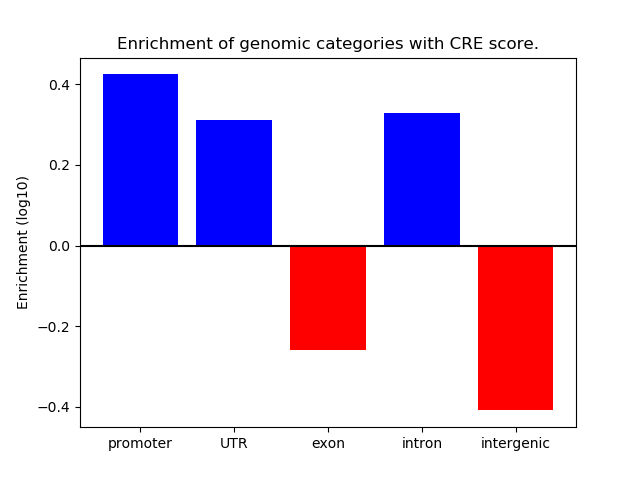
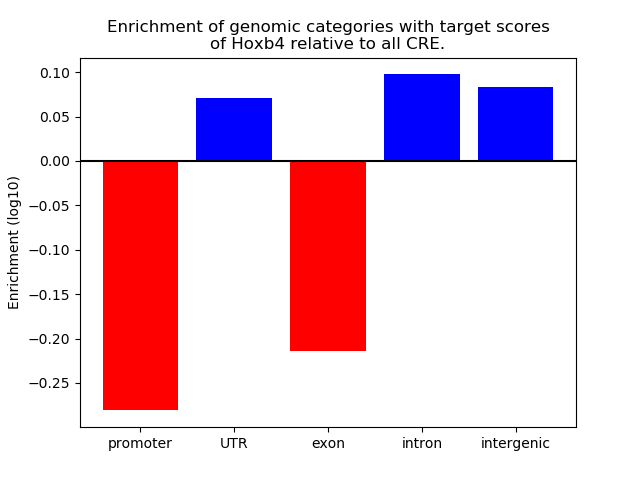
Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
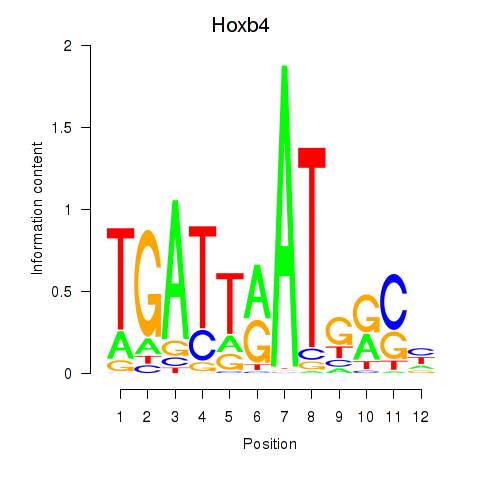
Results for Hoxb4
Z-value: 1.55
Transcription factors associated with Hoxb4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hoxb4
|
ENSMUSG00000038692.7 | homeobox B4 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr11_96318577_96318760 | Hoxb4 | 401 | 0.605874 | 0.53 | 2.8e-01 | Click! |
Activity of the Hoxb4 motif across conditions
Conditions sorted by the z-value of the Hoxb4 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
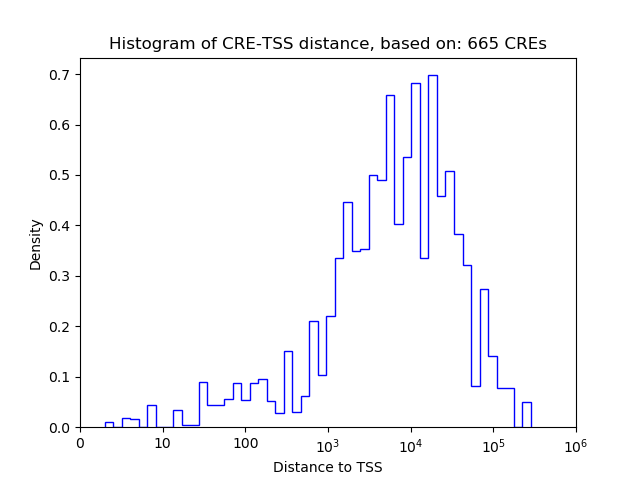
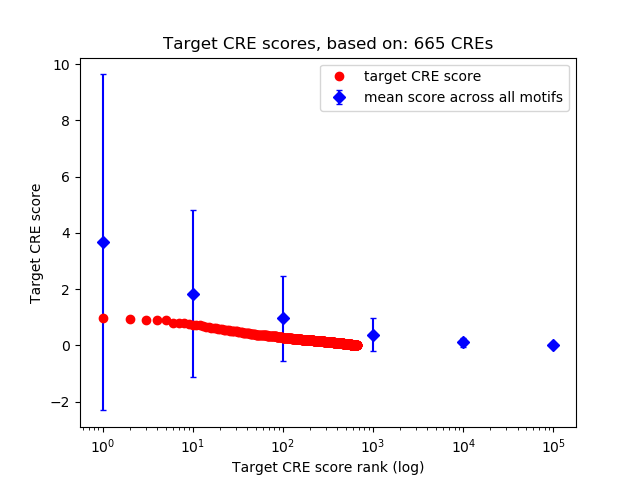
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr7_126260333_126260533 | 0.98 |
Sbk1 |
SH3-binding kinase 1 |
11571 |
0.11 |
| chr6_144895556_144895729 | 0.94 |
Gm22792 |
predicted gene, 22792 |
92010 |
0.07 |
| chr11_16866408_16866907 | 0.92 |
Egfr |
epidermal growth factor receptor |
11493 |
0.2 |
| chr1_162986828_162987029 | 0.91 |
Fmo3 |
flavin containing monooxygenase 3 |
2400 |
0.25 |
| chr7_140767632_140767811 | 0.91 |
Cyp2e1 |
cytochrome P450, family 2, subfamily e, polypeptide 1 |
3240 |
0.12 |
| chr18_62224841_62225012 | 0.81 |
Gm9949 |
predicted gene 9949 |
44800 |
0.14 |
| chr14_74794292_74794475 | 0.81 |
Lrch1 |
leucine-rich repeats and calponin homology (CH) domain containing 1 |
608 |
0.79 |
| chr11_16882054_16882234 | 0.80 |
Egfr |
epidermal growth factor receptor |
3994 |
0.24 |
| chr12_69759378_69759543 | 0.74 |
Mir681 |
microRNA 681 |
4484 |
0.15 |
| chr2_75564827_75565114 | 0.71 |
Gm13655 |
predicted gene 13655 |
68412 |
0.08 |
| chr12_35573403_35573591 | 0.71 |
Gm48237 |
predicted gene, 48237 |
27466 |
0.14 |
| chr13_52150386_52150766 | 0.71 |
Gm48199 |
predicted gene, 48199 |
29835 |
0.18 |
| chr18_31771523_31771682 | 0.70 |
Ammecr1l |
AMME chromosomal region gene 1-like |
9798 |
0.13 |
| chr13_109881168_109881343 | 0.67 |
Pde4d |
phosphodiesterase 4D, cAMP specific |
21847 |
0.23 |
| chr19_21919301_21919474 | 0.66 |
Ldhb-ps |
lactate dehydrogenase B, pseudogene |
17993 |
0.18 |
| chr13_80895235_80895839 | 0.62 |
Arrdc3 |
arrestin domain containing 3 |
5019 |
0.18 |
| chr10_95354659_95354831 | 0.61 |
2310039L15Rik |
RIKEN cDNA 2310039L15 gene |
143 |
0.94 |
| chr4_139875950_139876108 | 0.60 |
Gm1667 |
predicted gene 1667 |
34459 |
0.13 |
| chr6_51700821_51700988 | 0.60 |
Gm38811 |
predicted gene, 38811 |
10177 |
0.25 |
| chr2_72306157_72306335 | 0.60 |
Map3k20 |
mitogen-activated protein kinase kinase kinase 20 |
8345 |
0.2 |
| chr10_60933932_60934087 | 0.57 |
Gm26947 |
predicted gene, 26947 |
2018 |
0.24 |
| chr9_74886235_74886449 | 0.54 |
Onecut1 |
one cut domain, family member 1 |
19858 |
0.14 |
| chr9_13681735_13681905 | 0.53 |
Maml2 |
mastermind like transcriptional coactivator 2 |
19360 |
0.18 |
| chr2_13484483_13484816 | 0.53 |
Cubn |
cubilin (intrinsic factor-cobalamin receptor) |
7164 |
0.26 |
| chr11_79753212_79753391 | 0.53 |
Mir365-2 |
microRNA 365-2 |
26901 |
0.12 |
| chr9_83254483_83254659 | 0.52 |
Gm27216 |
predicted gene 27216 |
31 |
0.98 |
| chr1_71938584_71938854 | 0.51 |
Gm28818 |
predicted gene 28818 |
21594 |
0.17 |
| chr10_99314465_99314820 | 0.51 |
B530045E10Rik |
RIKEN cDNA B530045E10 gene |
15476 |
0.14 |
| chr4_111200292_111200482 | 0.51 |
Agbl4 |
ATP/GTP binding protein-like 4 |
81476 |
0.1 |
| chr8_40858142_40858306 | 0.50 |
Gm5345 |
predicted gene 5345 |
3197 |
0.19 |
| chr6_33958384_33958553 | 0.49 |
Gm13853 |
predicted gene 13853 |
33943 |
0.19 |
| chr1_181307865_181308016 | 0.48 |
Gm8146 |
predicted gene 8146 |
25894 |
0.14 |
| chr7_77888814_77888965 | 0.48 |
Gm23239 |
predicted gene, 23239 |
21431 |
0.28 |
| chr3_116969952_116970108 | 0.46 |
Palmd |
palmdelphin |
1043 |
0.39 |
| chr5_45534505_45534782 | 0.46 |
Fam184b |
family with sequence similarity 184, member B |
1899 |
0.22 |
| chr15_30285783_30285996 | 0.45 |
Ctnnd2 |
catenin (cadherin associated protein), delta 2 |
112751 |
0.06 |
| chr13_64388507_64388698 | 0.45 |
Ctsl |
cathepsin L |
17712 |
0.1 |
| chr5_145800258_145800417 | 0.45 |
Cyp3a44 |
cytochrome P450, family 3, subfamily a, polypeptide 44 |
5537 |
0.18 |
| chr4_7195720_7195887 | 0.44 |
Gm11804 |
predicted gene 11804 |
156289 |
0.04 |
| chr18_55107121_55107272 | 0.43 |
AC163347.1 |
novel transcript |
10836 |
0.17 |
| chr2_146099387_146099651 | 0.42 |
Cfap61 |
cilia and flagella associated protein 61 |
52268 |
0.15 |
| chr12_114053283_114053753 | 0.42 |
Ighv3-2 |
immunoglobulin heavy variable 3-2 |
19285 |
0.13 |
| chr15_77153726_77153986 | 0.42 |
Rbfox2 |
RNA binding protein, fox-1 homolog (C. elegans) 2 |
44 |
0.97 |
| chr1_43066043_43066369 | 0.41 |
Tgfbrap1 |
transforming growth factor, beta receptor associated protein 1 |
5273 |
0.14 |
| chr6_6279071_6279331 | 0.41 |
Gm20619 |
predicted gene 20619 |
20220 |
0.18 |
| chr12_15816769_15817061 | 0.40 |
Trib2 |
tribbles pseudokinase 2 |
7 |
0.56 |
| chr9_110676644_110676946 | 0.40 |
Gm35715 |
predicted gene, 35715 |
1841 |
0.21 |
| chr15_10713141_10713319 | 0.39 |
Rai14 |
retinoic acid induced 14 |
310 |
0.87 |
| chr1_9967919_9968070 | 0.39 |
Tcf24 |
transcription factor 24 |
62 |
0.9 |
| chr2_92380520_92380699 | 0.38 |
1700029I15Rik |
RIKEN cDNA 1700029I15 gene |
1929 |
0.18 |
| chr11_101669101_101669273 | 0.38 |
Arl4d |
ADP-ribosylation factor-like 4D |
3646 |
0.12 |
| chr10_28524334_28524485 | 0.38 |
Ptprk |
protein tyrosine phosphatase, receptor type, K |
35742 |
0.22 |
| chr2_78049569_78049722 | 0.37 |
4930440I19Rik |
RIKEN cDNA 4930440I19 gene |
1532 |
0.51 |
| chr2_163670938_163671176 | 0.37 |
Pkig |
protein kinase inhibitor, gamma |
9558 |
0.13 |
| chr6_116650512_116650679 | 0.37 |
Depp1 |
DEPP1 autophagy regulator |
14 |
0.95 |
| chr14_57107497_57107648 | 0.37 |
Gjb2 |
gap junction protein, beta 2 |
2870 |
0.2 |
| chr10_95942046_95942222 | 0.37 |
Eea1 |
early endosome antigen 1 |
1377 |
0.41 |
| chr13_101866951_101867131 | 0.36 |
Gm47007 |
predicted gene, 47007 |
11550 |
0.2 |
| chr9_116091537_116091709 | 0.36 |
Gm9385 |
predicted pseudogene 9385 |
50559 |
0.13 |
| chr11_6021664_6021840 | 0.35 |
Camk2b |
calcium/calmodulin-dependent protein kinase II, beta |
21304 |
0.15 |
| chr8_35920405_35920556 | 0.35 |
5430403N17Rik |
RIKEN cDNA 5430403N17 gene |
37523 |
0.14 |
| chr18_74805142_74805295 | 0.35 |
Acaa2 |
acetyl-Coenzyme A acyltransferase 2 (mitochondrial 3-oxoacyl-Coenzyme A thiolase) |
11835 |
0.15 |
| chr2_26477777_26477967 | 0.35 |
Notch1 |
notch 1 |
7374 |
0.1 |
| chr19_43971247_43971535 | 0.35 |
Cpn1 |
carboxypeptidase N, polypeptide 1 |
2698 |
0.21 |
| chr4_97960846_97961009 | 0.35 |
Nfia |
nuclear factor I/A |
49894 |
0.17 |
| chr14_18236888_18237050 | 0.35 |
Nr1d2 |
nuclear receptor subfamily 1, group D, member 2 |
2042 |
0.24 |
| chr10_127632738_127633305 | 0.35 |
Gm48815 |
predicted gene, 48815 |
3366 |
0.11 |
| chr12_113987570_113987864 | 0.35 |
Gm37961 |
predicted gene, 37961 |
18865 |
0.14 |
| chr6_95684732_95684897 | 0.35 |
Suclg2 |
succinate-Coenzyme A ligase, GDP-forming, beta subunit |
8068 |
0.32 |
| chrX_166678508_166678678 | 0.35 |
Gm1720 |
predicted gene 1720 |
16568 |
0.21 |
| chr9_94692147_94692428 | 0.35 |
Gm16262 |
predicted gene 16262 |
10032 |
0.17 |
| chr3_57435613_57435790 | 0.34 |
Tm4sf4 |
transmembrane 4 superfamily member 4 |
10387 |
0.22 |
| chr16_5662022_5662331 | 0.34 |
Rbfox1 |
RNA binding protein, fox-1 homolog (C. elegans) 1 |
223179 |
0.02 |
| chr19_42631725_42631876 | 0.34 |
Loxl4 |
lysyl oxidase-like 4 |
18987 |
0.18 |
| chr6_29538799_29539193 | 0.34 |
Irf5 |
interferon regulatory factor 5 |
2150 |
0.2 |
| chr16_36545463_36545634 | 0.33 |
Casr |
calcium-sensing receptor |
16058 |
0.11 |
| chr2_169390357_169390514 | 0.33 |
Gm14250 |
predicted gene 14250 |
1354 |
0.49 |
| chr3_101504490_101504690 | 0.33 |
Atp1a1 |
ATPase, Na+/K+ transporting, alpha 1 polypeptide |
72970 |
0.07 |
| chr19_47886740_47886936 | 0.32 |
Gsto2 |
glutathione S-transferase omega 2 |
16576 |
0.13 |
| chr2_160702528_160702695 | 0.32 |
Top1 |
topoisomerase (DNA) I |
468 |
0.81 |
| chr15_59055272_59055945 | 0.32 |
Mtss1 |
MTSS I-BAR domain containing 1 |
144 |
0.97 |
| chr6_101102390_101102554 | 0.32 |
Gm43942 |
predicted gene, 43942 |
41820 |
0.12 |
| chr17_84082154_84082324 | 0.31 |
4933433H22Rik |
RIKEN cDNA 4933433H22 gene |
3579 |
0.19 |
| chr8_88863011_88863356 | 0.31 |
Gm45504 |
predicted gene 45504 |
109 |
0.98 |
| chr9_69442354_69442630 | 0.31 |
Gm18587 |
predicted gene, 18587 |
2471 |
0.17 |
| chr17_31295551_31296040 | 0.31 |
Slc37a1 |
solute carrier family 37 (glycerol-3-phosphate transporter), member 1 |
31 |
0.94 |
| chr14_118635288_118635751 | 0.31 |
Abcc4 |
ATP-binding cassette, sub-family C (CFTR/MRP), member 4 |
41412 |
0.12 |
| chr3_107943742_107943913 | 0.31 |
Gstm6 |
glutathione S-transferase, mu 6 |
78 |
0.92 |
| chr19_32190045_32190211 | 0.31 |
Sgms1 |
sphingomyelin synthase 1 |
6300 |
0.24 |
| chr6_37537920_37538092 | 0.30 |
Akr1d1 |
aldo-keto reductase family 1, member D1 |
1675 |
0.4 |
| chr11_49247308_49247488 | 0.30 |
Mgat1 |
mannoside acetylglucosaminyltransferase 1 |
79 |
0.95 |
| chr10_51453986_51454193 | 0.29 |
Gm46189 |
predicted gene, 46189 |
1891 |
0.26 |
| chr9_58105721_58106037 | 0.29 |
Ccdc33 |
coiled-coil domain containing 33 |
3582 |
0.17 |
| chr10_110927526_110927725 | 0.29 |
Csrp2 |
cysteine and glycine-rich protein 2 |
3576 |
0.19 |
| chr12_77045836_77046012 | 0.28 |
Gm35189 |
predicted gene, 35189 |
3723 |
0.26 |
| chr6_148199306_148199711 | 0.28 |
Ergic2 |
ERGIC and golgi 2 |
2980 |
0.22 |
| chr4_88546855_88547012 | 0.28 |
Ifna15 |
interferon alpha 15 |
11312 |
0.1 |
| chr13_9445564_9445785 | 0.28 |
Gm48871 |
predicted gene, 48871 |
226 |
0.93 |
| chr17_78379801_78380005 | 0.28 |
Fez2 |
fasciculation and elongation protein zeta 2 (zygin II) |
21527 |
0.15 |
| chr6_142422023_142422193 | 0.28 |
Gys2 |
glycogen synthase 2 |
1511 |
0.33 |
| chr10_68868968_68869160 | 0.28 |
1700048P04Rik |
RIKEN cDNA 1700048P04 gene |
35436 |
0.21 |
| chr13_34242299_34242490 | 0.27 |
Slc22a23 |
solute carrier family 22, member 23 |
48863 |
0.11 |
| chr11_61092676_61092849 | 0.27 |
Kcnj12 |
potassium inwardly-rectifying channel, subfamily J, member 12 |
26958 |
0.13 |
| chr14_27102040_27102293 | 0.27 |
Arhgef3 |
Rho guanine nucleotide exchange factor (GEF) 3 |
12733 |
0.19 |
| chr18_80258103_80258267 | 0.27 |
Slc66a2 |
solute carrier family 66 member 2 |
1867 |
0.22 |
| chr4_40823217_40823652 | 0.26 |
Mir5123 |
microRNA 5123 |
26704 |
0.09 |
| chr2_60878102_60878265 | 0.26 |
Rbms1 |
RNA binding motif, single stranded interacting protein 1 |
3255 |
0.35 |
| chr4_34551560_34551735 | 0.26 |
Akirin2 |
akirin 2 |
696 |
0.68 |
| chr16_25877460_25877655 | 0.26 |
Trp63 |
transformation related protein 63 |
75641 |
0.1 |
| chr17_48039561_48039744 | 0.26 |
1700122O11Rik |
RIKEN cDNA 1700122O11 gene |
1359 |
0.33 |
| chr8_22414456_22414613 | 0.26 |
Mrps31 |
mitochondrial ribosomal protein S31 |
3136 |
0.13 |
| chr10_66631116_66631267 | 0.26 |
Gm28139 |
predicted gene 28139 |
9727 |
0.22 |
| chr12_84043890_84044041 | 0.25 |
Acot4 |
acyl-CoA thioesterase 4 |
1416 |
0.21 |
| chr7_89423273_89423442 | 0.25 |
AI314278 |
expressed sequence AI314278 |
2949 |
0.2 |
| chr4_3696728_3696981 | 0.25 |
Lyn |
LYN proto-oncogene, Src family tyrosine kinase |
18724 |
0.14 |
| chr9_63613110_63613389 | 0.25 |
Aagab |
alpha- and gamma-adaptin binding protein |
3739 |
0.23 |
| chr8_91657490_91657652 | 0.25 |
Gm45295 |
predicted gene 45295 |
14326 |
0.12 |
| chr9_102726405_102726576 | 0.25 |
Amotl2 |
angiomotin-like 2 |
1841 |
0.24 |
| chrX_60362413_60362564 | 0.25 |
Mir505 |
microRNA 505 |
32004 |
0.17 |
| chr8_25769504_25769685 | 0.25 |
Bag4 |
BCL2-associated athanogene 4 |
7988 |
0.1 |
| chr12_54205848_54206011 | 0.25 |
Egln3 |
egl-9 family hypoxia-inducible factor 3 |
2069 |
0.28 |
| chr6_142668760_142668977 | 0.24 |
Abcc9 |
ATP-binding cassette, sub-family C (CFTR/MRP), member 9 |
33350 |
0.16 |
| chr15_55317395_55317627 | 0.24 |
Col14a1 |
collagen, type XIV, alpha 1 |
9592 |
0.24 |
| chr1_10993153_10993537 | 0.24 |
Prex2 |
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 2 |
120 |
0.98 |
| chr18_84084702_84084863 | 0.24 |
Tshz1 |
teashirt zinc finger family member 1 |
293 |
0.88 |
| chr11_106486200_106486366 | 0.24 |
Ern1 |
endoplasmic reticulum (ER) to nucleus signalling 1 |
1513 |
0.34 |
| chr14_105253933_105254092 | 0.24 |
Gm22290 |
predicted gene, 22290 |
4002 |
0.17 |
| chr2_53063355_53063523 | 0.24 |
Prpf40a |
pre-mRNA processing factor 40A |
81870 |
0.1 |
| chr19_37648111_37648271 | 0.24 |
Cyp26c1 |
cytochrome P450, family 26, subfamily c, polypeptide 1 |
37390 |
0.14 |
| chr8_114117394_114117594 | 0.24 |
Nudt7 |
nudix (nucleoside diphosphate linked moiety X)-type motif 7 |
16063 |
0.27 |
| chr1_31094891_31095067 | 0.24 |
4931428L18Rik |
RIKEN cDNA 4931428L18 gene |
1475 |
0.35 |
| chr6_115884148_115884359 | 0.23 |
Ift122 |
intraflagellar transport 122 |
1655 |
0.26 |
| chr10_125327017_125327436 | 0.23 |
Slc16a7 |
solute carrier family 16 (monocarboxylic acid transporters), member 7 |
1309 |
0.51 |
| chr7_137697020_137697186 | 0.23 |
Gm45682 |
predicted gene 45682 |
37163 |
0.18 |
| chr1_191254506_191254830 | 0.23 |
Gm37074 |
predicted gene, 37074 |
5244 |
0.15 |
| chr19_12719350_12719546 | 0.23 |
Gm15962 |
predicted gene 15962 |
1755 |
0.22 |
| chr7_71370855_71371052 | 0.23 |
Gm29328 |
predicted gene 29328 |
620 |
0.73 |
| chr3_52338842_52339028 | 0.22 |
Gm38034 |
predicted gene, 38034 |
56694 |
0.1 |
| chr7_81250438_81250962 | 0.22 |
Gm7180 |
predicted pseudogene 7180 |
27826 |
0.13 |
| chr2_169634592_169634925 | 0.22 |
Tshz2 |
teashirt zinc finger family member 2 |
1082 |
0.55 |
| chr17_86470447_86470748 | 0.22 |
Prkce |
protein kinase C, epsilon |
19965 |
0.22 |
| chr3_101430547_101430727 | 0.22 |
Gm42538 |
predicted gene 42538 |
3701 |
0.2 |
| chr7_25987036_25987409 | 0.22 |
Gm45225 |
predicted gene 45225 |
5720 |
0.14 |
| chr4_124704877_124705041 | 0.22 |
Fhl3 |
four and a half LIM domains 3 |
4239 |
0.1 |
| chr4_60591701_60591858 | 0.22 |
Mup-ps5 |
major urinary protein, pseudogene 5 |
5818 |
0.16 |
| chr11_98784049_98784205 | 0.22 |
Nr1d1 |
nuclear receptor subfamily 1, group D, member 1 |
8794 |
0.1 |
| chr12_3867079_3867237 | 0.22 |
Dnmt3a |
DNA methyltransferase 3A |
967 |
0.52 |
| chr6_67102942_67103110 | 0.22 |
A430010J10Rik |
RIKEN cDNA A430010J10 gene |
12647 |
0.15 |
| chr11_16842718_16842956 | 0.22 |
Egfros |
epidermal growth factor receptor, opposite strand |
12135 |
0.2 |
| chr4_118123096_118123255 | 0.22 |
St3gal3 |
ST3 beta-galactoside alpha-2,3-sialyltransferase 3 |
11707 |
0.16 |
| chr6_145855503_145855668 | 0.22 |
Gm43909 |
predicted gene, 43909 |
7712 |
0.17 |
| chr16_34322645_34322806 | 0.22 |
Kalrn |
kalirin, RhoGEF kinase |
39074 |
0.22 |
| chr11_23101055_23101224 | 0.21 |
Gm12059 |
predicted gene 12059 |
33141 |
0.12 |
| chr12_118792020_118792190 | 0.21 |
Gm6171 |
predicted gene 6171 |
1727 |
0.38 |
| chr13_81368031_81368195 | 0.21 |
Adgrv1 |
adhesion G protein-coupled receptor V1 |
16487 |
0.26 |
| chr13_4143590_4143758 | 0.21 |
Akr1c18 |
aldo-keto reductase family 1, member C18 |
6954 |
0.13 |
| chr11_79305709_79305885 | 0.21 |
Gm9964 |
predicted gene 9964 |
8891 |
0.16 |
| chr11_106268502_106268656 | 0.21 |
Smarcd2 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 2 |
780 |
0.42 |
| chr6_27967716_27967883 | 0.21 |
Mir592 |
microRNA 592 |
31049 |
0.23 |
| chr14_69729701_69729901 | 0.21 |
Chmp7 |
charged multivesicular body protein 7 |
1074 |
0.4 |
| chr13_115090695_115090877 | 0.21 |
Pelo |
pelota mRNA surveillance and ribosome rescue factor |
600 |
0.58 |
| chr1_138510594_138510753 | 0.20 |
Gm28501 |
predicted gene 28501 |
4942 |
0.23 |
| chr9_78149872_78150023 | 0.20 |
Cilk1 |
ciliogenesis associated kinase 1 |
5376 |
0.13 |
| chr11_22893094_22893344 | 0.20 |
Gm24917 |
predicted gene, 24917 |
24318 |
0.11 |
| chr2_102706025_102706412 | 0.20 |
Slc1a2 |
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
138 |
0.97 |
| chr12_79642030_79642203 | 0.20 |
9430078K24Rik |
RIKEN cDNA 9430078K24 gene |
282617 |
0.01 |
| chr4_60149368_60149527 | 0.20 |
Mup2 |
major urinary protein 2 |
4842 |
0.19 |
| chr3_97631636_97631799 | 0.20 |
Fmo5 |
flavin containing monooxygenase 5 |
2834 |
0.19 |
| chr4_60299101_60299261 | 0.20 |
Mup-ps3 |
major urinary protein, pseudogene 3 |
61836 |
0.09 |
| chr5_63936466_63936784 | 0.20 |
Rell1 |
RELT-like 1 |
5711 |
0.18 |
| chr17_35242702_35242853 | 0.20 |
Gm22589 |
predicted gene, 22589 |
99 |
0.48 |
| chr11_16894642_16894943 | 0.20 |
Egfr |
epidermal growth factor receptor |
10393 |
0.2 |
| chr5_130024306_130024463 | 0.20 |
Asl |
argininosuccinate lyase |
3 |
0.96 |
| chr4_60231470_60231627 | 0.20 |
Mup-ps3 |
major urinary protein, pseudogene 3 |
5797 |
0.2 |
| chr2_24839386_24839554 | 0.20 |
Ehmt1 |
euchromatic histone methyltransferase 1 |
121 |
0.95 |
| chr12_73408646_73408959 | 0.19 |
D830013O20Rik |
RIKEN cDNA D830013O20 gene |
755 |
0.63 |
| chr14_11569795_11569971 | 0.19 |
Ptprg |
protein tyrosine phosphatase, receptor type, G |
16302 |
0.21 |
| chr13_104035617_104035778 | 0.19 |
Nln |
neurolysin (metallopeptidase M3 family) |
870 |
0.63 |
| chr3_95901553_95901705 | 0.19 |
Car14 |
carbonic anhydrase 14 |
2119 |
0.14 |
| chr4_144957116_144957269 | 0.19 |
Gm38074 |
predicted gene, 38074 |
1656 |
0.38 |
| chr16_62881474_62881642 | 0.19 |
Pros1 |
protein S (alpha) |
16591 |
0.19 |
| chr1_21228840_21229005 | 0.19 |
Gm38224 |
predicted gene, 38224 |
5800 |
0.13 |
| chr8_10439826_10440109 | 0.19 |
Myo16 |
myosin XVI |
167395 |
0.03 |
| chr7_99170677_99170840 | 0.19 |
Dgat2 |
diacylglycerol O-acyltransferase 2 |
652 |
0.61 |
| chr12_76431059_76431246 | 0.19 |
Ppp1r36 |
protein phosphatase 1, regulatory subunit 36 |
4561 |
0.12 |
| chr4_61844883_61845043 | 0.19 |
Mup5 |
major urinary protein 5 |
9730 |
0.12 |
| chr4_148616208_148616450 | 0.19 |
Tardbp |
TAR DNA binding protein |
191 |
0.9 |
| chr19_34745215_34745404 | 0.19 |
Slc16a12 |
solute carrier family 16 (monocarboxylic acid transporters), member 12 |
1980 |
0.23 |
| chr4_61312938_61313093 | 0.19 |
Mup-ps11 |
major urinary protein, pseudogene 11 |
5715 |
0.2 |
| chr16_70352273_70352442 | 0.19 |
Gbe1 |
glucan (1,4-alpha-), branching enzyme 1 |
38223 |
0.19 |
| chr16_13159523_13159853 | 0.19 |
Ercc4 |
excision repair cross-complementing rodent repair deficiency, complementation group 4 |
27297 |
0.22 |
| chr18_24054985_24055136 | 0.19 |
Gm18085 |
predicted gene, 18085 |
26864 |
0.14 |
| chr9_57723131_57723308 | 0.19 |
Edc3 |
enhancer of mRNA decapping 3 |
14652 |
0.12 |
| chr8_128350486_128350652 | 0.19 |
Nrp1 |
neuropilin 1 |
8035 |
0.23 |
| chr9_99353176_99353333 | 0.19 |
Esyt3 |
extended synaptotagmin-like protein 3 |
5264 |
0.13 |
| chr13_99667017_99667216 | 0.18 |
1700012J22Rik |
RIKEN cDNA 1700012J22 gene |
25142 |
0.18 |
| chr1_176982464_176982654 | 0.18 |
Gm37519 |
predicted gene, 37519 |
8323 |
0.15 |
| chr5_31199891_31200213 | 0.18 |
Zfp513 |
zinc finger protein 513 |
234 |
0.8 |
| chr2_64093669_64093820 | 0.18 |
Fign |
fidgetin |
4244 |
0.37 |
| chr17_84269883_84270034 | 0.18 |
Gm24492 |
predicted gene, 24492 |
17789 |
0.18 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.1 | 0.4 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.1 | 0.3 | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.1 | 0.2 | GO:0045074 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 0.1 | 0.2 | GO:0019676 | ammonia assimilation cycle(GO:0019676) |
| 0.1 | 0.1 | GO:1902809 | regulation of skeletal muscle fiber differentiation(GO:1902809) |
| 0.1 | 0.3 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.1 | 0.2 | GO:1901078 | negative regulation of relaxation of muscle(GO:1901078) |
| 0.0 | 0.2 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.0 | 0.1 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.0 | 0.1 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) |
| 0.0 | 0.2 | GO:0060023 | soft palate development(GO:0060023) |
| 0.0 | 0.1 | GO:0045869 | negative regulation of single stranded viral RNA replication via double stranded DNA intermediate(GO:0045869) |
| 0.0 | 0.1 | GO:1901475 | pyruvate transmembrane transport(GO:1901475) |
| 0.0 | 0.1 | GO:0002086 | diaphragm contraction(GO:0002086) |
| 0.0 | 0.1 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.0 | 0.3 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.0 | 0.3 | GO:0060613 | fat pad development(GO:0060613) |
| 0.0 | 0.3 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 0.2 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.0 | 0.1 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.0 | 0.1 | GO:0046499 | S-adenosylmethioninamine metabolic process(GO:0046499) |
| 0.0 | 0.1 | GO:0032789 | saturated monocarboxylic acid metabolic process(GO:0032788) unsaturated monocarboxylic acid metabolic process(GO:0032789) |
| 0.0 | 0.1 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.0 | 0.3 | GO:0006047 | UDP-N-acetylglucosamine metabolic process(GO:0006047) |
| 0.0 | 0.1 | GO:2000002 | negative regulation of DNA damage checkpoint(GO:2000002) |
| 0.0 | 0.1 | GO:2000645 | negative regulation of receptor catabolic process(GO:2000645) |
| 0.0 | 0.1 | GO:0003241 | growth involved in heart morphogenesis(GO:0003241) |
| 0.0 | 0.1 | GO:0015819 | lysine transport(GO:0015819) |
| 0.0 | 0.1 | GO:0032914 | positive regulation of transforming growth factor beta1 production(GO:0032914) |
| 0.0 | 0.0 | GO:0006549 | isoleucine metabolic process(GO:0006549) |
| 0.0 | 0.1 | GO:0016103 | diterpenoid catabolic process(GO:0016103) retinoic acid catabolic process(GO:0034653) |
| 0.0 | 0.1 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 0.1 | GO:0006104 | succinyl-CoA metabolic process(GO:0006104) |
| 0.0 | 0.4 | GO:0017144 | drug metabolic process(GO:0017144) |
| 0.0 | 0.1 | GO:1902109 | negative regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902109) |
| 0.0 | 0.1 | GO:1903553 | positive regulation of extracellular exosome assembly(GO:1903553) |
| 0.0 | 0.1 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.0 | GO:2000587 | regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000586) negative regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000587) |
| 0.0 | 0.0 | GO:0035609 | C-terminal protein deglutamylation(GO:0035609) |
| 0.0 | 0.0 | GO:0051562 | negative regulation of mitochondrial calcium ion concentration(GO:0051562) |
| 0.0 | 0.0 | GO:1900825 | regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900825) |
| 0.0 | 0.1 | GO:0060830 | ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) |
| 0.0 | 0.0 | GO:0006578 | amino-acid betaine biosynthetic process(GO:0006578) |
| 0.0 | 0.0 | GO:0017187 | peptidyl-glutamic acid carboxylation(GO:0017187) |
| 0.0 | 0.0 | GO:0021886 | cerebral cortex tangential migration using cell-axon interactions(GO:0021824) gonadotrophin-releasing hormone neuronal migration to the hypothalamus(GO:0021828) hypothalamic tangential migration using cell-axon interactions(GO:0021856) hypothalamus gonadotrophin-releasing hormone neuron differentiation(GO:0021886) hypothalamus gonadotrophin-releasing hormone neuron development(GO:0021888) sensory neuron axon guidance(GO:0097374) facioacoustic ganglion development(GO:1903375) |
| 0.0 | 0.1 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.1 | GO:0006987 | activation of signaling protein activity involved in unfolded protein response(GO:0006987) |
| 0.0 | 0.0 | GO:0006296 | nucleotide-excision repair, DNA incision, 5'-to lesion(GO:0006296) |
| 0.0 | 0.1 | GO:0010815 | bradykinin catabolic process(GO:0010815) |
| 0.0 | 0.0 | GO:0061368 | behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 0.0 | 0.1 | GO:0016127 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.0 | 0.1 | GO:0009597 | detection of virus(GO:0009597) |
| 0.0 | 0.1 | GO:0090427 | activation of meiosis(GO:0090427) |
| 0.0 | 0.1 | GO:0002513 | tolerance induction to self antigen(GO:0002513) |
| 0.0 | 0.1 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.0 | 0.1 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) |
| 0.0 | 0.1 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.0 | 0.0 | GO:0035771 | interleukin-4-mediated signaling pathway(GO:0035771) |
| 0.0 | 0.0 | GO:0021699 | cerebellar cortex maturation(GO:0021699) |
| 0.0 | 0.1 | GO:0016264 | gap junction assembly(GO:0016264) |
| 0.0 | 0.0 | GO:0016115 | terpenoid catabolic process(GO:0016115) |
| 0.0 | 0.0 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.0 | 0.0 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.0 | 0.0 | GO:0021943 | formation of radial glial scaffolds(GO:0021943) |
| 0.0 | 0.0 | GO:0006573 | valine metabolic process(GO:0006573) |
| 0.0 | 0.0 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.0 | 0.0 | GO:0071926 | cannabinoid signaling pathway(GO:0038171) endocannabinoid signaling pathway(GO:0071926) |
| 0.0 | 0.1 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.0 | 0.0 | GO:0046864 | retinol transport(GO:0034633) isoprenoid transport(GO:0046864) terpenoid transport(GO:0046865) |
| 0.0 | 0.0 | GO:0070889 | platelet alpha granule organization(GO:0070889) |
| 0.0 | 0.0 | GO:0008050 | female courtship behavior(GO:0008050) |
| 0.0 | 0.1 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.1 | 0.3 | GO:0001651 | dense fibrillar component(GO:0001651) |
| 0.0 | 0.1 | GO:0044308 | axonal spine(GO:0044308) |
| 0.0 | 0.1 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.0 | 0.1 | GO:1990597 | AIP1-IRE1 complex(GO:1990597) |
| 0.0 | 0.1 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.1 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.0 | 0.1 | GO:0030062 | mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 0.0 | 0.2 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.0 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.0 | 0.1 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 0.1 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.0 | 0.0 | GO:0097441 | basilar dendrite(GO:0097441) |
| 0.0 | 0.0 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.0 | 0.1 | GO:0002177 | manchette(GO:0002177) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.1 | 0.3 | GO:0015526 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.1 | 0.2 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.1 | 0.3 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.1 | 0.2 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.0 | 0.1 | GO:0050252 | retinol O-fatty-acyltransferase activity(GO:0050252) |
| 0.0 | 0.1 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.0 | 0.3 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.1 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) DNA (cytosine-5-)-methyltransferase activity, acting on CpG substrates(GO:0051718) |
| 0.0 | 0.1 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) |
| 0.0 | 0.1 | GO:0030621 | U4 snRNA binding(GO:0030621) |
| 0.0 | 0.1 | GO:0035671 | enone reductase activity(GO:0035671) |
| 0.0 | 0.1 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.0 | 0.1 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 0.1 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.0 | 0.1 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.1 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.0 | 0.3 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.4 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.2 | GO:0005313 | L-glutamate transmembrane transporter activity(GO:0005313) |
| 0.0 | 0.1 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.0 | 0.1 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.0 | 0.1 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.1 | GO:0050649 | testosterone 6-beta-hydroxylase activity(GO:0050649) |
| 0.0 | 0.5 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.2 | GO:0055106 | ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.0 | 0.0 | GO:0004487 | methenyltetrahydrofolate cyclohydrolase activity(GO:0004477) methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
| 0.0 | 0.1 | GO:0034802 | 3-(3-hydroxyphenyl)propionate hydroxylase activity(GO:0008688) 4-chlorobenzaldehyde oxidase activity(GO:0018471) 3,5-xylenol methylhydroxylase activity(GO:0018630) phenylacetate hydroxylase activity(GO:0018631) 4-nitrophenol 4-monooxygenase activity(GO:0018632) dimethyl sulfide monooxygenase activity(GO:0018633) alpha-pinene monooxygenase [NADH] activity(GO:0018634) 1-hydroxy-2-naphthoate hydroxylase activity(GO:0018637) toluene 4-monooxygenase activity(GO:0018638) xylene monooxygenase activity(GO:0018639) dibenzothiophene monooxygenase activity(GO:0018640) 6-hydroxy-3-methyl-2-oxo-1,2-dihydroquinoline 6-monooxygenase activity(GO:0018641) chlorophenol 4-monooxygenase activity(GO:0018642) carbon disulfide oxygenase activity(GO:0018643) toluene 2-monooxygenase activity(GO:0018644) 1-hydroxy-2-oxolimonene 1,2-monooxygenase activity(GO:0018646) phenanthrene 1,2-monooxygenase activity(GO:0018647) tetrahydrofuran hydroxylase activity(GO:0018649) styrene monooxygenase activity(GO:0018650) toluene-4-sulfonate monooxygenase activity(GO:0018651) toluene-sulfonate methyl-monooxygenase activity(GO:0018652) 3-methyl-2-oxo-1,2-dihydroquinoline 6-monooxygenase activity(GO:0018653) 2-hydroxy-phenylacetate hydroxylase activity(GO:0018654) 2-oxo-delta3-4,5,5-trimethylcyclopentenylacetyl-CoA 1,2-monooxygenase activity(GO:0018655) phenanthrene 3,4-monooxygenase activity(GO:0018656) toluene 3-monooxygenase activity(GO:0018657) 4-hydroxyphenylacetate,NADH:oxygen oxidoreductase (3-hydroxylating) activity(GO:0018660) limonene monooxygenase activity(GO:0019113) 2-methylnaphthalene hydroxylase activity(GO:0034526) 1-methylnaphthalene hydroxylase activity(GO:0034534) bisphenol A hydroxylase A activity(GO:0034560) salicylate 5-hydroxylase activity(GO:0034785) isobutylamine N-hydroxylase activity(GO:0034791) branched-chain dodecylbenzene sulfonate monooxygenase activity(GO:0034802) 3-HSA hydroxylase activity(GO:0034819) 4-hydroxypyridine-3-hydroxylase activity(GO:0034894) 2-octaprenyl-3-methyl-6-methoxy-1,4-benzoquinol hydroxylase activity(GO:0043719) 6-hydroxynicotinate 3-monooxygenase activity(GO:0043731) thalianol hydroxylase activity(GO:0080014) |
| 0.0 | 0.1 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.0 | GO:0003941 | L-serine ammonia-lyase activity(GO:0003941) |
| 0.0 | 0.1 | GO:1990239 | steroid hormone binding(GO:1990239) |
| 0.0 | 0.0 | GO:0051032 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.0 | 0.0 | GO:0043125 | ErbB-3 class receptor binding(GO:0043125) |
| 0.0 | 0.0 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 0.0 | 0.0 | GO:0030984 | kininogen binding(GO:0030984) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.0 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.0 | 0.5 | REACTOME SHC1 EVENTS IN EGFR SIGNALING | Genes involved in SHC1 events in EGFR signaling |
| 0.0 | 0.1 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.1 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |