like 4 |
825 |
0.63 |
| chr9_75033527_75033678 | 0.08 |
Arpp19 |
cAMP-regulated phosphoprotein 19 |
4012 |
0.2 |
| chr15_83452696_83452861 | 0.08 |
Pacsin2 |
protein kinase C and casein kinase substrate in neurons 2 |
11774 |
0.15 |
| chr1_121327051_121327452 | 0.08 |
Insig2 |
insulin induced gene 2 |
427 |
0.67 |
| chr3_101599190_101599434 | 0.08 |
Gm42941 |
predicted gene 42941 |
2182 |
0.28 |
| chr14_51063014_51063572 | 0.07 |
Rnase12 |
ribonuclease, RNase A family, 12 (non-active) |
5632 |
0.09 |
| chr4_120101853_120102132 | 0.07 |
Hivep3 |
human immunodeficiency virus type I enhancer binding protein 3 |
7503 |
0.23 |
| chr19_30158286_30158444 | 0.07 |
Rpl31-ps20 |
ribosomal protein L31, pseudogene 20 |
2255 |
0.3 |
| chr11_115536611_115536915 | 0.07 |
Sumo2 |
small ubiquitin-like modifier 2 |
487 |
0.64 |
| chr11_31718721_31719066 | 0.07 |
Gm38061 |
predicted gene, 38061 |
20245 |
0.21 |
| chr11_117955741_117955916 | 0.07 |
Socs3 |
suppressor of cytokine signaling 3 |
13358 |
0.13 |
| chr5_105793398_105793591 | 0.07 |
Rps15a-ps5 |
ribosomal protein S15A, pseudogene 5 |
4219 |
0.19 |
| chr1_73966372_73966602 | 0.07 |
Tns1 |
tensin 1 |
3444 |
0.29 |
| chr2_62643487_62643902 | 0.07 |
Ifih1 |
interferon induced with helicase C domain 1 |
2459 |
0.28 |
| chr15_62017519_62017928 | 0.07 |
Pvt1 |
Pvt1 oncogene |
20263 |
0.19 |
| chr9_83102256_83102418 | 0.07 |
Gm38398 |
predicted gene, 38398 |
4113 |
0.16 |
| chr16_29315105_29315271 | 0.07 |
Gm15752 |
predicted gene 15752 |
30829 |
0.19 |
| chr9_119154224_119154644 | 0.07 |
Acaa1b |
acetyl-Coenzyme A acyltransferase 1B |
2659 |
0.16 |
| chr6_71204039_71204219 | 0.07 |
Fabp1 |
fatty acid binding protein 1, liver |
4302 |
0.15 |
| chr9_46231796_46232011 | 0.07 |
Apoa1 |
apolipoprotein A-I |
3182 |
0.11 |
| chr1_183377966_183378118 | 0.07 |
Gm37339 |
predicted gene, 37339 |
6966 |
0.13 |
| chr19_47306885_47307217 | 0.07 |
Sh3pxd2a |
SH3 and PX domains 2A |
7700 |
0.17 |
| chr14_59533298_59533481 | 0.07 |
Cab39l |
calcium binding protein 39-like |
54573 |
0.1 |
| chr14_74952353_74952521 | 0.07 |
Lrch1 |
leucine-rich repeats and calponin homology (CH) domain containing 1 |
4561 |
0.21 |
| chr13_84514974_84515153 | 0.07 |
Gm26927 |
predicted gene, 26927 |
174950 |
0.03 |
| chr6_33417499_33417665 | 0.07 |
Gm43168 |
predicted gene 43168 |
15426 |
0.23 |
| chr13_73734744_73734913 | 0.07 |
Slc12a7 |
solute carrier family 12, member 7 |
1734 |
0.31 |
| chr4_7195720_7195887 | 0.07 |
Gm11804 |
predicted gene 11804 |
156289 |
0.04 |
| chr15_10308305_10308606 | 0.07 |
Prlr |
prolactin receptor |
5647 |
0.22 |
| chr11_59428179_59428372 | 0.07 |
Snap47 |
synaptosomal-associated protein, 47 |
19216 |
0.1 |
| chr13_51312028_51312200 | 0.07 |
Gm6056 |
predicted gene 6056 |
26807 |
0.16 |
| chr1_155259994_155260145 | 0.07 |
BC034090 |
cDNA sequence BC034090 |
15625 |
0.15 |
| chr5_117979162_117979338 | 0.07 |
Fbxo21 |
F-box protein 21 |
74 |
0.96 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0070460 | thyroid-stimulating hormone secretion(GO:0070460) |
| 0.0 | 0.1 | GO:1904688 | regulation of cap-independent translational initiation(GO:1903677) positive regulation of cap-independent translational initiation(GO:1903679) regulation of cytoplasmic translational initiation(GO:1904688) positive regulation of cytoplasmic translational initiation(GO:1904690) |
| 0.0 | 0.2 | GO:1902459 | positive regulation of stem cell population maintenance(GO:1902459) |
| 0.0 | 0.1 | GO:1904684 | negative regulation of metalloendopeptidase activity(GO:1904684) negative regulation of metallopeptidase activity(GO:1905049) |
| 0.0 | 0.1 | GO:0070973 | COPI-coated vesicle budding(GO:0035964) protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.1 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.0 | 0.1 | GO:0038108 | negative regulation of appetite by leptin-mediated signaling pathway(GO:0038108) |
| 0.0 | 0.1 | GO:0006553 | lysine metabolic process(GO:0006553) |
| 0.0 | 0.1 | GO:0046499 | S-adenosylmethioninamine metabolic process(GO:0046499) |
| 0.0 | 0.1 | GO:0072675 | osteoclast fusion(GO:0072675) |
| 0.0 | 0.0 | GO:0061144 | alveolar secondary septum development(GO:0061144) |
| 0.0 | 0.1 | GO:0019368 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.1 | GO:0032933 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.0 | 0.0 | GO:0006059 | hexitol metabolic process(GO:0006059) |
| 0.0 | 0.0 | GO:0002414 | immunoglobulin transcytosis in epithelial cells(GO:0002414) |
| 0.0 | 0.3 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.0 | 0.2 | GO:0021692 | cerebellar Purkinje cell layer morphogenesis(GO:0021692) |
| 0.0 | 0.0 | GO:0002086 | diaphragm contraction(GO:0002086) |
| 0.0 | 0.0 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.1 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.1 | GO:0044308 | axonal spine(GO:0044308) |
| 0.0 | 0.1 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.0 | 0.1 | GO:0034464 | BBSome(GO:0034464) |
| 0.0 | 0.1 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.0 | 0.1 | GO:0051718 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) DNA (cytosine-5-)-methyltransferase activity, acting on CpG substrates(GO:0051718) |
| 0.0 | 0.1 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.0 | 0.1 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.0 | 0.1 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.0 | 0.0 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.1 | GO:0000832 | inositol hexakisphosphate kinase activity(GO:0000828) inositol hexakisphosphate 5-kinase activity(GO:0000832) inositol hexakisphosphate 1-kinase activity(GO:0052723) inositol hexakisphosphate 3-kinase activity(GO:0052724) |
| 0.0 | 0.1 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.1 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.0 | 0.1 | GO:0102337 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.0 | GO:0030350 | iron-responsive element binding(GO:0030350) |
| 0.0 | 0.0 | GO:0003945 | N-acetyllactosamine synthase activity(GO:0003945) |
| 0.0 | 0.1 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.1 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.0 | 0.0 | GO:0031711 | bradykinin receptor binding(GO:0031711) |
| 0.0 | 0.0 | GO:0015038 | glutathione disulfide oxidoreductase activity(GO:0015038) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.1 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |