Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
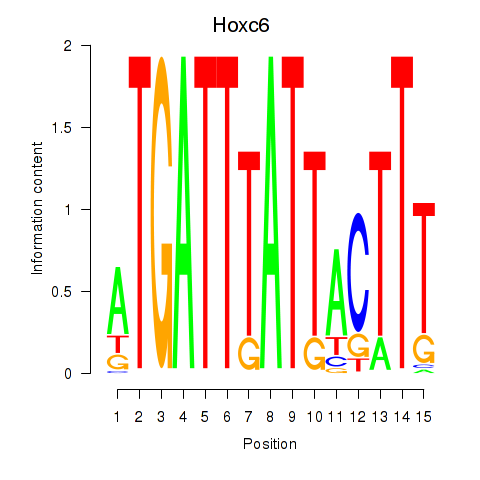
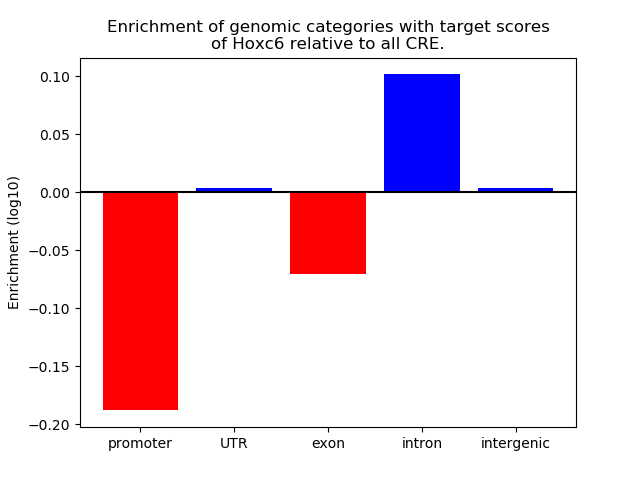
Results for Hoxc6
Z-value: 1.05
Transcription factors associated with Hoxc6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hoxc6
|
ENSMUSG00000001661.4 | homeobox C6 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr15_102999275_102999447 | Hoxc6 | 1104 | 0.276127 | -0.35 | 4.9e-01 | Click! |
Activity of the Hoxc6 motif across conditions
Conditions sorted by the z-value of the Hoxc6 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
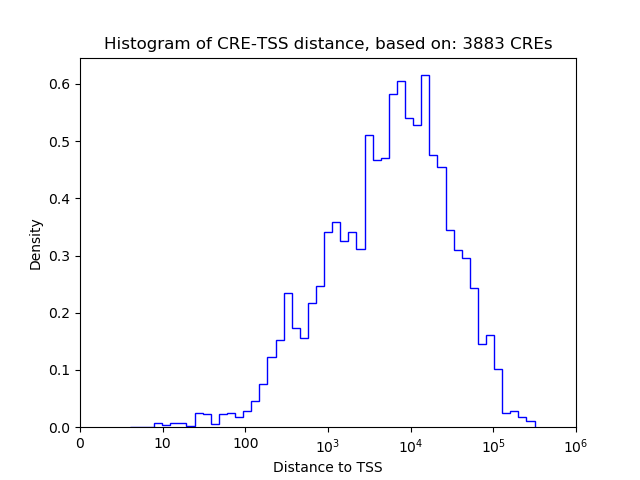
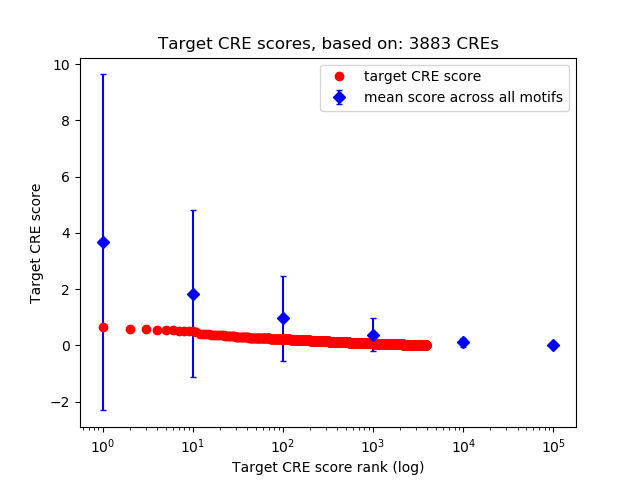
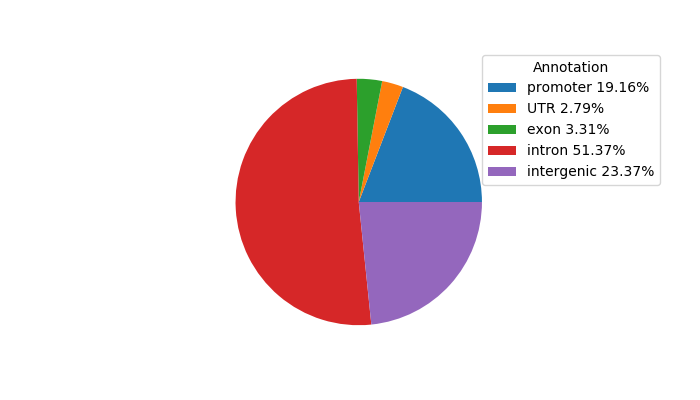
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr1_9968620_9968771 | 0.65 |
Tcf24 |
transcription factor 24 |
763 |
0.41 |
| chr19_39287276_39287531 | 0.58 |
Cyp2c29 |
cytochrome P450, family 2, subfamily c, polypeptide 29 |
329 |
0.91 |
| chr2_60118945_60119257 | 0.57 |
Gm13620 |
predicted gene 13620 |
5995 |
0.16 |
| chr7_104995535_104995703 | 0.54 |
Olfr672 |
olfactory receptor 672 |
1283 |
0.2 |
| chr5_29596501_29596658 | 0.53 |
Ube3c |
ubiquitin protein ligase E3C |
5750 |
0.22 |
| chr8_45341558_45341748 | 0.53 |
Cyp4v3 |
cytochrome P450, family 4, subfamily v, polypeptide 3 |
8437 |
0.15 |
| chr2_134491985_134492278 | 0.52 |
Hao1 |
hydroxyacid oxidase 1, liver |
62176 |
0.15 |
| chr4_47366562_47366857 | 0.52 |
Tgfbr1 |
transforming growth factor, beta receptor I |
13099 |
0.22 |
| chr6_108281132_108281339 | 0.52 |
Itpr1 |
inositol 1,4,5-trisphosphate receptor 1 |
28277 |
0.22 |
| chr11_28696545_28697113 | 0.50 |
2810471M01Rik |
RIKEN cDNA 2810471M01 gene |
15265 |
0.17 |
| chr2_131345745_131345918 | 0.47 |
Rnf24 |
ring finger protein 24 |
7031 |
0.14 |
| chr13_119432132_119432318 | 0.42 |
Paip1 |
polyadenylate binding protein-interacting protein 1 |
3067 |
0.2 |
| chr2_130932638_130932843 | 0.41 |
Atrn |
attractin |
25809 |
0.11 |
| chr13_34626488_34626639 | 0.41 |
Gm47127 |
predicted gene, 47127 |
14317 |
0.13 |
| chr3_117078471_117078622 | 0.39 |
1700061I17Rik |
RIKEN cDNA 1700061I17 gene |
781 |
0.64 |
| chr19_37323790_37323978 | 0.37 |
Ide |
insulin degrading enzyme |
6729 |
0.16 |
| chr10_88719373_88719562 | 0.36 |
Gm48144 |
predicted gene, 48144 |
3373 |
0.12 |
| chr1_188125975_188126247 | 0.36 |
Gm38315 |
predicted gene, 38315 |
34596 |
0.21 |
| chr13_102570134_102570306 | 0.36 |
Gm29927 |
predicted gene, 29927 |
20065 |
0.22 |
| chr5_21175987_21176254 | 0.35 |
Gsap |
gamma-secretase activating protein |
10135 |
0.15 |
| chr17_35590917_35591091 | 0.35 |
Sfta2 |
surfactant associated 2 |
10545 |
0.08 |
| chr1_100298746_100298897 | 0.35 |
Gm29667 |
predicted gene 29667 |
16030 |
0.18 |
| chr15_3533289_3533452 | 0.33 |
Ghr |
growth hormone receptor |
48472 |
0.16 |
| chr4_88893074_88893256 | 0.33 |
Ifne |
interferon epsilon |
12964 |
0.08 |
| chr9_42461424_42462009 | 0.33 |
Tbcel |
tubulin folding cofactor E-like |
255 |
0.91 |
| chr3_138287619_138288216 | 0.33 |
Adh1 |
alcohol dehydrogenase 1 (class I) |
10266 |
0.12 |
| chr6_100204562_100204726 | 0.33 |
Rybp |
RING1 and YY1 binding protein |
28487 |
0.15 |
| chr15_60694082_60694233 | 0.32 |
Gm23835 |
predicted gene, 23835 |
9966 |
0.19 |
| chr13_95772378_95772529 | 0.32 |
Iqgap2 |
IQ motif containing GTPase activating protein 2 |
8056 |
0.19 |
| chr11_115459206_115459357 | 0.31 |
Slc16a5 |
solute carrier family 16 (monocarboxylic acid transporters), member 5 |
3193 |
0.1 |
| chr7_120604351_120604519 | 0.31 |
E130201H02Rik |
RIKEN cDNA E130201H02 gene |
6810 |
0.11 |
| chr2_168734428_168734613 | 0.31 |
Atp9a |
ATPase, class II, type 9A |
181 |
0.95 |
| chr10_95354659_95354831 | 0.31 |
2310039L15Rik |
RIKEN cDNA 2310039L15 gene |
143 |
0.94 |
| chr16_93873807_93873958 | 0.30 |
Chaf1b |
chromatin assembly factor 1, subunit B (p60) |
10019 |
0.13 |
| chr19_12771696_12771894 | 0.30 |
Cntf |
ciliary neurotrophic factor |
6163 |
0.12 |
| chr15_32923240_32923575 | 0.30 |
Sdc2 |
syndecan 2 |
2684 |
0.39 |
| chr4_131882954_131883105 | 0.29 |
Srsf4 |
serine and arginine-rich splicing factor 4 |
1266 |
0.29 |
| chr3_60097884_60098081 | 0.29 |
Sucnr1 |
succinate receptor 1 |
16080 |
0.18 |
| chr16_56714341_56714503 | 0.29 |
Tfg |
Trk-fused gene |
1446 |
0.47 |
| chr13_9077339_9077490 | 0.29 |
Gm36264 |
predicted gene, 36264 |
963 |
0.5 |
| chr13_95811525_95811676 | 0.28 |
Iqgap2 |
IQ motif containing GTPase activating protein 2 |
47203 |
0.13 |
| chr11_43783562_43783743 | 0.28 |
Ttc1 |
tetratricopeptide repeat domain 1 |
35644 |
0.16 |
| chr2_155583504_155583661 | 0.28 |
Gss |
glutathione synthetase |
920 |
0.33 |
| chr12_36155790_36155945 | 0.27 |
Bzw2 |
basic leucine zipper and W2 domains 2 |
760 |
0.46 |
| chr4_10835624_10835785 | 0.27 |
Mir3471-1 |
microRNA 3471-1 |
9926 |
0.12 |
| chr19_41244661_41244826 | 0.27 |
Tm9sf3 |
transmembrane 9 superfamily member 3 |
19237 |
0.21 |
| chr5_7964334_7964518 | 0.27 |
Steap4 |
STEAP family member 4 |
3969 |
0.19 |
| chr9_108309327_108309485 | 0.27 |
Rhoa |
ras homolog family member A |
2577 |
0.11 |
| chr3_5425056_5425268 | 0.27 |
4930555M17Rik |
RIKEN cDNA 4930555M17 gene |
90171 |
0.09 |
| chr1_185370452_185370690 | 0.27 |
Eprs |
glutamyl-prolyl-tRNA synthetase |
2974 |
0.15 |
| chr13_41219562_41219715 | 0.27 |
Elovl2 |
elongation of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 2 |
524 |
0.68 |
| chr2_50284584_50284735 | 0.27 |
Mmadhc |
methylmalonic aciduria (cobalamin deficiency) cblD type, with homocystinuria |
4540 |
0.25 |
| chr2_35060101_35060327 | 0.26 |
Hc |
hemolytic complement |
1224 |
0.43 |
| chr5_90315666_90315817 | 0.26 |
Ankrd17 |
ankyrin repeat domain 17 |
11637 |
0.21 |
| chr4_106354190_106354378 | 0.26 |
Usp24 |
ubiquitin specific peptidase 24 |
38050 |
0.13 |
| chr17_81386388_81386573 | 0.26 |
Gm50044 |
predicted gene, 50044 |
15647 |
0.24 |
| chr17_47923114_47923463 | 0.26 |
Gm15556 |
predicted gene 15556 |
910 |
0.38 |
| chr16_30386539_30386690 | 0.25 |
Atp13a3 |
ATPase type 13A3 |
1871 |
0.37 |
| chr6_67268305_67268456 | 0.25 |
Serbp1 |
serpine1 mRNA binding protein 1 |
1050 |
0.44 |
| chr12_78218868_78219034 | 0.25 |
Gphn |
gephyrin |
7428 |
0.15 |
| chr1_72583626_72583816 | 0.25 |
Smarcal1 |
SWI/SNF related matrix associated, actin dependent regulator of chromatin, subfamily a-like 1 |
106 |
0.97 |
| chr1_139780384_139780535 | 0.25 |
Gm4788 |
predicted gene 4788 |
709 |
0.69 |
| chr2_64079562_64079736 | 0.25 |
Fign |
fidgetin |
18339 |
0.3 |
| chr13_101760313_101760479 | 0.25 |
Gm36638 |
predicted gene, 36638 |
7191 |
0.21 |
| chr3_75530671_75531016 | 0.25 |
Pdcd10 |
programmed cell death 10 |
9273 |
0.2 |
| chr2_48403972_48404299 | 0.25 |
Gm13472 |
predicted gene 13472 |
16098 |
0.2 |
| chr7_70483265_70483615 | 0.25 |
Gm7656 |
predicted gene 7656 |
7408 |
0.16 |
| chr18_43131844_43132003 | 0.24 |
Gm8181 |
predicted gene 8181 |
39236 |
0.13 |
| chr7_136914047_136914226 | 0.24 |
Gm45280 |
predicted gene 45280 |
6805 |
0.24 |
| chr12_73855812_73855997 | 0.24 |
Gm15283 |
predicted gene 15283 |
13708 |
0.17 |
| chr1_160996279_160996443 | 0.24 |
Gm37072 |
predicted gene, 37072 |
5484 |
0.07 |
| chr13_97927511_97927679 | 0.24 |
Arhgef28 |
Rho guanine nucleotide exchange factor (GEF) 28 |
19332 |
0.21 |
| chr11_53418931_53419109 | 0.24 |
Leap2 |
liver-expressed antimicrobial peptide 2 |
4150 |
0.1 |
| chr7_100907329_100907491 | 0.24 |
Arhgef17 |
Rho guanine nucleotide exchange factor (GEF) 17 |
12720 |
0.14 |
| chr10_53113782_53113941 | 0.24 |
Gm47622 |
predicted gene, 47622 |
11318 |
0.22 |
| chr2_132853961_132854112 | 0.24 |
Crls1 |
cardiolipin synthase 1 |
6298 |
0.14 |
| chr3_51437862_51438013 | 0.24 |
Naa15 |
N(alpha)-acetyltransferase 15, NatA auxiliary subunit |
2980 |
0.14 |
| chr8_116404224_116404375 | 0.24 |
1700018P08Rik |
RIKEN cDNA 1700018P08 gene |
47238 |
0.17 |
| chr13_117508625_117508817 | 0.24 |
Gm5200 |
predicted gene 5200 |
49646 |
0.17 |
| chr16_4772912_4773063 | 0.24 |
Cdip1 |
cell death inducing Trp53 target 1 |
1475 |
0.28 |
| chr2_71498544_71498972 | 0.24 |
Gm23253 |
predicted gene, 23253 |
3487 |
0.17 |
| chr10_108979346_108979586 | 0.24 |
Gm47477 |
predicted gene, 47477 |
22978 |
0.22 |
| chr19_56171122_56171273 | 0.23 |
Gm31912 |
predicted gene, 31912 |
64887 |
0.11 |
| chr14_114123652_114123812 | 0.23 |
Gm18369 |
predicted gene, 18369 |
48711 |
0.19 |
| chr2_145233540_145233706 | 0.23 |
Slc24a3 |
solute carrier family 24 (sodium/potassium/calcium exchanger), member 3 |
8988 |
0.28 |
| chr10_87831885_87832059 | 0.23 |
Igf1os |
insulin-like growth factor 1, opposite strand |
3610 |
0.27 |
| chr3_135510262_135510447 | 0.23 |
Manba |
mannosidase, beta A, lysosomal |
6796 |
0.13 |
| chr18_61422291_61422459 | 0.23 |
Gm8755 |
predicted gene 8755 |
11826 |
0.12 |
| chr12_40608115_40608266 | 0.23 |
Dock4 |
dedicator of cytokinesis 4 |
161854 |
0.03 |
| chr16_17340724_17340946 | 0.23 |
Gm24927 |
predicted gene, 24927 |
1177 |
0.36 |
| chr2_5968425_5968579 | 0.23 |
Upf2 |
UPF2 regulator of nonsense transcripts homolog (yeast) |
17033 |
0.17 |
| chr6_117059962_117060118 | 0.23 |
Gm43929 |
predicted gene, 43929 |
78021 |
0.08 |
| chr15_97247773_97248330 | 0.23 |
Amigo2 |
adhesion molecule with Ig like domain 2 |
764 |
0.48 |
| chr2_91378186_91378346 | 0.23 |
Gm22071 |
predicted gene, 22071 |
28870 |
0.13 |
| chr9_122138979_122139323 | 0.23 |
Gm47121 |
predicted gene, 47121 |
3298 |
0.16 |
| chr4_82378492_82378694 | 0.23 |
n-R5s188 |
nuclear encoded rRNA 5S 188 |
60817 |
0.14 |
| chr19_42150846_42151168 | 0.22 |
Marveld1 |
MARVEL (membrane-associating) domain containing 1 |
3299 |
0.14 |
| chr12_54967535_54967800 | 0.22 |
Gm27014 |
predicted gene, 27014 |
4603 |
0.15 |
| chr11_49260229_49260441 | 0.22 |
Mgat1 |
mannoside acetylglucosaminyltransferase 1 |
4091 |
0.14 |
| chr3_138430699_138430859 | 0.22 |
Adh5 |
alcohol dehydrogenase 5 (class III), chi polypeptide |
12314 |
0.12 |
| chr8_61273697_61273901 | 0.22 |
1700001D01Rik |
RIKEN cDNA 1700001D01 gene |
15073 |
0.2 |
| chr15_3482890_3483092 | 0.22 |
Ghr |
growth hormone receptor |
11347 |
0.28 |
| chr12_7816660_7817001 | 0.22 |
Gm32828 |
predicted gene, 32828 |
42861 |
0.14 |
| chr6_141613801_141613980 | 0.22 |
Slco1b2 |
solute carrier organic anion transporter family, member 1b2 |
15628 |
0.25 |
| chr19_40154709_40154860 | 0.22 |
Cyp2c70 |
cytochrome P450, family 2, subfamily c, polypeptide 70 |
32502 |
0.13 |
| chr10_124590350_124590536 | 0.22 |
4930503E24Rik |
RIKEN cDNA 4930503E24 gene |
67005 |
0.14 |
| chr11_72631760_72631911 | 0.22 |
Gm24143 |
predicted gene, 24143 |
15685 |
0.13 |
| chr2_15056785_15057135 | 0.21 |
Gm38257 |
predicted gene, 38257 |
247 |
0.86 |
| chr2_130908138_130908309 | 0.21 |
Atrn |
attractin |
1292 |
0.28 |
| chr1_102517589_102517767 | 0.21 |
Gm20281 |
predicted gene, 20281 |
58756 |
0.14 |
| chr17_33667328_33667604 | 0.21 |
Hnrnpm |
heterogeneous nuclear ribonucleoprotein M |
896 |
0.43 |
| chr10_115511796_115512121 | 0.21 |
Lgr5 |
leucine rich repeat containing G protein coupled receptor 5 |
48879 |
0.12 |
| chr1_57342831_57342992 | 0.21 |
4930558J18Rik |
RIKEN cDNA 4930558J18 gene |
34633 |
0.12 |
| chr8_41096211_41096362 | 0.21 |
Mtus1 |
mitochondrial tumor suppressor 1 |
13510 |
0.18 |
| chr12_7948991_7949142 | 0.21 |
Gm48633 |
predicted gene, 48633 |
995 |
0.56 |
| chr6_95003514_95003791 | 0.21 |
4930511E03Rik |
RIKEN cDNA 4930511E03 gene |
59818 |
0.12 |
| chr10_4333753_4333924 | 0.21 |
Akap12 |
A kinase (PRKA) anchor protein (gravin) 12 |
707 |
0.68 |
| chr4_72166936_72167097 | 0.21 |
Gm11250 |
predicted gene 11250 |
1752 |
0.4 |
| chr4_76363527_76363872 | 0.21 |
Ptprd |
protein tyrosine phosphatase, receptor type, D |
19456 |
0.22 |
| chr16_26702287_26702472 | 0.21 |
Il1rap |
interleukin 1 receptor accessory protein |
20055 |
0.24 |
| chr13_64099099_64099277 | 0.21 |
Slc35d2 |
solute carrier family 35, member D2 |
7975 |
0.15 |
| chr7_25448767_25449204 | 0.21 |
Gm15495 |
predicted gene 15495 |
5522 |
0.11 |
| chr10_29019588_29019757 | 0.21 |
Gm9824 |
predicted pseudogene 9824 |
10456 |
0.14 |
| chr18_33210537_33210688 | 0.20 |
Stard4 |
StAR-related lipid transfer (START) domain containing 4 |
3154 |
0.37 |
| chr13_44432624_44432823 | 0.20 |
1700029N11Rik |
RIKEN cDNA 1700029N11 gene |
6989 |
0.16 |
| chr19_31886956_31887132 | 0.20 |
A1cf |
APOBEC1 complementation factor |
18263 |
0.2 |
| chr9_100619847_100620182 | 0.20 |
Gm8661 |
predicted gene 8661 |
5951 |
0.15 |
| chr5_23441774_23441951 | 0.20 |
Kmt2e |
lysine (K)-specific methyltransferase 2E |
7413 |
0.15 |
| chr5_65431439_65431654 | 0.20 |
Gm43289 |
predicted gene 43289 |
34 |
0.95 |
| chr5_106573979_106574163 | 0.20 |
Gm28050 |
predicted gene, 28050 |
644 |
0.46 |
| chr15_3469696_3469847 | 0.20 |
Ghr |
growth hormone receptor |
1873 |
0.48 |
| chr3_18166354_18166574 | 0.20 |
Gm23686 |
predicted gene, 23686 |
11161 |
0.24 |
| chr13_4192623_4192780 | 0.20 |
Akr1c13 |
aldo-keto reductase family 1, member C13 |
1157 |
0.38 |
| chr14_14071519_14071682 | 0.20 |
Atxn7 |
ataxin 7 |
15869 |
0.17 |
| chr15_11190225_11190388 | 0.20 |
Adamts12 |
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 12 |
125488 |
0.05 |
| chr3_149318468_149318647 | 0.20 |
Gm30382 |
predicted gene, 30382 |
5317 |
0.21 |
| chr4_132930929_132931084 | 0.20 |
Gm24913 |
predicted gene, 24913 |
311 |
0.85 |
| chr16_42999237_42999415 | 0.20 |
Ndufs5-ps |
NADH:ubiquinone oxidoreductase core subunit S5, pseudogene |
43695 |
0.16 |
| chr2_40479361_40479517 | 0.19 |
Gm13456 |
predicted gene 13456 |
76313 |
0.11 |
| chr6_54035175_54035554 | 0.19 |
Chn2 |
chimerin 2 |
4190 |
0.24 |
| chr1_67091897_67092231 | 0.19 |
Cps1 |
carbamoyl-phosphate synthetase 1 |
30962 |
0.17 |
| chr18_51157676_51157865 | 0.19 |
Prr16 |
proline rich 16 |
40032 |
0.21 |
| chr12_7751611_7751762 | 0.19 |
Gm32828 |
predicted gene, 32828 |
108005 |
0.06 |
| chr7_14529256_14529446 | 0.19 |
Obox4-ps2 |
oocyte specific homeobox 4, pseudogene 2 |
715 |
0.53 |
| chr16_43403257_43403418 | 0.19 |
Gm15713 |
predicted gene 15713 |
16859 |
0.18 |
| chr7_100609531_100609682 | 0.19 |
Mrpl48 |
mitochondrial ribosomal protein L48 |
1305 |
0.26 |
| chr3_60515813_60516054 | 0.19 |
Mbnl1 |
muscleblind like splicing factor 1 |
12195 |
0.22 |
| chr15_3570914_3571065 | 0.19 |
Ghr |
growth hormone receptor |
10853 |
0.25 |
| chr6_47814259_47814426 | 0.19 |
Pdia4 |
protein disulfide isomerase associated 4 |
912 |
0.35 |
| chr18_39503638_39503872 | 0.19 |
Nr3c1 |
nuclear receptor subfamily 3, group C, member 1 |
12454 |
0.26 |
| chr11_52351256_52351613 | 0.19 |
Vdac1 |
voltage-dependent anion channel 1 |
9426 |
0.17 |
| chr2_130058269_130058427 | 0.19 |
Gm22424 |
predicted gene, 22424 |
13376 |
0.13 |
| chr12_79436402_79436553 | 0.19 |
Rad51b |
RAD51 paralog B |
109124 |
0.06 |
| chr1_21261382_21261702 | 0.19 |
Gsta3 |
glutathione S-transferase, alpha 3 |
8021 |
0.11 |
| chr12_57441896_57442047 | 0.19 |
Gm16246 |
predicted gene 16246 |
8149 |
0.23 |
| chr7_70365214_70365757 | 0.19 |
B130024G19Rik |
RIKEN cDNA B130024G19 gene |
326 |
0.78 |
| chr9_75037221_75037495 | 0.19 |
Arpp19 |
cAMP-regulated phosphoprotein 19 |
256 |
0.91 |
| chrX_93630327_93630494 | 0.19 |
Pola1 |
polymerase (DNA directed), alpha 1 |
1705 |
0.34 |
| chr9_72288048_72288199 | 0.19 |
Gm19353 |
predicted gene, 19353 |
1544 |
0.21 |
| chr13_95786097_95786248 | 0.19 |
Iqgap2 |
IQ motif containing GTPase activating protein 2 |
21775 |
0.18 |
| chr16_91678296_91678553 | 0.19 |
Donson |
downstream neighbor of SON |
2886 |
0.15 |
| chr11_95365489_95365701 | 0.19 |
Fam117a |
family with sequence similarity 117, member A |
9872 |
0.12 |
| chr15_36496930_36497165 | 0.19 |
Ankrd46 |
ankyrin repeat domain 46 |
227 |
0.91 |
| chr6_14897077_14897293 | 0.19 |
Foxp2 |
forkhead box P2 |
4164 |
0.36 |
| chr1_56623913_56624064 | 0.19 |
Hsfy2 |
heat shock transcription factor, Y-linked 2 |
13447 |
0.28 |
| chr5_42887251_42887423 | 0.18 |
Gm5554 |
predicted gene 5554 |
77272 |
0.11 |
| chr12_45069728_45070323 | 0.18 |
Stxbp6 |
syntaxin binding protein 6 (amisyn) |
4087 |
0.24 |
| chr9_111124362_111124513 | 0.18 |
Lrrfip2 |
leucine rich repeat (in FLII) interacting protein 2 |
4762 |
0.17 |
| chr7_72247872_72248068 | 0.18 |
Mctp2 |
multiple C2 domains, transmembrane 2 |
18610 |
0.26 |
| chr1_158541726_158541892 | 0.18 |
Gm37367 |
predicted gene, 37367 |
7002 |
0.15 |
| chr1_156558196_156558347 | 0.18 |
Abl2 |
v-abl Abelson murine leukemia viral oncogene 2 (arg, Abelson-related gene) |
515 |
0.76 |
| chr9_122148160_122148327 | 0.18 |
Gm47121 |
predicted gene, 47121 |
5794 |
0.13 |
| chr18_59092890_59093288 | 0.18 |
Minar2 |
membrane integral NOTCH2 associated receptor 2 |
30578 |
0.22 |
| chr10_44766057_44766218 | 0.18 |
4930431F10Rik |
RIKEN cDNA 4930431F10 gene |
27237 |
0.12 |
| chr10_69104876_69105058 | 0.18 |
Gm47107 |
predicted gene, 47107 |
7101 |
0.19 |
| chr5_134311252_134311463 | 0.18 |
Gtf2i |
general transcription factor II I |
370 |
0.8 |
| chr5_86888528_86888722 | 0.18 |
Ugt2b34 |
UDP glucuronosyltransferase 2 family, polypeptide B34 |
18312 |
0.1 |
| chr6_24610042_24610446 | 0.18 |
Lmod2 |
leiomodin 2 (cardiac) |
12482 |
0.14 |
| chr9_20581625_20581828 | 0.18 |
Zfp846 |
zinc finger protein 846 |
194 |
0.9 |
| chr8_68008523_68008836 | 0.18 |
Gm22018 |
predicted gene, 22018 |
1661 |
0.43 |
| chr7_118571834_118571985 | 0.18 |
Tmc7 |
transmembrane channel-like gene family 7 |
5282 |
0.16 |
| chr4_84545854_84546088 | 0.18 |
Bnc2 |
basonuclin 2 |
319 |
0.94 |
| chr4_6923149_6923300 | 0.18 |
Tox |
thymocyte selection-associated high mobility group box |
67259 |
0.13 |
| chr2_84727873_84728024 | 0.18 |
Clp1 |
CLP1, cleavage and polyadenylation factor I subunit |
598 |
0.5 |
| chr11_116501017_116501181 | 0.18 |
Rpl36-ps1 |
ribosomal protein L36, pseudogene 1 |
3437 |
0.12 |
| chr1_31097093_31097258 | 0.18 |
4931428L18Rik |
RIKEN cDNA 4931428L18 gene |
721 |
0.62 |
| chr17_4944661_4944832 | 0.18 |
Gm41517 |
predicted gene, 41517 |
2068 |
0.36 |
| chr11_28698214_28698534 | 0.18 |
2810471M01Rik |
RIKEN cDNA 2810471M01 gene |
16810 |
0.16 |
| chr18_43845253_43845404 | 0.18 |
Gm41715 |
predicted gene, 41715 |
5232 |
0.18 |
| chr10_41886623_41887088 | 0.17 |
Sesn1 |
sestrin 1 |
584 |
0.78 |
| chr14_117733261_117733423 | 0.17 |
Mir6239 |
microRNA 6239 |
220505 |
0.02 |
| chr6_27753809_27753967 | 0.17 |
Gm26310 |
predicted gene, 26310 |
85801 |
0.1 |
| chr5_121431395_121431558 | 0.17 |
Naa25 |
N(alpha)-acetyltransferase 25, NatB auxiliary subunit |
7942 |
0.1 |
| chr15_76314416_76314716 | 0.17 |
Oplah |
5-oxoprolinase (ATP-hydrolysing) |
4811 |
0.07 |
| chr10_67176584_67176792 | 0.17 |
Jmjd1c |
jumonji domain containing 1C |
9060 |
0.22 |
| chrX_152327737_152327888 | 0.17 |
Kantr |
Kdm5c adjacent non-coding transcript |
317 |
0.85 |
| chr4_70517804_70518135 | 0.17 |
Megf9 |
multiple EGF-like-domains 9 |
16959 |
0.29 |
| chrX_166458575_166458751 | 0.17 |
Trappc2 |
trafficking protein particle complex 2 |
13360 |
0.13 |
| chr1_9989244_9989423 | 0.17 |
Ppp1r42 |
protein phosphatase 1, regulatory subunit 42 |
3641 |
0.12 |
| chr18_6514098_6514308 | 0.17 |
Epc1 |
enhancer of polycomb homolog 1 |
1905 |
0.3 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0072338 | creatinine metabolic process(GO:0046449) cellular lactam metabolic process(GO:0072338) |
| 0.1 | 0.3 | GO:0000733 | DNA strand renaturation(GO:0000733) |
| 0.1 | 0.3 | GO:0034310 | primary alcohol catabolic process(GO:0034310) |
| 0.0 | 0.1 | GO:2000302 | positive regulation of synaptic vesicle exocytosis(GO:2000302) |
| 0.0 | 0.1 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.1 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.0 | 0.1 | GO:0060741 | prostate gland stromal morphogenesis(GO:0060741) |
| 0.0 | 0.1 | GO:0021593 | rhombomere morphogenesis(GO:0021593) |
| 0.0 | 0.2 | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.0 | 0.1 | GO:0006701 | progesterone biosynthetic process(GO:0006701) |
| 0.0 | 0.1 | GO:0001983 | baroreceptor response to increased systemic arterial blood pressure(GO:0001983) |
| 0.0 | 0.1 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.0 | 0.1 | GO:0014016 | neuroblast differentiation(GO:0014016) |
| 0.0 | 0.0 | GO:0006069 | ethanol metabolic process(GO:0006067) ethanol oxidation(GO:0006069) |
| 0.0 | 0.1 | GO:0015959 | diadenosine polyphosphate metabolic process(GO:0015959) |
| 0.0 | 0.1 | GO:0061724 | lipophagy(GO:0061724) |
| 0.0 | 0.1 | GO:0035523 | protein K29-linked deubiquitination(GO:0035523) protein K33-linked deubiquitination(GO:1990168) |
| 0.0 | 0.1 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.0 | 0.1 | GO:0060375 | regulation of mast cell differentiation(GO:0060375) |
| 0.0 | 0.2 | GO:0034626 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.1 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.0 | 0.1 | GO:1901078 | negative regulation of relaxation of muscle(GO:1901078) |
| 0.0 | 0.1 | GO:0030397 | membrane disassembly(GO:0030397) nuclear envelope disassembly(GO:0051081) |
| 0.0 | 0.1 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.0 | 0.1 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.0 | 0.2 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.0 | 0.1 | GO:1903334 | positive regulation of protein folding(GO:1903334) |
| 0.0 | 0.2 | GO:0006930 | substrate-dependent cell migration, cell extension(GO:0006930) |
| 0.0 | 0.2 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 0.0 | 0.1 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.0 | 0.1 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.0 | 0.1 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.0 | 0.6 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.0 | 0.1 | GO:1903599 | positive regulation of mitophagy(GO:1903599) |
| 0.0 | 0.0 | GO:0007529 | establishment of synaptic specificity at neuromuscular junction(GO:0007529) |
| 0.0 | 0.2 | GO:0021684 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.0 | 0.1 | GO:1900127 | positive regulation of hyaluronan biosynthetic process(GO:1900127) |
| 0.0 | 0.0 | GO:0019676 | ammonia assimilation cycle(GO:0019676) |
| 0.0 | 0.1 | GO:0019287 | isopentenyl diphosphate biosynthetic process(GO:0009240) isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.0 | 0.1 | GO:0030423 | targeting of mRNA for destruction involved in RNA interference(GO:0030423) |
| 0.0 | 0.0 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 0.0 | 0.1 | GO:0006982 | response to lipid hydroperoxide(GO:0006982) |
| 0.0 | 0.1 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.0 | 0.1 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.0 | 0.1 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.0 | 0.1 | GO:0032911 | negative regulation of transforming growth factor beta1 production(GO:0032911) |
| 0.0 | 0.1 | GO:1903774 | positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 0.0 | 0.1 | GO:1900016 | negative regulation of cytokine production involved in inflammatory response(GO:1900016) |
| 0.0 | 0.1 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 0.0 | 0.1 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.0 | 0.1 | GO:1903587 | regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903587) |
| 0.0 | 0.0 | GO:0042427 | serotonin biosynthetic process(GO:0042427) primary amino compound biosynthetic process(GO:1901162) |
| 0.0 | 0.1 | GO:0033216 | ferric iron import(GO:0033216) ferric iron import into cell(GO:0097461) |
| 0.0 | 0.0 | GO:0044333 | Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) |
| 0.0 | 0.0 | GO:0002282 | microglial cell activation involved in immune response(GO:0002282) |
| 0.0 | 0.0 | GO:0016080 | synaptic vesicle targeting(GO:0016080) |
| 0.0 | 0.0 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.0 | 0.0 | GO:0043366 | beta selection(GO:0043366) |
| 0.0 | 0.1 | GO:0021555 | midbrain-hindbrain boundary morphogenesis(GO:0021555) |
| 0.0 | 0.1 | GO:1901727 | positive regulation of histone deacetylase activity(GO:1901727) |
| 0.0 | 0.0 | GO:0044375 | regulation of peroxisome size(GO:0044375) |
| 0.0 | 0.0 | GO:0045608 | negative regulation of auditory receptor cell differentiation(GO:0045608) |
| 0.0 | 0.1 | GO:0097070 | ductus arteriosus closure(GO:0097070) |
| 0.0 | 0.0 | GO:0003331 | regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) |
| 0.0 | 0.0 | GO:0032789 | saturated monocarboxylic acid metabolic process(GO:0032788) unsaturated monocarboxylic acid metabolic process(GO:0032789) |
| 0.0 | 0.0 | GO:0021882 | regulation of transcription from RNA polymerase II promoter involved in forebrain neuron fate commitment(GO:0021882) |
| 0.0 | 0.0 | GO:0001827 | inner cell mass cell fate commitment(GO:0001827) |
| 0.0 | 0.1 | GO:0071447 | cellular response to hydroperoxide(GO:0071447) |
| 0.0 | 0.1 | GO:0003383 | apical constriction(GO:0003383) |
| 0.0 | 0.1 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.0 | 0.0 | GO:0009212 | dTTP biosynthetic process(GO:0006235) pyrimidine deoxyribonucleoside triphosphate biosynthetic process(GO:0009212) dTTP metabolic process(GO:0046075) |
| 0.0 | 0.2 | GO:0045722 | positive regulation of gluconeogenesis(GO:0045722) |
| 0.0 | 0.1 | GO:0006307 | DNA dealkylation involved in DNA repair(GO:0006307) |
| 0.0 | 0.0 | GO:1902809 | regulation of skeletal muscle fiber differentiation(GO:1902809) |
| 0.0 | 0.0 | GO:0045917 | positive regulation of complement activation(GO:0045917) positive regulation of protein activation cascade(GO:2000259) |
| 0.0 | 0.1 | GO:0070100 | negative regulation of chemokine-mediated signaling pathway(GO:0070100) |
| 0.0 | 0.0 | GO:1903371 | regulation of endoplasmic reticulum tubular network organization(GO:1903371) |
| 0.0 | 0.0 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.0 | 0.1 | GO:0042447 | hormone catabolic process(GO:0042447) |
| 0.0 | 0.1 | GO:1902966 | regulation of protein localization to early endosome(GO:1902965) positive regulation of protein localization to early endosome(GO:1902966) |
| 0.0 | 0.1 | GO:0098596 | vocal learning(GO:0042297) imitative learning(GO:0098596) learned vocalization behavior or vocal learning(GO:0098598) |
| 0.0 | 0.0 | GO:0055099 | response to high density lipoprotein particle(GO:0055099) |
| 0.0 | 0.1 | GO:2000370 | positive regulation of clathrin-mediated endocytosis(GO:2000370) |
| 0.0 | 0.1 | GO:1900086 | positive regulation of peptidyl-tyrosine autophosphorylation(GO:1900086) |
| 0.0 | 0.0 | GO:0015677 | copper ion import(GO:0015677) |
| 0.0 | 0.1 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 0.0 | 0.0 | GO:0009202 | deoxyribonucleoside triphosphate biosynthetic process(GO:0009202) |
| 0.0 | 0.0 | GO:0006768 | biotin metabolic process(GO:0006768) |
| 0.0 | 0.1 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.0 | 0.1 | GO:0097105 | presynaptic membrane assembly(GO:0097105) |
| 0.0 | 0.0 | GO:0010982 | regulation of high-density lipoprotein particle clearance(GO:0010982) |
| 0.0 | 0.0 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.0 | 0.1 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.0 | GO:0070989 | oxidative demethylation(GO:0070989) |
| 0.0 | 0.0 | GO:0010963 | regulation of L-arginine import(GO:0010963) |
| 0.0 | 0.1 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.0 | 0.0 | GO:0070948 | regulation of neutrophil mediated cytotoxicity(GO:0070948) regulation of neutrophil mediated killing of symbiont cell(GO:0070949) |
| 0.0 | 0.1 | GO:0060836 | lymphatic endothelial cell differentiation(GO:0060836) |
| 0.0 | 0.1 | GO:0044828 | negative regulation by host of viral genome replication(GO:0044828) |
| 0.0 | 0.1 | GO:0002756 | MyD88-independent toll-like receptor signaling pathway(GO:0002756) |
| 0.0 | 0.1 | GO:0090036 | regulation of protein kinase C signaling(GO:0090036) |
| 0.0 | 0.0 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.0 | 0.0 | GO:0036493 | positive regulation of translation in response to endoplasmic reticulum stress(GO:0036493) |
| 0.0 | 0.0 | GO:0070649 | formin-nucleated actin cable assembly(GO:0070649) |
| 0.0 | 0.0 | GO:0070782 | phosphatidylserine exposure on apoptotic cell surface(GO:0070782) |
| 0.0 | 0.0 | GO:1904995 | negative regulation of leukocyte adhesion to vascular endothelial cell(GO:1904995) |
| 0.0 | 0.3 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 0.0 | GO:0071638 | negative regulation of monocyte chemotactic protein-1 production(GO:0071638) |
| 0.0 | 0.0 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.0 | 0.0 | GO:1902309 | negative regulation of peptidyl-serine dephosphorylation(GO:1902309) |
| 0.0 | 0.1 | GO:0009404 | toxin metabolic process(GO:0009404) |
| 0.0 | 0.1 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0045293 | mRNA editing complex(GO:0045293) |
| 0.0 | 0.1 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.1 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.1 | GO:0016942 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) insulin-like growth factor ternary complex(GO:0042567) |
| 0.0 | 0.1 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.0 | 0.1 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.0 | 0.1 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 0.1 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.0 | 0.1 | GO:0034715 | pICln-Sm protein complex(GO:0034715) |
| 0.0 | 0.3 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 0.1 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.1 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.0 | 0.1 | GO:0031094 | platelet dense tubular network(GO:0031094) |
| 0.0 | 0.1 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.1 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.0 | 0.1 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.0 | 0.1 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.1 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.1 | GO:0045275 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 0.1 | GO:0000306 | extrinsic component of vacuolar membrane(GO:0000306) |
| 0.0 | 0.1 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.0 | 0.0 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.0 | 0.0 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.0 | 0.0 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.0 | 0.0 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.0 | 0.1 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.0 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0033695 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 0.1 | 0.2 | GO:0016899 | oxidoreductase activity, acting on the CH-OH group of donors, oxygen as acceptor(GO:0016899) |
| 0.1 | 0.2 | GO:0016743 | carboxyl- or carbamoyltransferase activity(GO:0016743) |
| 0.0 | 0.1 | GO:0070538 | oleic acid binding(GO:0070538) |
| 0.0 | 0.1 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 0.1 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.0 | 0.3 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.0 | 0.1 | GO:0005345 | purine nucleobase transmembrane transporter activity(GO:0005345) pyrimidine nucleobase transmembrane transporter activity(GO:0005350) nucleobase transmembrane transporter activity(GO:0015205) |
| 0.0 | 0.2 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 0.1 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.0 | 0.1 | GO:0046404 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) |
| 0.0 | 0.1 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) |
| 0.0 | 0.1 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.0 | 0.1 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.1 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.0 | 0.2 | GO:0102337 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.1 | GO:0016151 | nickel cation binding(GO:0016151) |
| 0.0 | 0.1 | GO:0016822 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.0 | 0.1 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.0 | 0.1 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.1 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.1 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.0 | 0.0 | GO:0035276 | ethanol binding(GO:0035276) |
| 0.0 | 0.1 | GO:0050656 | 3'-phosphoadenosine 5'-phosphosulfate binding(GO:0050656) |
| 0.0 | 0.1 | GO:0103116 | alpha-D-galactofuranose transporter activity(GO:0103116) |
| 0.0 | 0.1 | GO:1990239 | steroid hormone binding(GO:1990239) |
| 0.0 | 0.1 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.0 | 0.1 | GO:0043559 | insulin binding(GO:0043559) |
| 0.0 | 0.0 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.0 | 0.1 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.0 | 0.1 | GO:0052851 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.0 | 0.0 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.0 | 0.0 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.0 | 0.1 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.0 | 0.0 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.0 | 0.1 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.1 | GO:0004792 | thiosulfate sulfurtransferase activity(GO:0004792) |
| 0.0 | 0.0 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.0 | 0.1 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.2 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.1 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.0 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) |
| 0.0 | 0.0 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.0 | 0.1 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.0 | 0.1 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.1 | GO:0018631 | 3-(3-hydroxyphenyl)propionate hydroxylase activity(GO:0008688) 4-chlorobenzaldehyde oxidase activity(GO:0018471) 3,5-xylenol methylhydroxylase activity(GO:0018630) phenylacetate hydroxylase activity(GO:0018631) 4-nitrophenol 4-monooxygenase activity(GO:0018632) dimethyl sulfide monooxygenase activity(GO:0018633) alpha-pinene monooxygenase [NADH] activity(GO:0018634) 1-hydroxy-2-naphthoate hydroxylase activity(GO:0018637) toluene 4-monooxygenase activity(GO:0018638) xylene monooxygenase activity(GO:0018639) dibenzothiophene monooxygenase activity(GO:0018640) 6-hydroxy-3-methyl-2-oxo-1,2-dihydroquinoline 6-monooxygenase activity(GO:0018641) chlorophenol 4-monooxygenase activity(GO:0018642) carbon disulfide oxygenase activity(GO:0018643) toluene 2-monooxygenase activity(GO:0018644) 1-hydroxy-2-oxolimonene 1,2-monooxygenase activity(GO:0018646) phenanthrene 1,2-monooxygenase activity(GO:0018647) tetrahydrofuran hydroxylase activity(GO:0018649) styrene monooxygenase activity(GO:0018650) toluene-4-sulfonate monooxygenase activity(GO:0018651) toluene-sulfonate methyl-monooxygenase activity(GO:0018652) 3-methyl-2-oxo-1,2-dihydroquinoline 6-monooxygenase activity(GO:0018653) 2-hydroxy-phenylacetate hydroxylase activity(GO:0018654) 2-oxo-delta3-4,5,5-trimethylcyclopentenylacetyl-CoA 1,2-monooxygenase activity(GO:0018655) phenanthrene 3,4-monooxygenase activity(GO:0018656) toluene 3-monooxygenase activity(GO:0018657) 4-hydroxyphenylacetate,NADH:oxygen oxidoreductase (3-hydroxylating) activity(GO:0018660) limonene monooxygenase activity(GO:0019113) 2-methylnaphthalene hydroxylase activity(GO:0034526) 1-methylnaphthalene hydroxylase activity(GO:0034534) bisphenol A hydroxylase A activity(GO:0034560) salicylate 5-hydroxylase activity(GO:0034785) isobutylamine N-hydroxylase activity(GO:0034791) branched-chain dodecylbenzene sulfonate monooxygenase activity(GO:0034802) 3-HSA hydroxylase activity(GO:0034819) 4-hydroxypyridine-3-hydroxylase activity(GO:0034894) 2-octaprenyl-3-methyl-6-methoxy-1,4-benzoquinol hydroxylase activity(GO:0043719) 6-hydroxynicotinate 3-monooxygenase activity(GO:0043731) thalianol hydroxylase activity(GO:0080014) |
| 0.0 | 0.1 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.0 | 0.1 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 0.1 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.0 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.0 | 0.0 | GO:0038181 | bile acid receptor activity(GO:0038181) |
| 0.0 | 0.1 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 0.1 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.0 | 0.1 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.0 | 0.0 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 0.0 | 0.0 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.0 | 0.1 | GO:1990405 | protein antigen binding(GO:1990405) |
| 0.0 | 0.1 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.1 | GO:0019103 | pyrimidine nucleotide binding(GO:0019103) |
| 0.0 | 0.1 | GO:0017040 | ceramidase activity(GO:0017040) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.2 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.2 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.2 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.0 | 0.2 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.1 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.0 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.0 | 0.1 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |