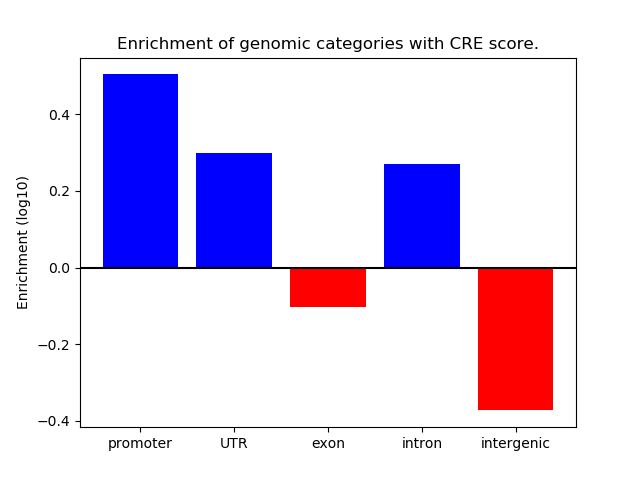
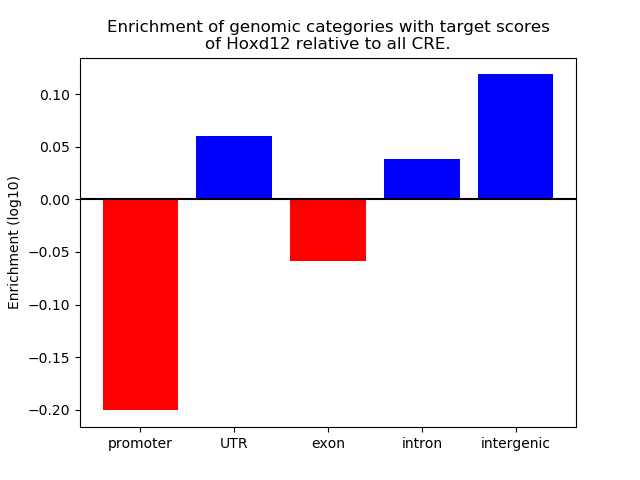
Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
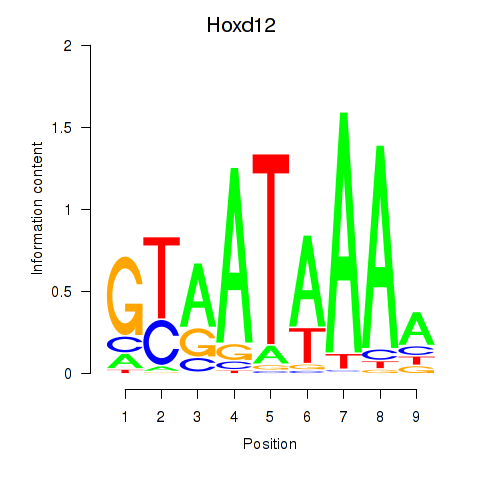
Results for Hoxd12
Z-value: 1.05
Transcription factors associated with Hoxd12
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hoxd12
|
ENSMUSG00000001823.4 | homeobox D12 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr2_74673098_74673252 | Hoxd12 | 1838 | 0.118513 | -0.55 | 2.6e-01 | Click! |
| chr2_74672857_74673036 | Hoxd12 | 2067 | 0.106046 | -0.09 | 8.7e-01 | Click! |
Activity of the Hoxd12 motif across conditions
Conditions sorted by the z-value of the Hoxd12 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
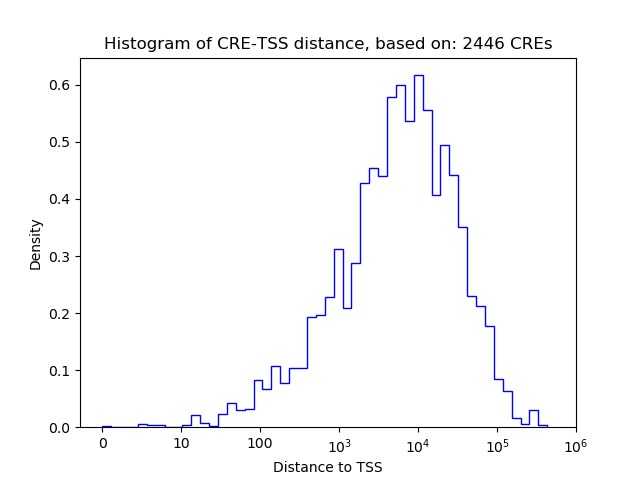
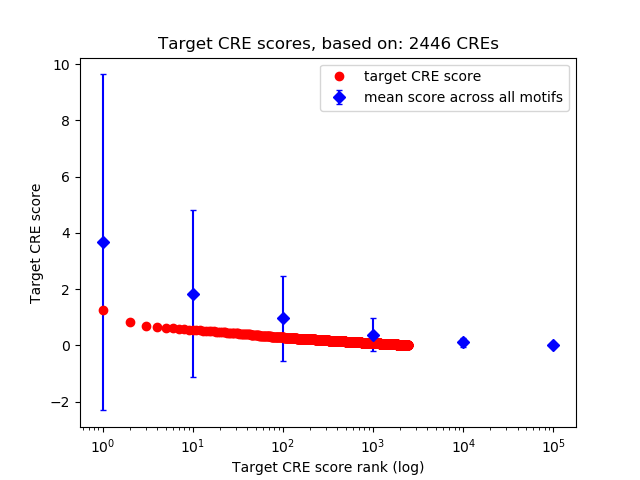
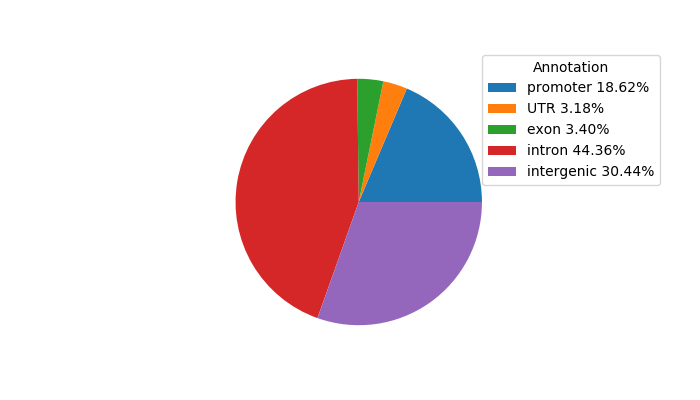
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr7_143779216_143779367 | 1.24 |
Gm22064 |
predicted gene, 22064 |
799 |
0.46 |
| chr14_46741092_46741246 | 0.82 |
Gm48924 |
predicted gene, 48924 |
5230 |
0.13 |
| chr18_70622479_70622643 | 0.68 |
Mbd2 |
methyl-CpG binding domain protein 2 |
4788 |
0.23 |
| chr13_59870307_59870458 | 0.67 |
Gm48384 |
predicted gene, 48384 |
3260 |
0.18 |
| chr6_15232040_15232214 | 0.60 |
Foxp2 |
forkhead box P2 |
22156 |
0.28 |
| chr8_114979772_114979943 | 0.60 |
Gm22556 |
predicted gene, 22556 |
73056 |
0.13 |
| chr6_145811271_145811882 | 0.59 |
Rassf8 |
Ras association (RalGDS/AF-6) domain family (N-terminal) member 8 |
3193 |
0.26 |
| chr15_101320859_101321043 | 0.58 |
Gm35853 |
predicted gene, 35853 |
1937 |
0.18 |
| chr17_63076780_63076958 | 0.55 |
Gm25348 |
predicted gene, 25348 |
20131 |
0.27 |
| chr15_55866367_55866558 | 0.54 |
Sntb1 |
syntrophin, basic 1 |
39848 |
0.15 |
| chr15_74872740_74872891 | 0.53 |
Ly6m |
lymphocyte antigen 6 complex, locus M |
8889 |
0.1 |
| chr8_128484406_128484701 | 0.53 |
Nrp1 |
neuropilin 1 |
125156 |
0.05 |
| chr19_23071512_23071878 | 0.52 |
C330002G04Rik |
RIKEN cDNA C330002G04 gene |
4158 |
0.22 |
| chr2_10156330_10156481 | 0.51 |
Itih5 |
inter-alpha (globulin) inhibitor H5 |
2834 |
0.18 |
| chr11_49615889_49616040 | 0.51 |
Flt4 |
FMS-like tyrosine kinase 4 |
6701 |
0.12 |
| chr2_80600719_80600876 | 0.50 |
Gm13688 |
predicted gene 13688 |
1936 |
0.21 |
| chr8_119445523_119446899 | 0.49 |
Necab2 |
N-terminal EF-hand calcium binding protein 2 |
508 |
0.74 |
| chr16_30257794_30257955 | 0.49 |
Gm49645 |
predicted gene, 49645 |
2722 |
0.2 |
| chr11_106492486_106492637 | 0.49 |
Ern1 |
endoplasmic reticulum (ER) to nucleus signalling 1 |
4709 |
0.17 |
| chr9_73050700_73050869 | 0.48 |
Rab27a |
RAB27A, member RAS oncogene family |
5800 |
0.09 |
| chr4_108166700_108166861 | 0.48 |
Echdc2 |
enoyl Coenzyme A hydratase domain containing 2 |
1262 |
0.4 |
| chr19_23066388_23066539 | 0.47 |
Gm50136 |
predicted gene, 50136 |
4990 |
0.21 |
| chr14_70338039_70338256 | 0.46 |
Slc39a14 |
solute carrier family 39 (zinc transporter), member 14 |
3949 |
0.15 |
| chr1_121332181_121332567 | 0.45 |
Insig2 |
insulin induced gene 2 |
176 |
0.94 |
| chr7_145056434_145056606 | 0.45 |
Gm45181 |
predicted gene 45181 |
106476 |
0.05 |
| chr9_65289050_65289226 | 0.43 |
Clpx |
caseinolytic mitochondrial matrix peptidase chaperone subunit |
5122 |
0.12 |
| chr19_21297468_21297791 | 0.43 |
Gm50255 |
predicted gene, 50255 |
15654 |
0.17 |
| chr3_83043842_83044694 | 0.42 |
Fgb |
fibrinogen beta chain |
5595 |
0.15 |
| chr1_72032135_72032300 | 0.42 |
4930556G22Rik |
RIKEN cDNA 4930556G22 gene |
6514 |
0.17 |
| chr8_124844006_124844208 | 0.42 |
Gm16163 |
predicted gene 16163 |
12409 |
0.11 |
| chr5_117293923_117294074 | 0.42 |
Gm3786 |
predicted gene 3786 |
408 |
0.73 |
| chr5_123865495_123865700 | 0.42 |
Hcar2 |
hydroxycarboxylic acid receptor 2 |
98 |
0.95 |
| chr11_45892970_45893144 | 0.42 |
Clint1 |
clathrin interactor 1 |
10660 |
0.16 |
| chr6_47533521_47533999 | 0.41 |
Ezh2 |
enhancer of zeste 2 polycomb repressive complex 2 subunit |
1365 |
0.42 |
| chr11_119130166_119130354 | 0.41 |
Gm11755 |
predicted gene 11755 |
8161 |
0.13 |
| chr12_84277793_84277988 | 0.41 |
Ptgr2 |
prostaglandin reductase 2 |
7342 |
0.12 |
| chr3_104359662_104359838 | 0.40 |
Gm5546 |
predicted gene 5546 |
6863 |
0.18 |
| chr4_46987889_46988055 | 0.40 |
Gabbr2 |
gamma-aminobutyric acid (GABA) B receptor, 2 |
3901 |
0.21 |
| chr15_28001392_28001573 | 0.40 |
Trio |
triple functional domain (PTPRF interacting) |
6119 |
0.24 |
| chr15_99865164_99865320 | 0.40 |
Lima1 |
LIM domain and actin binding 1 |
9429 |
0.08 |
| chr1_88975960_88976113 | 0.39 |
1700067G17Rik |
RIKEN cDNA 1700067G17 gene |
40077 |
0.14 |
| chr12_91929516_91929695 | 0.39 |
Rpl31-ps1 |
ribosomal protein L31, pseudogene 1 |
38332 |
0.12 |
| chr3_60079118_60079300 | 0.38 |
Sucnr1 |
succinate receptor 1 |
2660 |
0.27 |
| chr16_13258895_13259057 | 0.38 |
Mrtfb |
myocardin related transcription factor B |
2470 |
0.38 |
| chr2_114007839_114008018 | 0.38 |
A530058N18Rik |
RIKEN cDNA A530058N18 gene |
5635 |
0.18 |
| chr5_146114373_146114524 | 0.38 |
Cyp3a59 |
cytochrome P450, family 3, subfamily a, polypeptide 59 |
35181 |
0.09 |
| chr4_105105680_105105928 | 0.38 |
Prkaa2 |
protein kinase, AMP-activated, alpha 2 catalytic subunit |
4086 |
0.31 |
| chr8_45253213_45253379 | 0.37 |
F11 |
coagulation factor XI |
8735 |
0.18 |
| chr5_118332124_118332291 | 0.37 |
Gm28563 |
predicted gene 28563 |
65123 |
0.09 |
| chr2_63814449_63814614 | 0.37 |
Fign |
fidgetin |
283457 |
0.01 |
| chr14_34635884_34636046 | 0.36 |
Gm49024 |
predicted gene, 49024 |
25742 |
0.1 |
| chr7_132810815_132810966 | 0.36 |
Fam53b |
family with sequence similarity 53, member B |
2205 |
0.27 |
| chr16_76225574_76225868 | 0.35 |
Gm26915 |
predicted gene, 26915 |
80736 |
0.08 |
| chr1_93144926_93145077 | 0.35 |
Agxt |
alanine-glyoxylate aminotransferase |
5122 |
0.14 |
| chr1_165348964_165349116 | 0.35 |
Dcaf6 |
DDB1 and CUL4 associated factor 6 |
2958 |
0.22 |
| chr4_71839761_71839932 | 0.35 |
Gm11233 |
predicted gene 11233 |
28142 |
0.23 |
| chr9_48343199_48343563 | 0.35 |
Nxpe2 |
neurexophilin and PC-esterase domain family, member 2 |
2483 |
0.3 |
| chr8_26999709_26999860 | 0.35 |
Gm45370 |
predicted gene 45370 |
3534 |
0.12 |
| chr2_52928058_52928266 | 0.34 |
Fmnl2 |
formin-like 2 |
70294 |
0.12 |
| chr4_72049920_72050259 | 0.34 |
Gm11234 |
predicted gene 11234 |
69612 |
0.11 |
| chr4_46952731_46952882 | 0.34 |
Gabbr2 |
gamma-aminobutyric acid (GABA) B receptor, 2 |
39067 |
0.14 |
| chr6_145830100_145830412 | 0.34 |
Rassf8 |
Ras association (RalGDS/AF-6) domain family (N-terminal) member 8 |
10312 |
0.18 |
| chr3_108314174_108314346 | 0.34 |
Gm43099 |
predicted gene 43099 |
26379 |
0.07 |
| chr16_18430516_18430667 | 0.33 |
Txnrd2 |
thioredoxin reductase 2 |
1666 |
0.22 |
| chr12_117763341_117763497 | 0.33 |
Rapgef5 |
Rap guanine nucleotide exchange factor (GEF) 5 |
22824 |
0.18 |
| chr1_58120047_58120378 | 0.32 |
Aox3 |
aldehyde oxidase 3 |
7026 |
0.2 |
| chr3_51380110_51380295 | 0.32 |
Gm5103 |
predicted gene 5103 |
2126 |
0.17 |
| chr12_73270111_73270267 | 0.32 |
Trmt5 |
TRM5 tRNA methyltransferase 5 |
12482 |
0.19 |
| chr5_103783262_103783453 | 0.32 |
Aff1 |
AF4/FMR2 family, member 1 |
1002 |
0.59 |
| chr11_96862257_96862408 | 0.32 |
Gm11524 |
predicted gene 11524 |
7863 |
0.09 |
| chr8_108993830_108994020 | 0.32 |
Mir3108 |
microRNA 3108 |
57065 |
0.13 |
| chr7_140760788_140760954 | 0.31 |
Cyp2e1 |
cytochrome P450, family 2, subfamily e, polypeptide 1 |
2868 |
0.13 |
| chr1_180757526_180757875 | 0.31 |
Gm37768 |
predicted gene, 37768 |
1864 |
0.22 |
| chr4_141133226_141133487 | 0.31 |
Szrd1 |
SUZ RNA binding domain containing 1 |
6371 |
0.12 |
| chr17_30136017_30136188 | 0.31 |
Zfand3 |
zinc finger, AN1-type domain 3 |
88 |
0.97 |
| chr10_99269736_99269912 | 0.31 |
Gm34921 |
predicted gene, 34921 |
622 |
0.56 |
| chr18_23805749_23805916 | 0.31 |
Mapre2 |
microtubule-associated protein, RP/EB family, member 2 |
1729 |
0.33 |
| chr8_128394116_128394267 | 0.31 |
Nrp1 |
neuropilin 1 |
34794 |
0.18 |
| chr6_134614784_134614949 | 0.31 |
Gm38910 |
predicted gene, 38910 |
7185 |
0.14 |
| chr9_21424708_21424871 | 0.30 |
Dnm2 |
dynamin 2 |
119 |
0.93 |
| chr14_35119944_35120361 | 0.30 |
Gm49034 |
predicted gene, 49034 |
99286 |
0.08 |
| chr11_62487235_62487386 | 0.30 |
Gm12278 |
predicted gene 12278 |
4513 |
0.13 |
| chr3_37769304_37769512 | 0.30 |
Gm42921 |
predicted gene 42921 |
8371 |
0.13 |
| chr3_142300612_142300795 | 0.30 |
Pdlim5 |
PDZ and LIM domain 5 |
3276 |
0.34 |
| chr11_79229335_79229498 | 0.30 |
Wsb1 |
WD repeat and SOCS box-containing 1 |
13533 |
0.16 |
| chr14_118764159_118764325 | 0.29 |
Gm22379 |
predicted gene, 22379 |
10910 |
0.16 |
| chr4_138365532_138365692 | 0.29 |
Cda |
cytidine deaminase |
2380 |
0.19 |
| chr4_103386324_103386511 | 0.29 |
Gm12718 |
predicted gene 12718 |
3896 |
0.25 |
| chr12_98375326_98375477 | 0.29 |
5330409N07Rik |
RIKEN cDNA 5330409N07 gene |
72933 |
0.08 |
| chr6_145355079_145355230 | 0.29 |
Gm23498 |
predicted gene, 23498 |
11704 |
0.13 |
| chr1_58145328_58145479 | 0.29 |
Gm24548 |
predicted gene, 24548 |
9685 |
0.19 |
| chr11_26112294_26112697 | 0.29 |
5730522E02Rik |
RIKEN cDNA 5730522E02 gene |
30895 |
0.23 |
| chr6_129525955_129526168 | 0.29 |
Gm44120 |
predicted gene, 44120 |
3499 |
0.1 |
| chr12_25235200_25235794 | 0.29 |
Gm19340 |
predicted gene, 19340 |
47098 |
0.12 |
| chr6_17172773_17172927 | 0.28 |
Gm4876 |
predicted gene 4876 |
1017 |
0.58 |
| chr7_68696821_68697010 | 0.28 |
Gm44692 |
predicted gene 44692 |
29552 |
0.18 |
| chr19_55166765_55167101 | 0.28 |
Tectb |
tectorin beta |
13800 |
0.17 |
| chr13_114457557_114457872 | 0.28 |
Fst |
follistatin |
872 |
0.39 |
| chr13_21779387_21779554 | 0.28 |
H1f5 |
H1.5 linker histone, cluster member |
1155 |
0.16 |
| chr15_100679132_100679301 | 0.28 |
Cela1 |
chymotrypsin-like elastase family, member 1 |
150 |
0.91 |
| chr6_37633621_37633893 | 0.28 |
Ybx1-ps2 |
Y box protein 1, pseudogene 2 |
53438 |
0.14 |
| chr12_40034812_40034992 | 0.28 |
Arl4a |
ADP-ribosylation factor-like 4A |
2465 |
0.26 |
| chr11_52210469_52210623 | 0.28 |
Olfr1371 |
olfactory receptor 1371 |
3441 |
0.15 |
| chr1_185703517_185703677 | 0.28 |
Gm38093 |
predicted gene, 38093 |
41206 |
0.19 |
| chr4_107241963_107242114 | 0.28 |
Lrrc42 |
leucine rich repeat containing 42 |
1489 |
0.29 |
| chr9_89218115_89218458 | 0.27 |
Mthfs |
5, 10-methenyltetrahydrofolate synthetase |
168 |
0.93 |
| chr15_102245005_102245156 | 0.27 |
Rarg |
retinoic acid receptor, gamma |
151 |
0.91 |
| chr3_50989485_50989700 | 0.27 |
Gm37209 |
predicted gene, 37209 |
10781 |
0.22 |
| chr7_132578212_132578403 | 0.27 |
Oat |
ornithine aminotransferase |
1909 |
0.28 |
| chr5_115266968_115267119 | 0.27 |
Gm13831 |
predicted gene 13831 |
3950 |
0.1 |
| chr6_48666335_48666494 | 0.27 |
Gm44262 |
predicted gene, 44262 |
554 |
0.52 |
| chr8_36452309_36452599 | 0.26 |
Gm19140 |
predicted gene, 19140 |
2074 |
0.31 |
| chr7_6149176_6149327 | 0.26 |
Zfp787 |
zinc finger protein 787 |
1797 |
0.19 |
| chr5_146679213_146679653 | 0.26 |
4930573C15Rik |
RIKEN cDNA 4930573C15 gene |
27189 |
0.15 |
| chr6_122161223_122161580 | 0.26 |
Mug-ps1 |
murinoglobulin, pseudogene 1 |
14614 |
0.16 |
| chr2_48470032_48470196 | 0.26 |
Gm13481 |
predicted gene 13481 |
12869 |
0.26 |
| chr5_96162930_96163161 | 0.26 |
Cnot6l |
CCR4-NOT transcription complex, subunit 6-like |
1055 |
0.55 |
| chr17_13819577_13819728 | 0.26 |
Afdn |
afadin, adherens junction formation factor |
31310 |
0.13 |
| chr15_66945679_66946053 | 0.26 |
Ndrg1 |
N-myc downstream regulated gene 1 |
2613 |
0.24 |
| chr16_94694044_94694202 | 0.26 |
Gm41504 |
predicted gene, 41504 |
19780 |
0.16 |
| chr2_61138777_61138928 | 0.26 |
Gm13581 |
predicted gene 13581 |
85940 |
0.09 |
| chr19_47860475_47860640 | 0.26 |
Gsto1 |
glutathione S-transferase omega 1 |
2438 |
0.22 |
| chr6_5174317_5174636 | 0.26 |
Pon1 |
paraoxonase 1 |
19287 |
0.18 |
| chr2_61086233_61086389 | 0.25 |
Gm13579 |
predicted gene 13579 |
55341 |
0.15 |
| chr6_99506876_99507039 | 0.25 |
Foxp1 |
forkhead box P1 |
14040 |
0.19 |
| chr4_20032087_20032276 | 0.25 |
Ggh |
gamma-glutamyl hydrolase |
9871 |
0.2 |
| chr6_5389469_5389626 | 0.25 |
Asb4 |
ankyrin repeat and SOCS box-containing 4 |
880 |
0.69 |
| chr18_64326124_64326341 | 0.25 |
St8sia3os |
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 3, opposite strand |
7474 |
0.17 |
| chr6_48741844_48742003 | 0.25 |
Gimap1 |
GTPase, IMAP family member 1 |
259 |
0.61 |
| chr2_52298963_52299373 | 0.25 |
Gm22095 |
predicted gene, 22095 |
16091 |
0.19 |
| chr6_121872993_121873786 | 0.25 |
Mug1 |
murinoglobulin 1 |
12188 |
0.19 |
| chr4_11237010_11237175 | 0.25 |
Ints8 |
integrator complex subunit 8 |
1124 |
0.45 |
| chr16_94694268_94694544 | 0.25 |
Gm41504 |
predicted gene, 41504 |
19497 |
0.16 |
| chr3_81956990_81957439 | 0.25 |
Ctso |
cathepsin O |
6277 |
0.16 |
| chr5_21564099_21564250 | 0.25 |
Lrrc17 |
leucine rich repeat containing 17 |
20611 |
0.14 |
| chr3_138415171_138415339 | 0.25 |
Adh4 |
alcohol dehydrogenase 4 (class II), pi polypeptide |
211 |
0.91 |
| chr1_136911502_136911882 | 0.25 |
Nr5a2 |
nuclear receptor subfamily 5, group A, member 2 |
28891 |
0.18 |
| chr11_86705520_86705677 | 0.25 |
Cltc |
clathrin, heavy polypeptide (Hc) |
1901 |
0.3 |
| chr19_36677883_36678283 | 0.25 |
Hectd2os |
Hectd2, opposite strand |
11190 |
0.23 |
| chr7_80629460_80629678 | 0.25 |
Crtc3 |
CREB regulated transcription coactivator 3 |
57 |
0.97 |
| chr1_105903667_105904011 | 0.25 |
Gm37779 |
predicted gene, 37779 |
12827 |
0.17 |
| chr3_60437847_60438007 | 0.24 |
Mbnl1 |
muscleblind like splicing factor 1 |
34903 |
0.19 |
| chr10_99038868_99039040 | 0.24 |
Gm48761 |
predicted gene, 48761 |
518 |
0.77 |
| chr17_79068246_79068411 | 0.24 |
Qpct |
glutaminyl-peptide cyclotransferase (glutaminyl cyclase) |
8674 |
0.17 |
| chr3_106496064_106496222 | 0.24 |
Dennd2d |
DENN/MADD domain containing 2D |
2968 |
0.18 |
| chr1_72836877_72837059 | 0.24 |
Igfbp2 |
insulin-like growth factor binding protein 2 |
11646 |
0.21 |
| chr17_57224886_57225407 | 0.24 |
C3 |
complement component 3 |
2319 |
0.17 |
| chr7_132972226_132972386 | 0.24 |
Zranb1 |
zinc finger, RAN-binding domain containing 1 |
8553 |
0.15 |
| chr1_22996790_22997104 | 0.24 |
Gm7761 |
predicted gene 7761 |
9822 |
0.28 |
| chr1_134357391_134357543 | 0.24 |
Tmem183a |
transmembrane protein 183A |
4223 |
0.15 |
| chr4_35305303_35305463 | 0.24 |
C9orf72 |
C9orf72, member of C9orf72-SMCR8 complex |
79208 |
0.08 |
| chr10_84381402_84381782 | 0.24 |
Nuak1 |
NUAK family, SNF1-like kinase, 1 |
10796 |
0.2 |
| chr10_88507811_88508139 | 0.24 |
Chpt1 |
choline phosphotransferase 1 |
3902 |
0.18 |
| chr13_112237981_112238162 | 0.24 |
Gm37427 |
predicted gene, 37427 |
18878 |
0.17 |
| chr9_52003605_52003767 | 0.24 |
Gm6981 |
predicted pseudogene 6981 |
405 |
0.84 |
| chr7_110881871_110882036 | 0.24 |
Lyve1 |
lymphatic vessel endothelial hyaluronan receptor 1 |
18714 |
0.14 |
| chr6_17578467_17578785 | 0.24 |
Met |
met proto-oncogene |
31653 |
0.17 |
| chr17_77596575_77596938 | 0.24 |
Gm17538 |
predicted gene, 17538 |
77944 |
0.11 |
| chr11_70730299_70730756 | 0.24 |
Mir6925 |
microRNA 6925 |
24537 |
0.06 |
| chr4_104779771_104779980 | 0.24 |
C8b |
complement component 8, beta polypeptide |
13480 |
0.25 |
| chr6_5281010_5281176 | 0.23 |
Pon2 |
paraoxonase 2 |
759 |
0.54 |
| chr5_121220767_121220929 | 0.23 |
Hectd4 |
HECT domain E3 ubiquitin protein ligase 4 |
629 |
0.63 |
| chr9_69306275_69306692 | 0.23 |
Rora |
RAR-related orphan receptor alpha |
16801 |
0.21 |
| chr11_100837297_100837448 | 0.23 |
Stat5b |
signal transducer and activator of transcription 5B |
13166 |
0.12 |
| chr2_75723637_75723808 | 0.23 |
Gm13656 |
predicted gene 13656 |
6450 |
0.14 |
| chr6_128521281_128521434 | 0.23 |
Pzp |
PZP, alpha-2-macroglobulin like |
5346 |
0.1 |
| chr3_135456700_135456878 | 0.23 |
Ube2d3 |
ubiquitin-conjugating enzyme E2D 3 |
1859 |
0.21 |
| chr9_64234619_64234783 | 0.23 |
Uchl4 |
ubiquitin carboxyl-terminal esterase L4 |
500 |
0.63 |
| chr6_141635343_141635562 | 0.23 |
Slco1b2 |
solute carrier organic anion transporter family, member 1b2 |
994 |
0.67 |
| chr3_135675157_135675328 | 0.23 |
Nfkb1 |
nuclear factor of kappa light polypeptide gene enhancer in B cells 1, p105 |
5903 |
0.21 |
| chr2_10099902_10100053 | 0.23 |
Kin |
Kin17 DNA and RNA binding protein |
9680 |
0.11 |
| chr9_110661245_110661404 | 0.23 |
Ccdc12 |
coiled-coil domain containing 12 |
4712 |
0.11 |
| chr2_63184621_63184772 | 0.23 |
Kcnh7 |
potassium voltage-gated channel, subfamily H (eag-related), member 7 |
409 |
0.92 |
| chr5_90893727_90893898 | 0.23 |
Gm22816 |
predicted gene, 22816 |
432 |
0.73 |
| chr16_30064275_30064431 | 0.23 |
Hes1 |
hes family bHLH transcription factor 1 |
31 |
0.97 |
| chr3_96065817_96065981 | 0.23 |
Vps45 |
vacuolar protein sorting 45 |
7433 |
0.11 |
| chr2_155538152_155538306 | 0.23 |
Mipep-ps |
mitochondrial intermediate peptidase, pseudogene |
4418 |
0.1 |
| chr10_20421094_20421290 | 0.23 |
Pde7b |
phosphodiesterase 7B |
25699 |
0.16 |
| chr8_108706681_108707155 | 0.23 |
Zfhx3 |
zinc finger homeobox 3 |
3818 |
0.29 |
| chr14_122107635_122107897 | 0.23 |
Tm9sf2 |
transmembrane 9 superfamily member 2 |
498 |
0.55 |
| chr13_3893671_3893882 | 0.22 |
Net1 |
neuroepithelial cell transforming gene 1 |
198 |
0.91 |
| chr13_56571544_56571919 | 0.22 |
Lect2 |
leukocyte cell-derived chemotaxin 2 |
23229 |
0.14 |
| chr11_46583348_46583524 | 0.22 |
BC053393 |
cDNA sequence BC053393 |
11900 |
0.12 |
| chr1_24693656_24693810 | 0.22 |
Lmbrd1 |
LMBR1 domain containing 1 |
14807 |
0.17 |
| chr13_43954875_43955028 | 0.22 |
Gm2233 |
predicted gene 2233 |
6072 |
0.23 |
| chr6_100896186_100896354 | 0.22 |
4930595O18Rik |
RIKEN cDNA 4930595O18 gene |
26886 |
0.16 |
| chr11_75459071_75459222 | 0.22 |
Mir22hg |
Mir22 host gene (non-protein coding) |
2393 |
0.12 |
| chr8_127447720_127447900 | 0.22 |
Pard3 |
par-3 family cell polarity regulator |
64 |
0.99 |
| chr1_105178921_105179094 | 0.22 |
Gm29012 |
predicted gene 29012 |
62553 |
0.12 |
| chr14_8260306_8260457 | 0.22 |
Acox2 |
acyl-Coenzyme A oxidase 2, branched chain |
1028 |
0.55 |
| chr8_38723154_38723538 | 0.22 |
AC163680.1 |
NADH-ubiquinone oxidoreductase pseudogene |
15206 |
0.24 |
| chr3_51916312_51916511 | 0.22 |
Gm10728 |
predicted gene 10728 |
7595 |
0.13 |
| chr2_104121396_104121547 | 0.22 |
A930018P22Rik |
RIKEN cDNA A930018P22 gene |
1298 |
0.35 |
| chr19_55266780_55267136 | 0.22 |
Acsl5 |
acyl-CoA synthetase long-chain family member 5 |
5912 |
0.19 |
| chr10_107321805_107321969 | 0.22 |
Lin7a |
lin-7 homolog A (C. elegans) |
49493 |
0.15 |
| chr7_19025229_19025442 | 0.22 |
Sympk |
symplekin |
909 |
0.3 |
| chr11_95778633_95778791 | 0.22 |
Polr2k-ps |
polymerase (RNA) II (DNA directed) polypeptide K, pseudogene |
17221 |
0.11 |
| chr13_52976319_52976492 | 0.22 |
Nfil3 |
nuclear factor, interleukin 3, regulated |
4668 |
0.2 |
| chr2_69061493_69062160 | 0.22 |
Gm38377 |
predicted gene, 38377 |
43730 |
0.11 |
| chr8_106072684_106072835 | 0.22 |
Nfatc3 |
nuclear factor of activated T cells, cytoplasmic, calcineurin dependent 3 |
13126 |
0.11 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0033031 | positive regulation of neutrophil apoptotic process(GO:0033031) |
| 0.1 | 0.3 | GO:1903525 | regulation of membrane tubulation(GO:1903525) |
| 0.1 | 0.3 | GO:0003431 | growth plate cartilage chondrocyte development(GO:0003431) |
| 0.1 | 0.3 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.1 | 0.2 | GO:0035905 | ascending aorta development(GO:0035905) ascending aorta morphogenesis(GO:0035910) |
| 0.1 | 0.2 | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 0.1 | 0.3 | GO:0009115 | xanthine catabolic process(GO:0009115) |
| 0.1 | 0.5 | GO:0006991 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.1 | 0.1 | GO:0090259 | regulation of retinal ganglion cell axon guidance(GO:0090259) |
| 0.1 | 0.2 | GO:0046439 | cysteine catabolic process(GO:0009093) L-cysteine catabolic process(GO:0019448) L-cysteine metabolic process(GO:0046439) |
| 0.0 | 0.1 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.0 | 0.2 | GO:0035983 | response to trichostatin A(GO:0035983) cellular response to trichostatin A(GO:0035984) |
| 0.0 | 0.2 | GO:0021943 | formation of radial glial scaffolds(GO:0021943) |
| 0.0 | 0.2 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.0 | 0.1 | GO:0097503 | sialylation(GO:0097503) |
| 0.0 | 0.2 | GO:1903599 | positive regulation of mitophagy(GO:1903599) |
| 0.0 | 0.2 | GO:1902459 | positive regulation of stem cell population maintenance(GO:1902459) |
| 0.0 | 0.1 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.0 | 0.1 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.0 | 0.2 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.0 | 0.1 | GO:1901492 | positive regulation of lymphangiogenesis(GO:1901492) |
| 0.0 | 0.2 | GO:0098598 | vocal learning(GO:0042297) imitative learning(GO:0098596) learned vocalization behavior or vocal learning(GO:0098598) |
| 0.0 | 0.1 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.0 | 0.1 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.0 | 0.1 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.0 | 0.1 | GO:0002017 | regulation of blood volume by renal aldosterone(GO:0002017) |
| 0.0 | 0.2 | GO:0060613 | fat pad development(GO:0060613) |
| 0.0 | 0.1 | GO:1902744 | negative regulation of lamellipodium organization(GO:1902744) |
| 0.0 | 0.1 | GO:0003383 | apical constriction(GO:0003383) |
| 0.0 | 0.1 | GO:0006069 | ethanol metabolic process(GO:0006067) ethanol oxidation(GO:0006069) |
| 0.0 | 0.1 | GO:1902956 | regulation of mitochondrial electron transport, NADH to ubiquinone(GO:1902956) |
| 0.0 | 0.1 | GO:0061470 | T follicular helper cell differentiation(GO:0061470) |
| 0.0 | 0.1 | GO:0097477 | lateral motor column neuron migration(GO:0097477) |
| 0.0 | 0.2 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.0 | 0.1 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.2 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.0 | 0.2 | GO:2001214 | positive regulation of vasculogenesis(GO:2001214) |
| 0.0 | 0.2 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.1 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.0 | 0.1 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.0 | 0.2 | GO:0035234 | ectopic germ cell programmed cell death(GO:0035234) |
| 0.0 | 0.2 | GO:0021873 | forebrain neuroblast division(GO:0021873) |
| 0.0 | 0.1 | GO:0009092 | homoserine metabolic process(GO:0009092) transsulfuration(GO:0019346) |
| 0.0 | 0.2 | GO:0019852 | L-ascorbic acid metabolic process(GO:0019852) |
| 0.0 | 0.1 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.0 | 0.1 | GO:0045054 | constitutive secretory pathway(GO:0045054) |
| 0.0 | 0.1 | GO:2001280 | positive regulation of prostaglandin biosynthetic process(GO:0031394) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.0 | 0.1 | GO:1900454 | positive regulation of long term synaptic depression(GO:1900454) |
| 0.0 | 0.1 | GO:0046618 | drug export(GO:0046618) |
| 0.0 | 0.1 | GO:0016479 | negative regulation of transcription from RNA polymerase I promoter(GO:0016479) |
| 0.0 | 0.1 | GO:0035284 | central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.0 | 0.1 | GO:1904424 | regulation of GTP binding(GO:1904424) |
| 0.0 | 0.2 | GO:0043518 | negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) |
| 0.0 | 0.1 | GO:0034214 | protein hexamerization(GO:0034214) |
| 0.0 | 0.0 | GO:0070814 | hydrogen sulfide biosynthetic process(GO:0070814) |
| 0.0 | 0.1 | GO:0006285 | base-excision repair, AP site formation(GO:0006285) |
| 0.0 | 0.2 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 0.0 | 0.1 | GO:0051562 | negative regulation of mitochondrial calcium ion concentration(GO:0051562) |
| 0.0 | 0.0 | GO:2000619 | negative regulation of histone H4-K16 acetylation(GO:2000619) |
| 0.0 | 0.1 | GO:1904017 | response to Thyroglobulin triiodothyronine(GO:1904016) cellular response to Thyroglobulin triiodothyronine(GO:1904017) |
| 0.0 | 0.1 | GO:0010956 | negative regulation of calcidiol 1-monooxygenase activity(GO:0010956) negative regulation of vitamin D biosynthetic process(GO:0010957) |
| 0.0 | 0.1 | GO:0046087 | cytidine catabolic process(GO:0006216) cytidine deamination(GO:0009972) cytidine metabolic process(GO:0046087) |
| 0.0 | 0.0 | GO:1901668 | regulation of superoxide dismutase activity(GO:1901668) |
| 0.0 | 0.0 | GO:0010520 | regulation of reciprocal meiotic recombination(GO:0010520) |
| 0.0 | 0.1 | GO:0032348 | negative regulation of aldosterone metabolic process(GO:0032345) negative regulation of aldosterone biosynthetic process(GO:0032348) negative regulation of cortisol biosynthetic process(GO:2000065) |
| 0.0 | 0.2 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 0.0 | 0.0 | GO:0090210 | regulation of establishment of blood-brain barrier(GO:0090210) |
| 0.0 | 0.2 | GO:0035745 | T-helper 2 cell cytokine production(GO:0035745) |
| 0.0 | 0.1 | GO:0001905 | activation of membrane attack complex(GO:0001905) regulation of activation of membrane attack complex(GO:0001969) |
| 0.0 | 0.0 | GO:0007354 | zygotic determination of anterior/posterior axis, embryo(GO:0007354) |
| 0.0 | 0.0 | GO:0030576 | Cajal body organization(GO:0030576) |
| 0.0 | 0.0 | GO:2000721 | positive regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000721) |
| 0.0 | 0.1 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.0 | 0.0 | GO:0044375 | regulation of peroxisome size(GO:0044375) |
| 0.0 | 0.1 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 0.0 | 0.0 | GO:0008228 | opsonization(GO:0008228) |
| 0.0 | 0.0 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.0 | 0.0 | GO:0018199 | peptidyl-glutamine modification(GO:0018199) |
| 0.0 | 0.1 | GO:1900864 | mitochondrial tRNA modification(GO:0070900) mitochondrial RNA modification(GO:1900864) |
| 0.0 | 0.1 | GO:0002913 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.0 | 0.0 | GO:0046098 | guanine metabolic process(GO:0046098) |
| 0.0 | 0.1 | GO:0015760 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.0 | 0.1 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.0 | 0.0 | GO:2000850 | negative regulation of steroid hormone secretion(GO:2000832) negative regulation of corticosteroid hormone secretion(GO:2000847) negative regulation of glucocorticoid secretion(GO:2000850) |
| 0.0 | 0.1 | GO:0070126 | mitochondrial translational termination(GO:0070126) |
| 0.0 | 0.0 | GO:0006551 | leucine metabolic process(GO:0006551) |
| 0.0 | 0.1 | GO:0090232 | positive regulation of spindle checkpoint(GO:0090232) |
| 0.0 | 0.0 | GO:0034443 | negative regulation of lipoprotein oxidation(GO:0034443) |
| 0.0 | 0.0 | GO:1901069 | guanosine-containing compound catabolic process(GO:1901069) |
| 0.0 | 0.0 | GO:0098597 | observational learning(GO:0098597) |
| 0.0 | 0.0 | GO:0071139 | resolution of recombination intermediates(GO:0071139) |
| 0.0 | 0.0 | GO:0060486 | Clara cell differentiation(GO:0060486) |
| 0.0 | 0.0 | GO:0045914 | negative regulation of catecholamine metabolic process(GO:0045914) negative regulation of dopamine metabolic process(GO:0045963) |
| 0.0 | 0.0 | GO:0046813 | receptor-mediated virion attachment to host cell(GO:0046813) |
| 0.0 | 0.0 | GO:0048696 | regulation of collateral sprouting in absence of injury(GO:0048696) |
| 0.0 | 0.0 | GO:0061502 | early endosome to recycling endosome transport(GO:0061502) |
| 0.0 | 0.0 | GO:0002331 | pre-B cell allelic exclusion(GO:0002331) |
| 0.0 | 0.0 | GO:1990169 | detoxification of copper ion(GO:0010273) stress response to copper ion(GO:1990169) |
| 0.0 | 0.2 | GO:0043516 | regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043516) |
| 0.0 | 0.1 | GO:1904970 | brush border assembly(GO:1904970) |
| 0.0 | 0.0 | GO:0070889 | platelet alpha granule organization(GO:0070889) |
| 0.0 | 0.0 | GO:2000173 | negative regulation of branching morphogenesis of a nerve(GO:2000173) |
| 0.0 | 0.0 | GO:0015842 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.0 | 0.0 | GO:0050713 | negative regulation of interleukin-1 beta secretion(GO:0050713) |
| 0.0 | 0.1 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.0 | 0.0 | GO:0046469 | platelet activating factor biosynthetic process(GO:0006663) platelet activating factor metabolic process(GO:0046469) |
| 0.0 | 0.0 | GO:0000350 | generation of catalytic spliceosome for second transesterification step(GO:0000350) |
| 0.0 | 0.0 | GO:1904217 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) regulation of serine C-palmitoyltransferase activity(GO:1904220) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.0 | 0.0 | GO:0002337 | B-1a B cell differentiation(GO:0002337) |
| 0.0 | 0.0 | GO:0061043 | regulation of vascular wound healing(GO:0061043) |
| 0.0 | 0.0 | GO:1902109 | negative regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902109) |
| 0.0 | 0.1 | GO:0031000 | response to caffeine(GO:0031000) |
| 0.0 | 0.1 | GO:0051798 | positive regulation of hair follicle development(GO:0051798) |
| 0.0 | 0.1 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.1 | GO:0036289 | peptidyl-serine autophosphorylation(GO:0036289) |
| 0.0 | 0.1 | GO:0010746 | regulation of plasma membrane long-chain fatty acid transport(GO:0010746) |
| 0.0 | 0.0 | GO:0070460 | thyroid-stimulating hormone secretion(GO:0070460) |
| 0.0 | 0.1 | GO:0010944 | negative regulation of transcription by competitive promoter binding(GO:0010944) |
| 0.0 | 0.1 | GO:0070120 | ciliary neurotrophic factor-mediated signaling pathway(GO:0070120) |
| 0.0 | 0.1 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.0 | GO:1902534 | single-organism membrane invagination(GO:1902534) |
| 0.0 | 0.1 | GO:0033601 | positive regulation of mammary gland epithelial cell proliferation(GO:0033601) |
| 0.0 | 0.0 | GO:0070673 | response to interleukin-18(GO:0070673) |
| 0.0 | 0.0 | GO:1902075 | cellular response to salt(GO:1902075) |
| 0.0 | 0.0 | GO:0050847 | progesterone receptor signaling pathway(GO:0050847) |
| 0.0 | 0.1 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.0 | 0.0 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.0 | 0.1 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.0 | 0.0 | GO:0021827 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) postnatal olfactory bulb interneuron migration(GO:0021827) |
| 0.0 | 0.1 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.0 | 0.2 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 0.0 | 0.1 | GO:0097648 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor complex(GO:0097648) |
| 0.0 | 0.2 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.0 | 0.2 | GO:1990851 | Wnt-Frizzled-LRP5/6 complex(GO:1990851) |
| 0.0 | 0.1 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 0.2 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.1 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.1 | GO:1990597 | AIP1-IRE1 complex(GO:1990597) |
| 0.0 | 0.1 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.0 | 0.1 | GO:0005683 | U7 snRNP(GO:0005683) |
| 0.0 | 0.3 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.1 | GO:1990761 | growth cone lamellipodium(GO:1990761) |
| 0.0 | 0.1 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 0.1 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 0.0 | 0.2 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.2 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.1 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.1 | GO:0045261 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.0 | 0.0 | GO:0034679 | integrin alpha9-beta1 complex(GO:0034679) |
| 0.0 | 0.0 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.0 | 0.0 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.0 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.0 | 0.0 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.0 | 0.0 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.0 | 0.0 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.0 | 0.1 | GO:0044214 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.0 | 0.1 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 0.0 | GO:0002142 | stereocilia ankle link complex(GO:0002142) |
| 0.0 | 0.0 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.0 | 0.2 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0016882 | cyclo-ligase activity(GO:0016882) |
| 0.1 | 0.3 | GO:0102007 | lactonohydrolase activity(GO:0046573) acyl-L-homoserine-lactone lactonohydrolase activity(GO:0102007) |
| 0.1 | 0.3 | GO:0004854 | xanthine dehydrogenase activity(GO:0004854) oxidoreductase activity, acting on CH or CH2 groups, NAD or NADP as acceptor(GO:0016726) |
| 0.1 | 0.2 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.0 | 0.3 | GO:0031749 | D2 dopamine receptor binding(GO:0031749) |
| 0.0 | 0.2 | GO:0070878 | primary miRNA binding(GO:0070878) |
| 0.0 | 0.1 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 0.1 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 0.2 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.0 | 0.2 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.0 | 0.2 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.1 | GO:0015038 | glutathione disulfide oxidoreductase activity(GO:0015038) |
| 0.0 | 0.1 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 0.0 | 0.1 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 0.1 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.1 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.1 | GO:0070137 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.0 | 0.1 | GO:0004792 | thiosulfate sulfurtransferase activity(GO:0004792) |
| 0.0 | 0.1 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.1 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.0 | 0.1 | GO:0030375 | thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 0.1 | GO:0015526 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.0 | 0.2 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.2 | GO:0035197 | siRNA binding(GO:0035197) |
| 0.0 | 0.3 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 0.1 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.0 | 0.1 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.0 | 0.1 | GO:0015252 | hydrogen ion channel activity(GO:0015252) |
| 0.0 | 0.1 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.0 | 0.4 | GO:0016502 | purinergic nucleotide receptor activity(GO:0001614) nucleotide receptor activity(GO:0016502) |
| 0.0 | 0.1 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.1 | GO:0047023 | androsterone dehydrogenase activity(GO:0047023) |
| 0.0 | 0.1 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.0 | 0.1 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.0 | GO:0071209 | U7 snRNA binding(GO:0071209) |
| 0.0 | 0.1 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.0 | 0.1 | GO:0043426 | MRF binding(GO:0043426) |
| 0.0 | 0.1 | GO:0019534 | toxin transporter activity(GO:0019534) |
| 0.0 | 0.1 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.0 | 0.0 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.0 | 0.0 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.0 | 0.1 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.0 | 0.1 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.0 | 0.0 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.1 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.0 | 0.0 | GO:0070326 | very-low-density lipoprotein particle receptor binding(GO:0070326) |
| 0.0 | 0.0 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.0 | 0.0 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.0 | 0.0 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.0 | 0.0 | GO:0001226 | RNA polymerase II transcription corepressor binding(GO:0001226) |
| 0.0 | 0.2 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 0.1 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) |
| 0.0 | 0.1 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.0 | 0.0 | GO:0032139 | dinucleotide insertion or deletion binding(GO:0032139) |
| 0.0 | 0.1 | GO:0004528 | phosphodiesterase I activity(GO:0004528) |
| 0.0 | 0.3 | GO:0015238 | drug transmembrane transporter activity(GO:0015238) |
| 0.0 | 0.2 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.0 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.0 | 0.1 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.2 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.0 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.0 | 0.0 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.0 | 0.0 | GO:0090482 | vitamin transmembrane transporter activity(GO:0090482) |
| 0.0 | 0.1 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.1 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.1 | GO:0051880 | G-quadruplex DNA binding(GO:0051880) |
| 0.0 | 0.1 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 0.0 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.0 | 0.1 | GO:0038085 | vascular endothelial growth factor binding(GO:0038085) |
| 0.0 | 0.2 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.0 | 0.1 | GO:0016443 | bidentate ribonuclease III activity(GO:0016443) |
| 0.0 | 0.1 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.0 | 0.0 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.0 | 0.2 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.0 | 0.1 | GO:0050649 | testosterone 6-beta-hydroxylase activity(GO:0050649) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.0 | 0.3 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 0.0 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 0.0 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.1 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 0.3 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.5 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.3 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.2 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.1 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.2 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.0 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.0 | 0.1 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.2 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.0 | 0.0 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 0.0 | REACTOME CLASS A1 RHODOPSIN LIKE RECEPTORS | Genes involved in Class A/1 (Rhodopsin-like receptors) |
| 0.0 | 0.1 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.2 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.1 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.0 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.1 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.1 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |