Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
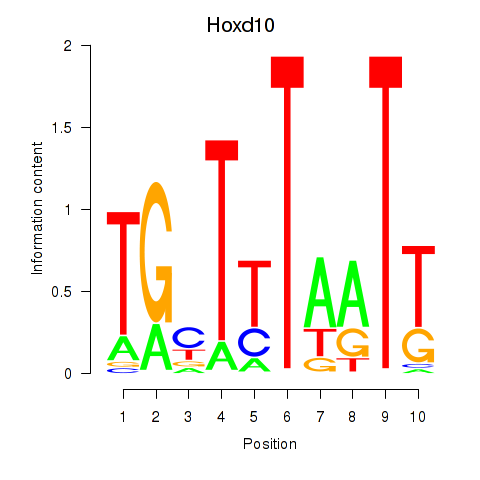
Results for Hoxd10
Z-value: 0.64
Transcription factors associated with Hoxd10
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hoxd10
|
ENSMUSG00000050368.3 | homeobox D10 |
Activity of the Hoxd10 motif across conditions
Conditions sorted by the z-value of the Hoxd10 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
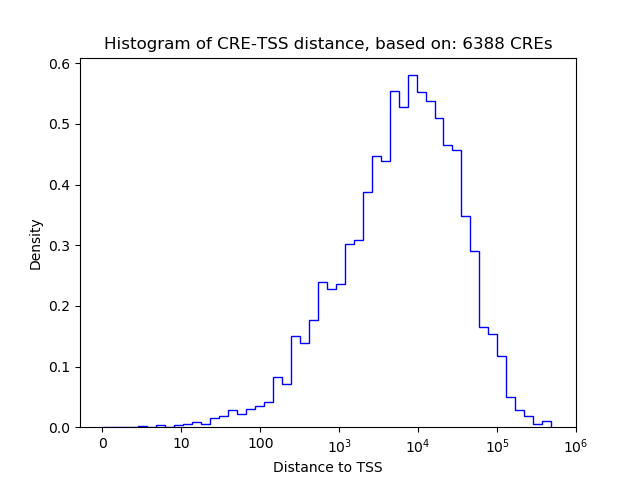
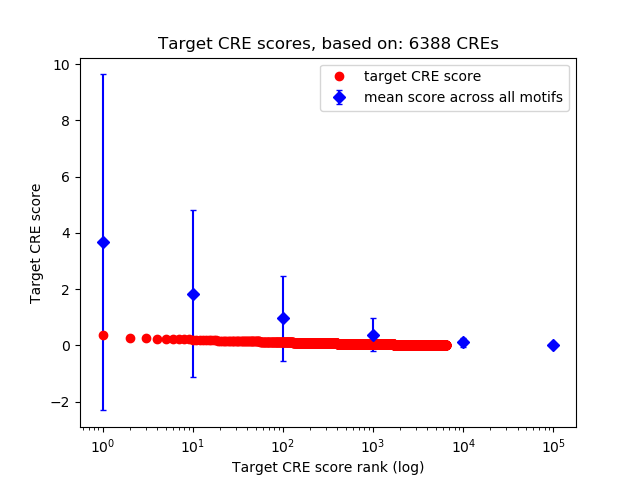
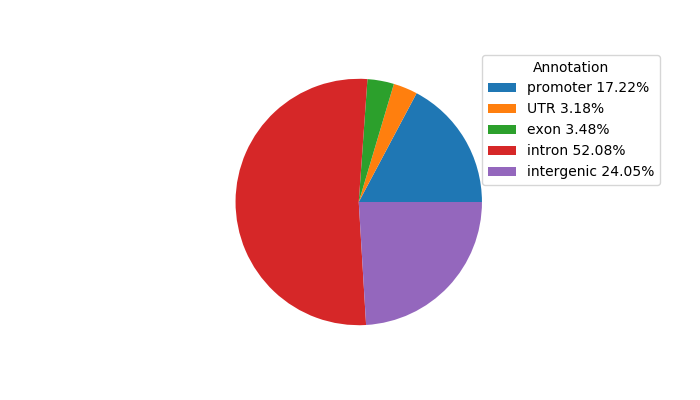
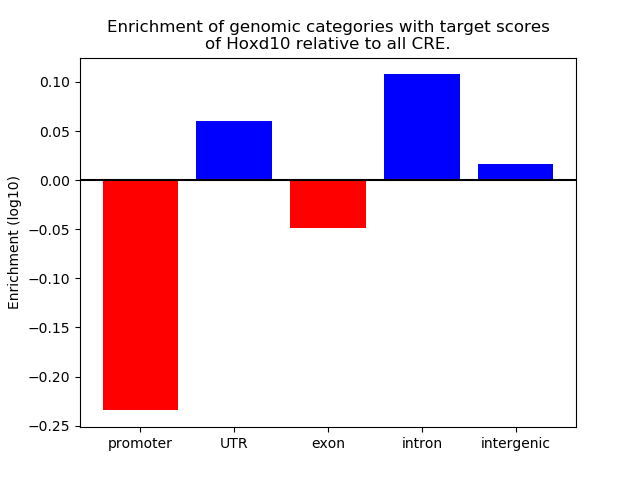
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr19_40191993_40192245 | 0.38 |
Gm5827 |
predicted gene 5827 |
1081 |
0.44 |
| chr3_5425056_5425268 | 0.27 |
4930555M17Rik |
RIKEN cDNA 4930555M17 gene |
90171 |
0.09 |
| chr13_112282869_112283044 | 0.25 |
Ankrd55 |
ankyrin repeat domain 55 |
5495 |
0.19 |
| chr19_29520832_29521011 | 0.23 |
A930007I19Rik |
RIKEN cDNA A930007I19 gene |
49 |
0.96 |
| chr1_162891657_162891835 | 0.23 |
Fmo2 |
flavin containing monooxygenase 2 |
5231 |
0.19 |
| chr17_64763760_64763987 | 0.23 |
Dreh |
down-regulated in hepatocellular carcinoma |
3238 |
0.25 |
| chr4_109317104_109317295 | 0.23 |
Eps15 |
epidermal growth factor receptor pathway substrate 15 |
8048 |
0.21 |
| chr7_97414019_97414355 | 0.23 |
Thrsp |
thyroid hormone responsive |
3332 |
0.16 |
| chr1_155035905_155036069 | 0.22 |
Gm29441 |
predicted gene 29441 |
4248 |
0.21 |
| chr14_68455613_68455772 | 0.21 |
Gm31227 |
predicted gene, 31227 |
5939 |
0.27 |
| chr11_16832225_16832426 | 0.20 |
Egfros |
epidermal growth factor receptor, opposite strand |
1623 |
0.42 |
| chr5_125479434_125479671 | 0.20 |
Gm27551 |
predicted gene, 27551 |
175 |
0.92 |
| chr11_16839257_16839592 | 0.20 |
Egfros |
epidermal growth factor receptor, opposite strand |
8722 |
0.22 |
| chr6_66996154_66996403 | 0.18 |
Gm36816 |
predicted gene, 36816 |
12135 |
0.12 |
| chr19_40174636_40174787 | 0.17 |
Cyp2c70 |
cytochrome P450, family 2, subfamily c, polypeptide 70 |
12575 |
0.15 |
| chr8_76152377_76152560 | 0.17 |
Gm45742 |
predicted gene 45742 |
35441 |
0.19 |
| chr11_16764973_16765411 | 0.17 |
Egfr |
epidermal growth factor receptor |
12962 |
0.2 |
| chr9_74791015_74791321 | 0.17 |
Gm22315 |
predicted gene, 22315 |
9098 |
0.19 |
| chr2_31519719_31520357 | 0.17 |
Ass1 |
argininosuccinate synthetase 1 |
1548 |
0.36 |
| chr2_177902871_177903175 | 0.17 |
Gm14327 |
predicted gene 14327 |
1262 |
0.38 |
| chr13_16575026_16575177 | 0.17 |
Gm48497 |
predicted gene, 48497 |
41280 |
0.17 |
| chr8_40895225_40895376 | 0.17 |
Slc7a2 |
solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 |
3138 |
0.22 |
| chr2_72753133_72753284 | 0.17 |
6430710C18Rik |
RIKEN cDNA 6430710C18 gene |
2355 |
0.34 |
| chr2_18066311_18066462 | 0.17 |
Mllt10 |
myeloid/lymphoid or mixed-lineage leukemia; translocated to, 10 |
1545 |
0.26 |
| chr10_24258031_24258221 | 0.17 |
Moxd1 |
monooxygenase, DBH-like 1 |
34581 |
0.16 |
| chr2_58773392_58773555 | 0.17 |
Upp2 |
uridine phosphorylase 2 |
8148 |
0.21 |
| chr5_89466890_89467069 | 0.16 |
Gc |
vitamin D binding protein |
9081 |
0.22 |
| chr4_76963069_76963220 | 0.16 |
Gm23159 |
predicted gene, 23159 |
4646 |
0.28 |
| chr9_74876222_74876428 | 0.16 |
Onecut1 |
one cut domain, family member 1 |
9841 |
0.15 |
| chr4_62704468_62704619 | 0.16 |
Gm11210 |
predicted gene 11210 |
1438 |
0.36 |
| chr2_72210584_72210735 | 0.16 |
Rapgef4 |
Rap guanine nucleotide exchange factor (GEF) 4 |
11860 |
0.17 |
| chr2_177471886_177472177 | 0.16 |
Zfp970 |
zinc finger protein 970 |
7185 |
0.16 |
| chr1_128080862_128081018 | 0.16 |
Gm37625 |
predicted gene, 37625 |
8391 |
0.12 |
| chr12_32693085_32693261 | 0.16 |
Gm18726 |
predicted gene, 18726 |
16988 |
0.2 |
| chr12_79501822_79502159 | 0.16 |
Rad51b |
RAD51 paralog B |
174637 |
0.03 |
| chr2_117026913_117027140 | 0.15 |
Gm13981 |
predicted gene 13981 |
28540 |
0.17 |
| chr4_141926137_141926288 | 0.15 |
Fhad1 |
forkhead-associated (FHA) phosphopeptide binding domain 1 |
3786 |
0.16 |
| chr3_67267572_67267743 | 0.15 |
Mlf1 |
myeloid leukemia factor 1 |
106440 |
0.06 |
| chr5_86938648_86938802 | 0.15 |
Gm24121 |
predicted gene, 24121 |
9717 |
0.09 |
| chr13_19395072_19395660 | 0.15 |
Gm42683 |
predicted gene 42683 |
87 |
0.82 |
| chr10_31956696_31956848 | 0.15 |
Gm18189 |
predicted gene, 18189 |
16897 |
0.27 |
| chr15_81249111_81249532 | 0.15 |
8430426J06Rik |
RIKEN cDNA 8430426J06 gene |
1353 |
0.36 |
| chr9_74894770_74895613 | 0.15 |
Onecut1 |
one cut domain, family member 1 |
28707 |
0.13 |
| chr13_109823681_109823902 | 0.15 |
Pde4d |
phosphodiesterase 4D, cAMP specific |
66570 |
0.13 |
| chr13_90464205_90464390 | 0.15 |
Gm47520 |
predicted gene, 47520 |
46197 |
0.16 |
| chr3_101578122_101578273 | 0.15 |
Atp1a1 |
ATPase, Na+/K+ transporting, alpha 1 polypeptide |
637 |
0.68 |
| chr11_16783062_16783213 | 0.15 |
Egfr |
epidermal growth factor receptor |
30907 |
0.16 |
| chr9_106273797_106274092 | 0.14 |
Poc1a |
POC1 centriolar protein A |
7117 |
0.12 |
| chr11_28684227_28684558 | 0.14 |
2810471M01Rik |
RIKEN cDNA 2810471M01 gene |
2828 |
0.27 |
| chr11_16890762_16890930 | 0.14 |
Egfr |
epidermal growth factor receptor |
12696 |
0.19 |
| chr3_114294859_114295010 | 0.14 |
Col11a1 |
collagen, type XI, alpha 1 |
129656 |
0.06 |
| chr19_44396602_44396753 | 0.14 |
Scd1 |
stearoyl-Coenzyme A desaturase 1 |
10013 |
0.14 |
| chr6_57647294_57647456 | 0.14 |
Gm19244 |
predicted gene, 19244 |
30098 |
0.11 |
| chr11_17651863_17652036 | 0.14 |
Gm12016 |
predicted gene 12016 |
12766 |
0.3 |
| chr5_92607049_92607221 | 0.14 |
Stbd1 |
starch binding domain 1 |
4067 |
0.19 |
| chr6_119883269_119883450 | 0.13 |
Gm15532 |
predicted gene 15532 |
9561 |
0.15 |
| chr1_162964214_162964365 | 0.13 |
Gm37273 |
predicted gene, 37273 |
18725 |
0.14 |
| chr12_16562529_16562714 | 0.13 |
Lpin1 |
lipin 1 |
177 |
0.97 |
| chr13_24359125_24359350 | 0.13 |
Gm11342 |
predicted gene 11342 |
16713 |
0.12 |
| chr11_16779122_16779339 | 0.13 |
Egfr |
epidermal growth factor receptor |
27000 |
0.17 |
| chr11_16761103_16761540 | 0.13 |
Egfr |
epidermal growth factor receptor |
9091 |
0.2 |
| chr2_134495277_134495573 | 0.13 |
Hao1 |
hydroxyacid oxidase 1, liver |
58882 |
0.16 |
| chr6_112635828_112636003 | 0.13 |
Gm5578 |
predicted pseudogene 5578 |
30371 |
0.14 |
| chr5_151018980_151019139 | 0.13 |
Gm8675 |
predicted gene 8675 |
9758 |
0.23 |
| chr5_90550918_90551069 | 0.13 |
Gm2602 |
predicted gene 2602 |
7856 |
0.13 |
| chr13_24358292_24358443 | 0.12 |
Gm11342 |
predicted gene 11342 |
17583 |
0.12 |
| chr11_69098275_69098686 | 0.12 |
Per1 |
period circadian clock 1 |
468 |
0.58 |
| chr9_94016628_94017010 | 0.12 |
Gm5369 |
predicted gene 5369 |
122643 |
0.06 |
| chr3_60515813_60516054 | 0.12 |
Mbnl1 |
muscleblind like splicing factor 1 |
12195 |
0.22 |
| chr6_113144373_113144524 | 0.12 |
Setd5 |
SET domain containing 5 |
15767 |
0.14 |
| chr5_104436788_104436985 | 0.12 |
Spp1 |
secreted phosphoprotein 1 |
694 |
0.6 |
| chr1_127898839_127898990 | 0.12 |
Rab3gap1 |
RAB3 GTPase activating protein subunit 1 |
1999 |
0.31 |
| chr11_16831950_16832169 | 0.12 |
Egfros |
epidermal growth factor receptor, opposite strand |
1357 |
0.48 |
| chr6_91752974_91753164 | 0.12 |
Slc6a6 |
solute carrier family 6 (neurotransmitter transporter, taurine), member 6 |
2885 |
0.21 |
| chr17_81175582_81175959 | 0.12 |
Gm50042 |
predicted gene, 50042 |
70968 |
0.11 |
| chr11_11831601_11831799 | 0.12 |
Ddc |
dopa decarboxylase |
4580 |
0.21 |
| chr8_40650786_40650937 | 0.12 |
Mtmr7 |
myotubularin related protein 7 |
16064 |
0.14 |
| chr13_28877494_28877645 | 0.12 |
2610307P16Rik |
RIKEN cDNA 2610307P16 gene |
5887 |
0.2 |
| chr11_94216406_94216835 | 0.12 |
Tob1 |
transducer of ErbB-2.1 |
5166 |
0.19 |
| chr3_107256063_107256233 | 0.11 |
Prok1 |
prokineticin 1 |
16441 |
0.12 |
| chr8_36283239_36283399 | 0.11 |
Lonrf1 |
LON peptidase N-terminal domain and ring finger 1 |
33803 |
0.16 |
| chr13_44819256_44819472 | 0.11 |
Gm22213 |
predicted gene, 22213 |
18919 |
0.2 |
| chr2_71874267_71874418 | 0.11 |
Pdk1 |
pyruvate dehydrogenase kinase, isoenzyme 1 |
599 |
0.75 |
| chr1_67169746_67169962 | 0.11 |
Cps1 |
carbamoyl-phosphate synthetase 1 |
46828 |
0.14 |
| chr8_60947880_60948031 | 0.11 |
Clcn3 |
chloride channel, voltage-sensitive 3 |
6793 |
0.17 |
| chr12_103996630_103996801 | 0.11 |
Gm28577 |
predicted gene 28577 |
136 |
0.92 |
| chr11_16892807_16893180 | 0.11 |
Egfr |
epidermal growth factor receptor |
12192 |
0.19 |
| chr7_140723099_140723551 | 0.11 |
Olfr542-ps1 |
olfactory receptor 542, pseudogene 1 |
2043 |
0.18 |
| chr15_56280670_56280835 | 0.11 |
Hba-ps3 |
hemoglobin alpha, pseudogene 3 |
108363 |
0.07 |
| chr19_21625734_21625885 | 0.11 |
1110059E24Rik |
RIKEN cDNA 1110059E24 gene |
4959 |
0.24 |
| chr11_16814706_16815122 | 0.11 |
Egfros |
epidermal growth factor receptor, opposite strand |
15788 |
0.21 |
| chr6_71999696_71999905 | 0.11 |
Gm26628 |
predicted gene, 26628 |
36025 |
0.1 |
| chr13_9057691_9057882 | 0.11 |
Gm36264 |
predicted gene, 36264 |
18665 |
0.13 |
| chr5_99251194_99251660 | 0.11 |
Rasgef1b |
RasGEF domain family, member 1B |
1500 |
0.46 |
| chr15_59048966_59049301 | 0.11 |
Mtss1 |
MTSS I-BAR domain containing 1 |
6331 |
0.24 |
| chr8_76147063_76147228 | 0.11 |
Gm45742 |
predicted gene 45742 |
30118 |
0.21 |
| chr5_66149579_66149965 | 0.11 |
Rbm47 |
RNA binding motif protein 47 |
1184 |
0.28 |
| chr19_26823384_26823535 | 0.11 |
4931403E22Rik |
RIKEN cDNA 4931403E22 gene |
448 |
0.84 |
| chr3_118487666_118487837 | 0.11 |
Gm26871 |
predicted gene, 26871 |
30092 |
0.12 |
| chr11_16901525_16901732 | 0.11 |
Egfr |
epidermal growth factor receptor |
3557 |
0.24 |
| chr14_116350280_116350464 | 0.11 |
Gm38045 |
predicted gene, 38045 |
399843 |
0.01 |
| chr4_76368191_76368359 | 0.11 |
Ptprd |
protein tyrosine phosphatase, receptor type, D |
24032 |
0.21 |
| chr15_85747646_85747958 | 0.11 |
Ppara |
peroxisome proliferator activated receptor alpha |
11671 |
0.14 |
| chr5_92503161_92503312 | 0.11 |
Scarb2 |
scavenger receptor class B, member 2 |
2401 |
0.25 |
| chr9_31589555_31589723 | 0.11 |
Gm18226 |
predicted gene, 18226 |
21393 |
0.16 |
| chr17_64759833_64759998 | 0.11 |
Gm17133 |
predicted gene 17133 |
5819 |
0.21 |
| chr14_67053186_67053337 | 0.11 |
Ppp2r2a |
protein phosphatase 2, regulatory subunit B, alpha |
9732 |
0.17 |
| chr4_130716973_130717173 | 0.11 |
Snord85 |
small nucleolar RNA, C/D box 85 |
32561 |
0.11 |
| chr9_116389320_116389471 | 0.11 |
D730003K21Rik |
RIKEN cDNA D730003K21 gene |
58227 |
0.15 |
| chr4_108060521_108060686 | 0.11 |
Scp2 |
sterol carrier protein 2, liver |
10760 |
0.13 |
| chr11_120560210_120560367 | 0.11 |
P4hb |
prolyl 4-hydroxylase, beta polypeptide |
948 |
0.26 |
| chrX_57836208_57836359 | 0.10 |
Gm14631 |
predicted gene 14631 |
82415 |
0.08 |
| chr7_114203967_114204169 | 0.10 |
Gm45454 |
predicted gene 45454 |
5020 |
0.23 |
| chr17_35808048_35808407 | 0.10 |
Ier3 |
immediate early response 3 |
13457 |
0.07 |
| chr11_58167864_58168078 | 0.10 |
Gemin5 |
gem nuclear organelle associated protein 5 |
568 |
0.6 |
| chr19_10103951_10104278 | 0.10 |
Fads2 |
fatty acid desaturase 2 |
2368 |
0.23 |
| chr3_116271828_116272000 | 0.10 |
Gpr88 |
G-protein coupled receptor 88 |
18411 |
0.18 |
| chr15_97187929_97188091 | 0.10 |
Gm32885 |
predicted gene, 32885 |
20766 |
0.21 |
| chr17_64644992_64645205 | 0.10 |
Man2a1 |
mannosidase 2, alpha 1 |
44362 |
0.16 |
| chr7_99193825_99193976 | 0.10 |
Gm45012 |
predicted gene 45012 |
8466 |
0.13 |
| chr19_10087853_10088030 | 0.10 |
Fads2 |
fatty acid desaturase 2 |
775 |
0.57 |
| chr19_60532004_60532219 | 0.10 |
Cacul1 |
CDK2 associated, cullin domain 1 |
5438 |
0.23 |
| chr19_40155227_40155577 | 0.10 |
Cyp2c70 |
cytochrome P450, family 2, subfamily c, polypeptide 70 |
31884 |
0.13 |
| chr1_24116155_24116306 | 0.10 |
Gm26607 |
predicted gene, 26607 |
9045 |
0.17 |
| chr18_90541359_90541533 | 0.10 |
Tmx3 |
thioredoxin-related transmembrane protein 3 |
4392 |
0.18 |
| chr6_24262961_24263227 | 0.10 |
Slc13a1 |
solute carrier family 13 (sodium/sulfate symporters), member 1 |
95002 |
0.08 |
| chr18_33377697_33377885 | 0.10 |
Gm5503 |
predicted gene 5503 |
7164 |
0.27 |
| chr6_114877506_114877696 | 0.10 |
Vgll4 |
vestigial like family member 4 |
2494 |
0.3 |
| chr7_68976071_68976222 | 0.10 |
Gm44691 |
predicted gene 44691 |
13258 |
0.21 |
| chr3_58235650_58235811 | 0.10 |
Gm26166 |
predicted gene, 26166 |
54627 |
0.11 |
| chr6_114888678_114888863 | 0.10 |
Vgll4 |
vestigial like family member 4 |
13663 |
0.2 |
| chr3_52011173_52011324 | 0.10 |
Gm37465 |
predicted gene, 37465 |
7223 |
0.13 |
| chr5_45903098_45903253 | 0.10 |
4930405L22Rik |
RIKEN cDNA 4930405L22 gene |
29234 |
0.16 |
| chr2_69724507_69724673 | 0.10 |
Ppig |
peptidyl-prolyl isomerase G (cyclophilin G) |
1264 |
0.33 |
| chr13_115162784_115162981 | 0.10 |
Gm18759 |
predicted gene, 18759 |
1938 |
0.33 |
| chr11_118265609_118265912 | 0.10 |
Usp36 |
ubiquitin specific peptidase 36 |
288 |
0.89 |
| chr11_63891944_63892129 | 0.10 |
Hs3st3b1 |
heparan sulfate (glucosamine) 3-O-sulfotransferase 3B1 |
30254 |
0.17 |
| chr6_103697460_103697614 | 0.10 |
Chl1 |
cell adhesion molecule L1-like |
213 |
0.95 |
| chr6_15930296_15930597 | 0.10 |
Gm43990 |
predicted gene, 43990 |
94357 |
0.08 |
| chr4_121163852_121164031 | 0.10 |
Rlf |
rearranged L-myc fusion sequence |
24593 |
0.11 |
| chr6_56907100_56907281 | 0.10 |
Gm3793 |
predicted gene 3793 |
608 |
0.64 |
| chr10_68056586_68056943 | 0.10 |
Rtkn2 |
rhotekin 2 |
12796 |
0.2 |
| chr3_60099135_60099299 | 0.10 |
Sucnr1 |
succinate receptor 1 |
17315 |
0.18 |
| chr16_93367939_93368144 | 0.10 |
1810044K17Rik |
RIKEN cDNA 1810044K17 gene |
198 |
0.89 |
| chr13_24359929_24360183 | 0.10 |
Gm11342 |
predicted gene 11342 |
15894 |
0.12 |
| chr2_129197178_129197487 | 0.09 |
Gm4430 |
predicted gene 4430 |
206 |
0.85 |
| chr17_71561749_71561923 | 0.09 |
Spdya |
speedy/RINGO cell cycle regulator family, member A |
5578 |
0.12 |
| chr3_129593850_129594054 | 0.09 |
Elovl6 |
ELOVL family member 6, elongation of long chain fatty acids (yeast) |
41572 |
0.12 |
| chr3_94710144_94710297 | 0.09 |
Selenbp2 |
selenium binding protein 2 |
16561 |
0.1 |
| chr6_108606639_108606790 | 0.09 |
0610040F04Rik |
RIKEN cDNA 0610040F04 gene |
7281 |
0.16 |
| chr1_67144557_67144947 | 0.09 |
Cps1 |
carbamoyl-phosphate synthetase 1 |
21726 |
0.21 |
| chr6_56904208_56904367 | 0.09 |
Gm3793 |
predicted gene 3793 |
2295 |
0.19 |
| chr10_53318105_53318261 | 0.09 |
Gm17823 |
predicted gene, 17823 |
18041 |
0.12 |
| chr15_59430248_59430452 | 0.09 |
Nsmce2 |
NSE2/MMS21 homolog, SMC5-SMC6 complex SUMO ligase |
56063 |
0.12 |
| chr9_74318679_74318830 | 0.09 |
Gm24141 |
predicted gene, 24141 |
43856 |
0.14 |
| chr8_93168995_93169226 | 0.09 |
Ces1d |
carboxylesterase 1D |
865 |
0.51 |
| chr1_67153015_67153221 | 0.09 |
Cps1 |
carbamoyl-phosphate synthetase 1 |
30092 |
0.19 |
| chr2_72223568_72223719 | 0.09 |
Rapgef4os2 |
Rap guanine nucleotide exchange factor (GEF) 4, opposite strand 2 |
8659 |
0.17 |
| chr4_44920931_44921086 | 0.09 |
Zcchc7 |
zinc finger, CCHC domain containing 7 |
2110 |
0.25 |
| chr2_12899690_12899932 | 0.09 |
Pter |
phosphotriesterase related |
24230 |
0.21 |
| chr11_84290556_84290741 | 0.09 |
Acaca |
acetyl-Coenzyme A carboxylase alpha |
10193 |
0.25 |
| chr10_87541080_87541552 | 0.09 |
Pah |
phenylalanine hydroxylase |
5357 |
0.23 |
| chr13_25015682_25015833 | 0.09 |
Mrs2 |
MRS2 magnesium transporter |
4019 |
0.19 |
| chr16_42910828_42911152 | 0.09 |
Gm19522 |
predicted gene, 19522 |
1075 |
0.5 |
| chr1_153739671_153739993 | 0.09 |
Rgs16 |
regulator of G-protein signaling 16 |
517 |
0.61 |
| chr16_56598260_56598451 | 0.09 |
Tfg |
Trk-fused gene |
107339 |
0.06 |
| chr6_144086911_144087062 | 0.09 |
Sox5os1 |
SRY (sex determining region Y)-box 5, opposite strand 1 |
65078 |
0.13 |
| chr2_28119005_28119156 | 0.09 |
F730016J06Rik |
RIKEN cDNA F730016J06 gene |
873 |
0.59 |
| chr10_80437858_80438035 | 0.09 |
Tcf3 |
transcription factor 3 |
4299 |
0.1 |
| chr4_134837509_134837670 | 0.09 |
Maco1 |
macoilin 1 |
15560 |
0.17 |
| chr15_7195034_7195185 | 0.09 |
Egflam |
EGF-like, fibronectin type III and laminin G domains |
27960 |
0.21 |
| chr17_69982645_69982841 | 0.09 |
Dlgap1 |
DLG associated protein 1 |
13322 |
0.24 |
| chr2_126622639_126622795 | 0.09 |
Hdc |
histidine decarboxylase |
3418 |
0.2 |
| chr5_8422645_8423469 | 0.09 |
Slc25a40 |
solute carrier family 25, member 40 |
158 |
0.72 |
| chr7_113961088_113961254 | 0.09 |
Gm45615 |
predicted gene 45615 |
125727 |
0.05 |
| chr10_95255137_95255325 | 0.09 |
Gm48880 |
predicted gene, 48880 |
59622 |
0.08 |
| chr2_27769095_27769412 | 0.09 |
Rxra |
retinoid X receptor alpha |
29052 |
0.21 |
| chr10_57301198_57301349 | 0.09 |
Gm7001 |
predicted gene 7001 |
10384 |
0.21 |
| chr18_56396921_56397088 | 0.09 |
Gramd3 |
GRAM domain containing 3 |
3333 |
0.25 |
| chr2_90583663_90583816 | 0.09 |
Ptprj |
protein tyrosine phosphatase, receptor type, J |
3092 |
0.29 |
| chr14_16572962_16573173 | 0.09 |
Rarb |
retinoic acid receptor, beta |
1978 |
0.34 |
| chr6_50561677_50561828 | 0.09 |
Cycs |
cytochrome c, somatic |
4197 |
0.12 |
| chr5_45437140_45437291 | 0.09 |
Gm42413 |
predicted gene, 42413 |
2826 |
0.18 |
| chr15_55042916_55043114 | 0.09 |
Taf2 |
TATA-box binding protein associated factor 2 |
3151 |
0.21 |
| chr14_45385956_45386177 | 0.09 |
Gnpnat1 |
glucosamine-phosphate N-acetyltransferase 1 |
582 |
0.65 |
| chr2_6328324_6328528 | 0.09 |
AL845275.1 |
novel protein |
5346 |
0.2 |
| chr10_40511959_40512217 | 0.09 |
Gm18671 |
predicted gene, 18671 |
36804 |
0.14 |
| chr3_141299865_141300028 | 0.09 |
Pdha2 |
pyruvate dehydrogenase E1 alpha 2 |
87591 |
0.1 |
| chr11_16882054_16882234 | 0.09 |
Egfr |
epidermal growth factor receptor |
3994 |
0.24 |
| chr19_57258284_57258443 | 0.09 |
4930449E18Rik |
RIKEN cDNA 4930449E18 gene |
15597 |
0.18 |
| chr4_134847461_134847666 | 0.09 |
Maco1 |
macoilin 1 |
5586 |
0.19 |
| chr6_39781272_39781654 | 0.09 |
Mrps33 |
mitochondrial ribosomal protein S33 |
24579 |
0.15 |
| chr3_98407172_98407364 | 0.09 |
Zfp697 |
zinc finger protein 697 |
24720 |
0.13 |
| chr12_30348373_30348544 | 0.09 |
Sntg2 |
syntrophin, gamma 2 |
9225 |
0.26 |
| chr17_46448212_46448499 | 0.09 |
Gm5093 |
predicted gene 5093 |
8258 |
0.1 |
| chr1_67201067_67201218 | 0.09 |
Gm15668 |
predicted gene 15668 |
48058 |
0.14 |
| chr10_89512416_89512679 | 0.09 |
Nr1h4 |
nuclear receptor subfamily 1, group H, member 4 |
5889 |
0.23 |
| chr13_58389633_58389784 | 0.09 |
Mir7-1 |
microRNA 7-1 |
3178 |
0.14 |
| chr8_128726356_128726507 | 0.09 |
Itgb1 |
integrin beta 1 (fibronectin receptor beta) |
2098 |
0.32 |
| chr12_29082462_29082658 | 0.09 |
4833405L11Rik |
RIKEN cDNA 4833405L11 gene |
24177 |
0.19 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.1 | 0.2 | GO:1901078 | negative regulation of relaxation of muscle(GO:1901078) |
| 0.0 | 0.3 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.0 | 0.1 | GO:0019676 | ammonia assimilation cycle(GO:0019676) |
| 0.0 | 0.1 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.1 | GO:1903416 | response to glycoside(GO:1903416) |
| 0.0 | 0.1 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.0 | 0.1 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.0 | 0.2 | GO:0034310 | primary alcohol catabolic process(GO:0034310) |
| 0.0 | 0.1 | GO:0072338 | creatinine metabolic process(GO:0046449) cellular lactam metabolic process(GO:0072338) |
| 0.0 | 0.1 | GO:0097167 | circadian regulation of translation(GO:0097167) |
| 0.0 | 0.1 | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.0 | 0.1 | GO:0060584 | regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.0 | 0.1 | GO:0007161 | calcium-independent cell-matrix adhesion(GO:0007161) |
| 0.0 | 0.0 | GO:1901187 | regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.0 | 0.1 | GO:0060620 | regulation of cholesterol import(GO:0060620) regulation of sterol import(GO:2000909) |
| 0.0 | 0.2 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.0 | 0.1 | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 0.0 | 0.1 | GO:0019065 | receptor-mediated endocytosis of virus by host cell(GO:0019065) endocytosis involved in viral entry into host cell(GO:0075509) |
| 0.0 | 0.1 | GO:0021699 | cerebellar cortex maturation(GO:0021699) |
| 0.0 | 0.0 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.0 | GO:2000619 | negative regulation of histone H4-K16 acetylation(GO:2000619) |
| 0.0 | 0.1 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.0 | 0.1 | GO:1903966 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.0 | 0.1 | GO:0061641 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.0 | 0.1 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.0 | 0.1 | GO:0030647 | polyketide metabolic process(GO:0030638) aminoglycoside antibiotic metabolic process(GO:0030647) daunorubicin metabolic process(GO:0044597) doxorubicin metabolic process(GO:0044598) |
| 0.0 | 0.1 | GO:0045908 | negative regulation of vasodilation(GO:0045908) |
| 0.0 | 0.0 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.0 | 0.1 | GO:1903347 | negative regulation of bicellular tight junction assembly(GO:1903347) |
| 0.0 | 0.0 | GO:0060166 | olfactory pit development(GO:0060166) |
| 0.0 | 0.0 | GO:0015959 | diadenosine polyphosphate metabolic process(GO:0015959) |
| 0.0 | 0.0 | GO:1901740 | negative regulation of myoblast fusion(GO:1901740) |
| 0.0 | 0.1 | GO:0010572 | positive regulation of platelet activation(GO:0010572) |
| 0.0 | 0.1 | GO:0006559 | L-phenylalanine catabolic process(GO:0006559) erythrose 4-phosphate/phosphoenolpyruvate family amino acid catabolic process(GO:1902222) |
| 0.0 | 0.1 | GO:0070508 | sterol import(GO:0035376) cholesterol import(GO:0070508) |
| 0.0 | 0.0 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.0 | 0.0 | GO:0018211 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.0 | 0.0 | GO:1901856 | negative regulation of cellular respiration(GO:1901856) |
| 0.0 | 0.0 | GO:0070672 | response to interleukin-15(GO:0070672) |
| 0.0 | 0.0 | GO:0006041 | glucosamine metabolic process(GO:0006041) |
| 0.0 | 0.0 | GO:1901162 | serotonin biosynthetic process(GO:0042427) primary amino compound biosynthetic process(GO:1901162) |
| 0.0 | 0.0 | GO:1902915 | negative regulation of protein K63-linked ubiquitination(GO:1900045) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.0 | 0.0 | GO:1903352 | ornithine transport(GO:0015822) L-ornithine transmembrane transport(GO:1903352) |
| 0.0 | 0.0 | GO:2000359 | regulation of binding of sperm to zona pellucida(GO:2000359) |
| 0.0 | 0.1 | GO:0060283 | negative regulation of oocyte development(GO:0060283) |
| 0.0 | 0.0 | GO:0061535 | glutamate secretion, neurotransmission(GO:0061535) |
| 0.0 | 0.1 | GO:0046618 | drug export(GO:0046618) |
| 0.0 | 0.0 | GO:0090219 | negative regulation of lipid kinase activity(GO:0090219) |
| 0.0 | 0.0 | GO:0002071 | glandular epithelial cell maturation(GO:0002071) |
| 0.0 | 0.0 | GO:0007089 | traversing start control point of mitotic cell cycle(GO:0007089) |
| 0.0 | 0.0 | GO:0010887 | negative regulation of cholesterol storage(GO:0010887) |
| 0.0 | 0.0 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.0 | 0.0 | GO:0040009 | regulation of growth rate(GO:0040009) |
| 0.0 | 0.0 | GO:2000259 | positive regulation of complement activation(GO:0045917) positive regulation of protein activation cascade(GO:2000259) |
| 0.0 | 0.1 | GO:0045714 | low-density lipoprotein particle receptor biosynthetic process(GO:0045713) regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045714) |
| 0.0 | 0.0 | GO:0009826 | unidimensional cell growth(GO:0009826) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.0 | 0.1 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.0 | 0.1 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.0 | 0.1 | GO:1990423 | RZZ complex(GO:1990423) |
| 0.0 | 0.1 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.0 | 0.1 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.0 | 0.1 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.0 | 0.1 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.0 | 0.1 | GO:0097433 | dense body(GO:0097433) |
| 0.0 | 0.0 | GO:0034666 | integrin alpha2-beta1 complex(GO:0034666) |
| 0.0 | 0.1 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.1 | 0.2 | GO:0016743 | carboxyl- or carbamoyltransferase activity(GO:0016743) |
| 0.0 | 0.1 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.0 | 0.1 | GO:0038181 | bile acid receptor activity(GO:0038181) |
| 0.0 | 0.1 | GO:0070538 | oleic acid binding(GO:0070538) |
| 0.0 | 0.1 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 0.0 | 0.2 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) |
| 0.0 | 0.2 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.1 | GO:1990239 | steroid hormone binding(GO:1990239) |
| 0.0 | 0.1 | GO:0016899 | oxidoreductase activity, acting on the CH-OH group of donors, oxygen as acceptor(GO:0016899) |
| 0.0 | 0.1 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.0 | 0.1 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.0 | 0.1 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.1 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.0 | 0.1 | GO:0015065 | uridine nucleotide receptor activity(GO:0015065) G-protein coupled pyrimidinergic nucleotide receptor activity(GO:0071553) |
| 0.0 | 0.0 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.0 | 0.0 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.0 | 0.0 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.0 | 0.0 | GO:0048030 | disaccharide binding(GO:0048030) |
| 0.0 | 0.1 | GO:0047023 | androsterone dehydrogenase activity(GO:0047023) |
| 0.0 | 0.0 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.0 | 0.1 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 0.0 | 0.0 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.0 | 0.0 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.0 | 0.0 | GO:0003846 | 2-acylglycerol O-acyltransferase activity(GO:0003846) |
| 0.0 | 0.0 | GO:0016215 | acyl-CoA desaturase activity(GO:0016215) |
| 0.0 | 0.1 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.3 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME SHC1 EVENTS IN EGFR SIGNALING | Genes involved in SHC1 events in EGFR signaling |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.1 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.0 | 0.1 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.1 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.1 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |