Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
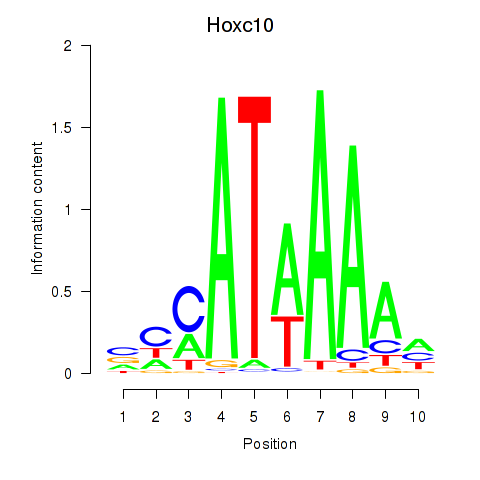
Results for Hoxc10
Z-value: 0.91
Transcription factors associated with Hoxc10
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hoxc10
|
ENSMUSG00000022484.7 | homeobox C10 |
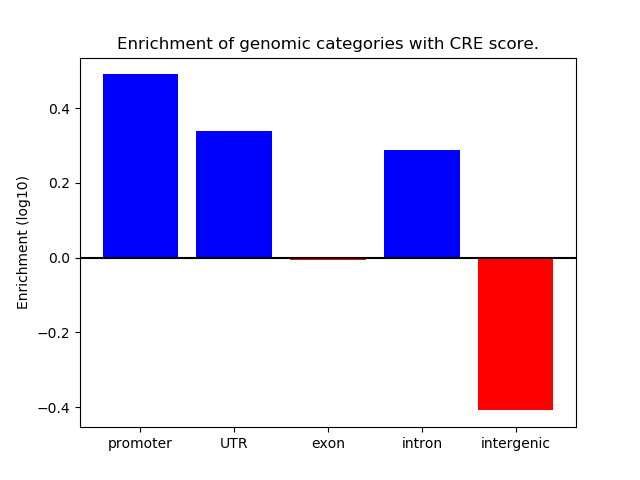
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr15_102963900_102964090 | Hoxc10 | 2801 | 0.109164 | -0.19 | 7.2e-01 | Click! |
Activity of the Hoxc10 motif across conditions
Conditions sorted by the z-value of the Hoxc10 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
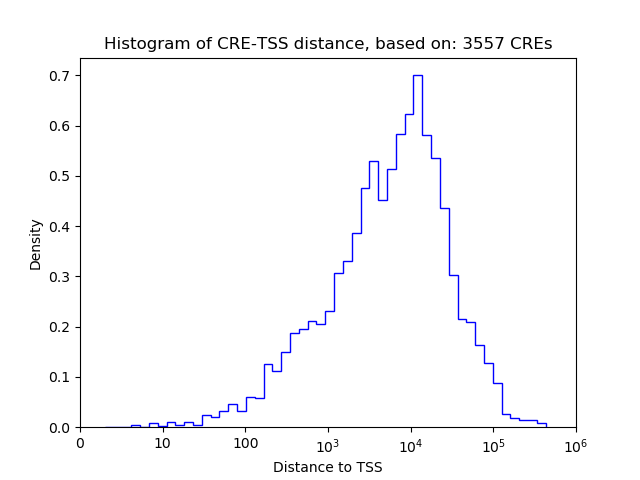
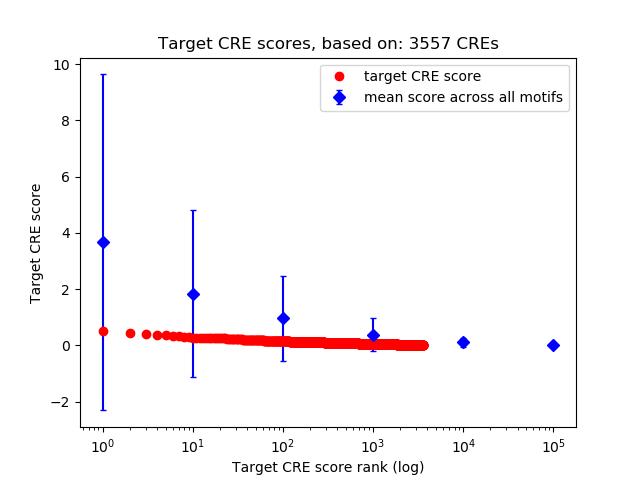
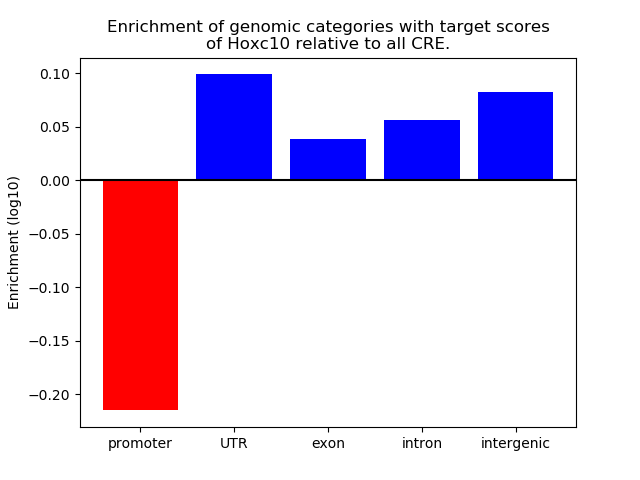
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr13_20205112_20205287 | 0.52 |
Elmo1 |
engulfment and cell motility 1 |
19992 |
0.25 |
| chr18_20988207_20988358 | 0.45 |
Rnf138 |
ring finger protein 138 |
13059 |
0.19 |
| chr8_123736768_123736919 | 0.39 |
Gm45781 |
predicted gene 45781 |
1701 |
0.16 |
| chr16_76237397_76237728 | 0.37 |
Nrip1 |
nuclear receptor interacting protein 1 |
86096 |
0.08 |
| chr2_103846113_103846314 | 0.35 |
Gm13879 |
predicted gene 13879 |
2557 |
0.13 |
| chr13_93622998_93623302 | 0.34 |
Gm15622 |
predicted gene 15622 |
2232 |
0.26 |
| chr12_21190031_21190182 | 0.32 |
AC156032.1 |
|
57217 |
0.09 |
| chr3_57605137_57605288 | 0.30 |
Mir6377 |
microRNA 6377 |
11397 |
0.12 |
| chr11_82138240_82138391 | 0.29 |
Ccl8 |
chemokine (C-C motif) ligand 8 |
23130 |
0.15 |
| chr16_24087119_24087285 | 0.27 |
Gm31583 |
predicted gene, 31583 |
2887 |
0.23 |
| chr1_46345449_46345605 | 0.27 |
Dnah7b |
dynein, axonemal, heavy chain 7B |
11093 |
0.21 |
| chr10_69160830_69160981 | 0.27 |
Rhobtb1 |
Rho-related BTB domain containing 1 |
9471 |
0.16 |
| chr3_84231506_84231924 | 0.27 |
Trim2 |
tripartite motif-containing 2 |
10826 |
0.24 |
| chr15_26388832_26388991 | 0.27 |
Marchf11 |
membrane associated ring-CH-type finger 11 |
1192 |
0.64 |
| chr9_65334133_65334359 | 0.26 |
Gm39363 |
predicted gene, 39363 |
1726 |
0.18 |
| chr19_47315824_47316178 | 0.26 |
Sh3pxd2a |
SH3 and PX domains 2A |
1250 |
0.45 |
| chr12_99342051_99342202 | 0.26 |
3300002A11Rik |
RIKEN cDNA 3300002A11 gene |
972 |
0.46 |
| chr9_57283258_57283868 | 0.26 |
1700017B05Rik |
RIKEN cDNA 1700017B05 gene |
20951 |
0.14 |
| chr11_120916572_120916723 | 0.25 |
Ccdc57 |
coiled-coil domain containing 57 |
16225 |
0.11 |
| chr16_24394028_24394581 | 0.25 |
Lpp |
LIM domain containing preferred translocation partner in lipoma |
566 |
0.73 |
| chr3_58583526_58583677 | 0.25 |
Selenot |
selenoprotein T |
6903 |
0.16 |
| chr15_102245005_102245156 | 0.25 |
Rarg |
retinoic acid receptor, gamma |
151 |
0.91 |
| chr13_93630762_93631092 | 0.24 |
Gm15622 |
predicted gene 15622 |
5545 |
0.17 |
| chr19_30096271_30096642 | 0.24 |
Uhrf2 |
ubiquitin-like, containing PHD and RING finger domains 2 |
4495 |
0.24 |
| chr5_30231710_30232189 | 0.24 |
Selenoi |
selenoprotein I |
632 |
0.63 |
| chr15_31528246_31529049 | 0.23 |
Marchf6 |
membrane associated ring-CH-type finger 6 |
2406 |
0.2 |
| chr5_110596813_110596964 | 0.23 |
Galnt9 |
polypeptide N-acetylgalactosaminyltransferase 9 |
6633 |
0.15 |
| chr10_24111238_24111394 | 0.22 |
Taar9 |
trace amine-associated receptor 9 |
1782 |
0.2 |
| chr18_20931708_20931938 | 0.22 |
Rnf125 |
ring finger protein 125 |
12802 |
0.2 |
| chr3_132887148_132887306 | 0.22 |
Gm29811 |
predicted gene, 29811 |
8631 |
0.16 |
| chr6_94783873_94784024 | 0.22 |
Gm43997 |
predicted gene, 43997 |
15484 |
0.16 |
| chr10_37195063_37195247 | 0.22 |
5930403N24Rik |
RIKEN cDNA 5930403N24 gene |
54066 |
0.12 |
| chr4_139791597_139791792 | 0.21 |
Pax7 |
paired box 7 |
41313 |
0.15 |
| chr13_32797231_32797395 | 0.21 |
Wrnip1 |
Werner helicase interacting protein 1 |
4725 |
0.15 |
| chr17_71251888_71252113 | 0.21 |
Emilin2 |
elastin microfibril interfacer 2 |
3406 |
0.2 |
| chr7_118304499_118304787 | 0.21 |
Gm23229 |
predicted gene, 23229 |
24782 |
0.12 |
| chr7_115868443_115868809 | 0.20 |
Sox6 |
SRY (sex determining region Y)-box 6 |
8774 |
0.29 |
| chr19_3831749_3831913 | 0.20 |
Gm19209 |
predicted gene, 19209 |
9442 |
0.1 |
| chr6_57726433_57726603 | 0.20 |
Lancl2 |
LanC (bacterial lantibiotic synthetase component C)-like 2 |
23431 |
0.12 |
| chr8_119414638_119414789 | 0.20 |
Osgin1 |
oxidative stress induced growth inhibitor 1 |
19411 |
0.13 |
| chr14_18234075_18234258 | 0.20 |
Nr1d2 |
nuclear receptor subfamily 1, group D, member 2 |
4845 |
0.16 |
| chr19_46983871_46984029 | 0.20 |
Nt5c2 |
5'-nucleotidase, cytosolic II |
14382 |
0.14 |
| chr3_98295816_98295967 | 0.19 |
Gm43189 |
predicted gene 43189 |
8903 |
0.15 |
| chr15_80673197_80673348 | 0.19 |
Fam83f |
family with sequence similarity 83, member F |
1425 |
0.31 |
| chr3_21479148_21479326 | 0.19 |
Gm43675 |
predicted gene 43675 |
124573 |
0.06 |
| chr5_139825857_139826025 | 0.19 |
Gm26938 |
predicted gene, 26938 |
466 |
0.61 |
| chr3_149086949_149087100 | 0.19 |
Gm25127 |
predicted gene, 25127 |
58092 |
0.12 |
| chr12_12320785_12320940 | 0.19 |
Fam49a |
family with sequence similarity 49, member A |
58673 |
0.14 |
| chr5_25086751_25086975 | 0.19 |
Prkag2 |
protein kinase, AMP-activated, gamma 2 non-catalytic subunit |
13779 |
0.15 |
| chr4_120986544_120986936 | 0.19 |
Smap2 |
small ArfGAP 2 |
11032 |
0.12 |
| chr1_66895610_66896410 | 0.19 |
Gm25832 |
predicted gene, 25832 |
29213 |
0.09 |
| chr6_145355079_145355230 | 0.18 |
Gm23498 |
predicted gene, 23498 |
11704 |
0.13 |
| chr8_46540344_46540495 | 0.18 |
Acsl1 |
acyl-CoA synthetase long-chain family member 1 |
9355 |
0.15 |
| chr13_90922459_90922719 | 0.18 |
Rps23 |
ribosomal protein S23 |
369 |
0.88 |
| chr15_22789821_22790146 | 0.18 |
Hnrnpa1l2-ps2 |
heterogeneous nuclear ribonucleoprotein A1-like 2, pseudogene 2 |
75153 |
0.12 |
| chr15_36681116_36681275 | 0.18 |
Gm10385 |
predicted gene 10385 |
8908 |
0.14 |
| chr17_30010480_30010631 | 0.18 |
Zfand3 |
zinc finger, AN1-type domain 3 |
2758 |
0.19 |
| chr16_95709296_95709447 | 0.18 |
Ets2 |
E26 avian leukemia oncogene 2, 3' domain |
3032 |
0.28 |
| chr8_106882742_106882961 | 0.18 |
Chtf8 |
CTF8, chromosome transmission fidelity factor 8 |
10364 |
0.12 |
| chr13_20115696_20115876 | 0.17 |
Elmo1 |
engulfment and cell motility 1 |
4274 |
0.33 |
| chr11_99160094_99160256 | 0.17 |
Ccr7 |
chemokine (C-C motif) receptor 7 |
5098 |
0.18 |
| chr15_36471981_36472313 | 0.17 |
Ankrd46 |
ankyrin repeat domain 46 |
24568 |
0.12 |
| chr11_70648552_70648949 | 0.17 |
Rnf167 |
ring finger protein 167 |
372 |
0.59 |
| chr9_108669991_108670142 | 0.17 |
Slc25a20 |
solute carrier family 25 (mitochondrial carnitine/acylcarnitine translocase), member 20 |
2430 |
0.15 |
| chr1_82261807_82261976 | 0.17 |
Gm9747 |
predicted gene 9747 |
28779 |
0.15 |
| chr15_74922383_74922543 | 0.17 |
Gm6610 |
predicted gene 6610 |
1981 |
0.17 |
| chr15_100112289_100112692 | 0.17 |
4930478M13Rik |
RIKEN cDNA 4930478M13 gene |
4329 |
0.17 |
| chr4_66018173_66018346 | 0.16 |
Gm11484 |
predicted gene 11484 |
335590 |
0.01 |
| chr2_35338418_35338609 | 0.16 |
Stom |
stomatin |
1537 |
0.27 |
| chr2_36091025_36091199 | 0.16 |
Lhx6 |
LIM homeobox protein 6 |
3161 |
0.17 |
| chr6_58832026_58832202 | 0.16 |
Herc3 |
hect domain and RLD 3 |
253 |
0.93 |
| chr9_108314669_108314825 | 0.16 |
Rhoa |
ras homolog family member A |
7918 |
0.07 |
| chr19_26731899_26732052 | 0.16 |
Smarca2 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
1976 |
0.4 |
| chr4_62966144_62966295 | 0.16 |
Zfp618 |
zinc finger protein 618 |
645 |
0.69 |
| chr5_146703840_146704008 | 0.16 |
4930573C15Rik |
RIKEN cDNA 4930573C15 gene |
2698 |
0.26 |
| chr4_138348310_138348502 | 0.16 |
Cda |
cytidine deaminase |
4757 |
0.13 |
| chr10_109973597_109973769 | 0.16 |
Nav3 |
neuron navigator 3 |
26536 |
0.21 |
| chr15_38471890_38472111 | 0.16 |
G930009F23Rik |
RIKEN cDNA G930009F23 gene |
16866 |
0.13 |
| chr3_121982194_121982345 | 0.16 |
Arhgap29 |
Rho GTPase activating protein 29 |
719 |
0.68 |
| chr1_162301492_162301643 | 0.15 |
Dnm3 |
dynamin 3 |
5931 |
0.2 |
| chr7_66077810_66077998 | 0.15 |
Gm45081 |
predicted gene 45081 |
1569 |
0.23 |
| chr10_122384642_122385270 | 0.15 |
Gm36041 |
predicted gene, 36041 |
1936 |
0.39 |
| chr19_58311318_58311494 | 0.15 |
Gm16277 |
predicted gene 16277 |
107267 |
0.07 |
| chr6_50176334_50176508 | 0.15 |
Mpp6 |
membrane protein, palmitoylated 6 (MAGUK p55 subfamily member 6) |
3879 |
0.27 |
| chr16_48303590_48303752 | 0.15 |
Dppa2 |
developmental pluripotency associated 2 |
237 |
0.92 |
| chr8_105825543_105825721 | 0.15 |
Ranbp10 |
RAN binding protein 10 |
1573 |
0.19 |
| chr2_156175837_156176038 | 0.15 |
Rbm39 |
RNA binding motif protein 39 |
1446 |
0.28 |
| chr2_76673306_76673477 | 0.15 |
Fkbp7 |
FK506 binding protein 7 |
275 |
0.87 |
| chr10_31956966_31957141 | 0.15 |
Gm18189 |
predicted gene, 18189 |
16616 |
0.27 |
| chr13_34345499_34345841 | 0.15 |
Slc22a23 |
solute carrier family 22, member 23 |
488 |
0.8 |
| chr4_141708335_141708559 | 0.15 |
Ddi2 |
DNA-damage inducible protein 2 |
14972 |
0.12 |
| chr5_65608952_65609103 | 0.15 |
Pds5a |
PDS5 cohesin associated factor A |
2746 |
0.14 |
| chr3_83000200_83000478 | 0.15 |
Gm30097 |
predicted gene, 30097 |
4754 |
0.17 |
| chr9_32648459_32648646 | 0.15 |
Ets1 |
E26 avian leukemia oncogene 1, 5' domain |
12304 |
0.16 |
| chr2_145813125_145813276 | 0.15 |
Rin2 |
Ras and Rab interactor 2 |
8950 |
0.22 |
| chr8_36452309_36452599 | 0.15 |
Gm19140 |
predicted gene, 19140 |
2074 |
0.31 |
| chr3_60437847_60438007 | 0.15 |
Mbnl1 |
muscleblind like splicing factor 1 |
34903 |
0.19 |
| chr3_65527351_65527507 | 0.15 |
4931440P22Rik |
RIKEN cDNA 4931440P22 gene |
755 |
0.39 |
| chr10_85161219_85161370 | 0.15 |
Cry1 |
cryptochrome 1 (photolyase-like) |
9938 |
0.18 |
| chr4_61303822_61304309 | 0.14 |
Mup14 |
major urinary protein 14 |
65 |
0.97 |
| chr19_3883590_3883781 | 0.14 |
Chka |
choline kinase alpha |
8046 |
0.08 |
| chr1_170667297_170667480 | 0.14 |
Olfml2b |
olfactomedin-like 2B |
22856 |
0.16 |
| chr19_42535855_42536113 | 0.14 |
R3hcc1l |
R3H domain and coiled-coil containing 1 like |
17162 |
0.18 |
| chr13_21395579_21395832 | 0.14 |
Mir6942 |
microRNA 6942 |
933 |
0.25 |
| chr12_28905051_28905477 | 0.14 |
Gm31508 |
predicted gene, 31508 |
4965 |
0.21 |
| chr12_26125627_26125778 | 0.14 |
Gm47750 |
predicted gene, 47750 |
24614 |
0.21 |
| chr12_98375326_98375477 | 0.14 |
5330409N07Rik |
RIKEN cDNA 5330409N07 gene |
72933 |
0.08 |
| chr19_30095981_30096172 | 0.14 |
Uhrf2 |
ubiquitin-like, containing PHD and RING finger domains 2 |
4115 |
0.24 |
| chr3_100785704_100785863 | 0.14 |
Vtcn1 |
V-set domain containing T cell activation inhibitor 1 |
39676 |
0.14 |
| chr8_35413438_35413609 | 0.14 |
Gm45301 |
predicted gene 45301 |
4017 |
0.2 |
| chr11_67516560_67516876 | 0.14 |
Gas7 |
growth arrest specific 7 |
30436 |
0.19 |
| chr2_157556257_157556418 | 0.14 |
Blcap |
bladder cancer associated protein |
3670 |
0.14 |
| chr15_10712881_10713032 | 0.14 |
Rai14 |
retinoic acid induced 14 |
584 |
0.71 |
| chr7_113778913_113779066 | 0.14 |
Spon1 |
spondin 1, (f-spondin) extracellular matrix protein |
12815 |
0.22 |
| chr4_33223953_33224209 | 0.14 |
Srsf12 |
serine and arginine-rich splicing factor 12 |
14818 |
0.14 |
| chr14_20315223_20315409 | 0.14 |
Nudt13 |
nudix (nucleoside diphosphate linked moiety X)-type motif 13 |
3887 |
0.15 |
| chr2_145727998_145728180 | 0.14 |
Gm11763 |
predicted gene 11763 |
26364 |
0.18 |
| chr16_6592226_6592401 | 0.14 |
Rbfox1 |
RNA binding protein, fox-1 homolog (C. elegans) 1 |
216909 |
0.02 |
| chr15_78913574_78913725 | 0.13 |
1700027A07Rik |
RIKEN cDNA 1700027A07 gene |
11 |
0.77 |
| chr18_9632745_9632896 | 0.13 |
Gm4834 |
predicted gene 4834 |
3958 |
0.16 |
| chr17_45566033_45566184 | 0.13 |
Slc35b2 |
solute carrier family 35, member B2 |
1369 |
0.23 |
| chr19_33099137_33099698 | 0.13 |
Gm29946 |
predicted gene, 29946 |
23230 |
0.18 |
| chr8_128363089_128363260 | 0.13 |
Nrp1 |
neuropilin 1 |
3777 |
0.26 |
| chr13_17717656_17717837 | 0.13 |
Gm48621 |
predicted gene, 48621 |
7882 |
0.12 |
| chr6_114936968_114937119 | 0.13 |
Vgll4 |
vestigial like family member 4 |
15222 |
0.21 |
| chr9_22314126_22314292 | 0.13 |
Zfp810 |
zinc finger protein 810 |
6561 |
0.09 |
| chr11_45872012_45872204 | 0.13 |
Clint1 |
clathrin interactor 1 |
10289 |
0.16 |
| chr1_191480561_191480712 | 0.13 |
Gm37688 |
predicted gene, 37688 |
3305 |
0.18 |
| chr16_26733286_26733437 | 0.13 |
Il1rap |
interleukin 1 receptor accessory protein |
6319 |
0.26 |
| chr5_89012654_89012806 | 0.13 |
Slc4a4 |
solute carrier family 4 (anion exchanger), member 4 |
15236 |
0.27 |
| chr12_99537214_99537482 | 0.13 |
Foxn3 |
forkhead box N3 |
26160 |
0.12 |
| chr1_121332181_121332567 | 0.13 |
Insig2 |
insulin induced gene 2 |
176 |
0.94 |
| chr15_91608960_91609149 | 0.13 |
4933438A12Rik |
RIKEN cDNA 4933438A12 gene |
11471 |
0.18 |
| chr5_36177248_36177418 | 0.13 |
Psapl1 |
prosaposin-like 1 |
26688 |
0.22 |
| chr4_90616833_90616984 | 0.13 |
Gm12636 |
predicted gene 12636 |
72538 |
0.12 |
| chr18_20671827_20672076 | 0.13 |
Ttr |
transthyretin |
6671 |
0.17 |
| chr5_135741186_135741337 | 0.13 |
Tmem120a |
transmembrane protein 120A |
1138 |
0.32 |
| chr18_38312030_38312222 | 0.13 |
Rnf14 |
ring finger protein 14 |
12374 |
0.1 |
| chr19_29370028_29370202 | 0.13 |
Cd274 |
CD274 antigen |
2660 |
0.18 |
| chr8_45529825_45529981 | 0.13 |
Gm45458 |
predicted gene 45458 |
10212 |
0.18 |
| chr19_21824245_21824555 | 0.13 |
Gm50130 |
predicted gene, 50130 |
9356 |
0.22 |
| chr19_17159808_17159970 | 0.13 |
Prune2 |
prune homolog 2 |
21150 |
0.22 |
| chr16_35919055_35919394 | 0.13 |
Gm10237 |
predicted gene 10237 |
1240 |
0.32 |
| chr11_77728722_77728873 | 0.13 |
Cryba1 |
crystallin, beta A1 |
3514 |
0.17 |
| chr1_24743279_24743468 | 0.13 |
Lmbrd1 |
LMBR1 domain containing 1 |
1500 |
0.46 |
| chr9_111209525_111209710 | 0.13 |
Lrrfip2 |
leucine rich repeat (in FLII) interacting protein 2 |
4576 |
0.2 |
| chr19_23961632_23961790 | 0.13 |
Fam189a2 |
family with sequence similarity 189, member A2 |
14724 |
0.16 |
| chr4_139974069_139974220 | 0.13 |
Klhdc7a |
kelch domain containing 7A |
6118 |
0.17 |
| chr11_101369874_101370035 | 0.13 |
G6pc |
glucose-6-phosphatase, catalytic |
2393 |
0.1 |
| chr6_98954486_98954663 | 0.13 |
Foxp1 |
forkhead box P1 |
19029 |
0.26 |
| chr2_122887240_122887411 | 0.13 |
Sqor |
sulfide quinone oxidoreductase |
87563 |
0.08 |
| chr7_49488722_49488898 | 0.13 |
Gm38059 |
predicted gene, 38059 |
19098 |
0.22 |
| chr3_79549707_79549864 | 0.13 |
Gm3513 |
predicted gene 3513 |
2408 |
0.21 |
| chr17_50515527_50515699 | 0.13 |
Plcl2 |
phospholipase C-like 2 |
6210 |
0.31 |
| chr4_33292661_33293055 | 0.13 |
Pnrc1 |
proline-rich nuclear receptor coactivator 1 |
2695 |
0.24 |
| chr2_170128598_170128786 | 0.13 |
Zfp217 |
zinc finger protein 217 |
2528 |
0.4 |
| chr14_121181940_121182118 | 0.13 |
B930095G15Rik |
RIKEN cDNA B930095G15 gene |
66677 |
0.11 |
| chr11_20829675_20830062 | 0.12 |
Lgalsl |
lectin, galactoside binding-like |
887 |
0.49 |
| chr4_20032087_20032276 | 0.12 |
Ggh |
gamma-glutamyl hydrolase |
9871 |
0.2 |
| chr5_131017810_131017961 | 0.12 |
Gm43165 |
predicted gene 43165 |
25085 |
0.21 |
| chrX_170004028_170004179 | 0.12 |
Erdr1 |
erythroid differentiation regulator 1 |
5556 |
0.17 |
| chr4_104530575_104530756 | 0.12 |
Dab1 |
disabled 1 |
18592 |
0.29 |
| chr10_59786230_59786381 | 0.12 |
Gm17059 |
predicted gene 17059 |
13949 |
0.14 |
| chr11_102744260_102744413 | 0.12 |
Gm16342 |
predicted gene 16342 |
11931 |
0.11 |
| chr15_25650294_25650463 | 0.12 |
Gm48996 |
predicted gene, 48996 |
15751 |
0.17 |
| chr14_22817406_22817579 | 0.12 |
Gm7473 |
predicted gene 7473 |
42248 |
0.2 |
| chr4_86686025_86686176 | 0.12 |
Plin2 |
perilipin 2 |
16040 |
0.18 |
| chrY_90775341_90775665 | 0.12 |
Gm47283 |
predicted gene, 47283 |
9235 |
0.17 |
| chr4_88184310_88184469 | 0.12 |
Gm12645 |
predicted gene 12645 |
32693 |
0.16 |
| chr15_81798335_81798552 | 0.12 |
Tef |
thyrotroph embryonic factor |
3978 |
0.12 |
| chr10_91170398_91170640 | 0.12 |
Tmpo |
thymopoietin |
57 |
0.97 |
| chr11_31897693_31897844 | 0.12 |
Cpeb4 |
cytoplasmic polyadenylation element binding protein 4 |
24493 |
0.18 |
| chr5_41572654_41572805 | 0.12 |
Rab28 |
RAB28, member RAS oncogene family |
54773 |
0.16 |
| chr2_30163705_30163866 | 0.12 |
Slc39a1-ps |
solute carrier family 39 (zinc transporter), member 1, pseudogene |
4387 |
0.11 |
| chr13_32823696_32823847 | 0.12 |
Wrnip1 |
Werner helicase interacting protein 1 |
3005 |
0.19 |
| chr11_106982284_106982435 | 0.12 |
Gm11707 |
predicted gene 11707 |
9269 |
0.13 |
| chr17_63491587_63491744 | 0.12 |
Fbxl17 |
F-box and leucine-rich repeat protein 17 |
1088 |
0.61 |
| chr8_128687218_128687546 | 0.12 |
Itgb1 |
integrin beta 1 (fibronectin receptor beta) |
1512 |
0.39 |
| chr2_103801242_103801527 | 0.12 |
Caprin1 |
cell cycle associated protein 1 |
3735 |
0.12 |
| chr7_127973702_127973919 | 0.12 |
Fus |
fused in sarcoma |
333 |
0.72 |
| chrY_90764765_90764916 | 0.12 |
Gm21860 |
predicted gene, 21860 |
9373 |
0.17 |
| chr12_21183631_21184095 | 0.12 |
AC156032.1 |
|
63460 |
0.08 |
| chrX_169990489_169990640 | 0.12 |
Gm15247 |
predicted gene 15247 |
3625 |
0.18 |
| chr4_44997538_44997819 | 0.12 |
1700055D18Rik |
RIKEN cDNA 1700055D18 gene |
14009 |
0.1 |
| chrX_170001003_170001326 | 0.12 |
Erdr1 |
erythroid differentiation regulator 1 |
8495 |
0.15 |
| chr4_53414447_53414950 | 0.12 |
Gm12496 |
predicted gene 12496 |
10668 |
0.21 |
| chr11_95726300_95726491 | 0.12 |
Zfp652 |
zinc finger protein 652 |
13384 |
0.12 |
| chr4_153239041_153239351 | 0.12 |
Gm13174 |
predicted gene 13174 |
21020 |
0.26 |
| chr16_18430516_18430667 | 0.12 |
Txnrd2 |
thioredoxin reductase 2 |
1666 |
0.22 |
| chr1_134580810_134580961 | 0.12 |
Kdm5b |
lysine (K)-specific demethylase 5B |
7092 |
0.15 |
| chr4_139169377_139169633 | 0.12 |
Gm16287 |
predicted gene 16287 |
11150 |
0.13 |
| chr4_145215459_145215632 | 0.12 |
Vps13d |
vacuolar protein sorting 13D |
20540 |
0.19 |
| chr1_88975960_88976113 | 0.12 |
1700067G17Rik |
RIKEN cDNA 1700067G17 gene |
40077 |
0.14 |
| chr13_95794713_95795054 | 0.12 |
Iqgap2 |
IQ motif containing GTPase activating protein 2 |
30486 |
0.16 |
| chr6_54772634_54772785 | 0.12 |
Znrf2 |
zinc and ring finger 2 |
44207 |
0.14 |
| chr5_92867642_92867801 | 0.12 |
Shroom3 |
shroom family member 3 |
11250 |
0.22 |
| chr11_94383447_94383613 | 0.12 |
Abcc3 |
ATP-binding cassette, sub-family C (CFTR/MRP), member 3 |
9390 |
0.16 |
| chr15_102245839_102245990 | 0.12 |
Rarg |
retinoic acid receptor, gamma |
586 |
0.56 |
| chr18_46711768_46711921 | 0.11 |
Cdo1 |
cysteine dioxygenase 1, cytosolic |
16185 |
0.12 |
| chr1_121320571_121320766 | 0.11 |
Insig2 |
insulin induced gene 2 |
940 |
0.52 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0070384 | Harderian gland development(GO:0070384) |
| 0.0 | 0.1 | GO:0090230 | regulation of centromere complex assembly(GO:0090230) |
| 0.0 | 0.1 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.0 | 0.1 | GO:0035441 | cell migration involved in vasculogenesis(GO:0035441) |
| 0.0 | 0.1 | GO:0014873 | response to muscle activity involved in regulation of muscle adaptation(GO:0014873) |
| 0.0 | 0.1 | GO:0002337 | B-1a B cell differentiation(GO:0002337) |
| 0.0 | 0.2 | GO:0006991 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.0 | 0.1 | GO:0072675 | osteoclast fusion(GO:0072675) |
| 0.0 | 0.1 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.0 | 0.2 | GO:0071267 | amino acid salvage(GO:0043102) L-methionine biosynthetic process(GO:0071265) L-methionine salvage(GO:0071267) |
| 0.0 | 0.1 | GO:0018199 | peptidyl-glutamine modification(GO:0018199) |
| 0.0 | 0.1 | GO:0019464 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.0 | 0.1 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.0 | 0.1 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.1 | GO:0006481 | C-terminal protein methylation(GO:0006481) |
| 0.0 | 0.1 | GO:0009972 | cytidine catabolic process(GO:0006216) cytidine deamination(GO:0009972) cytidine metabolic process(GO:0046087) |
| 0.0 | 0.1 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.0 | 0.1 | GO:0009093 | cysteine catabolic process(GO:0009093) L-cysteine catabolic process(GO:0019448) L-cysteine metabolic process(GO:0046439) |
| 0.0 | 0.1 | GO:0040038 | polar body extrusion after meiotic divisions(GO:0040038) |
| 0.0 | 0.1 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.0 | 0.1 | GO:0008050 | female courtship behavior(GO:0008050) |
| 0.0 | 0.1 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.0 | 0.1 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.0 | 0.1 | GO:0007161 | calcium-independent cell-matrix adhesion(GO:0007161) |
| 0.0 | 0.1 | GO:0033159 | negative regulation of protein import into nucleus, translocation(GO:0033159) |
| 0.0 | 0.0 | GO:2000410 | thymocyte migration(GO:0072679) regulation of thymocyte migration(GO:2000410) positive regulation of thymocyte migration(GO:2000412) |
| 0.0 | 0.1 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.0 | 0.0 | GO:0034499 | late endosome to Golgi transport(GO:0034499) |
| 0.0 | 0.0 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.0 | 0.1 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.0 | 0.0 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.0 | 0.0 | GO:0061470 | T follicular helper cell differentiation(GO:0061470) |
| 0.0 | 0.1 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.0 | 0.1 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.0 | 0.0 | GO:2000850 | negative regulation of steroid hormone secretion(GO:2000832) negative regulation of corticosteroid hormone secretion(GO:2000847) negative regulation of glucocorticoid secretion(GO:2000850) |
| 0.0 | 0.1 | GO:0090308 | regulation of methylation-dependent chromatin silencing(GO:0090308) |
| 0.0 | 0.1 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.0 | 0.0 | GO:0048294 | negative regulation of isotype switching to IgE isotypes(GO:0048294) |
| 0.0 | 0.0 | GO:0061642 | chemoattraction of axon(GO:0061642) |
| 0.0 | 0.0 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.0 | 0.1 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 | 0.0 | GO:0050713 | negative regulation of interleukin-1 beta secretion(GO:0050713) |
| 0.0 | 0.1 | GO:0001712 | ectodermal cell fate commitment(GO:0001712) |
| 0.0 | 0.0 | GO:0070346 | positive regulation of fat cell proliferation(GO:0070346) |
| 0.0 | 0.1 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.0 | 0.0 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 0.0 | 0.1 | GO:1901727 | positive regulation of histone deacetylase activity(GO:1901727) |
| 0.0 | 0.1 | GO:0030953 | astral microtubule organization(GO:0030953) |
| 0.0 | 0.0 | GO:0071879 | positive regulation of adrenergic receptor signaling pathway(GO:0071879) |
| 0.0 | 0.0 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.0 | GO:0006172 | ADP biosynthetic process(GO:0006172) |
| 0.0 | 0.1 | GO:0060484 | lung-associated mesenchyme development(GO:0060484) |
| 0.0 | 0.0 | GO:0010909 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.0 | 0.1 | GO:0019695 | choline metabolic process(GO:0019695) |
| 0.0 | 0.0 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.0 | 0.0 | GO:1903525 | regulation of membrane tubulation(GO:1903525) |
| 0.0 | 0.0 | GO:0010835 | regulation of protein ADP-ribosylation(GO:0010835) |
| 0.0 | 0.0 | GO:0006982 | response to lipid hydroperoxide(GO:0006982) |
| 0.0 | 0.1 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.0 | 0.0 | GO:0071930 | negative regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071930) |
| 0.0 | 0.0 | GO:0090154 | positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 0.0 | 0.0 | GO:0097296 | activation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097296) |
| 0.0 | 0.1 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.0 | 0.1 | GO:0048133 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.0 | 0.1 | GO:0032464 | positive regulation of protein homooligomerization(GO:0032464) |
| 0.0 | 0.0 | GO:1902075 | cellular response to salt(GO:1902075) |
| 0.0 | 0.0 | GO:0097477 | lateral motor column neuron migration(GO:0097477) |
| 0.0 | 0.0 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.1 | GO:0030174 | regulation of DNA-dependent DNA replication initiation(GO:0030174) |
| 0.0 | 0.0 | GO:0007290 | spermatid nucleus elongation(GO:0007290) |
| 0.0 | 0.0 | GO:0021914 | negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021914) |
| 0.0 | 0.1 | GO:0010886 | positive regulation of cholesterol storage(GO:0010886) |
| 0.0 | 0.0 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 0.0 | 0.0 | GO:0036135 | Schwann cell migration(GO:0036135) regulation of Schwann cell migration(GO:1900147) |
| 0.0 | 0.1 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.0 | GO:2000467 | positive regulation of glycogen (starch) synthase activity(GO:2000467) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.0 | 0.1 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.2 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.1 | GO:0034666 | integrin alpha2-beta1 complex(GO:0034666) |
| 0.0 | 0.1 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.0 | 0.1 | GO:0009279 | cell outer membrane(GO:0009279) cell envelope(GO:0030313) external encapsulating structure part(GO:0044462) |
| 0.0 | 0.1 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.1 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.0 | 0.0 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.0 | 0.0 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.0 | 0.0 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.0 | 0.1 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 0.1 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.0 | 0.0 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0008898 | S-adenosylmethionine-homocysteine S-methyltransferase activity(GO:0008898) |
| 0.0 | 0.3 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.0 | 0.1 | GO:0004104 | cholinesterase activity(GO:0004104) |
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.1 | GO:0004346 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.0 | 0.1 | GO:0033142 | progesterone receptor binding(GO:0033142) |
| 0.0 | 0.1 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.1 | GO:0030899 | calcium-dependent ATPase activity(GO:0030899) |
| 0.0 | 0.1 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 0.1 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.0 | 0.1 | GO:0002094 | polyprenyltransferase activity(GO:0002094) |
| 0.0 | 0.1 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.1 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.0 | 0.0 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 0.1 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.0 | 0.0 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.0 | 0.0 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.0 | 0.1 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 0.0 | 0.1 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.0 | 0.1 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.0 | 0.0 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 0.0 | 0.1 | GO:0035473 | lipase binding(GO:0035473) |
| 0.0 | 0.0 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.0 | 0.1 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.0 | 0.1 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.0 | GO:0052834 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.0 | 0.0 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.0 | 0.0 | GO:0019961 | interferon binding(GO:0019961) |
| 0.0 | 0.0 | GO:0000171 | ribonuclease MRP activity(GO:0000171) |
| 0.0 | 0.1 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 0.1 | GO:0042910 | xenobiotic transporter activity(GO:0042910) |
| 0.0 | 0.1 | GO:0019960 | C-X3-C chemokine binding(GO:0019960) |
| 0.0 | 0.0 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.0 | 0.1 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.0 | 0.1 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.0 | 0.0 | REACTOME CELL DEATH SIGNALLING VIA NRAGE NRIF AND NADE | Genes involved in Cell death signalling via NRAGE, NRIF and NADE |
| 0.0 | 0.0 | REACTOME ARMS MEDIATED ACTIVATION | Genes involved in ARMS-mediated activation |
| 0.0 | 0.6 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |