Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
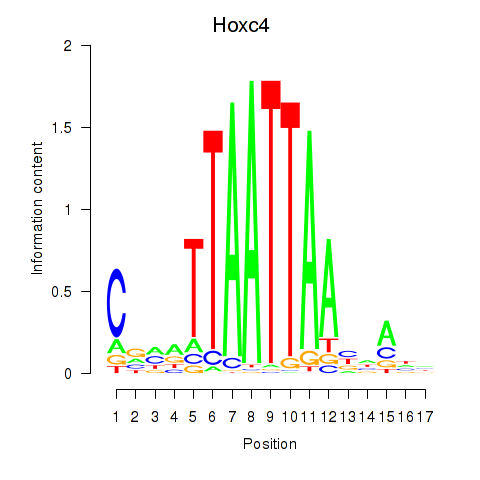
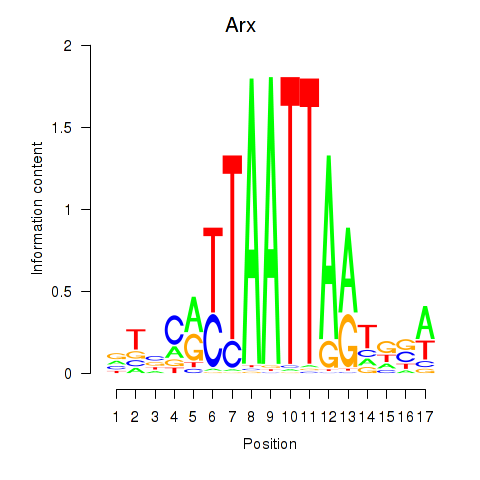
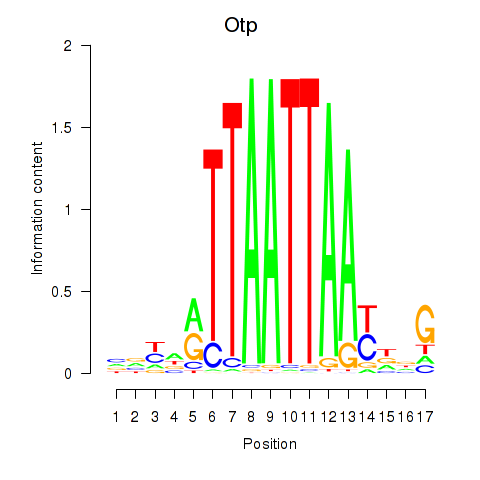
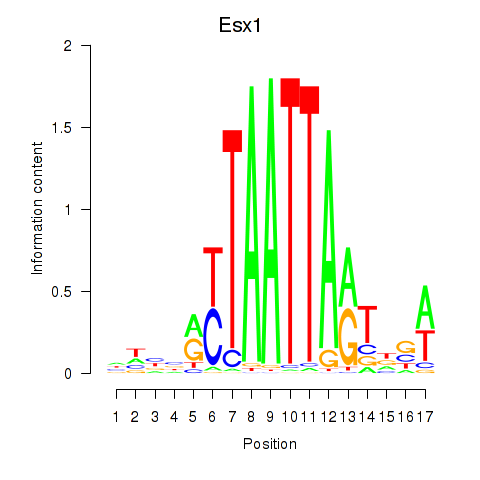
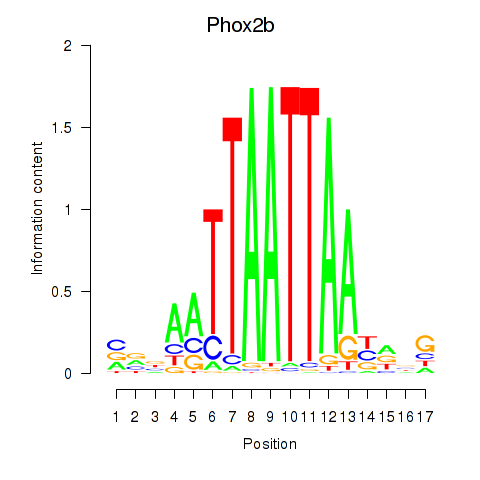
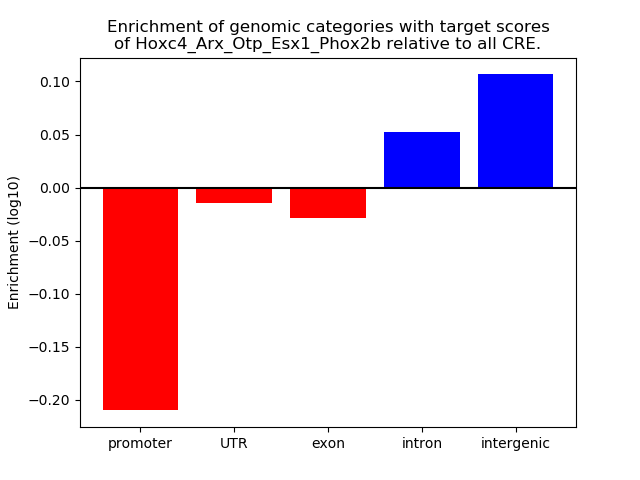
Results for Hoxc4_Arx_Otp_Esx1_Phox2b
Z-value: 0.88
Transcription factors associated with Hoxc4_Arx_Otp_Esx1_Phox2b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hoxc4
|
ENSMUSG00000075394.3 | homeobox C4 |
|
Arx
|
ENSMUSG00000035277.9 | aristaless related homeobox |
|
Otp
|
ENSMUSG00000021685.10 | orthopedia homeobox |
|
Esx1
|
ENSMUSG00000023443.7 | extraembryonic, spermatogenesis, homeobox 1 |
|
Phox2b
|
ENSMUSG00000012520.8 | paired-like homeobox 2b |
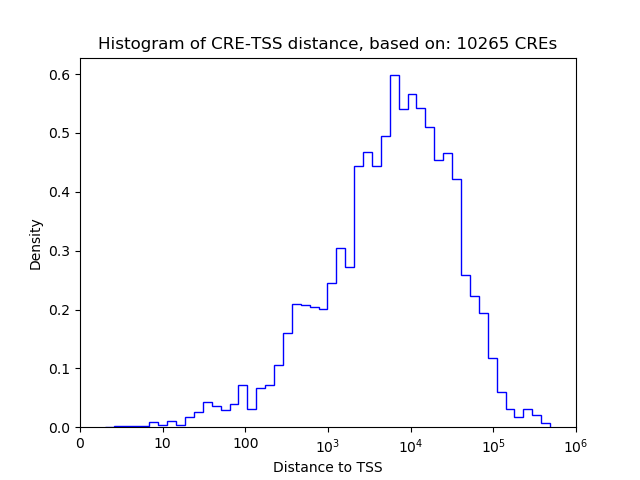
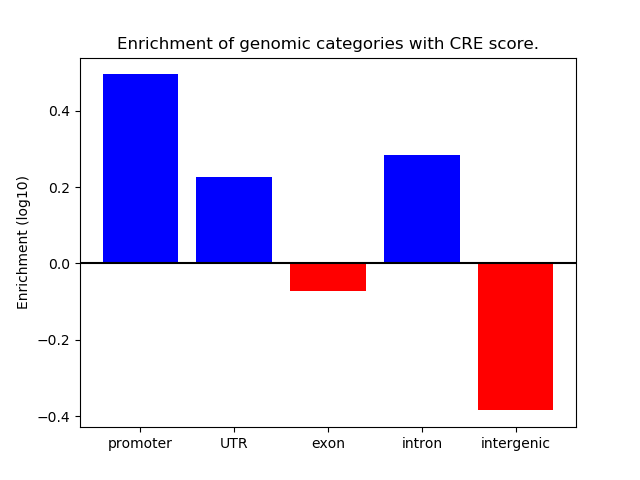
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chrX_93281375_93281550 | Arx | 4983 | 0.249545 | -0.75 | 8.5e-02 | Click! |
| chrX_93305904_93306060 | Arx | 19472 | 0.219806 | -0.54 | 2.6e-01 | Click! |
| chr13_94929247_94929398 | Otp | 46722 | 0.112046 | -0.39 | 4.5e-01 | Click! |
| chr13_94868720_94868871 | Otp | 6807 | 0.208618 | -0.14 | 7.9e-01 | Click! |
Activity of the Hoxc4_Arx_Otp_Esx1_Phox2b motif across conditions
Conditions sorted by the z-value of the Hoxc4_Arx_Otp_Esx1_Phox2b motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
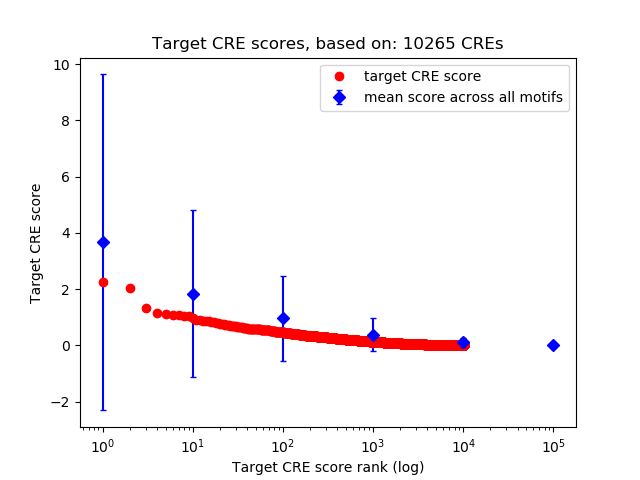
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr5_138080510_138080792 | 2.25 |
Zkscan1 |
zinc finger with KRAB and SCAN domains 1 |
4433 |
0.1 |
| chr12_21142640_21142894 | 2.05 |
Asap2 |
ArfGAP with SH3 domain, ankyrin repeat and PH domain 2 |
30813 |
0.16 |
| chr19_43971247_43971535 | 1.32 |
Cpn1 |
carboxypeptidase N, polypeptide 1 |
2698 |
0.21 |
| chr13_64229004_64229155 | 1.16 |
Cdc14b |
CDC14 cell division cycle 14B |
16797 |
0.1 |
| chr3_133765600_133766533 | 1.11 |
Gm6135 |
prediticted gene 6135 |
25438 |
0.2 |
| chr10_115337191_115337351 | 1.07 |
Tmem19 |
transmembrane protein 19 |
11793 |
0.14 |
| chr13_59875722_59876043 | 1.06 |
Gm48384 |
predicted gene, 48384 |
2240 |
0.23 |
| chr13_117745874_117746186 | 1.04 |
4933413L06Rik |
RIKEN cDNA 4933413L06 gene |
26019 |
0.26 |
| chr5_150228070_150228236 | 1.02 |
Gm36378 |
predicted gene, 36378 |
15710 |
0.19 |
| chr2_165921904_165922055 | 0.97 |
Gm11461 |
predicted gene 11461 |
3862 |
0.16 |
| chr11_86917162_86917343 | 0.90 |
Ypel2 |
yippee like 2 |
54772 |
0.11 |
| chr10_31644816_31644968 | 0.90 |
Gm8793 |
predicted gene 8793 |
5734 |
0.23 |
| chr16_24394028_24394581 | 0.88 |
Lpp |
LIM domain containing preferred translocation partner in lipoma |
566 |
0.73 |
| chr6_121878156_121878361 | 0.87 |
Mug1 |
murinoglobulin 1 |
7319 |
0.2 |
| chr11_112218841_112219019 | 0.85 |
Gm11680 |
predicted gene 11680 |
111492 |
0.07 |
| chr13_58058115_58058280 | 0.85 |
Mir874 |
microRNA 874 |
34997 |
0.13 |
| chr1_181000299_181000450 | 0.82 |
Ephx1 |
epoxide hydrolase 1, microsomal |
4204 |
0.11 |
| chr12_78248706_78249176 | 0.81 |
Gm18899 |
predicted gene, 18899 |
7494 |
0.15 |
| chr15_31279226_31279377 | 0.79 |
Gm49296 |
predicted gene, 49296 |
2788 |
0.19 |
| chr6_121874198_121874349 | 0.76 |
Mug1 |
murinoglobulin 1 |
11304 |
0.19 |
| chr2_136986571_136986744 | 0.75 |
Slx4ip |
SLX4 interacting protein |
81114 |
0.08 |
| chr6_121890040_121890777 | 0.75 |
Mug1 |
murinoglobulin 1 |
4831 |
0.21 |
| chrX_48193537_48193988 | 0.74 |
Zdhhc9 |
zinc finger, DHHC domain containing 9 |
14566 |
0.16 |
| chr14_50991070_50991230 | 0.73 |
Gm49038 |
predicted gene, 49038 |
5925 |
0.08 |
| chr1_119253363_119253514 | 0.71 |
Gm3508 |
predicted gene 3508 |
10685 |
0.22 |
| chr4_54875588_54875760 | 0.70 |
Gm12479 |
predicted gene 12479 |
22546 |
0.23 |
| chr12_4526866_4527037 | 0.69 |
Gm31938 |
predicted gene, 31938 |
2838 |
0.19 |
| chr3_94338287_94338715 | 0.69 |
Gm43743 |
predicted gene 43743 |
3169 |
0.1 |
| chr8_36706136_36706391 | 0.69 |
Dlc1 |
deleted in liver cancer 1 |
26791 |
0.24 |
| chr16_10952807_10952965 | 0.69 |
Gm26268 |
predicted gene, 26268 |
12838 |
0.11 |
| chr19_38356216_38356572 | 0.66 |
Gm50150 |
predicted gene, 50150 |
13780 |
0.14 |
| chr3_107949519_107949837 | 0.65 |
Gm12494 |
predicted gene 12494 |
3443 |
0.09 |
| chr13_43232894_43233157 | 0.65 |
Tbc1d7 |
TBC1 domain family, member 7 |
61524 |
0.1 |
| chr10_28095778_28096012 | 0.64 |
Ptprk |
protein tyrosine phosphatase, receptor type, K |
20885 |
0.21 |
| chr9_105878487_105878719 | 0.64 |
Col6a5 |
collagen, type VI, alpha 5 |
3203 |
0.27 |
| chr13_93629307_93629685 | 0.63 |
Gm15622 |
predicted gene 15622 |
4114 |
0.18 |
| chr2_165559022_165559181 | 0.63 |
Eya2 |
EYA transcriptional coactivator and phosphatase 2 |
35931 |
0.12 |
| chr14_20319722_20320198 | 0.62 |
Nudt13 |
nudix (nucleoside diphosphate linked moiety X)-type motif 13 |
8531 |
0.12 |
| chr2_45443502_45443687 | 0.61 |
Gm13479 |
predicted gene 13479 |
86329 |
0.09 |
| chr4_148085280_148085458 | 0.61 |
Agtrap |
angiotensin II, type I receptor-associated protein |
593 |
0.56 |
| chr10_40095200_40095431 | 0.60 |
Gm25613 |
predicted gene, 25613 |
14147 |
0.14 |
| chr9_57391313_57391472 | 0.59 |
Ppcdc |
phosphopantothenoylcysteine decarboxylase |
29926 |
0.12 |
| chr3_119368468_119368634 | 0.59 |
Gm23432 |
predicted gene, 23432 |
270323 |
0.02 |
| chr11_49088297_49088883 | 0.58 |
Gm12188 |
predicted gene 12188 |
47 |
0.79 |
| chr13_43500025_43500247 | 0.58 |
Gm47683 |
predicted gene, 47683 |
31 |
0.97 |
| chr6_148139673_148139824 | 0.58 |
Ergic2 |
ERGIC and golgi 2 |
43460 |
0.13 |
| chr1_119189619_119189770 | 0.58 |
Gm8321 |
predicted gene 8321 |
32108 |
0.17 |
| chr11_81374417_81374616 | 0.58 |
4930527B05Rik |
RIKEN cDNA 4930527B05 gene |
10497 |
0.3 |
| chr18_46336206_46336490 | 0.58 |
4930415P13Rik |
RIKEN cDNA 4930415P13 gene |
4081 |
0.17 |
| chr5_45476407_45476609 | 0.58 |
Lap3 |
leucine aminopeptidase 3 |
16866 |
0.11 |
| chr15_38661347_38661720 | 0.57 |
Atp6v1c1 |
ATPase, H+ transporting, lysosomal V1 subunit C1 |
400 |
0.79 |
| chr10_96854885_96855043 | 0.57 |
Gm24587 |
predicted gene, 24587 |
706 |
0.71 |
| chr18_65149682_65149843 | 0.57 |
Nedd4l |
neural precursor cell expressed, developmentally down-regulated gene 4-like |
5972 |
0.25 |
| chr9_7571293_7571444 | 0.56 |
Mmp27 |
matrix metallopeptidase 27 |
28 |
0.97 |
| chr9_44591082_44591233 | 0.56 |
Gm47231 |
predicted gene, 47231 |
6861 |
0.08 |
| chr1_165609107_165609258 | 0.56 |
Mpzl1 |
myelin protein zero-like 1 |
364 |
0.78 |
| chr17_18321520_18321874 | 0.56 |
Vmn2r93 |
vomeronasal 2, receptor 93 |
23598 |
0.19 |
| chr11_100836746_100836921 | 0.55 |
Stat5b |
signal transducer and activator of transcription 5B |
13705 |
0.12 |
| chr4_55245568_55245866 | 0.55 |
Gm12508 |
predicted gene 12508 |
9023 |
0.17 |
| chr17_30632040_30632191 | 0.55 |
Dnah8 |
dynein, axonemal, heavy chain 8 |
3641 |
0.14 |
| chr10_43378887_43379063 | 0.55 |
Rps19-ps11 |
ribosomal protein S19, pseudogene 11 |
5055 |
0.15 |
| chr1_24612181_24612351 | 0.55 |
Gm28439 |
predicted gene 28439 |
144 |
0.72 |
| chr5_114553798_114553965 | 0.54 |
Gm13790 |
predicted gene 13790 |
2592 |
0.24 |
| chr2_164158645_164158808 | 0.54 |
n-R5s207 |
nuclear encoded rRNA 5S 207 |
1205 |
0.29 |
| chr12_8643213_8643385 | 0.54 |
Pum2 |
pumilio RNA-binding family member 2 |
30835 |
0.16 |
| chr8_128687947_128688299 | 0.54 |
Itgb1 |
integrin beta 1 (fibronectin receptor beta) |
2253 |
0.29 |
| chr19_36629275_36629447 | 0.54 |
Hectd2os |
Hectd2, opposite strand |
3337 |
0.28 |
| chr11_95711926_95712160 | 0.53 |
Zfp652 |
zinc finger protein 652 |
630 |
0.43 |
| chr3_119194156_119194307 | 0.53 |
Gm43410 |
predicted gene 43410 |
325987 |
0.01 |
| chr6_65569857_65570008 | 0.53 |
Tnip3 |
TNFAIP3 interacting protein 3 |
20466 |
0.22 |
| chr1_183979753_183980270 | 0.53 |
Dusp10 |
dual specificity phosphatase 10 |
33291 |
0.16 |
| chr12_85291673_85291832 | 0.52 |
Zc2hc1c |
zinc finger, C2HC-type containing 1C |
3161 |
0.14 |
| chr11_70713419_70713657 | 0.52 |
Mir6925 |
microRNA 6925 |
7548 |
0.08 |
| chr8_4313834_4314030 | 0.52 |
Elavl1 |
ELAV (embryonic lethal, abnormal vision)-like 1 (Hu antigen R) |
1313 |
0.29 |
| chr3_80058876_80059038 | 0.51 |
A330069K06Rik |
RIKEN cDNA A330069K06 gene |
4065 |
0.25 |
| chr3_146778044_146778430 | 0.51 |
Prkacb |
protein kinase, cAMP dependent, catalytic, beta |
3179 |
0.23 |
| chr7_122666197_122666401 | 0.51 |
Cacng3 |
calcium channel, voltage-dependent, gamma subunit 3 |
4193 |
0.22 |
| chr12_28636385_28636550 | 0.50 |
Rps7 |
ribosomal protein S7 |
514 |
0.71 |
| chr3_119261070_119261221 | 0.50 |
Gm23432 |
predicted gene, 23432 |
377729 |
0.01 |
| chr11_116021292_116021891 | 0.50 |
H3f3b |
H3.3 histone B |
2882 |
0.14 |
| chr2_179292139_179292587 | 0.49 |
Gm14293 |
predicted gene 14293 |
51873 |
0.14 |
| chr15_64309233_64309403 | 0.49 |
Asap1 |
ArfGAP with SH3 domain, ankyrin repeat and PH domain1 |
2371 |
0.28 |
| chr2_126562513_126562799 | 0.49 |
Slc27a2 |
solute carrier family 27 (fatty acid transporter), member 2 |
2104 |
0.32 |
| chr4_6539560_6539735 | 0.48 |
Gm11801 |
predicted gene 11801 |
86 |
0.98 |
| chr18_57571362_57571554 | 0.48 |
Ccdc192 |
coiled-coil domain containing 192 |
8282 |
0.17 |
| chr11_109475236_109475399 | 0.48 |
Slc16a6 |
solute carrier family 16 (monocarboxylic acid transporters), member 6 |
1719 |
0.22 |
| chr3_108030847_108030998 | 0.48 |
Gm12499 |
predicted gene 12499 |
439 |
0.6 |
| chr3_81233539_81233911 | 0.48 |
Gm37300 |
predicted gene, 37300 |
171847 |
0.03 |
| chr11_23954708_23954885 | 0.48 |
Gm12062 |
predicted gene 12062 |
25942 |
0.16 |
| chr1_59381588_59381854 | 0.48 |
Gm29016 |
predicted gene 29016 |
2791 |
0.27 |
| chr9_116291010_116291488 | 0.48 |
D730003K21Rik |
RIKEN cDNA D730003K21 gene |
36088 |
0.18 |
| chr12_71840957_71841139 | 0.47 |
Gm7985 |
predicted gene 7985 |
6243 |
0.21 |
| chr1_40117818_40118037 | 0.47 |
Gm37347 |
predicted gene, 37347 |
29416 |
0.14 |
| chr4_116019975_116020341 | 0.47 |
Faah |
fatty acid amide hydrolase |
2232 |
0.22 |
| chr15_100400715_100400907 | 0.47 |
Slc11a2 |
solute carrier family 11 (proton-coupled divalent metal ion transporters), member 2 |
2432 |
0.17 |
| chr13_8591659_8591833 | 0.46 |
Gm48262 |
predicted gene, 48262 |
38726 |
0.19 |
| chr13_30242706_30242889 | 0.46 |
Mboat1 |
membrane bound O-acyltransferase domain containing 1 |
10944 |
0.22 |
| chr12_111454567_111454858 | 0.46 |
Tnfaip2 |
tumor necrosis factor, alpha-induced protein 2 |
4193 |
0.14 |
| chr2_132654710_132655013 | 0.46 |
Rpl23a-ps4 |
ribosomal protein L23A, pseudogene 4 |
7381 |
0.12 |
| chr19_5840987_5841190 | 0.46 |
Neat1 |
nuclear paraspeckle assembly transcript 1 (non-protein coding) |
4171 |
0.09 |
| chr19_56821183_56821529 | 0.46 |
Ccdc186 |
coiled-coil domain containing 186 |
834 |
0.58 |
| chr18_61398040_61398397 | 0.46 |
Mir378a |
microRNA 378a |
318 |
0.54 |
| chr19_46138051_46138212 | 0.45 |
Pitx3 |
paired-like homeodomain transcription factor 3 |
842 |
0.48 |
| chr9_70935063_70935240 | 0.45 |
Lipc |
lipase, hepatic |
343 |
0.89 |
| chr14_100132110_100132261 | 0.45 |
Klf12 |
Kruppel-like factor 12 |
17579 |
0.24 |
| chr19_61057720_61057890 | 0.45 |
Gm22520 |
predicted gene, 22520 |
44260 |
0.12 |
| chr17_24007025_24007189 | 0.45 |
Gm50066 |
predicted gene, 50066 |
2495 |
0.11 |
| chr17_29381584_29381926 | 0.45 |
Fgd2 |
FYVE, RhoGEF and PH domain containing 2 |
5254 |
0.14 |
| chr13_93653363_93653698 | 0.45 |
Bhmt |
betaine-homocysteine methyltransferase |
15569 |
0.13 |
| chr4_116689870_116690150 | 0.45 |
Prdx1 |
peroxiredoxin 1 |
2698 |
0.15 |
| chr5_147469423_147469844 | 0.44 |
Pan3 |
PAN3 poly(A) specific ribonuclease subunit |
16344 |
0.12 |
| chr9_36794772_36794937 | 0.44 |
Ei24 |
etoposide induced 2.4 mRNA |
2166 |
0.21 |
| chr16_76349002_76349359 | 0.44 |
Nrip1 |
nuclear receptor interacting protein 1 |
23857 |
0.18 |
| chr14_74866246_74866397 | 0.43 |
Lrch1 |
leucine-rich repeats and calponin homology (CH) domain containing 1 |
18934 |
0.21 |
| chr7_38187215_38187367 | 0.43 |
1600014C10Rik |
RIKEN cDNA 1600014C10 gene |
1020 |
0.43 |
| chr19_30096271_30096642 | 0.43 |
Uhrf2 |
ubiquitin-like, containing PHD and RING finger domains 2 |
4495 |
0.24 |
| chr3_18054195_18054346 | 0.43 |
Bhlhe22 |
basic helix-loop-helix family, member e22 |
96 |
0.97 |
| chr14_66552999_66553150 | 0.43 |
Gm23899 |
predicted gene, 23899 |
38621 |
0.18 |
| chr4_33229770_33229937 | 0.42 |
Pnrc1 |
proline-rich nuclear receptor coactivator 1 |
17722 |
0.14 |
| chr2_169724086_169724281 | 0.42 |
Tshz2 |
teashirt zinc finger family member 2 |
90507 |
0.08 |
| chr14_60635934_60636102 | 0.42 |
Spata13 |
spermatogenesis associated 13 |
1263 |
0.52 |
| chr15_54947760_54948020 | 0.42 |
Gm26684 |
predicted gene, 26684 |
4180 |
0.19 |
| chr6_5496841_5497314 | 0.42 |
Pdk4 |
pyruvate dehydrogenase kinase, isoenzyme 4 |
768 |
0.76 |
| chr6_17255791_17255960 | 0.42 |
Cav2 |
caveolin 2 |
25310 |
0.17 |
| chr15_77208076_77208227 | 0.41 |
Rbfox2 |
RNA binding protein, fox-1 homolog (C. elegans) 2 |
4715 |
0.18 |
| chrX_161939930_161940295 | 0.41 |
Gm15202 |
predicted gene 15202 |
31889 |
0.21 |
| chr11_30792834_30792985 | 0.41 |
Psme4 |
proteasome (prosome, macropain) activator subunit 4 |
18628 |
0.14 |
| chr4_132129267_132129447 | 0.41 |
Oprd1 |
opioid receptor, delta 1 |
15129 |
0.1 |
| chr3_138426007_138426180 | 0.41 |
Adh4 |
alcohol dehydrogenase 4 (class II), pi polypeptide |
10598 |
0.13 |
| chr4_104869752_104870593 | 0.41 |
C8a |
complement component 8, alpha polypeptide |
6211 |
0.21 |
| chr7_75470307_75470471 | 0.41 |
Gm44834 |
predicted gene 44834 |
11610 |
0.16 |
| chr17_56470014_56470183 | 0.41 |
Ptprs |
protein tyrosine phosphatase, receptor type, S |
4529 |
0.17 |
| chr10_41627379_41627538 | 0.41 |
Ccdc162 |
coiled-coil domain containing 162 |
14621 |
0.15 |
| chr8_71405347_71405498 | 0.41 |
Ankle1 |
ankyrin repeat and LEM domain containing 1 |
588 |
0.54 |
| chr6_121872993_121873786 | 0.40 |
Mug1 |
murinoglobulin 1 |
12188 |
0.19 |
| chr7_127666140_127666314 | 0.40 |
Gm44760 |
predicted gene 44760 |
7993 |
0.08 |
| chr7_84171490_84171660 | 0.40 |
Gm22177 |
predicted gene, 22177 |
1010 |
0.47 |
| chr14_119831250_119831463 | 0.40 |
4930404K13Rik |
RIKEN cDNA 4930404K13 gene |
32378 |
0.19 |
| chr2_36092387_36092643 | 0.40 |
Lhx6 |
LIM homeobox protein 6 |
1758 |
0.26 |
| chr13_54566023_54566188 | 0.40 |
4833439L19Rik |
RIKEN cDNA 4833439L19 gene |
670 |
0.53 |
| chr13_20090144_20090306 | 0.40 |
Elmo1 |
engulfment and cell motility 1 |
371 |
0.92 |
| chr15_82290409_82290560 | 0.39 |
Septin3 |
septin 3 |
6054 |
0.09 |
| chr6_35874026_35874908 | 0.39 |
Gm43442 |
predicted gene 43442 |
52244 |
0.17 |
| chr2_66712021_66712193 | 0.39 |
Scn7a |
sodium channel, voltage-gated, type VII, alpha |
72807 |
0.1 |
| chr1_136229022_136229181 | 0.39 |
Inava |
innate immunity activator |
918 |
0.4 |
| chr10_25563128_25563279 | 0.39 |
Gm29571 |
predicted gene 29571 |
26817 |
0.14 |
| chr12_3575421_3575577 | 0.39 |
Dtnb |
dystrobrevin, beta |
2515 |
0.31 |
| chr2_14099979_14100130 | 0.39 |
Stam |
signal transducing adaptor molecule (SH3 domain and ITAM motif) 1 |
25627 |
0.12 |
| chr3_81676672_81676850 | 0.39 |
Gm43346 |
predicted gene 43346 |
44915 |
0.18 |
| chr5_113041682_113041833 | 0.38 |
Gm22740 |
predicted gene, 22740 |
2387 |
0.2 |
| chr8_12804734_12804948 | 0.38 |
Atp11a |
ATPase, class VI, type 11A |
6575 |
0.17 |
| chr1_180211878_180212203 | 0.38 |
Gm37336 |
predicted gene, 37336 |
296 |
0.87 |
| chr9_77921035_77921417 | 0.38 |
Elovl5 |
ELOVL family member 5, elongation of long chain fatty acids (yeast) |
706 |
0.62 |
| chr8_71699244_71699423 | 0.38 |
B3gnt3 |
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 3 |
2456 |
0.13 |
| chr6_39241464_39242040 | 0.38 |
Gm43479 |
predicted gene 43479 |
1638 |
0.32 |
| chr7_34485188_34485527 | 0.37 |
Gm12780 |
predicted gene 12780 |
10708 |
0.16 |
| chr10_81407875_81408026 | 0.37 |
Nfic |
nuclear factor I/C |
309 |
0.68 |
| chr9_70942553_70942713 | 0.37 |
Lipc |
lipase, hepatic |
7825 |
0.21 |
| chr6_134743647_134743846 | 0.37 |
Dusp16 |
dual specificity phosphatase 16 |
80 |
0.96 |
| chr15_34493251_34493646 | 0.37 |
Rida |
reactive intermediate imine deaminase A homolog |
1723 |
0.21 |
| chr12_118887325_118887612 | 0.37 |
Abcb5 |
ATP-binding cassette, sub-family B (MDR/TAP), member 5 |
3291 |
0.3 |
| chr1_75652903_75653256 | 0.37 |
Gm5257 |
predicted gene 5257 |
16689 |
0.16 |
| chr19_42126675_42126826 | 0.36 |
Avpi1 |
arginine vasopressin-induced 1 |
2239 |
0.19 |
| chr9_8134789_8134974 | 0.36 |
Cep126 |
centrosomal protein 126 |
587 |
0.72 |
| chr9_66414274_66414486 | 0.36 |
Gm28379 |
predicted gene 28379 |
6261 |
0.21 |
| chr10_68276454_68276805 | 0.36 |
Arid5b |
AT rich interactive domain 5B (MRF1-like) |
2092 |
0.36 |
| chr9_58242475_58242626 | 0.36 |
Pml |
promyelocytic leukemia |
7073 |
0.12 |
| chr6_120242684_120242877 | 0.36 |
B4galnt3 |
beta-1,4-N-acetyl-galactosaminyl transferase 3 |
137 |
0.97 |
| chr19_3864646_3865022 | 0.35 |
Gm50383 |
predicted gene, 50383 |
2839 |
0.13 |
| chr12_8217539_8217881 | 0.35 |
Gm33037 |
predicted gene, 33037 |
8545 |
0.19 |
| chr10_20622440_20622618 | 0.35 |
Gm17230 |
predicted gene 17230 |
3106 |
0.32 |
| chr13_102905828_102906396 | 0.35 |
Mast4 |
microtubule associated serine/threonine kinase family member 4 |
6 |
0.99 |
| chr13_74920338_74920722 | 0.35 |
Hnrnpa1l2-ps |
heterogeneous nuclear ribonucleoprotein A1-like 2, pseudogene |
56225 |
0.14 |
| chr10_115298774_115299175 | 0.35 |
Rab21 |
RAB21, member RAS oncogene family |
16617 |
0.14 |
| chr4_116695063_116695819 | 0.35 |
Prdx1 |
peroxiredoxin 1 |
8129 |
0.1 |
| chr11_45943515_45943691 | 0.35 |
Lsm11 |
U7 snRNP-specific Sm-like protein LSM11 |
1332 |
0.38 |
| chr5_74623982_74624188 | 0.35 |
Lnx1 |
ligand of numb-protein X 1 |
564 |
0.75 |
| chr9_71073997_71074158 | 0.35 |
Gm47268 |
predicted gene, 47268 |
52765 |
0.13 |
| chr3_118507933_118508602 | 0.35 |
Gm37773 |
predicted gene, 37773 |
17884 |
0.14 |
| chr19_24270105_24270479 | 0.35 |
Fxn |
frataxin |
7879 |
0.18 |
| chr10_53348308_53348525 | 0.35 |
Cep85l |
centrosomal protein 85-like |
1407 |
0.3 |
| chr12_25061032_25061199 | 0.35 |
Gm47701 |
predicted gene, 47701 |
3577 |
0.2 |
| chr17_84997546_84997697 | 0.35 |
Ppm1b |
protein phosphatase 1B, magnesium dependent, beta isoform |
3941 |
0.22 |
| chr19_21784354_21784521 | 0.34 |
Cemip2 |
cell migration inducing hyaluronidase 2 |
6049 |
0.23 |
| chr12_25161386_25161720 | 0.34 |
Gm36287 |
predicted gene, 36287 |
10638 |
0.17 |
| chr17_4196946_4197109 | 0.34 |
4930548J01Rik |
RIKEN cDNA 4930548J01 gene |
74914 |
0.12 |
| chr3_132917768_132917922 | 0.34 |
Npnt |
nephronectin |
11486 |
0.16 |
| chr5_9059681_9059838 | 0.34 |
Gm40264 |
predicted gene, 40264 |
24635 |
0.13 |
| chr14_22783164_22783347 | 0.34 |
Gm7473 |
predicted gene 7473 |
8011 |
0.31 |
| chr9_108291116_108291267 | 0.34 |
Nicn1 |
nicolin 1 |
736 |
0.43 |
| chr2_103490980_103491171 | 0.34 |
Cat |
catalase |
5915 |
0.2 |
| chr13_17716892_17717054 | 0.34 |
Gm48621 |
predicted gene, 48621 |
7109 |
0.12 |
| chr15_26355371_26355533 | 0.34 |
Marchf11 |
membrane associated ring-CH-type finger 11 |
32267 |
0.24 |
| chr7_49458254_49458469 | 0.34 |
Nav2 |
neuron navigator 2 |
7601 |
0.23 |
| chr13_42342253_42342427 | 0.34 |
Gm47118 |
predicted gene, 47118 |
37349 |
0.17 |
| chr4_32507009_32507324 | 0.34 |
Bach2 |
BTB and CNC homology, basic leucine zipper transcription factor 2 |
5661 |
0.25 |
| chr5_151137806_151137957 | 0.34 |
Stard13 |
StAR-related lipid transfer (START) domain containing 13 |
10338 |
0.25 |
| chr18_9786838_9787153 | 0.34 |
Gm23350 |
predicted gene, 23350 |
23733 |
0.11 |
| chr11_3208642_3208934 | 0.34 |
Eif4enif1 |
eukaryotic translation initiation factor 4E nuclear import factor 1 |
5158 |
0.13 |
| chr13_14192509_14192814 | 0.33 |
Arid4b |
AT rich interactive domain 4B (RBP1-like) |
1484 |
0.37 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | GO:0046473 | phosphatidic acid metabolic process(GO:0046473) |
| 0.1 | 0.6 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.1 | 0.3 | GO:0070827 | chromatin maintenance(GO:0070827) |
| 0.1 | 0.2 | GO:0015744 | succinate transport(GO:0015744) |
| 0.1 | 0.3 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.1 | 0.6 | GO:0002489 | antigen processing and presentation of endogenous peptide antigen via MHC class Ib via ER pathway(GO:0002488) antigen processing and presentation of endogenous peptide antigen via MHC class Ib via ER pathway, TAP-dependent(GO:0002489) |
| 0.1 | 0.2 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.1 | 0.2 | GO:0061470 | T follicular helper cell differentiation(GO:0061470) |
| 0.1 | 0.2 | GO:0007161 | calcium-independent cell-matrix adhesion(GO:0007161) |
| 0.1 | 0.1 | GO:0072309 | mesenchymal stem cell maintenance involved in metanephric nephron morphogenesis(GO:0072309) |
| 0.1 | 0.2 | GO:0070458 | detoxification of nitrogen compound(GO:0051410) cellular detoxification of nitrogen compound(GO:0070458) |
| 0.1 | 0.2 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.1 | 0.2 | GO:0061502 | early endosome to recycling endosome transport(GO:0061502) |
| 0.1 | 0.4 | GO:0071267 | amino acid salvage(GO:0043102) L-methionine biosynthetic process(GO:0071265) L-methionine salvage(GO:0071267) |
| 0.1 | 0.2 | GO:0010911 | regulation of isomerase activity(GO:0010911) positive regulation of isomerase activity(GO:0010912) |
| 0.1 | 0.2 | GO:0030997 | regulation of centriole-centriole cohesion(GO:0030997) |
| 0.1 | 0.2 | GO:0019464 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.1 | 0.2 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.1 | 0.2 | GO:0036258 | multivesicular body assembly(GO:0036258) |
| 0.1 | 0.5 | GO:0010216 | maintenance of DNA methylation(GO:0010216) |
| 0.1 | 0.1 | GO:0014873 | response to muscle activity involved in regulation of muscle adaptation(GO:0014873) |
| 0.1 | 0.2 | GO:0090230 | regulation of centromere complex assembly(GO:0090230) |
| 0.1 | 0.1 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.1 | 0.2 | GO:0061073 | ciliary body morphogenesis(GO:0061073) |
| 0.1 | 0.3 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.1 | 0.1 | GO:0060585 | regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.1 | 0.1 | GO:1902071 | regulation of hypoxia-inducible factor-1alpha signaling pathway(GO:1902071) |
| 0.1 | 0.2 | GO:1904672 | regulation of mesenchymal cell apoptotic process involved in nephron morphogenesis(GO:0072039) negative regulation of mesenchymal cell apoptotic process involved in nephron morphogenesis(GO:0072040) mesenchymal cell apoptotic process involved in nephron morphogenesis(GO:1901145) regulation of somatic stem cell population maintenance(GO:1904672) negative regulation of somatic stem cell population maintenance(GO:1904673) |
| 0.1 | 0.4 | GO:0021540 | corpus callosum morphogenesis(GO:0021540) |
| 0.1 | 0.2 | GO:0035553 | oxidative RNA demethylation(GO:0035513) oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.1 | 0.2 | GO:1903223 | positive regulation of oxidative stress-induced neuron death(GO:1903223) |
| 0.1 | 0.3 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.1 | 0.2 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.1 | 0.1 | GO:0002019 | regulation of renal output by angiotensin(GO:0002019) |
| 0.1 | 0.1 | GO:0019448 | cysteine catabolic process(GO:0009093) L-cysteine catabolic process(GO:0019448) L-cysteine metabolic process(GO:0046439) |
| 0.0 | 0.2 | GO:0045578 | negative regulation of B cell differentiation(GO:0045578) |
| 0.0 | 0.1 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.0 | 0.2 | GO:0070885 | negative regulation of calcineurin-NFAT signaling cascade(GO:0070885) |
| 0.0 | 0.1 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.0 | 0.1 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.0 | 0.2 | GO:0009115 | xanthine catabolic process(GO:0009115) |
| 0.0 | 0.2 | GO:0070836 | caveola assembly(GO:0070836) |
| 0.0 | 0.2 | GO:0072675 | osteoclast fusion(GO:0072675) |
| 0.0 | 0.2 | GO:0052805 | histidine catabolic process(GO:0006548) imidazole-containing compound catabolic process(GO:0052805) |
| 0.0 | 0.1 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.0 | 0.1 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.0 | 0.2 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.3 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) |
| 0.0 | 0.3 | GO:0071501 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.0 | 0.2 | GO:0034773 | histone H4-K20 trimethylation(GO:0034773) |
| 0.0 | 0.2 | GO:0071286 | cellular response to magnesium ion(GO:0071286) |
| 0.0 | 0.0 | GO:0032345 | negative regulation of aldosterone metabolic process(GO:0032345) negative regulation of aldosterone biosynthetic process(GO:0032348) negative regulation of cortisol biosynthetic process(GO:2000065) |
| 0.0 | 0.4 | GO:0006544 | glycine metabolic process(GO:0006544) |
| 0.0 | 0.3 | GO:0002756 | MyD88-independent toll-like receptor signaling pathway(GO:0002756) |
| 0.0 | 0.1 | GO:1990791 | dorsal root ganglion development(GO:1990791) |
| 0.0 | 0.3 | GO:0060613 | fat pad development(GO:0060613) |
| 0.0 | 0.2 | GO:0010815 | bradykinin catabolic process(GO:0010815) |
| 0.0 | 0.3 | GO:0071257 | cellular response to electrical stimulus(GO:0071257) |
| 0.0 | 0.2 | GO:0015691 | cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.0 | 0.1 | GO:2000049 | positive regulation of cell-cell adhesion mediated by cadherin(GO:2000049) |
| 0.0 | 0.2 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.0 | 0.1 | GO:0045626 | negative regulation of T-helper 1 cell differentiation(GO:0045626) |
| 0.0 | 0.1 | GO:0003213 | cardiac right atrium morphogenesis(GO:0003213) |
| 0.0 | 0.2 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.0 | 0.4 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.3 | GO:2001214 | positive regulation of vasculogenesis(GO:2001214) |
| 0.0 | 0.4 | GO:0038166 | angiotensin-activated signaling pathway(GO:0038166) |
| 0.0 | 0.1 | GO:0006933 | negative regulation of cell adhesion involved in substrate-bound cell migration(GO:0006933) |
| 0.0 | 0.1 | GO:0048633 | positive regulation of skeletal muscle tissue growth(GO:0048633) |
| 0.0 | 0.1 | GO:1904504 | regulation of lipophagy(GO:1904502) positive regulation of lipophagy(GO:1904504) |
| 0.0 | 0.1 | GO:1900041 | negative regulation of interleukin-2 secretion(GO:1900041) |
| 0.0 | 0.1 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) |
| 0.0 | 0.1 | GO:0048807 | female genitalia morphogenesis(GO:0048807) |
| 0.0 | 0.0 | GO:2000670 | positive regulation of dendritic cell apoptotic process(GO:2000670) |
| 0.0 | 0.1 | GO:1900108 | negative regulation of nodal signaling pathway(GO:1900108) |
| 0.0 | 0.1 | GO:0090071 | negative regulation of ribosome biogenesis(GO:0090071) |
| 0.0 | 0.1 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 0.0 | 0.3 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 0.1 | GO:0051791 | medium-chain fatty acid metabolic process(GO:0051791) |
| 0.0 | 0.1 | GO:0033024 | mast cell homeostasis(GO:0033023) mast cell apoptotic process(GO:0033024) regulation of mast cell apoptotic process(GO:0033025) |
| 0.0 | 0.1 | GO:0060066 | oviduct development(GO:0060066) |
| 0.0 | 0.2 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.0 | 0.1 | GO:0050705 | regulation of interleukin-1 alpha secretion(GO:0050705) |
| 0.0 | 0.1 | GO:0060744 | thelarche(GO:0042695) mammary gland branching involved in thelarche(GO:0060744) |
| 0.0 | 0.1 | GO:0032485 | regulation of Ral protein signal transduction(GO:0032485) |
| 0.0 | 0.1 | GO:0001712 | ectodermal cell fate commitment(GO:0001712) |
| 0.0 | 0.2 | GO:0010886 | positive regulation of cholesterol storage(GO:0010886) |
| 0.0 | 0.0 | GO:0018916 | nitrobenzene metabolic process(GO:0018916) |
| 0.0 | 0.1 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.0 | 0.2 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.1 | GO:0051295 | establishment of meiotic spindle localization(GO:0051295) |
| 0.0 | 0.1 | GO:0071442 | positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.0 | 0.1 | GO:0086023 | adrenergic receptor signaling pathway involved in heart process(GO:0086023) |
| 0.0 | 0.1 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.0 | 0.1 | GO:0016479 | negative regulation of transcription from RNA polymerase I promoter(GO:0016479) |
| 0.0 | 0.3 | GO:0021702 | cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.0 | 0.2 | GO:0035811 | negative regulation of urine volume(GO:0035811) |
| 0.0 | 0.1 | GO:0046098 | guanine metabolic process(GO:0046098) |
| 0.0 | 0.1 | GO:0035860 | glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.0 | 0.1 | GO:0009642 | response to light intensity(GO:0009642) |
| 0.0 | 0.3 | GO:0051895 | negative regulation of focal adhesion assembly(GO:0051895) |
| 0.0 | 0.1 | GO:0042732 | D-xylose metabolic process(GO:0042732) |
| 0.0 | 0.1 | GO:0035336 | long-chain fatty-acyl-CoA metabolic process(GO:0035336) |
| 0.0 | 0.1 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.0 | 0.2 | GO:1900246 | positive regulation of RIG-I signaling pathway(GO:1900246) |
| 0.0 | 0.1 | GO:0046501 | protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.0 | 0.1 | GO:1902564 | negative regulation of neutrophil activation(GO:1902564) |
| 0.0 | 0.1 | GO:0034627 | 'de novo' NAD biosynthetic process(GO:0034627) |
| 0.0 | 0.1 | GO:0042045 | epithelial fluid transport(GO:0042045) |
| 0.0 | 0.1 | GO:0097503 | sialylation(GO:0097503) |
| 0.0 | 0.1 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.0 | 0.1 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.0 | 0.1 | GO:0010166 | wax biosynthetic process(GO:0010025) wax metabolic process(GO:0010166) |
| 0.0 | 0.1 | GO:0097195 | pilomotor reflex(GO:0097195) |
| 0.0 | 0.1 | GO:0006681 | galactosylceramide metabolic process(GO:0006681) |
| 0.0 | 0.1 | GO:0071504 | response to heparin(GO:0071503) cellular response to heparin(GO:0071504) |
| 0.0 | 0.1 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.0 | 0.0 | GO:0060594 | mammary gland specification(GO:0060594) |
| 0.0 | 0.1 | GO:0015888 | thiamine transport(GO:0015888) |
| 0.0 | 0.1 | GO:0016078 | tRNA catabolic process(GO:0016078) |
| 0.0 | 0.1 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.0 | 0.0 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.0 | 0.1 | GO:1904263 | positive regulation of TORC1 signaling(GO:1904263) |
| 0.0 | 0.2 | GO:0046185 | aldehyde catabolic process(GO:0046185) |
| 0.0 | 0.1 | GO:1902953 | positive regulation of ER to Golgi vesicle-mediated transport(GO:1902953) |
| 0.0 | 0.1 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.0 | 0.1 | GO:0070681 | glutaminyl-tRNAGln biosynthesis via transamidation(GO:0070681) |
| 0.0 | 0.2 | GO:0046040 | IMP metabolic process(GO:0046040) |
| 0.0 | 0.1 | GO:1990086 | lens fiber cell apoptotic process(GO:1990086) |
| 0.0 | 0.1 | GO:0060762 | regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 0.0 | 0.1 | GO:1904668 | positive regulation of ubiquitin protein ligase activity(GO:1904668) |
| 0.0 | 0.1 | GO:0010744 | positive regulation of macrophage derived foam cell differentiation(GO:0010744) |
| 0.0 | 0.1 | GO:0042360 | vitamin E metabolic process(GO:0042360) |
| 0.0 | 0.1 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.0 | 0.2 | GO:2000811 | negative regulation of anoikis(GO:2000811) |
| 0.0 | 0.1 | GO:0006499 | N-terminal protein myristoylation(GO:0006499) |
| 0.0 | 0.2 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 | 0.1 | GO:0042891 | antibiotic transport(GO:0042891) |
| 0.0 | 0.1 | GO:0042997 | negative regulation of Golgi to plasma membrane protein transport(GO:0042997) |
| 0.0 | 0.1 | GO:1902093 | positive regulation of sperm motility(GO:1902093) |
| 0.0 | 0.7 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.0 | 0.0 | GO:2001274 | negative regulation of glucose import in response to insulin stimulus(GO:2001274) |
| 0.0 | 0.1 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.0 | 0.2 | GO:0061000 | negative regulation of dendritic spine development(GO:0061000) |
| 0.0 | 0.1 | GO:2000323 | negative regulation of glucocorticoid receptor signaling pathway(GO:2000323) |
| 0.0 | 0.1 | GO:0061623 | galactose catabolic process via UDP-galactose(GO:0033499) glycolytic process from galactose(GO:0061623) |
| 0.0 | 0.1 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 0.0 | 0.1 | GO:0060355 | positive regulation of cell adhesion molecule production(GO:0060355) |
| 0.0 | 0.1 | GO:1900242 | regulation of synaptic vesicle endocytosis(GO:1900242) |
| 0.0 | 0.1 | GO:0007529 | establishment of synaptic specificity at neuromuscular junction(GO:0007529) |
| 0.0 | 0.1 | GO:0015817 | histidine transport(GO:0015817) |
| 0.0 | 0.1 | GO:2000169 | regulation of peptidyl-cysteine S-nitrosylation(GO:2000169) |
| 0.0 | 0.0 | GO:0043974 | histone H3-K27 acetylation(GO:0043974) regulation of histone H3-K27 acetylation(GO:1901674) |
| 0.0 | 0.0 | GO:0021914 | negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021914) |
| 0.0 | 0.1 | GO:0044332 | Wnt signaling pathway involved in dorsal/ventral axis specification(GO:0044332) |
| 0.0 | 0.0 | GO:0071608 | macrophage inflammatory protein-1 alpha production(GO:0071608) |
| 0.0 | 0.2 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.0 | 0.2 | GO:0007097 | nuclear migration(GO:0007097) |
| 0.0 | 0.1 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 | 0.0 | GO:1900126 | negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.0 | 0.1 | GO:0046337 | phosphatidylethanolamine metabolic process(GO:0046337) |
| 0.0 | 0.0 | GO:0045590 | negative regulation of regulatory T cell differentiation(GO:0045590) |
| 0.0 | 0.0 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.0 | 0.1 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.0 | 0.1 | GO:0060789 | hair follicle placode formation(GO:0060789) |
| 0.0 | 0.1 | GO:0051365 | cellular response to potassium ion starvation(GO:0051365) |
| 0.0 | 0.1 | GO:1902231 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.0 | 0.1 | GO:0045651 | positive regulation of macrophage differentiation(GO:0045651) |
| 0.0 | 0.4 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.0 | 0.3 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.1 | GO:0051466 | positive regulation of corticotropin-releasing hormone secretion(GO:0051466) |
| 0.0 | 0.0 | GO:1902630 | regulation of membrane hyperpolarization(GO:1902630) |
| 0.0 | 0.1 | GO:0021853 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.0 | 0.0 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.0 | 0.1 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.0 | 0.1 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.0 | 0.1 | GO:0034116 | positive regulation of heterotypic cell-cell adhesion(GO:0034116) |
| 0.0 | 0.1 | GO:0007039 | protein catabolic process in the vacuole(GO:0007039) |
| 0.0 | 0.1 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 0.0 | 0.1 | GO:1901069 | guanosine-containing compound catabolic process(GO:1901069) |
| 0.0 | 0.1 | GO:0071494 | cellular response to UV-C(GO:0071494) |
| 0.0 | 0.1 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.9 | GO:0007566 | embryo implantation(GO:0007566) |
| 0.0 | 0.0 | GO:2000389 | regulation of neutrophil extravasation(GO:2000389) positive regulation of neutrophil extravasation(GO:2000391) |
| 0.0 | 0.0 | GO:2000416 | regulation of eosinophil migration(GO:2000416) positive regulation of eosinophil migration(GO:2000418) |
| 0.0 | 0.0 | GO:0019230 | proprioception(GO:0019230) |
| 0.0 | 0.0 | GO:0002378 | immunoglobulin biosynthetic process(GO:0002378) |
| 0.0 | 0.1 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.0 | GO:0031296 | B cell costimulation(GO:0031296) |
| 0.0 | 0.0 | GO:0051771 | negative regulation of nitric-oxide synthase biosynthetic process(GO:0051771) |
| 0.0 | 0.0 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.0 | 0.1 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.0 | 0.0 | GO:2000173 | negative regulation of branching morphogenesis of a nerve(GO:2000173) |
| 0.0 | 0.0 | GO:0065001 | specification of axis polarity(GO:0065001) |
| 0.0 | 0.1 | GO:0097062 | dendritic spine maintenance(GO:0097062) |
| 0.0 | 0.1 | GO:0060057 | apoptotic process involved in mammary gland involution(GO:0060057) positive regulation of apoptotic process involved in mammary gland involution(GO:0060058) positive regulation of apoptotic process involved in morphogenesis(GO:1902339) regulation of mammary gland involution(GO:1903519) positive regulation of mammary gland involution(GO:1903521) positive regulation of apoptotic process involved in development(GO:1904747) |
| 0.0 | 0.1 | GO:0071404 | cellular response to low-density lipoprotein particle stimulus(GO:0071404) |
| 0.0 | 0.0 | GO:0038108 | negative regulation of appetite by leptin-mediated signaling pathway(GO:0038108) |
| 0.0 | 0.1 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.0 | 0.0 | GO:0002606 | positive regulation of dendritic cell antigen processing and presentation(GO:0002606) |
| 0.0 | 0.1 | GO:0044336 | canonical Wnt signaling pathway involved in negative regulation of apoptotic process(GO:0044336) |
| 0.0 | 0.1 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.0 | 0.0 | GO:0035262 | gonad morphogenesis(GO:0035262) |
| 0.0 | 0.1 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.0 | 0.0 | GO:0021937 | cerebellar Purkinje cell-granule cell precursor cell signaling involved in regulation of granule cell precursor cell proliferation(GO:0021937) smoothened signaling pathway involved in regulation of cerebellar granule cell precursor cell proliferation(GO:0021938) |
| 0.0 | 0.1 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.0 | 0.2 | GO:0035278 | miRNA mediated inhibition of translation(GO:0035278) |
| 0.0 | 0.1 | GO:1900113 | negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.0 | 0.0 | GO:0051890 | regulation of cardioblast differentiation(GO:0051890) |
| 0.0 | 0.0 | GO:0021912 | regulation of transcription from RNA polymerase II promoter involved in spinal cord motor neuron fate specification(GO:0021912) |
| 0.0 | 0.0 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.0 | GO:0048294 | negative regulation of isotype switching to IgE isotypes(GO:0048294) |
| 0.0 | 0.0 | GO:0030070 | insulin processing(GO:0030070) |
| 0.0 | 0.0 | GO:0006393 | termination of mitochondrial transcription(GO:0006393) |
| 0.0 | 0.1 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 0.1 | GO:0060670 | branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 0.0 | 0.1 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.0 | 0.4 | GO:0048873 | homeostasis of number of cells within a tissue(GO:0048873) |
| 0.0 | 0.1 | GO:0030497 | fatty acid elongation(GO:0030497) |
| 0.0 | 0.0 | GO:0097029 | mature conventional dendritic cell differentiation(GO:0097029) |
| 0.0 | 0.1 | GO:0022038 | corpus callosum development(GO:0022038) |
| 0.0 | 0.0 | GO:0070889 | platelet alpha granule organization(GO:0070889) |
| 0.0 | 0.0 | GO:0010993 | regulation of ubiquitin homeostasis(GO:0010993) free ubiquitin chain polymerization(GO:0010994) |
| 0.0 | 0.0 | GO:0043134 | hindgut contraction(GO:0043133) regulation of hindgut contraction(GO:0043134) |
| 0.0 | 0.1 | GO:0021942 | radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.0 | 0.0 | GO:0032074 | negative regulation of nuclease activity(GO:0032074) |
| 0.0 | 0.2 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 0.0 | GO:0021586 | pons maturation(GO:0021586) |
| 0.0 | 0.0 | GO:1900157 | regulation of bone mineralization involved in bone maturation(GO:1900157) |
| 0.0 | 0.1 | GO:0071481 | cellular response to X-ray(GO:0071481) |
| 0.0 | 0.0 | GO:0061110 | dense core granule biogenesis(GO:0061110) |
| 0.0 | 0.1 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.0 | 0.0 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.0 | 0.1 | GO:0046881 | positive regulation of follicle-stimulating hormone secretion(GO:0046881) |
| 0.0 | 0.1 | GO:0042256 | mature ribosome assembly(GO:0042256) |
| 0.0 | 0.1 | GO:0001765 | membrane raft assembly(GO:0001765) |
| 0.0 | 0.1 | GO:0060971 | embryonic heart tube left/right pattern formation(GO:0060971) |
| 0.0 | 0.0 | GO:0006435 | threonyl-tRNA aminoacylation(GO:0006435) |
| 0.0 | 0.0 | GO:2001023 | regulation of response to drug(GO:2001023) |
| 0.0 | 0.0 | GO:1903011 | negative regulation of bone development(GO:1903011) |
| 0.0 | 0.0 | GO:1901535 | regulation of DNA demethylation(GO:1901535) negative regulation of DNA demethylation(GO:1901536) |
| 0.0 | 0.0 | GO:2000121 | regulation of removal of superoxide radicals(GO:2000121) |
| 0.0 | 0.2 | GO:0020027 | hemoglobin metabolic process(GO:0020027) |
| 0.0 | 0.0 | GO:0072674 | multinuclear osteoclast differentiation(GO:0072674) regulation of osteoclast proliferation(GO:0090289) positive regulation of osteoclast proliferation(GO:0090290) |
| 0.0 | 0.0 | GO:0072282 | metanephric nephron tubule morphogenesis(GO:0072282) |
| 0.0 | 0.0 | GO:1901656 | glycoside transport(GO:1901656) |
| 0.0 | 0.0 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.0 | 0.1 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.0 | 0.0 | GO:2000040 | regulation of planar cell polarity pathway involved in axis elongation(GO:2000040) negative regulation of planar cell polarity pathway involved in axis elongation(GO:2000041) |
| 0.0 | 0.0 | GO:1902202 | regulation of hepatocyte growth factor receptor signaling pathway(GO:1902202) |
| 0.0 | 0.1 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.0 | 0.1 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.0 | 0.0 | GO:0001757 | somite specification(GO:0001757) |
| 0.0 | 0.2 | GO:0061014 | positive regulation of mRNA catabolic process(GO:0061014) |
| 0.0 | 0.4 | GO:0090662 | energy coupled proton transmembrane transport, against electrochemical gradient(GO:0015988) ATP hydrolysis coupled proton transport(GO:0015991) ATP hydrolysis coupled transmembrane transport(GO:0090662) |
| 0.0 | 0.0 | GO:0006203 | dGTP catabolic process(GO:0006203) |
| 0.0 | 0.1 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.0 | 0.1 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.1 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.0 | 0.0 | GO:0072718 | response to cisplatin(GO:0072718) |
| 0.0 | 0.1 | GO:0090232 | positive regulation of spindle checkpoint(GO:0090232) |
| 0.0 | 0.0 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.0 | 0.1 | GO:0090051 | negative regulation of cell migration involved in sprouting angiogenesis(GO:0090051) |
| 0.0 | 0.0 | GO:0097296 | activation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097296) |
| 0.0 | 0.0 | GO:0018171 | peptidyl-cysteine oxidation(GO:0018171) |
| 0.0 | 0.1 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 0.0 | GO:0061687 | detoxification of copper ion(GO:0010273) detoxification of inorganic compound(GO:0061687) stress response to copper ion(GO:1990169) |
| 0.0 | 0.1 | GO:0098719 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.1 | GO:0006670 | sphingosine metabolic process(GO:0006670) |
| 0.0 | 0.0 | GO:0021932 | hindbrain radial glia guided cell migration(GO:0021932) |
| 0.0 | 0.0 | GO:0002432 | granuloma formation(GO:0002432) |
| 0.0 | 0.1 | GO:0006104 | succinyl-CoA metabolic process(GO:0006104) |
| 0.0 | 0.0 | GO:0045041 | protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.0 | 0.1 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.0 | 0.0 | GO:0006106 | fumarate metabolic process(GO:0006106) |
| 0.0 | 0.1 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.0 | 0.0 | GO:0097680 | double-strand break repair via classical nonhomologous end joining(GO:0097680) |
| 0.0 | 0.7 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 0.0 | GO:0061038 | uterus morphogenesis(GO:0061038) |
| 0.0 | 0.0 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.0 | 0.0 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.0 | 0.2 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.0 | 0.0 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.0 | 0.2 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.0 | 0.0 | GO:1901317 | regulation of sperm motility(GO:1901317) |
| 0.0 | 0.0 | GO:0051799 | negative regulation of hair follicle development(GO:0051799) |
| 0.0 | 0.0 | GO:0014827 | intestine smooth muscle contraction(GO:0014827) |
| 0.0 | 0.1 | GO:0048102 | autophagic cell death(GO:0048102) |
| 0.0 | 0.0 | GO:0010940 | positive regulation of necrotic cell death(GO:0010940) |
| 0.0 | 0.0 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 | 0.0 | GO:0051775 | response to redox state(GO:0051775) |
| 0.0 | 0.0 | GO:1902237 | positive regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway(GO:1902237) |
| 0.0 | 0.1 | GO:0035428 | hexose transmembrane transport(GO:0035428) |
| 0.0 | 0.0 | GO:0036112 | medium-chain fatty-acyl-CoA metabolic process(GO:0036112) |
| 0.0 | 0.1 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.0 | 0.1 | GO:0051647 | nucleus localization(GO:0051647) |
| 0.0 | 0.1 | GO:0045717 | negative regulation of fatty acid biosynthetic process(GO:0045717) |
| 0.0 | 0.0 | GO:0021648 | vestibulocochlear nerve morphogenesis(GO:0021648) |
| 0.0 | 0.1 | GO:0086073 | bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 0.0 | 0.1 | GO:0051292 | nuclear pore complex assembly(GO:0051292) |
| 0.0 | 0.0 | GO:0051541 | elastin metabolic process(GO:0051541) |
| 0.0 | 0.0 | GO:0055098 | response to low-density lipoprotein particle(GO:0055098) |
| 0.0 | 0.0 | GO:0019244 | lactate biosynthetic process from pyruvate(GO:0019244) lactate biosynthetic process(GO:0019249) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.1 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.1 | 0.3 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.1 | 0.5 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.1 | 0.2 | GO:0034666 | integrin alpha2-beta1 complex(GO:0034666) |
| 0.1 | 0.3 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.1 | 0.2 | GO:0010009 | cytoplasmic side of endosome membrane(GO:0010009) |
| 0.1 | 0.4 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.1 | 0.2 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 0.1 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.0 | 0.2 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.1 | GO:0031523 | Myb complex(GO:0031523) |
| 0.0 | 0.2 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 0.1 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.0 | 0.4 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.3 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.1 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.2 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.3 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 0.1 | GO:0090498 | extrinsic component of Golgi membrane(GO:0090498) |
| 0.0 | 0.1 | GO:0072558 | NLRP1 inflammasome complex(GO:0072558) |
| 0.0 | 0.2 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.0 | 0.1 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 0.3 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.6 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 0.1 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 0.1 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.3 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.3 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) |
| 0.0 | 0.2 | GO:0032797 | SMN complex(GO:0032797) |
| 0.0 | 0.1 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.0 | 0.1 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.0 | 0.1 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.0 | 0.1 | GO:0030956 | glutamyl-tRNA(Gln) amidotransferase complex(GO:0030956) |
| 0.0 | 0.2 | GO:0090543 | Flemming body(GO:0090543) |
| 0.0 | 0.3 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.0 | 0.1 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.0 | 0.3 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 0.1 | GO:0031417 | NatC complex(GO:0031417) |
| 0.0 | 0.1 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.0 | 0.2 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.2 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.0 | 0.0 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.1 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.0 | 0.4 | GO:0097546 | ciliary base(GO:0097546) |
| 0.0 | 0.1 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.0 | 0.5 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 0.1 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.1 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.0 | 0.1 | GO:0030137 | COPI-coated vesicle(GO:0030137) |
| 0.0 | 0.1 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.8 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.1 | GO:0048500 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.0 | 0.2 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.0 | 0.0 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 0.1 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.0 | 0.0 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.0 | 0.1 | GO:0031010 | ISWI-type complex(GO:0031010) |
| 0.0 | 0.2 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 0.1 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.0 | 0.0 | GO:0005712 | chiasma(GO:0005712) |
| 0.0 | 0.0 | GO:0005745 | m-AAA complex(GO:0005745) |
| 0.0 | 0.1 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 0.0 | 0.0 | GO:0035838 | growing cell tip(GO:0035838) |
| 0.0 | 0.0 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.0 | 0.1 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.0 | GO:0043511 | inhibin complex(GO:0043511) |
| 0.0 | 0.2 | GO:0043189 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.0 | 0.0 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.0 | 0.0 | GO:0042627 | chylomicron(GO:0042627) |
| 0.0 | 0.0 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.0 | 0.1 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.0 | 0.1 | GO:0001931 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 0.1 | GO:1990851 | Wnt-Frizzled-LRP5/6 complex(GO:1990851) |
| 0.0 | 0.1 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.0 | 0.2 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.0 | 0.1 | GO:0060091 | kinocilium(GO:0060091) |
| 0.0 | 0.0 | GO:0071942 | XPC complex(GO:0071942) |
| 0.0 | 0.0 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.0 | 0.4 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.0 | 0.1 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.0 | 0.1 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.0 | GO:0034750 | Scrib-APC-beta-catenin complex(GO:0034750) |
| 0.0 | 0.0 | GO:0000178 | exosome (RNase complex)(GO:0000178) |
| 0.0 | 0.0 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.0 | 0.1 | GO:0071546 | pi-body(GO:0071546) |
| 0.0 | 0.0 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.0 | 0.0 | GO:0030891 | VCB complex(GO:0030891) |
| 0.0 | 0.0 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 0.0 | 0.1 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.0 | 0.1 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.1 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.1 | GO:0071437 | invadopodium(GO:0071437) |
| 0.0 | 0.0 | GO:0000938 | GARP complex(GO:0000938) |
| 0.0 | 0.0 | GO:0031094 | platelet dense tubular network(GO:0031094) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.2 | 0.5 | GO:0008898 | S-adenosylmethionine-homocysteine S-methyltransferase activity(GO:0008898) |
| 0.1 | 0.5 | GO:0004945 | angiotensin type II receptor activity(GO:0004945) |
| 0.1 | 0.3 | GO:0017153 | sodium:dicarboxylate symporter activity(GO:0017153) |
| 0.1 | 0.3 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.1 | 0.1 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.1 | 0.2 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.1 | 0.3 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.1 | 0.3 | GO:0030151 | molybdenum ion binding(GO:0030151) |
| 0.1 | 0.2 | GO:0035515 | oxidative RNA demethylase activity(GO:0035515) |
| 0.1 | 0.3 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.0 | 0.1 | GO:0000171 | ribonuclease MRP activity(GO:0000171) |
| 0.0 | 0.3 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.0 | 0.1 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.0 | 0.1 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 0.4 | GO:0034783 | 3-oxo-2-(2'-pentenyl)cyclopentane-1-octanoic acid CoA ligase activity(GO:0010435) 3-isopropenyl-6-oxoheptanoyl-CoA synthetase activity(GO:0018854) 2-oxo-delta3-4,5,5-trimethylcyclopentenylacetyl-CoA synthetase activity(GO:0018855) benzoyl acetate-CoA ligase activity(GO:0018856) 2,4-dichlorobenzoate-CoA ligase activity(GO:0018857) pivalate-CoA ligase activity(GO:0034783) cyclopropanecarboxylate-CoA ligase activity(GO:0034793) adipate-CoA ligase activity(GO:0034796) citronellyl-CoA ligase activity(GO:0034823) mentha-1,3-dione-CoA ligase activity(GO:0034841) thiophene-2-carboxylate-CoA ligase activity(GO:0034842) 2,4,4-trimethylpentanoate-CoA ligase activity(GO:0034865) cis-2-methyl-5-isopropylhexa-2,5-dienoate-CoA ligase activity(GO:0034942) trans-2-methyl-5-isopropylhexa-2,5-dienoate-CoA ligase activity(GO:0034943) branched-chain acyl-CoA synthetase (ADP-forming) activity(GO:0043759) aryl-CoA synthetase (ADP-forming) activity(GO:0043762) 3-hydroxypropionyl-CoA synthetase activity(GO:0043955) perillic acid:CoA ligase (ADP-forming) activity(GO:0052685) perillic acid:CoA ligase (AMP-forming) activity(GO:0052686) (3R)-3-isopropenyl-6-oxoheptanoate:CoA ligase (ADP-forming) activity(GO:0052687) (3R)-3-isopropenyl-6-oxoheptanoate:CoA ligase (AMP-forming) activity(GO:0052688) pristanate-CoA ligase activity(GO:0070251) malonyl-CoA synthetase activity(GO:0090409) |
| 0.0 | 0.1 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.0 | 0.1 | GO:0030350 | iron-responsive element binding(GO:0030350) |
| 0.0 | 0.2 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
| 0.0 | 0.2 | GO:0019960 | C-X3-C chemokine binding(GO:0019960) |
| 0.0 | 0.1 | GO:0071209 | U7 snRNA binding(GO:0071209) |
| 0.0 | 0.1 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.1 | GO:0031800 | type 3 metabotropic glutamate receptor binding(GO:0031800) |
| 0.0 | 0.1 | GO:0003985 | acetyl-CoA C-acetyltransferase activity(GO:0003985) C-acetyltransferase activity(GO:0016453) |
| 0.0 | 0.3 | GO:0004445 | inositol-polyphosphate 5-phosphatase activity(GO:0004445) |
| 0.0 | 0.5 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.3 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.0 | 0.1 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) DNA (cytosine-5-)-methyltransferase activity, acting on CpG substrates(GO:0051718) |
| 0.0 | 0.1 | GO:0050309 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.0 | 0.1 | GO:0016401 | palmitoyl-CoA oxidase activity(GO:0016401) |
| 0.0 | 0.1 | GO:0015563 | uptake transmembrane transporter activity(GO:0015563) |
| 0.0 | 0.6 | GO:0016646 | oxidoreductase activity, acting on the CH-NH group of donors, NAD or NADP as acceptor(GO:0016646) |
| 0.0 | 0.2 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.1 | GO:0008410 | CoA-transferase activity(GO:0008410) |
| 0.0 | 0.2 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 0.1 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.0 | 0.1 | GO:0015205 | purine nucleobase transmembrane transporter activity(GO:0005345) pyrimidine nucleobase transmembrane transporter activity(GO:0005350) nucleobase transmembrane transporter activity(GO:0015205) |
| 0.0 | 0.1 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.0 | 0.1 | GO:0015087 | cobalt ion transmembrane transporter activity(GO:0015087) |
| 0.0 | 0.1 | GO:0019145 | aminobutyraldehyde dehydrogenase activity(GO:0019145) 4-trimethylammoniobutyraldehyde dehydrogenase activity(GO:0047105) |
| 0.0 | 0.1 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 0.1 | GO:0047391 | alkylglycerophosphoethanolamine phosphodiesterase activity(GO:0047391) |
| 0.0 | 0.1 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.0 | 0.2 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.0 | 0.2 | GO:0102338 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.1 | GO:0004104 | cholinesterase activity(GO:0004104) |
| 0.0 | 0.3 | GO:0016645 | oxidoreductase activity, acting on the CH-NH group of donors(GO:0016645) |
| 0.0 | 0.2 | GO:0018642 | 3-(3-hydroxyphenyl)propionate hydroxylase activity(GO:0008688) 4-chlorobenzaldehyde oxidase activity(GO:0018471) 3,5-xylenol methylhydroxylase activity(GO:0018630) phenylacetate hydroxylase activity(GO:0018631) 4-nitrophenol 4-monooxygenase activity(GO:0018632) dimethyl sulfide monooxygenase activity(GO:0018633) alpha-pinene monooxygenase [NADH] activity(GO:0018634) 1-hydroxy-2-naphthoate hydroxylase activity(GO:0018637) toluene 4-monooxygenase activity(GO:0018638) xylene monooxygenase activity(GO:0018639) dibenzothiophene monooxygenase activity(GO:0018640) 6-hydroxy-3-methyl-2-oxo-1,2-dihydroquinoline 6-monooxygenase activity(GO:0018641) chlorophenol 4-monooxygenase activity(GO:0018642) carbon disulfide oxygenase activity(GO:0018643) toluene 2-monooxygenase activity(GO:0018644) 1-hydroxy-2-oxolimonene 1,2-monooxygenase activity(GO:0018646) phenanthrene 1,2-monooxygenase activity(GO:0018647) tetrahydrofuran hydroxylase activity(GO:0018649) styrene monooxygenase activity(GO:0018650) toluene-4-sulfonate monooxygenase activity(GO:0018651) toluene-sulfonate methyl-monooxygenase activity(GO:0018652) 3-methyl-2-oxo-1,2-dihydroquinoline 6-monooxygenase activity(GO:0018653) 2-hydroxy-phenylacetate hydroxylase activity(GO:0018654) 2-oxo-delta3-4,5,5-trimethylcyclopentenylacetyl-CoA 1,2-monooxygenase activity(GO:0018655) phenanthrene 3,4-monooxygenase activity(GO:0018656) toluene 3-monooxygenase activity(GO:0018657) 4-hydroxyphenylacetate,NADH:oxygen oxidoreductase (3-hydroxylating) activity(GO:0018660) limonene monooxygenase activity(GO:0019113) 2-methylnaphthalene hydroxylase activity(GO:0034526) 1-methylnaphthalene hydroxylase activity(GO:0034534) bisphenol A hydroxylase A activity(GO:0034560) salicylate 5-hydroxylase activity(GO:0034785) isobutylamine N-hydroxylase activity(GO:0034791) branched-chain dodecylbenzene sulfonate monooxygenase activity(GO:0034802) 3-HSA hydroxylase activity(GO:0034819) 4-hydroxypyridine-3-hydroxylase activity(GO:0034894) 2-octaprenyl-3-methyl-6-methoxy-1,4-benzoquinol hydroxylase activity(GO:0043719) 6-hydroxynicotinate 3-monooxygenase activity(GO:0043731) thalianol hydroxylase activity(GO:0080014) |
| 0.0 | 0.1 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 0.1 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.2 | GO:0034889 | alkyl sulfatase activity(GO:0018741) endosulfan hemisulfate sulfatase activity(GO:0034889) endosulfan sulfate hydrolase activity(GO:0034902) |
| 0.0 | 0.1 | GO:0050252 | retinol O-fatty-acyltransferase activity(GO:0050252) |
| 0.0 | 0.1 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.2 | GO:0004438 | phosphatidylinositol-3-phosphatase activity(GO:0004438) |
| 0.0 | 0.2 | GO:0050308 | sugar-phosphatase activity(GO:0050308) |
| 0.0 | 0.1 | GO:0016822 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.0 | 0.1 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.0 | 0.1 | GO:0018685 | alkane 1-monooxygenase activity(GO:0018685) |
| 0.0 | 0.1 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.0 | 0.1 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.0 | 0.1 | GO:0030519 | snoRNP binding(GO:0030519) |
| 0.0 | 0.2 | GO:0034784 | pivalyl-CoA mutase activity(GO:0034784) o-hydroxylaminobenzoate mutase activity(GO:0034951) lupeol synthase activity(GO:0042299) beta-amyrin synthase activity(GO:0042300) baruol synthase activity(GO:0080011) |
| 0.0 | 0.1 | GO:0050567 | glutaminyl-tRNA synthase (glutamine-hydrolyzing) activity(GO:0050567) |
| 0.0 | 0.4 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.1 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.1 | GO:0035473 | lipase binding(GO:0035473) |
| 0.0 | 0.0 | GO:0004774 | succinate-CoA ligase activity(GO:0004774) |
| 0.0 | 0.2 | GO:0050649 | testosterone 6-beta-hydroxylase activity(GO:0050649) |
| 0.0 | 0.3 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.2 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.0 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.0 | 0.3 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.1 | GO:0031750 | D3 dopamine receptor binding(GO:0031750) |
| 0.0 | 0.0 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.0 | 0.1 | GO:0003696 | satellite DNA binding(GO:0003696) |
| 0.0 | 0.1 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.0 | 0.1 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.0 | 0.1 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.0 | 0.1 | GO:0019966 | interleukin-1 binding(GO:0019966) |
| 0.0 | 0.0 | GO:0016742 | hydroxymethyl-, formyl- and related transferase activity(GO:0016742) |
| 0.0 | 0.1 | GO:0035651 | AP-3 adaptor complex binding(GO:0035651) |
| 0.0 | 0.0 | GO:0035673 | oligopeptide transmembrane transporter activity(GO:0035673) |
| 0.0 | 0.1 | GO:0034648 | histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.0 | 0.1 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.0 | 0.0 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 0.1 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.1 | GO:0008312 | 7S RNA binding(GO:0008312) |
| 0.0 | 0.1 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.0 | 0.1 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.3 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.0 | 0.1 | GO:0042895 | antibiotic transporter activity(GO:0042895) |
| 0.0 | 0.3 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.1 | GO:0005290 | L-histidine transmembrane transporter activity(GO:0005290) azole transporter activity(GO:0045118) azole transmembrane transporter activity(GO:1901474) |
| 0.0 | 0.0 | GO:0005146 | leukemia inhibitory factor receptor binding(GO:0005146) |
| 0.0 | 0.0 | GO:0016972 | thiol oxidase activity(GO:0016972) |
| 0.0 | 0.0 | GO:0033142 | progesterone receptor binding(GO:0033142) |
| 0.0 | 0.0 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.0 | 0.1 | GO:0016892 | endoribonuclease activity, producing 3'-phosphomonoesters(GO:0016892) |
| 0.0 | 0.1 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.0 | 0.0 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.1 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 0.0 | GO:0004611 | phosphoenolpyruvate carboxykinase activity(GO:0004611) |
| 0.0 | 0.1 | GO:0018812 | 3-hydroxyacyl-CoA dehydratase activity(GO:0018812) |
| 0.0 | 0.1 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.0 | 0.1 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.0 | 0.1 | GO:0005534 | galactose binding(GO:0005534) |
| 0.0 | 0.1 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.0 | 0.1 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.0 | 0.1 | GO:0031748 | D1 dopamine receptor binding(GO:0031748) |
| 0.0 | 0.1 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.0 | 0.0 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.0 | 0.2 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.0 | 0.1 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 0.0 | GO:0017098 | sulfonylurea receptor binding(GO:0017098) |
| 0.0 | 0.2 | GO:0070717 | poly-purine tract binding(GO:0070717) |
| 0.0 | 0.1 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) |
| 0.0 | 0.1 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.0 | 0.1 | GO:0022820 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.0 | 0.1 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.0 | 0.0 | GO:1904680 | peptide transmembrane transporter activity(GO:1904680) |
| 0.0 | 0.2 | GO:0008198 | ferrous iron binding(GO:0008198) |
| 0.0 | 0.0 | GO:0004829 | threonine-tRNA ligase activity(GO:0004829) |
| 0.0 | 0.0 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.0 | 0.1 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) |
| 0.0 | 0.1 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 0.1 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.2 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.2 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.0 | 0.1 | GO:0050897 | cobalt ion binding(GO:0050897) |
| 0.0 | 0.0 | GO:0042284 | sphingolipid delta-4 desaturase activity(GO:0042284) |
| 0.0 | 0.0 | GO:0046978 | TAP1 binding(GO:0046978) TAP2 binding(GO:0046979) |
| 0.0 | 0.1 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 0.1 | GO:0038085 | vascular endothelial growth factor binding(GO:0038085) |
| 0.0 | 0.1 | GO:0001091 | RNA polymerase II basal transcription factor binding(GO:0001091) |
| 0.0 | 0.0 | GO:0000405 | bubble DNA binding(GO:0000405) |
| 0.0 | 0.1 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 0.4 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.1 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.0 | GO:0008390 | testosterone 16-alpha-hydroxylase activity(GO:0008390) |
| 0.0 | 0.1 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.0 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 0.0 | 0.1 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 0.0 | 0.2 | GO:0016594 | glycine binding(GO:0016594) |
| 0.0 | 0.1 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.0 | 0.1 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 0.0 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.0 | 0.2 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.0 | 0.0 | GO:0016882 | cyclo-ligase activity(GO:0016882) |
| 0.0 | 0.3 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.1 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.0 | 0.0 | GO:0016149 | translation release factor activity, codon specific(GO:0016149) |
| 0.0 | 0.1 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.0 | GO:0008732 | threonine aldolase activity(GO:0004793) L-allo-threonine aldolase activity(GO:0008732) |
| 0.0 | 0.1 | GO:0070513 | death domain binding(GO:0070513) |
| 0.0 | 0.0 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) |
| 0.0 | 0.1 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.0 | 0.1 | GO:0003918 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.0 | 0.0 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.0 | 0.1 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 0.1 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 0.1 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.2 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 0.3 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.6 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.0 | 0.8 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.2 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.3 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.4 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 0.2 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.0 | 0.2 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.4 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 0.1 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.0 | 0.2 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.2 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.3 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.1 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.0 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 0.1 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 0.2 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 0.1 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.5 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.3 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.3 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.3 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 0.0 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.3 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 0.3 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.1 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.4 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.0 | 0.5 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.0 | REACTOME CTLA4 INHIBITORY SIGNALING | Genes involved in CTLA4 inhibitory signaling |
| 0.0 | 0.4 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 0.2 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.4 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.0 | 0.2 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 0.0 | REACTOME P53 DEPENDENT G1 DNA DAMAGE RESPONSE | Genes involved in p53-Dependent G1 DNA Damage Response |
| 0.0 | 0.2 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.0 | 0.0 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.0 | 0.1 | REACTOME TRANSPORT OF MATURE MRNA DERIVED FROM AN INTRONLESS TRANSCRIPT | Genes involved in Transport of Mature mRNA Derived from an Intronless Transcript |
| 0.0 | 0.1 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.0 | 0.0 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.0 | 0.0 | REACTOME CYTOKINE SIGNALING IN IMMUNE SYSTEM | Genes involved in Cytokine Signaling in Immune system |
| 0.0 | 0.3 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.1 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 0.1 | REACTOME TRAF6 MEDIATED NFKB ACTIVATION | Genes involved in TRAF6 mediated NF-kB activation |
| 0.0 | 0.1 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.0 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.0 | 0.0 | REACTOME P75NTR SIGNALS VIA NFKB | Genes involved in p75NTR signals via NF-kB |
| 0.0 | 0.2 | REACTOME APC CDC20 MEDIATED DEGRADATION OF NEK2A | Genes involved in APC-Cdc20 mediated degradation of Nek2A |
| 0.0 | 0.1 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.1 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.1 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.3 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.2 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.1 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.1 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.2 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 0.2 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 0.2 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.3 | REACTOME SEMA4D IN SEMAPHORIN SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.0 | 0.1 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.1 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.0 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 0.1 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.1 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |