Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
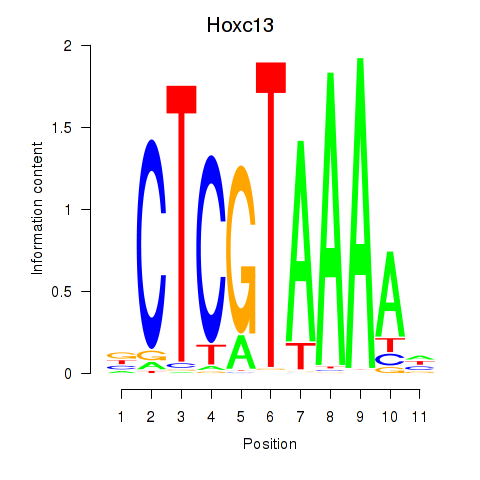
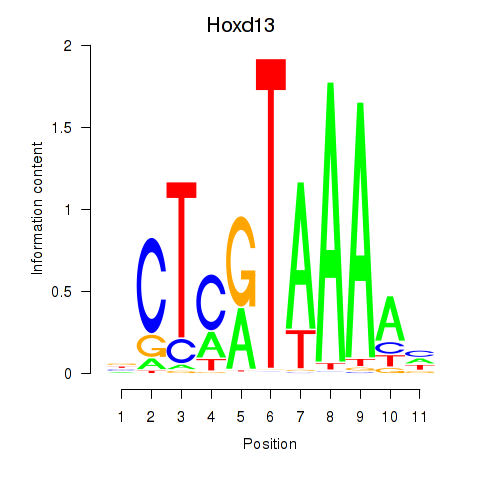
Results for Hoxc13_Hoxd13
Z-value: 0.59
Transcription factors associated with Hoxc13_Hoxd13
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hoxc13
|
ENSMUSG00000001655.6 | homeobox C13 |
|
Hoxd13
|
ENSMUSG00000001819.4 | homeobox D13 |
Activity of the Hoxc13_Hoxd13 motif across conditions
Conditions sorted by the z-value of the Hoxc13_Hoxd13 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
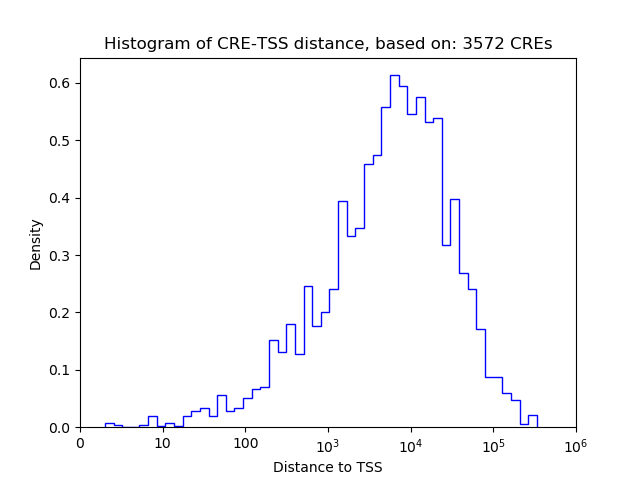
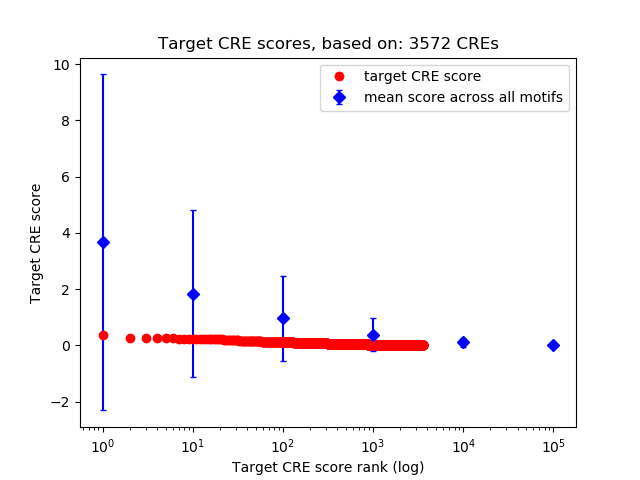
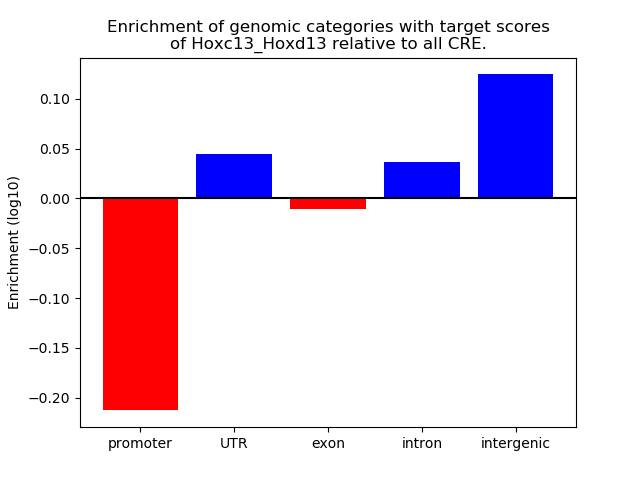
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr15_100112289_100112692 | 0.37 |
4930478M13Rik |
RIKEN cDNA 4930478M13 gene |
4329 |
0.17 |
| chr10_37195063_37195247 | 0.26 |
5930403N24Rik |
RIKEN cDNA 5930403N24 gene |
54066 |
0.12 |
| chrY_90775341_90775665 | 0.26 |
Gm47283 |
predicted gene, 47283 |
9235 |
0.17 |
| chrX_170001003_170001326 | 0.25 |
Erdr1 |
erythroid differentiation regulator 1 |
8495 |
0.15 |
| chrX_93159224_93159387 | 0.25 |
1700003E24Rik |
RIKEN cDNA 1700003E24 gene |
3151 |
0.24 |
| chr3_60053807_60054248 | 0.25 |
Aadac |
arylacetamide deacetylase |
22151 |
0.14 |
| chr10_68168093_68168385 | 0.24 |
Arid5b |
AT rich interactive domain 5B (MRF1-like) |
31613 |
0.21 |
| chr16_52043832_52044015 | 0.24 |
Cblb |
Casitas B-lineage lymphoma b |
5730 |
0.27 |
| chr15_66814050_66814231 | 0.23 |
Sla |
src-like adaptor |
1547 |
0.4 |
| chr17_78992754_78992910 | 0.23 |
Prkd3 |
protein kinase D3 |
7384 |
0.16 |
| chrX_170004028_170004179 | 0.23 |
Erdr1 |
erythroid differentiation regulator 1 |
5556 |
0.17 |
| chr13_21322712_21322863 | 0.22 |
Gpx6 |
glutathione peroxidase 6 |
5529 |
0.11 |
| chr10_10408279_10408449 | 0.22 |
Adgb |
androglobin |
3380 |
0.27 |
| chr4_149887083_149887262 | 0.22 |
Gm13070 |
predicted gene 13070 |
16834 |
0.13 |
| chrY_90778372_90778523 | 0.22 |
Gm47283 |
predicted gene, 47283 |
6291 |
0.19 |
| chr19_41257848_41258010 | 0.22 |
Tm9sf3 |
transmembrane 9 superfamily member 3 |
6051 |
0.26 |
| chr6_135552531_135552703 | 0.22 |
Gm25136 |
predicted gene, 25136 |
31575 |
0.2 |
| chr9_48987994_48988145 | 0.22 |
Usp28 |
ubiquitin specific peptidase 28 |
2604 |
0.25 |
| chr4_154705761_154705932 | 0.21 |
Actrt2 |
actin-related protein T2 |
37979 |
0.12 |
| chr8_11055813_11056042 | 0.21 |
9530052E02Rik |
RIKEN cDNA 9530052E02 gene |
6349 |
0.16 |
| chrY_90781351_90781502 | 0.21 |
Gm47283 |
predicted gene, 47283 |
3312 |
0.23 |
| chr1_72244368_72244519 | 0.21 |
Gm24497 |
predicted gene, 24497 |
230 |
0.88 |
| chr4_8714834_8715372 | 0.20 |
Chd7 |
chromodomain helicase DNA binding protein 7 |
4763 |
0.29 |
| chr9_35050174_35050386 | 0.20 |
St3gal4 |
ST3 beta-galactoside alpha-2,3-sialyltransferase 4 |
5514 |
0.2 |
| chr11_82138240_82138391 | 0.20 |
Ccl8 |
chemokine (C-C motif) ligand 8 |
23130 |
0.15 |
| chr7_100042239_100042447 | 0.19 |
Chrdl2 |
chordin-like 2 |
20244 |
0.14 |
| chr10_75049052_75049276 | 0.19 |
Rab36 |
RAB36, member RAS oncogene family |
10941 |
0.15 |
| chr18_56715593_56715797 | 0.19 |
Lmnb1 |
lamin B1 |
7882 |
0.19 |
| chr3_127769267_127769430 | 0.19 |
Alpk1 |
alpha-kinase 1 |
11142 |
0.11 |
| chr8_36668727_36668937 | 0.19 |
Dlc1 |
deleted in liver cancer 1 |
54889 |
0.16 |
| chr15_91608960_91609149 | 0.19 |
4933438A12Rik |
RIKEN cDNA 4933438A12 gene |
11471 |
0.18 |
| chr10_83369091_83369272 | 0.18 |
D10Wsu102e |
DNA segment, Chr 10, Wayne State University 102, expressed |
8960 |
0.18 |
| chr12_26125627_26125778 | 0.17 |
Gm47750 |
predicted gene, 47750 |
24614 |
0.21 |
| chr7_16052532_16052691 | 0.17 |
Bicra |
BRD4 interacting chromatin remodeling complex associated protein |
4690 |
0.15 |
| chr6_66981477_66981645 | 0.17 |
Gm36816 |
predicted gene, 36816 |
26852 |
0.11 |
| chr4_155566693_155567032 | 0.16 |
Nadk |
NAD kinase |
2790 |
0.15 |
| chr10_83032672_83032849 | 0.16 |
Gm10773 |
predicted gene 10773 |
1065 |
0.55 |
| chr10_69160830_69160981 | 0.16 |
Rhobtb1 |
Rho-related BTB domain containing 1 |
9471 |
0.16 |
| chr5_90431005_90431358 | 0.16 |
Alb |
albumin |
29716 |
0.13 |
| chr17_53650458_53650615 | 0.16 |
Kat2b |
K(lysine) acetyltransferase 2B |
5839 |
0.15 |
| chr10_118581468_118581731 | 0.16 |
Ifngas1 |
Ifng antisense RNA 1 |
25074 |
0.2 |
| chr7_19866622_19866883 | 0.15 |
Gm44659 |
predicted gene 44659 |
4686 |
0.08 |
| chr10_73140222_73140487 | 0.15 |
Pcdh15 |
protocadherin 15 |
40901 |
0.14 |
| chr4_66018173_66018346 | 0.15 |
Gm11484 |
predicted gene 11484 |
335590 |
0.01 |
| chr11_31672407_31672562 | 0.15 |
Bod1 |
biorientation of chromosomes in cell division 1 |
599 |
0.79 |
| chr7_19006605_19006762 | 0.15 |
Irf2bp1 |
interferon regulatory factor 2 binding protein 1 |
2639 |
0.11 |
| chr10_87572875_87573031 | 0.15 |
Pah |
phenylalanine hydroxylase |
26280 |
0.2 |
| chr1_192103290_192103784 | 0.15 |
Rcor3 |
REST corepressor 3 |
5314 |
0.13 |
| chr2_127845443_127845611 | 0.14 |
Acoxl |
acyl-Coenzyme A oxidase-like |
6651 |
0.17 |
| chr17_44061582_44061733 | 0.14 |
Enpp5 |
ectonucleotide pyrophosphatase/phosphodiesterase 5 |
17156 |
0.19 |
| chr7_101155027_101155182 | 0.14 |
Gm6341 |
predicted pseudogene 6341 |
49 |
0.97 |
| chr3_14957442_14957760 | 0.14 |
A930001A20Rik |
RIKEN cDNA A930001A20 gene |
13600 |
0.19 |
| chr11_62934808_62934982 | 0.14 |
Tvp23bos |
trans-golgi network vesicle protein 23B, opposite strand |
13303 |
0.13 |
| chr10_80173409_80173669 | 0.14 |
Fam174c |
family with sequence similarity 174, member C |
595 |
0.48 |
| chr12_100201612_100201850 | 0.14 |
Calm1 |
calmodulin 1 |
1587 |
0.29 |
| chr12_75664068_75664229 | 0.14 |
Wdr89 |
WD repeat domain 89 |
5379 |
0.22 |
| chr18_16745346_16745693 | 0.14 |
Gm15485 |
predicted gene 15485 |
16786 |
0.23 |
| chr8_115423133_115423517 | 0.13 |
4930488N15Rik |
RIKEN cDNA 4930488N15 gene |
162067 |
0.04 |
| chr4_11047537_11047728 | 0.13 |
Ndufaf6 |
NADH:ubiquinone oxidoreductase complex assembly factor 6 |
11451 |
0.17 |
| chr16_43524279_43524457 | 0.13 |
Zbtb20 |
zinc finger and BTB domain containing 20 |
14060 |
0.24 |
| chr5_146114890_146115041 | 0.13 |
Cyp3a59 |
cytochrome P450, family 3, subfamily a, polypeptide 59 |
35698 |
0.09 |
| chr16_48303590_48303752 | 0.13 |
Dppa2 |
developmental pluripotency associated 2 |
237 |
0.92 |
| chr2_102406798_102406949 | 0.13 |
Trim44 |
tripartite motif-containing 44 |
955 |
0.64 |
| chr10_17411439_17411645 | 0.13 |
Gm47760 |
predicted gene, 47760 |
78665 |
0.09 |
| chr16_26348570_26348826 | 0.13 |
Cldn1 |
claudin 1 |
12096 |
0.29 |
| chr10_84803969_84804150 | 0.13 |
Gm24226 |
predicted gene, 24226 |
7124 |
0.23 |
| chr8_108519534_108519698 | 0.13 |
Gm39244 |
predicted gene, 39244 |
17331 |
0.25 |
| chr2_18937664_18937815 | 0.13 |
Pip4k2a |
phosphatidylinositol-5-phosphate 4-kinase, type II, alpha |
31465 |
0.19 |
| chr6_108212621_108212836 | 0.13 |
Itpr1 |
inositol 1,4,5-trisphosphate receptor 1 |
368 |
0.89 |
| chr8_70695342_70695652 | 0.13 |
Jund |
jun D proto-oncogene |
3452 |
0.09 |
| chr4_107006336_107006624 | 0.13 |
Gm12786 |
predicted gene 12786 |
16019 |
0.15 |
| chr12_39952688_39952943 | 0.13 |
Gm18939 |
predicted gene, 18939 |
2183 |
0.32 |
| chr5_42970531_42970682 | 0.13 |
Gm5554 |
predicted gene 5554 |
5997 |
0.25 |
| chr2_103696259_103696442 | 0.13 |
Abtb2 |
ankyrin repeat and BTB (POZ) domain containing 2 |
18685 |
0.16 |
| chr17_4004285_4004445 | 0.13 |
4930470H14Rik |
RIKEN cDNA 4930470H14 gene |
78630 |
0.11 |
| chr1_53823867_53824094 | 0.13 |
Hecw2 |
HECT, C2 and WW domain containing E3 ubiquitin protein ligase 2 |
17191 |
0.16 |
| chr1_128174385_128174568 | 0.12 |
R3hdm1 |
R3H domain containing 1 |
217 |
0.89 |
| chr12_102469832_102469989 | 0.12 |
Golga5 |
golgi autoantigen, golgin subfamily a, 5 |
718 |
0.65 |
| chr6_121871287_121871502 | 0.12 |
Mug1 |
murinoglobulin 1 |
14183 |
0.19 |
| chr15_36478571_36478739 | 0.12 |
Ankrd46 |
ankyrin repeat domain 46 |
18060 |
0.14 |
| chr3_107654890_107655041 | 0.12 |
Ahcyl1 |
S-adenosylhomocysteine hydrolase-like 1 |
10631 |
0.14 |
| chr10_93749819_93749974 | 0.12 |
Gm15963 |
predicted gene 15963 |
21689 |
0.13 |
| chr2_49541398_49541585 | 0.12 |
Epc2 |
enhancer of polycomb homolog 2 |
12616 |
0.26 |
| chr10_118867636_118867819 | 0.12 |
Dyrk2 |
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 2 |
572 |
0.66 |
| chr19_29040294_29040634 | 0.12 |
1700018L02Rik |
RIKEN cDNA 1700018L02 gene |
5781 |
0.12 |
| chr8_115694479_115694630 | 0.12 |
Gm15655 |
predicted gene 15655 |
10524 |
0.23 |
| chr17_7884908_7885059 | 0.12 |
Tagap |
T cell activation Rho GTPase activating protein |
41017 |
0.13 |
| chr19_6273750_6273958 | 0.12 |
Gm14963 |
predicted gene 14963 |
2751 |
0.1 |
| chr14_121323111_121323262 | 0.12 |
Stk24 |
serine/threonine kinase 24 |
19711 |
0.2 |
| chr18_46558992_46559143 | 0.12 |
Ticam2 |
toll-like receptor adaptor molecule 2 |
15466 |
0.13 |
| chr13_28773702_28773903 | 0.12 |
Gm17528 |
predicted gene, 17528 |
53321 |
0.12 |
| chr5_145983671_145983967 | 0.12 |
Cyp3a25 |
cytochrome P450, family 3, subfamily a, polypeptide 25 |
7824 |
0.13 |
| chr19_43510885_43511105 | 0.12 |
Got1 |
glutamic-oxaloacetic transaminase 1, soluble |
4870 |
0.14 |
| chr6_145355079_145355230 | 0.12 |
Gm23498 |
predicted gene, 23498 |
11704 |
0.13 |
| chr16_30953629_30953810 | 0.12 |
Gm46565 |
predicted gene, 46565 |
18090 |
0.15 |
| chr5_28042416_28042567 | 0.12 |
Gm43611 |
predicted gene 43611 |
6085 |
0.17 |
| chr7_19131903_19132185 | 0.12 |
Fbxo46 |
F-box protein 46 |
351 |
0.71 |
| chr8_77034183_77034467 | 0.12 |
Nr3c2 |
nuclear receptor subfamily 3, group C, member 2 |
93688 |
0.08 |
| chr15_101299611_101299792 | 0.12 |
Smim41 |
small integral membrane protein 41 |
6469 |
0.1 |
| chr8_64728299_64728453 | 0.12 |
Msmo1 |
methylsterol monoxygenase 1 |
2683 |
0.23 |
| chr3_54731795_54731993 | 0.12 |
Exosc8 |
exosome component 8 |
1329 |
0.33 |
| chr5_90470187_90471180 | 0.11 |
Alb |
albumin |
7983 |
0.15 |
| chr7_19956116_19956267 | 0.11 |
Rpl7a-ps8 |
ribosomal protein L7A, pseudogene 8 |
2408 |
0.13 |
| chr3_57516324_57516475 | 0.11 |
Gm16016 |
predicted gene 16016 |
11833 |
0.19 |
| chr5_45513758_45514082 | 0.11 |
Lap3 |
leucine aminopeptidase 3 |
4711 |
0.13 |
| chr8_105825543_105825721 | 0.11 |
Ranbp10 |
RAN binding protein 10 |
1573 |
0.19 |
| chr17_12419000_12419151 | 0.11 |
Plg |
plasminogen |
40416 |
0.12 |
| chrX_36183753_36183920 | 0.11 |
Zcchc12 |
zinc finger, CCHC domain containing 12 |
12068 |
0.17 |
| chr14_105732464_105732657 | 0.11 |
Gm10076 |
predicted gene 10076 |
50731 |
0.13 |
| chr9_121376171_121376353 | 0.11 |
Trak1 |
trafficking protein, kinesin binding 1 |
6532 |
0.19 |
| chr11_106710560_106710711 | 0.11 |
Pecam1 |
platelet/endothelial cell adhesion molecule 1 |
4311 |
0.17 |
| chr6_47257138_47257289 | 0.11 |
Cntnap2 |
contactin associated protein-like 2 |
12760 |
0.28 |
| chr13_102886312_102886463 | 0.11 |
Mast4 |
microtubule associated serine/threonine kinase family member 4 |
19399 |
0.26 |
| chr14_103087898_103088058 | 0.11 |
Fbxl3 |
F-box and leucine-rich repeat protein 3 |
1839 |
0.28 |
| chr16_24081029_24081180 | 0.11 |
Gm46545 |
predicted gene, 46545 |
1586 |
0.36 |
| chr3_149935228_149935581 | 0.11 |
Rpsa-ps10 |
ribosomal protein SA, pseudogene 10 |
138138 |
0.05 |
| chr8_22628523_22628708 | 0.10 |
Dkk4 |
dickkopf WNT signaling pathway inhibitor 4 |
4572 |
0.16 |
| chr5_63966948_63967158 | 0.10 |
Rell1 |
RELT-like 1 |
1770 |
0.3 |
| chr10_51595230_51595419 | 0.10 |
Fam162b |
family with sequence similarity 162, member B |
4807 |
0.14 |
| chr13_58960442_58960601 | 0.10 |
Ntrk2 |
neurotrophic tyrosine kinase, receptor, type 2 |
88786 |
0.07 |
| chr10_128005224_128005731 | 0.10 |
Hsd17b6 |
hydroxysteroid (17-beta) dehydrogenase 6 |
2031 |
0.16 |
| chr3_146122899_146123050 | 0.10 |
Mcoln3 |
mucolipin 3 |
1302 |
0.37 |
| chr3_144617682_144617841 | 0.10 |
Rpl9-ps8 |
ribosomal protein L9, pseudogene 8 |
8203 |
0.15 |
| chr2_69224824_69225015 | 0.10 |
G6pc2 |
glucose-6-phosphatase, catalytic, 2 |
4891 |
0.16 |
| chr10_96822242_96822623 | 0.10 |
Gm48521 |
predicted gene, 48521 |
21393 |
0.15 |
| chr2_137156125_137156303 | 0.10 |
Jag1 |
jagged 1 |
39570 |
0.17 |
| chr1_136137177_136137371 | 0.10 |
Kif21b |
kinesin family member 21B |
5820 |
0.12 |
| chr5_146114373_146114524 | 0.10 |
Cyp3a59 |
cytochrome P450, family 3, subfamily a, polypeptide 59 |
35181 |
0.09 |
| chr2_180256342_180256726 | 0.10 |
Rps21 |
ribosomal protein S21 |
843 |
0.36 |
| chr9_50533781_50533973 | 0.10 |
Gm47077 |
predicted gene, 47077 |
5110 |
0.11 |
| chr13_98945834_98945985 | 0.10 |
Gm35215 |
predicted gene, 35215 |
7 |
0.97 |
| chr10_33847261_33847432 | 0.10 |
Sult3a1 |
sulfotransferase family 3A, member 1 |
10375 |
0.13 |
| chr3_81981051_81981260 | 0.10 |
Asic5 |
acid-sensing (proton-gated) ion channel family member 5 |
1135 |
0.45 |
| chr8_128483208_128483439 | 0.10 |
Nrp1 |
neuropilin 1 |
123926 |
0.05 |
| chr9_105878487_105878719 | 0.10 |
Col6a5 |
collagen, type VI, alpha 5 |
3203 |
0.27 |
| chr6_72428123_72428281 | 0.10 |
Ggcx |
gamma-glutamyl carboxylase |
1585 |
0.25 |
| chr6_129113449_129113636 | 0.10 |
Klrb1-ps1 |
killer cell lectin-like receptor subfamily B member 1, pseudogene 1 |
2976 |
0.16 |
| chr4_141893509_141893832 | 0.10 |
Fhad1os2 |
forkhead-associated (FHA) phosphopeptide binding domain 1, opposite strand 2 |
8016 |
0.12 |
| chr18_11915892_11916043 | 0.10 |
Gm49968 |
predicted gene, 49968 |
2836 |
0.3 |
| chr16_45608261_45608423 | 0.10 |
Gcsam |
germinal center associated, signaling and motility |
2038 |
0.27 |
| chr8_114555361_114555569 | 0.10 |
Gm16116 |
predicted gene 16116 |
39435 |
0.17 |
| chr11_6474966_6475143 | 0.10 |
Gm11973 |
predicted gene 11973 |
371 |
0.61 |
| chr17_12327944_12328104 | 0.10 |
Gm49964 |
predicted gene, 49964 |
7296 |
0.13 |
| chr17_43249770_43249932 | 0.10 |
Adgrf1 |
adhesion G protein-coupled receptor F1 |
20478 |
0.24 |
| chr19_42539449_42539629 | 0.09 |
R3hcc1l |
R3H domain and coiled-coil containing 1 like |
20717 |
0.17 |
| chr4_118132972_118133132 | 0.09 |
St3gal3 |
ST3 beta-galactoside alpha-2,3-sialyltransferase 3 |
1830 |
0.3 |
| chr7_67380279_67380446 | 0.09 |
Mef2a |
myocyte enhancer factor 2A |
7504 |
0.19 |
| chr4_6262876_6263048 | 0.09 |
Gm11798 |
predicted gene 11798 |
1997 |
0.33 |
| chr9_69291273_69291476 | 0.09 |
Rora |
RAR-related orphan receptor alpha |
1692 |
0.45 |
| chr3_9610870_9611024 | 0.09 |
Zfp704 |
zinc finger protein 704 |
862 |
0.67 |
| chr3_107982528_107982704 | 0.09 |
Gm12497 |
predicted pseudogene 12497 |
2984 |
0.1 |
| chr13_30346992_30347295 | 0.09 |
Agtr1a |
angiotensin II receptor, type 1a |
1602 |
0.41 |
| chr4_44046062_44046313 | 0.09 |
Gne |
glucosamine (UDP-N-acetyl)-2-epimerase/N-acetylmannosamine kinase |
9110 |
0.14 |
| chr14_45472712_45472872 | 0.09 |
Fermt2 |
fermitin family member 2 |
5069 |
0.13 |
| chr2_45573006_45573165 | 0.09 |
Gm13478 |
predicted gene 13478 |
74106 |
0.11 |
| chr17_86906233_86906392 | 0.09 |
Tmem247 |
transmembrane protein 247 |
11036 |
0.14 |
| chr6_86480319_86480711 | 0.09 |
A430078I02Rik |
RIKEN cDNA A430078I02 gene |
255 |
0.81 |
| chr7_132931708_132932726 | 0.09 |
1500002F19Rik |
RIKEN cDNA 1500002F19 gene |
1020 |
0.34 |
| chr4_150080632_150080823 | 0.09 |
Gpr157 |
G protein-coupled receptor 157 |
6638 |
0.11 |
| chr3_116080235_116080422 | 0.09 |
Gm26344 |
predicted gene, 26344 |
14409 |
0.14 |
| chr7_130016072_130016266 | 0.09 |
Gm23847 |
predicted gene, 23847 |
18925 |
0.24 |
| chr11_48819928_48820131 | 0.09 |
Trim41 |
tripartite motif-containing 41 |
2676 |
0.13 |
| chr1_181286269_181286427 | 0.09 |
Gm8146 |
predicted gene 8146 |
4302 |
0.19 |
| chr10_80622428_80622591 | 0.09 |
Csnk1g2 |
casein kinase 1, gamma 2 |
329 |
0.73 |
| chr4_65548543_65548702 | 0.09 |
Trim32 |
tripartite motif-containing 32 |
56364 |
0.18 |
| chr8_84743171_84743345 | 0.09 |
G430095P16Rik |
RIKEN cDNA G430095P16 gene |
20251 |
0.09 |
| chr11_55455186_55455561 | 0.09 |
Atox1 |
antioxidant 1 copper chaperone |
5824 |
0.12 |
| chr2_167351419_167351615 | 0.09 |
B4galt5 |
UDP-Gal:betaGlcNAc beta 1,4-galactosyltransferase, polypeptide 5 |
2334 |
0.3 |
| chr5_104399237_104399791 | 0.09 |
Spp1 |
secreted phosphoprotein 1 |
35604 |
0.12 |
| chr8_11012184_11012401 | 0.09 |
Irs2 |
insulin receptor substrate 2 |
3834 |
0.16 |
| chr8_107396318_107396469 | 0.09 |
Nqo1 |
NAD(P)H dehydrogenase, quinone 1 |
6813 |
0.15 |
| chr18_38312030_38312222 | 0.09 |
Rnf14 |
ring finger protein 14 |
12374 |
0.1 |
| chr12_79798610_79798778 | 0.09 |
9430078K24Rik |
RIKEN cDNA 9430078K24 gene |
126039 |
0.05 |
| chr8_72416870_72417051 | 0.09 |
Eps15l1 |
epidermal growth factor receptor pathway substrate 15-like 1 |
4478 |
0.11 |
| chr3_122417274_122417702 | 0.09 |
Bcar3 |
breast cancer anti-estrogen resistance 3 |
2269 |
0.24 |
| chr18_11807467_11807624 | 0.09 |
Cables1 |
CDK5 and Abl enzyme substrate 1 |
31675 |
0.13 |
| chr12_71718820_71718971 | 0.09 |
Gm47555 |
predicted gene, 47555 |
35490 |
0.17 |
| chr15_67420967_67421299 | 0.09 |
1700012I11Rik |
RIKEN cDNA 1700012I11 gene |
194364 |
0.03 |
| chrX_139807214_139807374 | 0.09 |
Cldn2 |
claudin 2 |
1213 |
0.4 |
| chr5_147894276_147894456 | 0.08 |
Slc46a3 |
solute carrier family 46, member 3 |
449 |
0.8 |
| chr9_65314578_65314742 | 0.08 |
Clpx |
caseinolytic mitochondrial matrix peptidase chaperone subunit |
4508 |
0.11 |
| chr15_96674208_96674363 | 0.08 |
Gm22045 |
predicted gene, 22045 |
2492 |
0.26 |
| chr14_21174635_21174786 | 0.08 |
Adk |
adenosine kinase |
98558 |
0.07 |
| chr16_23472139_23472320 | 0.08 |
Gm49514 |
predicted gene, 49514 |
42041 |
0.1 |
| chr3_115815549_115815774 | 0.08 |
Dph5 |
diphthamide biosynthesis 5 |
72176 |
0.07 |
| chr4_133751634_133751971 | 0.08 |
Arid1a |
AT rich interactive domain 1A (SWI-like) |
1809 |
0.28 |
| chr6_71172799_71172983 | 0.08 |
Gm44427 |
predicted gene, 44427 |
8092 |
0.13 |
| chr13_12550644_12550996 | 0.08 |
Gm30239 |
predicted gene, 30239 |
7420 |
0.14 |
| chr19_47536758_47536981 | 0.08 |
Stn1 |
STN1, CST complex subunit |
27 |
0.97 |
| chr14_105941342_105941672 | 0.08 |
Spry2 |
sprouty RTK signaling antagonist 2 |
44688 |
0.16 |
| chr3_79525252_79525403 | 0.08 |
Fnip2 |
folliculin interacting protein 2 |
6549 |
0.17 |
| chr2_92426557_92426708 | 0.08 |
Cry2 |
cryptochrome 2 (photolyase-like) |
5760 |
0.12 |
| chr11_95412110_95412292 | 0.08 |
Spop |
speckle-type BTB/POZ protein |
1879 |
0.25 |
| chr14_20969178_20969340 | 0.08 |
Vcl |
vinculin |
39861 |
0.15 |
| chr4_60066252_60066431 | 0.08 |
Mup7 |
major urinary protein 7 |
4070 |
0.21 |
| chr11_64588556_64588707 | 0.08 |
Gm24275 |
predicted gene, 24275 |
2009 |
0.47 |
| chr3_36862749_36862934 | 0.08 |
4932438A13Rik |
RIKEN cDNA 4932438A13 gene |
263 |
0.93 |
| chr14_41098877_41099452 | 0.08 |
Mat1a |
methionine adenosyltransferase I, alpha |
5871 |
0.13 |
| chr8_107425180_107425410 | 0.08 |
Nob1 |
NIN1/RPN12 binding protein 1 homolog |
244 |
0.89 |
| chr9_121672421_121672607 | 0.08 |
Vipr1 |
vasoactive intestinal peptide receptor 1 |
3801 |
0.14 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0003347 | epicardial cell to mesenchymal cell transition(GO:0003347) |
| 0.0 | 0.1 | GO:0042045 | epithelial fluid transport(GO:0042045) |
| 0.0 | 0.1 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) |
| 0.0 | 0.1 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.1 | GO:0002669 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.0 | 0.0 | GO:0032074 | negative regulation of nuclease activity(GO:0032074) |
| 0.0 | 0.0 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.0 | GO:0003431 | growth plate cartilage chondrocyte development(GO:0003431) |
| 0.0 | 0.1 | GO:2000402 | negative regulation of lymphocyte migration(GO:2000402) |
| 0.0 | 0.1 | GO:2000138 | positive regulation of cell proliferation involved in heart morphogenesis(GO:2000138) |
| 0.0 | 0.1 | GO:0006548 | histidine catabolic process(GO:0006548) imidazole-containing compound catabolic process(GO:0052805) |
| 0.0 | 0.1 | GO:0010273 | detoxification of copper ion(GO:0010273) stress response to copper ion(GO:1990169) |
| 0.0 | 0.0 | GO:1902075 | cellular response to salt(GO:1902075) |
| 0.0 | 0.0 | GO:0010956 | negative regulation of calcidiol 1-monooxygenase activity(GO:0010956) |
| 0.0 | 0.0 | GO:0034473 | U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) |
| 0.0 | 0.1 | GO:0060484 | lung-associated mesenchyme development(GO:0060484) |
| 0.0 | 0.0 | GO:0007161 | calcium-independent cell-matrix adhesion(GO:0007161) |
| 0.0 | 0.0 | GO:0002019 | regulation of renal output by angiotensin(GO:0002019) |
| 0.0 | 0.0 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.0 | 0.1 | GO:0016081 | synaptic vesicle docking(GO:0016081) |
| 0.0 | 0.0 | GO:0010835 | regulation of protein ADP-ribosylation(GO:0010835) |
| 0.0 | 0.0 | GO:0021914 | negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021914) |
| 0.0 | 0.1 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.0 | 0.0 | GO:0002667 | lymphocyte anergy(GO:0002249) regulation of T cell anergy(GO:0002667) T cell anergy(GO:0002870) regulation of lymphocyte anergy(GO:0002911) |
| 0.0 | 0.0 | GO:0006868 | glutamine transport(GO:0006868) |
| 0.0 | 0.0 | GO:0010748 | negative regulation of plasma membrane long-chain fatty acid transport(GO:0010748) |
| 0.0 | 0.0 | GO:0001907 | killing by symbiont of host cells(GO:0001907) disruption by symbiont of host cell(GO:0044004) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.0 | 0.1 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.0 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.0 | 0.1 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.0 | 0.0 | GO:0034666 | integrin alpha2-beta1 complex(GO:0034666) |
| 0.0 | 0.0 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 0.0 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.0 | GO:0042585 | germinal vesicle(GO:0042585) |
| 0.0 | 0.0 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 0.1 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.1 | GO:0031800 | type 3 metabotropic glutamate receptor binding(GO:0031800) |
| 0.0 | 0.1 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.0 | 0.0 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 0.0 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) |
| 0.0 | 0.1 | GO:0008390 | testosterone 16-alpha-hydroxylase activity(GO:0008390) |
| 0.0 | 0.0 | GO:0061513 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.0 | 0.0 | GO:0031711 | bradykinin receptor binding(GO:0031711) |
| 0.0 | 0.0 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.0 | 0.1 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 0.0 | GO:0015186 | L-asparagine transmembrane transporter activity(GO:0015182) L-glutamine transmembrane transporter activity(GO:0015186) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |