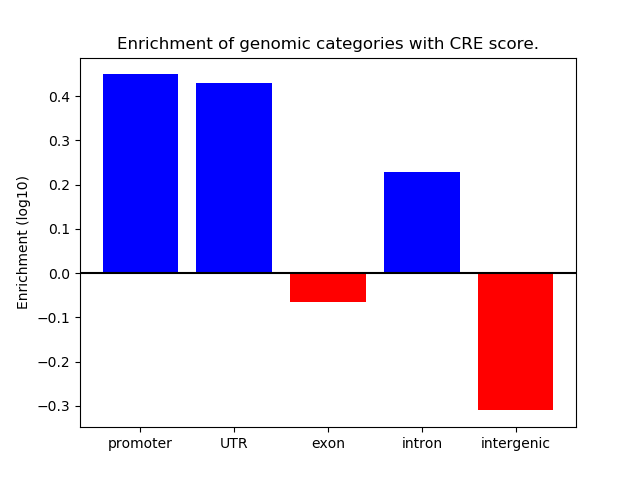
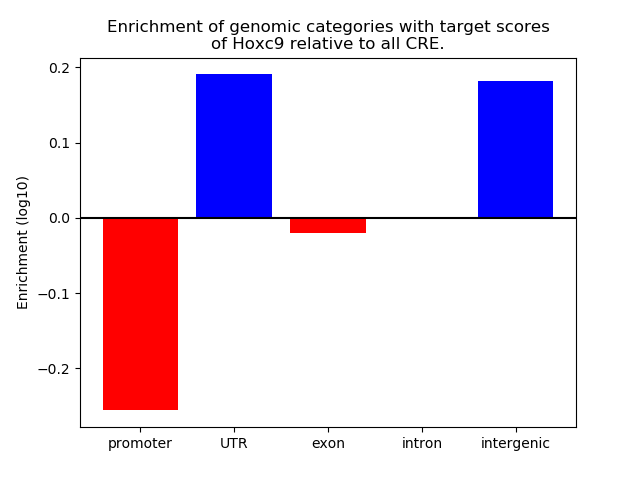
Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
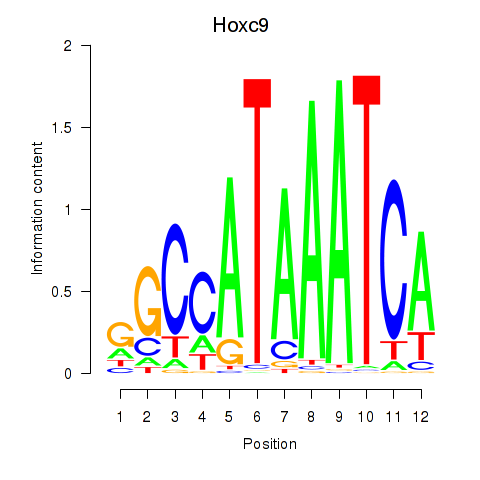
Results for Hoxc9
Z-value: 1.88
Transcription factors associated with Hoxc9
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hoxc9
|
ENSMUSG00000036139.6 | homeobox C9 |
Activity of the Hoxc9 motif across conditions
Conditions sorted by the z-value of the Hoxc9 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
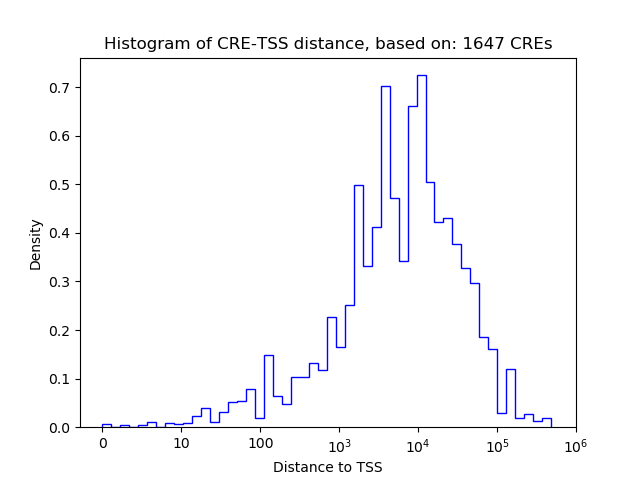
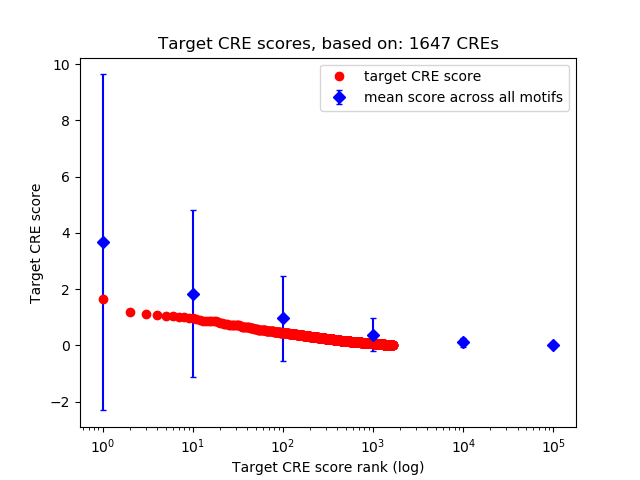
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr19_47699832_47699983 | 1.65 |
Gm50281 |
predicted gene, 50281 |
3374 |
0.13 |
| chr15_103305267_103305425 | 1.19 |
Gm49482 |
predicted gene, 49482 |
4281 |
0.11 |
| chr2_26487761_26487912 | 1.12 |
Notch1 |
notch 1 |
15986 |
0.09 |
| chr13_103544276_103544472 | 1.09 |
Gm24870 |
predicted gene, 24870 |
97552 |
0.07 |
| chr6_129526176_129526360 | 1.04 |
Gm44120 |
predicted gene, 44120 |
3706 |
0.1 |
| chr11_101669101_101669273 | 1.03 |
Arl4d |
ADP-ribosylation factor-like 4D |
3646 |
0.12 |
| chr16_32906853_32907017 | 1.02 |
Fyttd1 |
forty-two-three domain containing 1 |
4836 |
0.15 |
| chr1_88291560_88291711 | 1.02 |
Trpm8 |
transient receptor potential cation channel, subfamily M, member 8 |
11648 |
0.11 |
| chr2_36201411_36201590 | 0.99 |
Gm13429 |
predicted gene 13429 |
814 |
0.51 |
| chr13_109881168_109881343 | 0.98 |
Pde4d |
phosphodiesterase 4D, cAMP specific |
21847 |
0.23 |
| chr7_126260333_126260533 | 0.93 |
Sbk1 |
SH3-binding kinase 1 |
11571 |
0.11 |
| chr2_28401752_28401907 | 0.89 |
Ppp1r26 |
protein phosphatase 1, regulatory subunit 26 |
44971 |
0.1 |
| chr19_31530752_31530903 | 0.87 |
Prkg1 |
protein kinase, cGMP-dependent, type I |
133543 |
0.05 |
| chr18_67649043_67649260 | 0.87 |
Psmg2 |
proteasome (prosome, macropain) assembly chaperone 2 |
7543 |
0.16 |
| chr8_26984786_26984951 | 0.87 |
Gm45371 |
predicted gene 45371 |
4867 |
0.11 |
| chr1_184276149_184276321 | 0.87 |
Gm37223 |
predicted gene, 37223 |
82094 |
0.09 |
| chr2_137165742_137165931 | 0.85 |
Gm28214 |
predicted gene 28214 |
33147 |
0.2 |
| chr14_74794292_74794475 | 0.85 |
Lrch1 |
leucine-rich repeats and calponin homology (CH) domain containing 1 |
608 |
0.79 |
| chr6_119405432_119405608 | 0.83 |
Adipor2 |
adiponectin receptor 2 |
11955 |
0.2 |
| chr3_136671194_136671345 | 0.81 |
Ppp3ca |
protein phosphatase 3, catalytic subunit, alpha isoform |
499 |
0.84 |
| chr2_179292139_179292587 | 0.79 |
Gm14293 |
predicted gene 14293 |
51873 |
0.14 |
| chr10_66631116_66631267 | 0.77 |
Gm28139 |
predicted gene 28139 |
9727 |
0.22 |
| chr16_32427729_32427880 | 0.77 |
Pcyt1a |
phosphate cytidylyltransferase 1, choline, alpha isoform |
3117 |
0.15 |
| chr6_143530873_143531067 | 0.74 |
4930579D09Rik |
RIKEN cDNA 4930579D09 gene |
4538 |
0.3 |
| chr4_108362669_108362965 | 0.74 |
Shisal2a |
shisa like 2A |
20532 |
0.11 |
| chr8_3402101_3402252 | 0.73 |
Arhgef18 |
rho/rac guanine nucleotide exchange factor (GEF) 18 |
9103 |
0.17 |
| chr8_23255218_23255374 | 0.73 |
Gm45412 |
predicted gene 45412 |
1339 |
0.29 |
| chr11_22636888_22637044 | 0.72 |
Gm12053 |
predicted gene 12053 |
10302 |
0.19 |
| chr13_55613448_55613599 | 0.72 |
Gm15911 |
predicted gene 15911 |
8740 |
0.1 |
| chr1_71938584_71938854 | 0.71 |
Gm28818 |
predicted gene 28818 |
21594 |
0.17 |
| chr2_120518757_120518908 | 0.71 |
Zfp106 |
zinc finger protein 106 |
1714 |
0.3 |
| chr2_159732589_159732753 | 0.71 |
Gm11445 |
predicted gene 11445 |
45536 |
0.19 |
| chr9_33067630_33067799 | 0.71 |
Gm27166 |
predicted gene 27166 |
35923 |
0.17 |
| chr6_98920465_98920616 | 0.67 |
Foxp1 |
forkhead box P1 |
9245 |
0.27 |
| chr17_73845744_73845921 | 0.67 |
Gm4948 |
predicted gene 4948 |
25518 |
0.16 |
| chr17_5012388_5012845 | 0.66 |
Arid1b |
AT rich interactive domain 1B (SWI-like) |
16187 |
0.23 |
| chr13_110455904_110456069 | 0.66 |
Plk2 |
polo like kinase 2 |
58189 |
0.14 |
| chr10_84803969_84804150 | 0.65 |
Gm24226 |
predicted gene, 24226 |
7124 |
0.23 |
| chr13_113163450_113163622 | 0.65 |
Gm49564 |
predicted gene, 49564 |
12086 |
0.12 |
| chr1_184605964_184606231 | 0.65 |
Rpl21-ps1 |
ribosomal protein 21, pseudogene 1 |
11151 |
0.15 |
| chr2_146099387_146099651 | 0.64 |
Cfap61 |
cilia and flagella associated protein 61 |
52268 |
0.15 |
| chr17_8983661_8983825 | 0.64 |
1700010I14Rik |
RIKEN cDNA 1700010I14 gene |
4590 |
0.27 |
| chr6_34640326_34640484 | 0.62 |
Gm13861 |
predicted gene 13861 |
13489 |
0.18 |
| chr4_123633972_123634145 | 0.62 |
Gm12926 |
predicted gene 12926 |
3763 |
0.18 |
| chr5_45534505_45534782 | 0.62 |
Fam184b |
family with sequence similarity 184, member B |
1899 |
0.22 |
| chr14_40830166_40830336 | 0.61 |
Sh2d4b |
SH2 domain containing 4B |
23054 |
0.21 |
| chr3_54358525_54358723 | 0.61 |
Postn |
periostin, osteoblast specific factor |
2485 |
0.4 |
| chr17_47628875_47629038 | 0.60 |
Usp49 |
ubiquitin specific peptidase 49 |
1734 |
0.19 |
| chr3_85819269_85819420 | 0.59 |
Fam160a1 |
family with sequence similarity 160, member A1 |
2053 |
0.31 |
| chr18_62224841_62225012 | 0.59 |
Gm9949 |
predicted gene 9949 |
44800 |
0.14 |
| chr7_112270651_112270950 | 0.58 |
Mical2 |
microtubule associated monooxygenase, calponin and LIM domain containing 2 |
499 |
0.87 |
| chr3_79559741_79559912 | 0.57 |
Gm3513 |
predicted gene 3513 |
7633 |
0.12 |
| chr7_45509194_45509417 | 0.56 |
Nucb1 |
nucleobindin 1 |
1010 |
0.23 |
| chr11_103102697_103103091 | 0.56 |
Acbd4 |
acyl-Coenzyme A binding domain containing 4 |
127 |
0.92 |
| chr13_34242299_34242490 | 0.56 |
Slc22a23 |
solute carrier family 22, member 23 |
48863 |
0.11 |
| chr11_55406169_55406386 | 0.56 |
Sparc |
secreted acidic cysteine rich glycoprotein |
4112 |
0.19 |
| chr6_134043668_134043819 | 0.55 |
Etv6 |
ets variant 6 |
7774 |
0.2 |
| chr14_46017259_46017572 | 0.54 |
Gm6580 |
predicted gene 6580 |
10013 |
0.17 |
| chr16_77392160_77392548 | 0.54 |
Gm21816 |
predicted gene, 21816 |
7676 |
0.13 |
| chr16_91565059_91565210 | 0.54 |
Ifngr2 |
interferon gamma receptor 2 |
3811 |
0.13 |
| chr3_104531244_104531395 | 0.54 |
Lrig2 |
leucine-rich repeats and immunoglobulin-like domains 2 |
19401 |
0.12 |
| chr11_49247308_49247488 | 0.54 |
Mgat1 |
mannoside acetylglucosaminyltransferase 1 |
79 |
0.95 |
| chr5_74660839_74660990 | 0.53 |
Lnx1 |
ligand of numb-protein X 1 |
16715 |
0.17 |
| chr8_36031619_36031770 | 0.53 |
Rps12-ps24 |
ribosomal protein S12, pseudogene 24 |
5941 |
0.22 |
| chr4_120599080_120599244 | 0.52 |
Gm8439 |
predicted gene 8439 |
10416 |
0.14 |
| chr18_38310613_38310764 | 0.52 |
Rnf14 |
ring finger protein 14 |
10936 |
0.1 |
| chr8_123097696_123097880 | 0.52 |
Spg7 |
SPG7, paraplegin matrix AAA peptidase subunit |
3267 |
0.11 |
| chr2_26409405_26409631 | 0.51 |
Inpp5e |
inositol polyphosphate-5-phosphatase E |
315 |
0.75 |
| chr5_140852570_140852762 | 0.51 |
Gna12 |
guanine nucleotide binding protein, alpha 12 |
22235 |
0.18 |
| chr7_37208227_37208420 | 0.51 |
Gm28075 |
predicted gene 28075 |
67782 |
0.1 |
| chr6_84623037_84623198 | 0.51 |
Cyp26b1 |
cytochrome P450, family 26, subfamily b, polypeptide 1 |
29209 |
0.22 |
| chr2_26477777_26477967 | 0.51 |
Notch1 |
notch 1 |
7374 |
0.1 |
| chr1_39015594_39015753 | 0.50 |
Pdcl3 |
phosducin-like 3 |
20513 |
0.15 |
| chr3_148810176_148810327 | 0.50 |
Adgrl2 |
adhesion G protein-coupled receptor L2 |
12728 |
0.29 |
| chr6_58968401_58968557 | 0.50 |
BB365896 |
expressed sequence BB365896 |
2824 |
0.19 |
| chr18_31771523_31771682 | 0.50 |
Ammecr1l |
AMME chromosomal region gene 1-like |
9798 |
0.13 |
| chr6_94231502_94231666 | 0.50 |
Magi1 |
membrane associated guanylate kinase, WW and PDZ domain containing 1 |
51441 |
0.15 |
| chr9_69687494_69687666 | 0.49 |
B230323A14Rik |
RIKEN cDNA B230323A14 gene |
71160 |
0.09 |
| chr7_19818513_19818676 | 0.49 |
Gm16174 |
predicted gene 16174 |
2041 |
0.13 |
| chr7_140767632_140767811 | 0.49 |
Cyp2e1 |
cytochrome P450, family 2, subfamily e, polypeptide 1 |
3240 |
0.12 |
| chr3_75910357_75910514 | 0.49 |
Golim4 |
golgi integral membrane protein 4 |
7989 |
0.22 |
| chr3_127809212_127809380 | 0.48 |
Ap1ar |
adaptor-related protein complex 1 associated regulatory protein |
432 |
0.73 |
| chr5_64802893_64803289 | 0.48 |
Klf3 |
Kruppel-like factor 3 (basic) |
297 |
0.69 |
| chr5_67260097_67260248 | 0.48 |
Tmem33 |
transmembrane protein 33 |
393 |
0.75 |
| chr19_41838995_41839153 | 0.48 |
Frat2 |
frequently rearranged in advanced T cell lymphomas 2 |
9058 |
0.14 |
| chr11_112821671_112821851 | 0.48 |
4933434M16Rik |
RIKEN cDNA 4933434M16 gene |
3393 |
0.25 |
| chr1_170815750_170815913 | 0.48 |
Gm25235 |
predicted gene, 25235 |
17040 |
0.13 |
| chr5_129974844_129975008 | 0.47 |
Vkorc1l1 |
vitamin K epoxide reductase complex, subunit 1-like 1 |
2068 |
0.19 |
| chr4_135870496_135870942 | 0.47 |
Pnrc2 |
proline-rich nuclear receptor coactivator 2 |
2342 |
0.17 |
| chr16_22698076_22698372 | 0.47 |
Gm8118 |
predicted gene 8118 |
12030 |
0.18 |
| chr11_83852736_83852913 | 0.47 |
Hnf1b |
HNF1 homeobox B |
136 |
0.94 |
| chr2_70474959_70475123 | 0.46 |
Sp5 |
trans-acting transcription factor 5 |
118 |
0.95 |
| chr1_15610696_15611021 | 0.46 |
Gm37138 |
predicted gene, 37138 |
54609 |
0.13 |
| chr7_28957205_28957624 | 0.45 |
Actn4 |
actinin alpha 4 |
4809 |
0.11 |
| chr12_76578699_76578851 | 0.45 |
Sptb |
spectrin beta, erythrocytic |
5311 |
0.17 |
| chrX_73097052_73097231 | 0.45 |
Xlr3a |
X-linked lymphocyte-regulated 3A |
46 |
0.49 |
| chr8_106138930_106139081 | 0.45 |
1810019D21Rik |
RIKEN cDNA 1810019D21 gene |
1940 |
0.19 |
| chr9_116091537_116091709 | 0.45 |
Gm9385 |
predicted pseudogene 9385 |
50559 |
0.13 |
| chr15_82670462_82670884 | 0.44 |
Cyp2d36-ps |
cytochrome P450, family 2, subfamily d, polypeptide 36, pseudogene |
12584 |
0.08 |
| chr3_101922485_101922654 | 0.44 |
Slc22a15 |
solute carrier family 22 (organic anion/cation transporter), member 15 |
1810 |
0.41 |
| chr8_126734848_126735025 | 0.44 |
Gm45805 |
predicted gene 45805 |
23398 |
0.22 |
| chr2_181257756_181258013 | 0.44 |
Gmeb2 |
glucocorticoid modulatory element binding protein 2 |
1753 |
0.2 |
| chr5_45110945_45111100 | 0.44 |
D5Ertd615e |
DNA segment, Chr 5, ERATO Doi 615, expressed |
58542 |
0.13 |
| chr3_107943742_107943913 | 0.44 |
Gstm6 |
glutathione S-transferase, mu 6 |
78 |
0.92 |
| chr6_129113449_129113636 | 0.44 |
Klrb1-ps1 |
killer cell lectin-like receptor subfamily B member 1, pseudogene 1 |
2976 |
0.16 |
| chr4_60066252_60066431 | 0.44 |
Mup7 |
major urinary protein 7 |
4070 |
0.21 |
| chr7_26023934_26024181 | 0.44 |
Cyp2b27-ps |
cytochrome P450, family 2, subfamily b, polypeptide 27, pseudogene |
4849 |
0.14 |
| chr2_103870054_103870389 | 0.44 |
Gm13876 |
predicted gene 13876 |
18103 |
0.09 |
| chr2_72306157_72306335 | 0.43 |
Map3k20 |
mitogen-activated protein kinase kinase kinase 20 |
8345 |
0.2 |
| chr7_26143665_26143899 | 0.43 |
Cyp2b26-ps |
cytochrome P450, family 2, subfamily b, polypeptide 26, pseudogene |
4810 |
0.17 |
| chr1_121301989_121302163 | 0.43 |
Gm38283 |
predicted gene, 38283 |
1245 |
0.42 |
| chr11_69781883_69782061 | 0.43 |
Zbtb4 |
zinc finger and BTB domain containing 4 |
6566 |
0.06 |
| chr1_170485088_170485365 | 0.42 |
Nos1ap |
nitric oxide synthase 1 (neuronal) adaptor protein |
30312 |
0.19 |
| chr17_57344446_57344660 | 0.42 |
Adgre1 |
adhesion G protein-coupled receptor E1 |
14138 |
0.17 |
| chr17_68312521_68312883 | 0.42 |
L3mbtl4 |
L3MBTL4 histone methyl-lysine binding protein |
38905 |
0.18 |
| chr4_53082761_53082912 | 0.42 |
Abca1 |
ATP-binding cassette, sub-family A (ABC1), member 1 |
41910 |
0.13 |
| chr10_42278852_42279003 | 0.42 |
Foxo3 |
forkhead box O3 |
2172 |
0.38 |
| chr7_72345699_72346292 | 0.42 |
Mctp2 |
multiple C2 domains, transmembrane 2 |
39387 |
0.21 |
| chr2_102197430_102197592 | 0.41 |
Ldlrad3 |
low density lipoprotein receptor class A domain containing 3 |
11126 |
0.18 |
| chr4_149062510_149062918 | 0.41 |
Gm9506 |
predicted gene 9506 |
33669 |
0.11 |
| chr11_16882054_16882234 | 0.41 |
Egfr |
epidermal growth factor receptor |
3994 |
0.24 |
| chr10_122455472_122455951 | 0.41 |
Gm48878 |
predicted gene, 48878 |
1579 |
0.39 |
| chr2_3356420_3356581 | 0.41 |
Gm37525 |
predicted gene, 37525 |
2496 |
0.19 |
| chr13_31894655_31894821 | 0.41 |
Gm11380 |
predicted gene 11380 |
10513 |
0.21 |
| chr1_24614263_24614420 | 0.41 |
Gm10925 |
predicted gene 10925 |
310 |
0.44 |
| chr7_127009850_127010117 | 0.41 |
Mvp |
major vault protein |
1882 |
0.11 |
| chr3_133765600_133766533 | 0.40 |
Gm6135 |
prediticted gene 6135 |
25438 |
0.2 |
| chr4_139875950_139876108 | 0.40 |
Gm1667 |
predicted gene 1667 |
34459 |
0.13 |
| chr3_27859393_27859548 | 0.40 |
Gm26040 |
predicted gene, 26040 |
8643 |
0.24 |
| chr4_60737134_60737406 | 0.40 |
Mup12 |
major urinary protein 12 |
4056 |
0.21 |
| chr4_136143189_136143532 | 0.40 |
Id3 |
inhibitor of DNA binding 3 |
137 |
0.94 |
| chr11_69716247_69716398 | 0.40 |
Gm12307 |
predicted gene 12307 |
14262 |
0.05 |
| chr10_86161840_86162013 | 0.40 |
Gm15990 |
predicted gene 15990 |
36099 |
0.14 |
| chr13_52150386_52150766 | 0.40 |
Gm48199 |
predicted gene, 48199 |
29835 |
0.18 |
| chr4_60497810_60497972 | 0.40 |
Mup1 |
major urinary protein 1 |
1441 |
0.3 |
| chr3_150365886_150366049 | 0.39 |
Gm6439 |
predicted gene 6439 |
150114 |
0.05 |
| chr6_116650512_116650679 | 0.39 |
Depp1 |
DEPP1 autophagy regulator |
14 |
0.95 |
| chr10_27133710_27134037 | 0.39 |
Lama2 |
laminin, alpha 2 |
77228 |
0.1 |
| chr1_160040201_160040352 | 0.39 |
Gm37294 |
predicted gene, 37294 |
4055 |
0.19 |
| chr6_5517592_5517751 | 0.39 |
Pdk4 |
pyruvate dehydrogenase kinase, isoenzyme 4 |
21362 |
0.26 |
| chr6_12448993_12449152 | 0.39 |
Thsd7a |
thrombospondin, type I, domain containing 7A |
29711 |
0.24 |
| chr4_61777961_61778274 | 0.38 |
Mup19 |
major urinary protein 19 |
4107 |
0.15 |
| chr6_52479304_52479475 | 0.38 |
1700094M24Rik |
RIKEN cDNA 1700094M24 gene |
13062 |
0.16 |
| chr9_121677227_121677378 | 0.38 |
Vipr1 |
vasoactive intestinal peptide receptor 1 |
8589 |
0.11 |
| chr9_69442354_69442630 | 0.38 |
Gm18587 |
predicted gene, 18587 |
2471 |
0.17 |
| chr5_88796583_88796899 | 0.38 |
Gm42912 |
predicted gene 42912 |
2845 |
0.21 |
| chr10_80130534_80130701 | 0.38 |
Stk11 |
serine/threonine kinase 11 |
3935 |
0.1 |
| chr5_123203458_123203883 | 0.38 |
Gm43409 |
predicted gene 43409 |
12082 |
0.1 |
| chr13_70831157_70831325 | 0.38 |
Gm36529 |
predicted gene, 36529 |
9210 |
0.2 |
| chr19_12457466_12457644 | 0.38 |
Mpeg1 |
macrophage expressed gene 1 |
3224 |
0.15 |
| chr15_57889538_57889699 | 0.37 |
Derl1 |
Der1-like domain family, member 1 |
776 |
0.67 |
| chr3_104610491_104610642 | 0.37 |
Gm26091 |
predicted gene, 26091 |
9327 |
0.11 |
| chrX_36864362_36864513 | 0.37 |
C330007P06Rik |
RIKEN cDNA C330007P06 gene |
191 |
0.91 |
| chr7_49025943_49026104 | 0.37 |
Gm45207 |
predicted gene 45207 |
36558 |
0.14 |
| chr19_5688905_5689088 | 0.37 |
Map3k11 |
mitogen-activated protein kinase kinase kinase 11 |
41 |
0.53 |
| chr11_86954758_86954966 | 0.37 |
Ypel2 |
yippee like 2 |
17162 |
0.18 |
| chr2_84650666_84651104 | 0.37 |
Ctnnd1 |
catenin (cadherin associated protein), delta 1 |
120 |
0.92 |
| chr10_122384642_122385270 | 0.37 |
Gm36041 |
predicted gene, 36041 |
1936 |
0.39 |
| chr16_91736910_91737079 | 0.36 |
Itsn1 |
intersectin 1 (SH3 domain protein 1A) |
6760 |
0.15 |
| chr9_72988766_72988933 | 0.36 |
Ccpg1 |
cell cycle progression 1 |
3230 |
0.1 |
| chr1_60178803_60178972 | 0.36 |
Nbeal1 |
neurobeachin like 1 |
1712 |
0.36 |
| chr2_169634592_169634925 | 0.36 |
Tshz2 |
teashirt zinc finger family member 2 |
1082 |
0.55 |
| chr8_76985045_76985216 | 0.36 |
Nr3c2 |
nuclear receptor subfamily 3, group C, member 2 |
76858 |
0.1 |
| chr8_82940163_82940321 | 0.35 |
Gm18212 |
predicted gene, 18212 |
12270 |
0.24 |
| chr3_60532827_60533127 | 0.35 |
Gm37589 |
predicted gene, 37589 |
2774 |
0.28 |
| chr2_104684894_104685441 | 0.35 |
Tcp11l1 |
t-complex 11 like 1 |
901 |
0.52 |
| chr15_74785868_74786026 | 0.35 |
Gm17189 |
predicted gene 17189 |
1882 |
0.15 |
| chr1_93567830_93567992 | 0.35 |
Gm37250 |
predicted gene, 37250 |
36096 |
0.11 |
| chr8_10439826_10440109 | 0.35 |
Myo16 |
myosin XVI |
167395 |
0.03 |
| chr1_186182710_186182892 | 0.35 |
Gm37272 |
predicted gene, 37272 |
544 |
0.77 |
| chr11_50290912_50291474 | 0.34 |
Maml1 |
mastermind like transcriptional coactivator 1 |
611 |
0.62 |
| chr14_68447677_68447874 | 0.34 |
Gm31227 |
predicted gene, 31227 |
13856 |
0.25 |
| chr11_53763211_53763413 | 0.34 |
Mir7671 |
microRNA 7671 |
342 |
0.78 |
| chr2_163007366_163007517 | 0.34 |
Sgk2 |
serum/glucocorticoid regulated kinase 2 |
9329 |
0.12 |
| chr2_91763068_91763259 | 0.34 |
Ambra1 |
autophagy/beclin 1 regulator 1 |
3282 |
0.21 |
| chr10_95942046_95942222 | 0.34 |
Eea1 |
early endosome antigen 1 |
1377 |
0.41 |
| chrX_12081574_12081758 | 0.34 |
Bcor |
BCL6 interacting corepressor |
1113 |
0.62 |
| chr15_38661135_38661289 | 0.34 |
Atp6v1c1 |
ATPase, H+ transporting, lysosomal V1 subunit C1 |
721 |
0.55 |
| chr4_61299711_61299917 | 0.34 |
Mup14 |
major urinary protein 14 |
3551 |
0.24 |
| chr6_121992123_121992633 | 0.34 |
Mug2 |
murinoglobulin 2 |
14383 |
0.19 |
| chr13_90092035_90092214 | 0.34 |
Tmem167 |
transmembrane protein 167 |
2362 |
0.24 |
| chr11_79753212_79753391 | 0.34 |
Mir365-2 |
microRNA 365-2 |
26901 |
0.12 |
| chr8_35144585_35144741 | 0.33 |
Gm45626 |
predicted gene 45626 |
5783 |
0.16 |
| chr4_123718182_123718341 | 0.33 |
Ndufs5 |
NADH:ubiquinone oxidoreductase core subunit S5 |
59 |
0.96 |
| chr2_50625194_50625394 | 0.33 |
Gm13484 |
predicted gene 13484 |
32773 |
0.19 |
| chr4_60135275_60135767 | 0.33 |
Mup2 |
major urinary protein 2 |
4336 |
0.2 |
| chrX_164978971_164979139 | 0.33 |
Mospd2 |
motile sperm domain containing 2 |
1242 |
0.39 |
| chr6_94065091_94065371 | 0.33 |
Magi1 |
membrane associated guanylate kinase, WW and PDZ domain containing 1 |
151541 |
0.04 |
| chr4_45523336_45523508 | 0.33 |
Shb |
src homology 2 domain-containing transforming protein B |
6908 |
0.16 |
| chr3_143339999_143340399 | 0.33 |
Gm43613 |
predicted gene 43613 |
9779 |
0.21 |
| chr13_98595452_98595618 | 0.33 |
Gm4815 |
predicted gene 4815 |
17966 |
0.12 |
| chr1_43066043_43066369 | 0.32 |
Tgfbrap1 |
transforming growth factor, beta receptor associated protein 1 |
5273 |
0.14 |
| chr11_113725569_113725747 | 0.32 |
Cdc42ep4 |
CDC42 effector protein (Rho GTPase binding) 4 |
13664 |
0.13 |
| chr16_45608261_45608423 | 0.32 |
Gcsam |
germinal center associated, signaling and motility |
2038 |
0.27 |
| chr15_89477372_89477550 | 0.32 |
Arsa |
arylsulfatase A |
36 |
0.77 |
| chr3_38115997_38116148 | 0.32 |
Rpl21-ps10 |
ribosomal protein L21, pseudogene 10 |
8150 |
0.2 |
| chr19_34863787_34864179 | 0.32 |
Pank1 |
pantothenate kinase 1 |
13866 |
0.15 |
| chr1_145972338_145972489 | 0.32 |
Gm5263 |
predicted gene 5263 |
448023 |
0.01 |
| chr4_129000922_129001084 | 0.32 |
Ak2 |
adenylate kinase 2 |
7650 |
0.16 |
| chr15_10834414_10834584 | 0.32 |
Gm19276 |
predicted gene, 19276 |
19672 |
0.2 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.4 | GO:0003162 | atrioventricular node development(GO:0003162) |
| 0.2 | 0.5 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.1 | 0.4 | GO:1903899 | positive regulation of PERK-mediated unfolded protein response(GO:1903899) |
| 0.1 | 0.5 | GO:0035564 | regulation of kidney size(GO:0035564) |
| 0.1 | 0.7 | GO:0014744 | positive regulation of muscle adaptation(GO:0014744) |
| 0.1 | 0.3 | GO:0010912 | regulation of isomerase activity(GO:0010911) positive regulation of isomerase activity(GO:0010912) |
| 0.1 | 0.3 | GO:0032290 | peripheral nervous system myelin formation(GO:0032290) |
| 0.1 | 0.4 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.1 | 0.2 | GO:1901078 | negative regulation of relaxation of muscle(GO:1901078) |
| 0.1 | 0.3 | GO:0051572 | negative regulation of histone H3-K4 methylation(GO:0051572) |
| 0.1 | 0.3 | GO:1900086 | positive regulation of peptidyl-tyrosine autophosphorylation(GO:1900086) |
| 0.1 | 0.1 | GO:1902809 | regulation of skeletal muscle fiber differentiation(GO:1902809) |
| 0.1 | 0.2 | GO:0061470 | T follicular helper cell differentiation(GO:0061470) |
| 0.1 | 0.2 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.1 | GO:0002018 | renin-angiotensin regulation of aldosterone production(GO:0002018) |
| 0.0 | 0.1 | GO:0070649 | formin-nucleated actin cable assembly(GO:0070649) |
| 0.0 | 0.2 | GO:1904970 | brush border assembly(GO:1904970) |
| 0.0 | 0.2 | GO:0045875 | negative regulation of sister chromatid cohesion(GO:0045875) |
| 0.0 | 0.2 | GO:0021914 | negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021914) |
| 0.0 | 0.2 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.0 | 0.5 | GO:0006047 | UDP-N-acetylglucosamine metabolic process(GO:0006047) |
| 0.0 | 0.1 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.0 | 0.2 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.0 | 0.2 | GO:0060023 | soft palate development(GO:0060023) |
| 0.0 | 0.2 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.1 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.0 | 0.5 | GO:0046855 | inositol phosphate dephosphorylation(GO:0046855) |
| 0.0 | 0.1 | GO:0071879 | positive regulation of adrenergic receptor signaling pathway(GO:0071879) |
| 0.0 | 0.2 | GO:2000343 | positive regulation of chemokine (C-X-C motif) ligand 2 production(GO:2000343) |
| 0.0 | 0.1 | GO:0070889 | platelet alpha granule organization(GO:0070889) |
| 0.0 | 0.2 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.1 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.0 | 0.1 | GO:0042997 | negative regulation of Golgi to plasma membrane protein transport(GO:0042997) |
| 0.0 | 0.2 | GO:1903599 | positive regulation of mitophagy(GO:1903599) |
| 0.0 | 0.2 | GO:0072310 | glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.0 | 0.1 | GO:0014901 | regulation of satellite cell activation involved in skeletal muscle regeneration(GO:0014717) satellite cell activation involved in skeletal muscle regeneration(GO:0014901) |
| 0.0 | 0.1 | GO:0071316 | cellular response to nicotine(GO:0071316) |
| 0.0 | 0.3 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 0.0 | 0.1 | GO:0002125 | maternal aggressive behavior(GO:0002125) |
| 0.0 | 0.1 | GO:0034080 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.0 | 0.1 | GO:1903334 | positive regulation of protein folding(GO:1903334) |
| 0.0 | 0.1 | GO:0050955 | thermoception(GO:0050955) |
| 0.0 | 0.1 | GO:0006106 | fumarate metabolic process(GO:0006106) |
| 0.0 | 0.1 | GO:0017187 | peptidyl-glutamic acid carboxylation(GO:0017187) |
| 0.0 | 0.1 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.0 | 0.1 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.0 | 0.1 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.0 | 0.1 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.0 | 0.1 | GO:1902071 | regulation of hypoxia-inducible factor-1alpha signaling pathway(GO:1902071) |
| 0.0 | 0.1 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.0 | 0.1 | GO:0002572 | pro-T cell differentiation(GO:0002572) |
| 0.0 | 0.1 | GO:2000402 | negative regulation of lymphocyte migration(GO:2000402) |
| 0.0 | 0.0 | GO:2000645 | negative regulation of receptor catabolic process(GO:2000645) |
| 0.0 | 0.1 | GO:1904154 | positive regulation of retrograde protein transport, ER to cytosol(GO:1904154) |
| 0.0 | 0.1 | GO:1901300 | positive regulation of hydrogen peroxide-mediated programmed cell death(GO:1901300) positive regulation of hydrogen peroxide-induced cell death(GO:1905206) |
| 0.0 | 0.1 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.0 | 0.1 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.0 | 0.2 | GO:0046599 | regulation of centriole replication(GO:0046599) |
| 0.0 | 0.1 | GO:1902202 | regulation of hepatocyte growth factor receptor signaling pathway(GO:1902202) |
| 0.0 | 0.4 | GO:0010257 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.1 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.0 | 0.1 | GO:0032489 | regulation of Cdc42 protein signal transduction(GO:0032489) |
| 0.0 | 0.1 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) |
| 0.0 | 0.2 | GO:0016127 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.0 | 0.0 | GO:0061368 | behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 0.0 | 0.1 | GO:0070640 | vitamin D3 metabolic process(GO:0070640) |
| 0.0 | 0.1 | GO:0002838 | negative regulation of response to tumor cell(GO:0002835) negative regulation of immune response to tumor cell(GO:0002838) |
| 0.0 | 0.1 | GO:0042091 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 0.0 | 0.1 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.0 | 0.1 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.0 | 0.1 | GO:0051387 | negative regulation of neurotrophin TRK receptor signaling pathway(GO:0051387) |
| 0.0 | 0.1 | GO:0090160 | Golgi to lysosome transport(GO:0090160) |
| 0.0 | 0.3 | GO:0006098 | pentose-phosphate shunt(GO:0006098) |
| 0.0 | 0.1 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.0 | 0.1 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.0 | 0.1 | GO:0019676 | ammonia assimilation cycle(GO:0019676) |
| 0.0 | 0.1 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.1 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.2 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.1 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.0 | 0.1 | GO:0032933 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.0 | 0.1 | GO:2000169 | regulation of peptidyl-cysteine S-nitrosylation(GO:2000169) |
| 0.0 | 0.0 | GO:0015819 | lysine transport(GO:0015819) |
| 0.0 | 0.0 | GO:0021775 | smoothened signaling pathway involved in ventral spinal cord interneuron specification(GO:0021775) smoothened signaling pathway involved in spinal cord motor neuron cell fate specification(GO:0021776) |
| 0.0 | 0.0 | GO:1902608 | regulation of large conductance calcium-activated potassium channel activity(GO:1902606) positive regulation of large conductance calcium-activated potassium channel activity(GO:1902608) |
| 0.0 | 0.1 | GO:0051660 | establishment of centrosome localization(GO:0051660) |
| 0.0 | 0.1 | GO:0034242 | negative regulation of syncytium formation by plasma membrane fusion(GO:0034242) |
| 0.0 | 0.0 | GO:0032074 | negative regulation of nuclease activity(GO:0032074) |
| 0.0 | 0.1 | GO:0043985 | histone H4-R3 methylation(GO:0043985) |
| 0.0 | 0.0 | GO:0021773 | striatal medium spiny neuron differentiation(GO:0021773) |
| 0.0 | 0.0 | GO:0006172 | ADP biosynthetic process(GO:0006172) |
| 0.0 | 0.1 | GO:0001880 | Mullerian duct regression(GO:0001880) |
| 0.0 | 0.1 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.0 | 0.1 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.0 | 0.0 | GO:1902512 | positive regulation of apoptotic DNA fragmentation(GO:1902512) positive regulation of DNA catabolic process(GO:1903626) |
| 0.0 | 0.0 | GO:0044375 | regulation of peroxisome size(GO:0044375) |
| 0.0 | 0.0 | GO:0061030 | epithelial cell differentiation involved in mammary gland alveolus development(GO:0061030) |
| 0.0 | 0.1 | GO:0036498 | IRE1-mediated unfolded protein response(GO:0036498) |
| 0.0 | 0.4 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.0 | 0.0 | GO:0006578 | amino-acid betaine biosynthetic process(GO:0006578) |
| 0.0 | 0.1 | GO:1902897 | regulation of postsynaptic density protein 95 clustering(GO:1902897) |
| 0.0 | 0.0 | GO:0090154 | positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 0.0 | 0.1 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.0 | 0.0 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.0 | 0.1 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.1 | GO:1902035 | positive regulation of hematopoietic stem cell proliferation(GO:1902035) |
| 0.0 | 0.1 | GO:0006548 | histidine catabolic process(GO:0006548) imidazole-containing compound catabolic process(GO:0052805) |
| 0.0 | 0.0 | GO:0060266 | negative regulation of respiratory burst involved in inflammatory response(GO:0060266) |
| 0.0 | 0.1 | GO:0035279 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.0 | 0.0 | GO:0035771 | interleukin-4-mediated signaling pathway(GO:0035771) |
| 0.0 | 0.1 | GO:0033601 | positive regulation of mammary gland epithelial cell proliferation(GO:0033601) |
| 0.0 | 0.0 | GO:1903265 | positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 0.0 | 0.0 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.0 | 0.2 | GO:0006656 | phosphatidylcholine biosynthetic process(GO:0006656) |
| 0.0 | 0.1 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.0 | 0.1 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.0 | 0.1 | GO:0032986 | nucleosome disassembly(GO:0006337) protein-DNA complex disassembly(GO:0032986) |
| 0.0 | 0.0 | GO:2000017 | positive regulation of determination of dorsal identity(GO:2000017) |
| 0.0 | 0.0 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.0 | 0.0 | GO:1902510 | regulation of apoptotic DNA fragmentation(GO:1902510) |
| 0.0 | 0.3 | GO:0036342 | post-anal tail morphogenesis(GO:0036342) |
| 0.0 | 0.1 | GO:0071699 | olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 0.0 | 0.0 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.0 | 0.0 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.0 | 0.0 | GO:1990705 | cholangiocyte proliferation(GO:1990705) |
| 0.0 | 0.1 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 0.0 | GO:0002220 | innate immune response activating cell surface receptor signaling pathway(GO:0002220) |
| 0.0 | 0.0 | GO:0016561 | protein import into peroxisome matrix, translocation(GO:0016561) |
| 0.0 | 0.1 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.0 | 0.0 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.0 | 0.1 | GO:0033152 | immunoglobulin V(D)J recombination(GO:0033152) |
| 0.0 | 0.0 | GO:0060948 | cardiac vascular smooth muscle cell development(GO:0060948) |
| 0.0 | 0.0 | GO:0002331 | pre-B cell allelic exclusion(GO:0002331) |
| 0.0 | 0.0 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) |
| 0.0 | 0.1 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.0 | 0.2 | GO:0043011 | myeloid dendritic cell differentiation(GO:0043011) |
| 0.0 | 0.0 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.1 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.0 | GO:0050812 | regulation of acyl-CoA biosynthetic process(GO:0050812) |
| 0.0 | 0.0 | GO:0016480 | negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 0.0 | 0.0 | GO:0048296 | isotype switching to IgA isotypes(GO:0048290) regulation of isotype switching to IgA isotypes(GO:0048296) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.1 | 0.4 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.1 | 0.4 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.1 | 0.2 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 0.1 | 0.2 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.0 | 0.1 | GO:0090498 | extrinsic component of Golgi membrane(GO:0090498) |
| 0.0 | 0.3 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 0.2 | GO:0031983 | vesicle lumen(GO:0031983) |
| 0.0 | 0.1 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.3 | GO:0070578 | RISC-loading complex(GO:0070578) |
| 0.0 | 0.3 | GO:0045179 | apical cortex(GO:0045179) |
| 0.0 | 0.3 | GO:0036513 | Derlin-1 retrotranslocation complex(GO:0036513) |
| 0.0 | 0.1 | GO:0020016 | ciliary pocket(GO:0020016) ciliary pocket membrane(GO:0020018) |
| 0.0 | 0.1 | GO:0005745 | m-AAA complex(GO:0005745) |
| 0.0 | 0.2 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 0.3 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.0 | 0.1 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 0.0 | 0.2 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 0.1 | GO:0044308 | axonal spine(GO:0044308) |
| 0.0 | 0.1 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.0 | 0.1 | GO:0033061 | DNA recombinase mediator complex(GO:0033061) |
| 0.0 | 0.1 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.1 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.0 | 0.1 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.1 | GO:0030313 | cell outer membrane(GO:0009279) cell envelope(GO:0030313) external encapsulating structure part(GO:0044462) |
| 0.0 | 0.1 | GO:0000811 | GINS complex(GO:0000811) |
| 0.0 | 0.0 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.0 | GO:0043511 | inhibin complex(GO:0043511) |
| 0.0 | 0.0 | GO:1990597 | AIP1-IRE1 complex(GO:1990597) |
| 0.0 | 0.1 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.0 | 0.1 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 0.1 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.1 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.0 | 0.1 | GO:0044352 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.0 | 0.0 | GO:0005775 | vacuolar lumen(GO:0005775) |
| 0.0 | 0.0 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.0 | 0.0 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.3 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.0 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 0.0 | 0.0 | GO:0030868 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.0 | 0.0 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 0.1 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.1 | GO:0005869 | dynactin complex(GO:0005869) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.1 | 0.4 | GO:0035650 | AP-1 adaptor complex binding(GO:0035650) |
| 0.1 | 0.6 | GO:0018656 | 3-(3-hydroxyphenyl)propionate hydroxylase activity(GO:0008688) 4-chlorobenzaldehyde oxidase activity(GO:0018471) 3,5-xylenol methylhydroxylase activity(GO:0018630) phenylacetate hydroxylase activity(GO:0018631) 4-nitrophenol 4-monooxygenase activity(GO:0018632) dimethyl sulfide monooxygenase activity(GO:0018633) alpha-pinene monooxygenase [NADH] activity(GO:0018634) 1-hydroxy-2-naphthoate hydroxylase activity(GO:0018637) toluene 4-monooxygenase activity(GO:0018638) xylene monooxygenase activity(GO:0018639) dibenzothiophene monooxygenase activity(GO:0018640) 6-hydroxy-3-methyl-2-oxo-1,2-dihydroquinoline 6-monooxygenase activity(GO:0018641) chlorophenol 4-monooxygenase activity(GO:0018642) carbon disulfide oxygenase activity(GO:0018643) toluene 2-monooxygenase activity(GO:0018644) 1-hydroxy-2-oxolimonene 1,2-monooxygenase activity(GO:0018646) phenanthrene 1,2-monooxygenase activity(GO:0018647) tetrahydrofuran hydroxylase activity(GO:0018649) styrene monooxygenase activity(GO:0018650) toluene-4-sulfonate monooxygenase activity(GO:0018651) toluene-sulfonate methyl-monooxygenase activity(GO:0018652) 3-methyl-2-oxo-1,2-dihydroquinoline 6-monooxygenase activity(GO:0018653) 2-hydroxy-phenylacetate hydroxylase activity(GO:0018654) 2-oxo-delta3-4,5,5-trimethylcyclopentenylacetyl-CoA 1,2-monooxygenase activity(GO:0018655) phenanthrene 3,4-monooxygenase activity(GO:0018656) toluene 3-monooxygenase activity(GO:0018657) 4-hydroxyphenylacetate,NADH:oxygen oxidoreductase (3-hydroxylating) activity(GO:0018660) limonene monooxygenase activity(GO:0019113) 2-methylnaphthalene hydroxylase activity(GO:0034526) 1-methylnaphthalene hydroxylase activity(GO:0034534) bisphenol A hydroxylase A activity(GO:0034560) salicylate 5-hydroxylase activity(GO:0034785) isobutylamine N-hydroxylase activity(GO:0034791) branched-chain dodecylbenzene sulfonate monooxygenase activity(GO:0034802) 3-HSA hydroxylase activity(GO:0034819) 4-hydroxypyridine-3-hydroxylase activity(GO:0034894) 2-octaprenyl-3-methyl-6-methoxy-1,4-benzoquinol hydroxylase activity(GO:0043719) 6-hydroxynicotinate 3-monooxygenase activity(GO:0043731) thalianol hydroxylase activity(GO:0080014) |
| 0.1 | 0.2 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.1 | 0.4 | GO:0004439 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) |
| 0.1 | 0.2 | GO:0031711 | bradykinin receptor binding(GO:0031711) |
| 0.1 | 0.3 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.1 | 0.2 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.1 | 0.3 | GO:0070883 | pre-miRNA binding(GO:0070883) |
| 0.0 | 0.0 | GO:0034211 | GTP-dependent protein kinase activity(GO:0034211) |
| 0.0 | 0.1 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.0 | 0.1 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.0 | 0.2 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.0 | 0.1 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.0 | 0.1 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.0 | 0.1 | GO:0047023 | androsterone dehydrogenase activity(GO:0047023) |
| 0.0 | 0.2 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.0 | 0.3 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.0 | 0.2 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.0 | 0.1 | GO:0055100 | adiponectin binding(GO:0055100) |
| 0.0 | 0.1 | GO:0008184 | glycogen phosphorylase activity(GO:0008184) |
| 0.0 | 0.1 | GO:0031893 | vasopressin receptor binding(GO:0031893) |
| 0.0 | 0.1 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) |
| 0.0 | 0.1 | GO:0035671 | enone reductase activity(GO:0035671) |
| 0.0 | 0.3 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 0.1 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.0 | 0.2 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.0 | 0.1 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.1 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.0 | 0.1 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.0 | 0.1 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.3 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.4 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.0 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 0.1 | GO:0004445 | inositol-polyphosphate 5-phosphatase activity(GO:0004445) |
| 0.0 | 0.1 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.0 | 0.2 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.0 | 0.2 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.1 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.1 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.0 | 0.1 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.0 | GO:0008898 | S-adenosylmethionine-homocysteine S-methyltransferase activity(GO:0008898) |
| 0.0 | 0.1 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.1 | GO:0044020 | histone methyltransferase activity (H4-R3 specific)(GO:0044020) |
| 0.0 | 0.1 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 0.0 | 0.1 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.0 | 0.1 | GO:0035473 | lipase binding(GO:0035473) |
| 0.0 | 0.0 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.0 | 0.1 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.0 | 0.2 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.0 | 0.1 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 0.0 | 0.0 | GO:0047045 | testosterone 17-beta-dehydrogenase (NADP+) activity(GO:0047045) |
| 0.0 | 0.0 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.0 | 0.4 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.0 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.0 | 0.0 | GO:0004103 | choline kinase activity(GO:0004103) |
| 0.0 | 0.3 | GO:0004119 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) cGMP-inhibited cyclic-nucleotide phosphodiesterase activity(GO:0004119) photoreceptor cyclic-nucleotide phosphodiesterase activity(GO:0004120) 7,8-dihydro-D-neopterin 2',3'-cyclic phosphate phosphodiesterase activity(GO:0044688) inositol phosphosphingolipid phospholipase activity(GO:0052712) inositol phosphorylceramide phospholipase activity(GO:0052713) mannosyl-inositol phosphorylceramide phospholipase activity(GO:0052714) mannosyl-diinositol phosphorylceramide phospholipase activity(GO:0052715) |
| 0.0 | 0.1 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.2 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.0 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.0 | GO:0016882 | cyclo-ligase activity(GO:0016882) |
| 0.0 | 0.4 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.0 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.0 | 0.1 | GO:0070991 | medium-chain-acyl-CoA dehydrogenase activity(GO:0070991) |
| 0.0 | 0.1 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.0 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.0 | 0.0 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.0 | 0.0 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 0.0 | GO:0023029 | MHC class Ib protein binding(GO:0023029) |
| 0.0 | 0.1 | GO:0048273 | mitogen-activated protein kinase p38 binding(GO:0048273) |
| 0.0 | 0.1 | GO:0016443 | bidentate ribonuclease III activity(GO:0016443) |
| 0.0 | 0.1 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 0.1 | GO:0016623 | aldehyde oxidase activity(GO:0004031) oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) |
| 0.0 | 0.1 | GO:0042979 | ornithine decarboxylase regulator activity(GO:0042979) |
| 0.0 | 0.1 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.0 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.0 | 0.4 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.7 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 0.2 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.9 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.0 | REACTOME SIGNALING BY EGFR IN CANCER | Genes involved in Signaling by EGFR in Cancer |
| 0.0 | 0.4 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 0.2 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.0 | REACTOME FRS2 MEDIATED CASCADE | Genes involved in FRS2-mediated cascade |
| 0.0 | 0.2 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.0 | REACTOME S PHASE | Genes involved in S Phase |
| 0.0 | 0.1 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 0.0 | REACTOME VIF MEDIATED DEGRADATION OF APOBEC3G | Genes involved in Vif-mediated degradation of APOBEC3G |
| 0.0 | 0.2 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.0 | 0.0 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.0 | 0.2 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.4 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 0.3 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.3 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 0.2 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.0 | 0.1 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.0 | REACTOME PLATELET ACTIVATION SIGNALING AND AGGREGATION | Genes involved in Platelet activation, signaling and aggregation |
| 0.0 | 0.0 | REACTOME REGULATORY RNA PATHWAYS | Genes involved in Regulatory RNA pathways |
| 0.0 | 0.1 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 0.1 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 0.2 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 0.1 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |