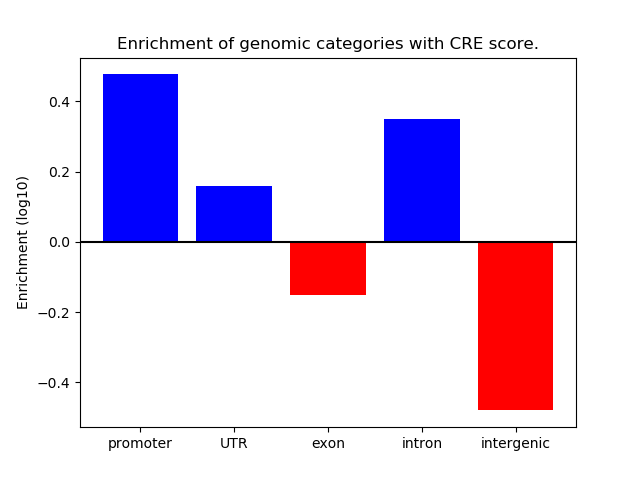
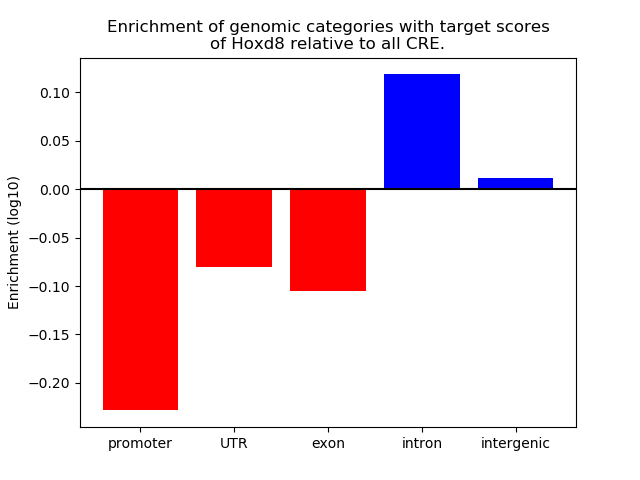
Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
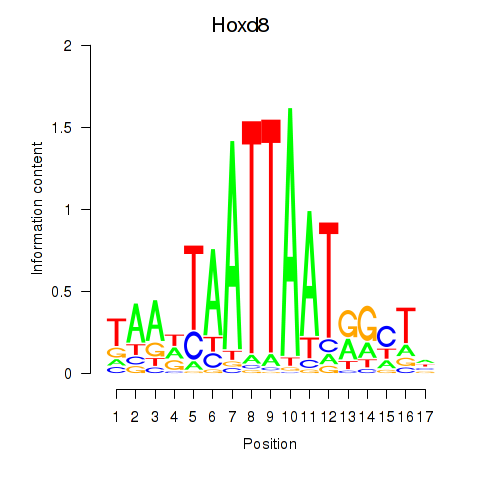
Results for Hoxd8
Z-value: 1.11
Transcription factors associated with Hoxd8
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hoxd8
|
ENSMUSG00000027102.4 | homeobox D8 |
Activity of the Hoxd8 motif across conditions
Conditions sorted by the z-value of the Hoxd8 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
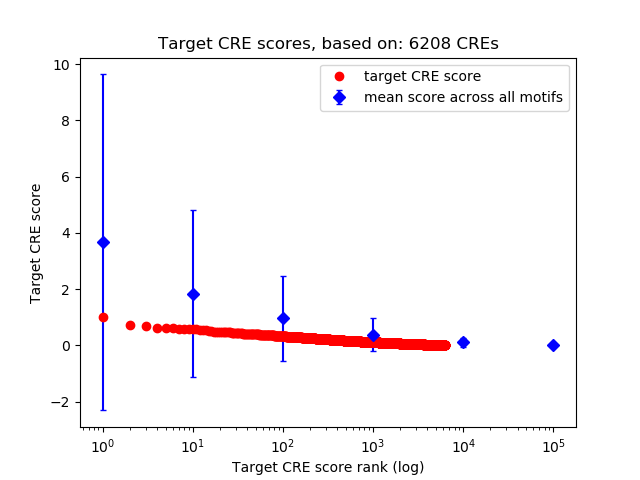
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr13_16574847_16575001 | 1.02 |
Gm48497 |
predicted gene, 48497 |
41103 |
0.17 |
| chr4_106354190_106354378 | 0.72 |
Usp24 |
ubiquitin specific peptidase 24 |
38050 |
0.13 |
| chr19_31884467_31884670 | 0.69 |
A1cf |
APOBEC1 complementation factor |
15787 |
0.21 |
| chr16_42999869_43000020 | 0.62 |
Ndufs5-ps |
NADH:ubiquinone oxidoreductase core subunit S5, pseudogene |
44313 |
0.16 |
| chr3_14787462_14787685 | 0.61 |
Car1 |
carbonic anhydrase 1 |
9114 |
0.18 |
| chr2_154875946_154876097 | 0.61 |
Eif2s2 |
eukaryotic translation initiation factor 2, subunit 2 (beta) |
16761 |
0.19 |
| chr6_29571610_29572022 | 0.59 |
Tnpo3 |
transportin 3 |
297 |
0.87 |
| chr16_30027841_30027992 | 0.59 |
9030404E10Rik |
RIKEN cDNA 9030404E10 gene |
11563 |
0.15 |
| chr8_93183570_93183871 | 0.59 |
Gm45909 |
predicted gene 45909 |
7638 |
0.14 |
| chr12_78268546_78268712 | 0.58 |
Gm48225 |
predicted gene, 48225 |
7336 |
0.17 |
| chr4_123990670_123991070 | 0.58 |
Gm12902 |
predicted gene 12902 |
64636 |
0.08 |
| chr3_136048131_136048315 | 0.55 |
Gm5281 |
predicted gene 5281 |
3465 |
0.25 |
| chr7_132941355_132941506 | 0.54 |
Zranb1 |
zinc finger, RAN-binding domain containing 1 |
6738 |
0.15 |
| chr6_28587807_28587958 | 0.53 |
Gm37978 |
predicted gene, 37978 |
19810 |
0.16 |
| chr9_122826708_122826859 | 0.50 |
Gm35549 |
predicted gene, 35549 |
17103 |
0.1 |
| chr8_56281836_56281987 | 0.50 |
Hpgd |
hydroxyprostaglandin dehydrogenase 15 (NAD) |
12674 |
0.23 |
| chr12_83905337_83905488 | 0.49 |
Numb |
NUMB endocytic adaptor protein |
16321 |
0.11 |
| chr11_100485778_100486208 | 0.49 |
Acly |
ATP citrate lyase |
6482 |
0.09 |
| chr9_109565625_109565786 | 0.48 |
Fbxw15 |
F-box and WD-40 domain protein 15 |
2543 |
0.17 |
| chr10_87040962_87041138 | 0.48 |
1700113H08Rik |
RIKEN cDNA 1700113H08 gene |
16995 |
0.16 |
| chr9_48723477_48723685 | 0.47 |
Zbtb16 |
zinc finger and BTB domain containing 16 |
112364 |
0.06 |
| chr5_40811186_40811352 | 0.47 |
Gm23022 |
predicted gene, 23022 |
405340 |
0.01 |
| chr3_66982581_66983014 | 0.46 |
Shox2 |
short stature homeobox 2 |
1026 |
0.42 |
| chr12_12940858_12941058 | 0.46 |
Mycn |
v-myc avian myelocytomatosis viral related oncogene, neuroblastoma derived |
342 |
0.83 |
| chr19_48883437_48883771 | 0.46 |
Gm50436 |
predicted gene, 50436 |
72468 |
0.11 |
| chr2_78656286_78656827 | 0.46 |
Gm14463 |
predicted gene 14463 |
873 |
0.71 |
| chr1_162867938_162868165 | 0.45 |
Fmo1 |
flavin containing monooxygenase 1 |
1441 |
0.39 |
| chr9_7858246_7858397 | 0.45 |
Birc3 |
baculoviral IAP repeat-containing 3 |
2005 |
0.3 |
| chr2_38823684_38823849 | 0.44 |
Nr6a1os |
nuclear receptor subfamily 6, group A, member 1, opposite strand |
425 |
0.74 |
| chr8_90953330_90953683 | 0.44 |
Chd9 |
chromodomain helicase DNA binding protein 9 |
1452 |
0.39 |
| chr2_112609858_112610011 | 0.44 |
Gm44374 |
predicted gene, 44374 |
4096 |
0.23 |
| chr4_136402761_136402948 | 0.43 |
Htr1d |
5-hydroxytryptamine (serotonin) receptor 1D |
20670 |
0.14 |
| chr3_138223407_138223558 | 0.43 |
Adh7 |
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
2011 |
0.23 |
| chr3_133544844_133545488 | 0.43 |
Tet2 |
tet methylcytosine dioxygenase 2 |
27 |
0.53 |
| chr3_34076650_34076843 | 0.42 |
Dnajc19 |
DnaJ heat shock protein family (Hsp40) member C19 |
3434 |
0.16 |
| chr6_108666051_108666462 | 0.42 |
Bhlhe40 |
basic helix-loop-helix family, member e40 |
3210 |
0.21 |
| chr17_80022007_80022158 | 0.41 |
Gm22215 |
predicted gene, 22215 |
11432 |
0.14 |
| chr18_45582508_45582659 | 0.41 |
Kcnn2 |
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 2 |
7963 |
0.24 |
| chr1_100298746_100298897 | 0.41 |
Gm29667 |
predicted gene 29667 |
16030 |
0.18 |
| chr12_73861884_73862127 | 0.41 |
Gm15283 |
predicted gene 15283 |
7607 |
0.18 |
| chr4_6885681_6885832 | 0.40 |
Tox |
thymocyte selection-associated high mobility group box |
104727 |
0.07 |
| chr17_64607771_64607970 | 0.40 |
Man2a1 |
mannosidase 2, alpha 1 |
7134 |
0.26 |
| chr6_37510078_37510463 | 0.40 |
Akr1d1 |
aldo-keto reductase family 1, member D1 |
19903 |
0.21 |
| chr3_8963133_8963313 | 0.40 |
Tpd52 |
tumor protein D52 |
823 |
0.64 |
| chr16_90199492_90199754 | 0.40 |
Gm49704 |
predicted gene, 49704 |
1475 |
0.35 |
| chr3_97649987_97650151 | 0.40 |
Prkab2 |
protein kinase, AMP-activated, beta 2 non-catalytic subunit |
8124 |
0.13 |
| chr4_97102666_97103047 | 0.40 |
Gm27521 |
predicted gene, 27521 |
185836 |
0.03 |
| chr14_75243267_75243429 | 0.39 |
Cpb2 |
carboxypeptidase B2 (plasma) |
1061 |
0.44 |
| chr14_32179471_32179639 | 0.39 |
Ncoa4 |
nuclear receptor coactivator 4 |
2823 |
0.15 |
| chr16_78490552_78490715 | 0.39 |
Gm49604 |
predicted gene, 49604 |
953 |
0.54 |
| chr4_63057046_63057197 | 0.39 |
Zfp618 |
zinc finger protein 618 |
63279 |
0.1 |
| chr9_15301981_15302132 | 0.39 |
4931406C07Rik |
RIKEN cDNA 4931406C07 gene |
500 |
0.46 |
| chr2_132859056_132859242 | 0.39 |
Crls1 |
cardiolipin synthase 1 |
11411 |
0.13 |
| chr12_75531904_75532183 | 0.39 |
Ppp2r5e |
protein phosphatase 2, regulatory subunit B', epsilon |
17631 |
0.18 |
| chr19_31886956_31887132 | 0.38 |
A1cf |
APOBEC1 complementation factor |
18263 |
0.2 |
| chr9_106264996_106265377 | 0.38 |
Poc1a |
POC1 centriolar protein A |
15875 |
0.1 |
| chr8_34164335_34164526 | 0.38 |
Mir6395 |
microRNA 6395 |
2282 |
0.18 |
| chr7_110538178_110538390 | 0.37 |
Gm10087 |
predicted gene 10087 |
11492 |
0.18 |
| chr11_115167087_115167259 | 0.37 |
Slc9a3r1 |
solute carrier family 9 (sodium/hydrogen exchanger), member 3 regulator 1 |
3832 |
0.13 |
| chr6_141994065_141994238 | 0.37 |
Gm6614 |
predicted gene 6614 |
14594 |
0.21 |
| chr14_30530847_30531018 | 0.37 |
Dcp1a |
decapping mRNA 1A |
7225 |
0.15 |
| chr5_147545326_147545477 | 0.37 |
Pan3 |
PAN3 poly(A) specific ribonuclease subunit |
6124 |
0.21 |
| chr7_100610040_100610203 | 0.37 |
Mrpl48 |
mitochondrial ribosomal protein L48 |
1820 |
0.19 |
| chr2_121507198_121507349 | 0.37 |
Wdr76 |
WD repeat domain 76 |
490 |
0.46 |
| chr10_84914599_84914750 | 0.37 |
Ric8b |
RIC8 guanine nucleotide exchange factor B |
2942 |
0.33 |
| chr1_127787130_127787281 | 0.36 |
Ccnt2 |
cyclin T2 |
9366 |
0.16 |
| chr2_115450732_115450919 | 0.36 |
3110099E03Rik |
RIKEN cDNA 3110099E03 gene |
61376 |
0.12 |
| chr14_55468841_55469329 | 0.36 |
Dhrs4 |
dehydrogenase/reductase (SDR family) member 4 |
9673 |
0.1 |
| chr14_65740190_65740357 | 0.36 |
Scara5 |
scavenger receptor class A, member 5 |
63796 |
0.1 |
| chr19_55111923_55112097 | 0.36 |
Gpam |
glycerol-3-phosphate acyltransferase, mitochondrial |
8919 |
0.2 |
| chr16_42998828_42999221 | 0.36 |
Ndufs5-ps |
NADH:ubiquinone oxidoreductase core subunit S5, pseudogene |
43393 |
0.16 |
| chr6_8862762_8862913 | 0.36 |
Ica1 |
islet cell autoantigen 1 |
84349 |
0.1 |
| chr15_59231191_59231350 | 0.36 |
Gm7691 |
predicted gene 7691 |
7219 |
0.19 |
| chr19_60570173_60570324 | 0.36 |
Gm25238 |
predicted gene, 25238 |
10281 |
0.18 |
| chr15_39146655_39146819 | 0.35 |
n-R5s39 |
nuclear encoded rRNA 5S 39 |
250 |
0.87 |
| chr5_101758048_101758202 | 0.35 |
Cds1 |
CDP-diacylglycerol synthase 1 |
7005 |
0.17 |
| chr2_19519895_19520058 | 0.35 |
Gm13345 |
predicted gene 13345 |
10659 |
0.16 |
| chr15_3721208_3721477 | 0.35 |
Gm4823 |
predicted gene 4823 |
25533 |
0.23 |
| chr8_36246396_36246684 | 0.35 |
Lonrf1 |
LON peptidase N-terminal domain and ring finger 1 |
2976 |
0.27 |
| chr16_30941233_30941532 | 0.35 |
Gm46565 |
predicted gene, 46565 |
5753 |
0.18 |
| chr13_4438641_4438805 | 0.35 |
Akr1c6 |
aldo-keto reductase family 1, member C6 |
2577 |
0.26 |
| chr4_97810121_97810272 | 0.34 |
E130114P18Rik |
RIKEN cDNA E130114P18 gene |
32118 |
0.17 |
| chr14_8425861_8426012 | 0.34 |
Gm8416 |
predicted gene 8416 |
17019 |
0.2 |
| chr3_18210346_18210497 | 0.34 |
Gm23686 |
predicted gene, 23686 |
32796 |
0.17 |
| chr15_44689672_44690007 | 0.34 |
Sybu |
syntabulin (syntaxin-interacting) |
57949 |
0.11 |
| chr13_92050387_92050727 | 0.34 |
Rasgrf2 |
RAS protein-specific guanine nucleotide-releasing factor 2 |
19653 |
0.26 |
| chr8_93253165_93253392 | 0.34 |
Gm45727 |
predicted gene 45727 |
2103 |
0.24 |
| chr10_87896267_87896418 | 0.33 |
Igf1os |
insulin-like growth factor 1, opposite strand |
32961 |
0.15 |
| chr6_88046142_88046293 | 0.33 |
Rab7 |
RAB7, member RAS oncogene family |
947 |
0.41 |
| chr6_26491486_26491715 | 0.33 |
Gm24691 |
predicted gene, 24691 |
31170 |
0.26 |
| chr8_18713888_18714465 | 0.33 |
Angpt2 |
angiopoietin 2 |
14629 |
0.17 |
| chr11_28702228_28702389 | 0.33 |
2810471M01Rik |
RIKEN cDNA 2810471M01 gene |
20744 |
0.15 |
| chr7_84113279_84113524 | 0.33 |
Cemip |
cell migration inducing protein, hyaluronan binding |
26899 |
0.14 |
| chr5_124228161_124228330 | 0.33 |
Pitpnm2os1 |
phosphatidylinositol transfer protein, membrane-associated 2, opposite strand 1 |
1480 |
0.26 |
| chr19_26058038_26058213 | 0.33 |
1700048O14Rik |
RIKEN cDNA 1700048O14 gene |
11658 |
0.29 |
| chr2_103790393_103790544 | 0.32 |
Caprin1 |
cell cycle associated protein 1 |
6076 |
0.11 |
| chr3_131485378_131485553 | 0.32 |
Sgms2 |
sphingomyelin synthase 2 |
5014 |
0.28 |
| chr5_86942001_86942153 | 0.32 |
Gm24121 |
predicted gene, 24121 |
13069 |
0.09 |
| chr2_36159195_36159603 | 0.32 |
Mrrf |
mitochondrial ribosome recycling factor |
12296 |
0.12 |
| chr10_111367147_111367312 | 0.32 |
Gm40761 |
predicted gene, 40761 |
30105 |
0.16 |
| chr7_70356055_70356242 | 0.32 |
Nr2f2 |
nuclear receptor subfamily 2, group F, member 2 |
911 |
0.45 |
| chr5_123865713_123865874 | 0.32 |
Hcar2 |
hydroxycarboxylic acid receptor 2 |
294 |
0.86 |
| chr11_38683198_38683743 | 0.32 |
Gm23520 |
predicted gene, 23520 |
114807 |
0.07 |
| chr13_100152373_100152603 | 0.32 |
Gm18413 |
predicted gene, 18413 |
2017 |
0.25 |
| chr13_4512002_4512170 | 0.32 |
Akr1c20 |
aldo-keto reductase family 1, member C20 |
11227 |
0.18 |
| chr15_50017534_50017718 | 0.31 |
Gm49186 |
predicted gene, 49186 |
104244 |
0.08 |
| chr5_40908767_40908928 | 0.31 |
4930519E07Rik |
RIKEN cDNA 4930519E07 gene |
372389 |
0.01 |
| chr6_100204562_100204726 | 0.31 |
Rybp |
RING1 and YY1 binding protein |
28487 |
0.15 |
| chr10_4605236_4605428 | 0.31 |
Esr1 |
estrogen receptor 1 (alpha) |
6261 |
0.23 |
| chr3_18167606_18167757 | 0.31 |
Gm23686 |
predicted gene, 23686 |
9944 |
0.24 |
| chr19_45902358_45902683 | 0.31 |
Gm6813 |
predicted gene 6813 |
24394 |
0.14 |
| chrX_140545006_140545195 | 0.31 |
Tsc22d3 |
TSC22 domain family, member 3 |
1913 |
0.35 |
| chr2_34049474_34049625 | 0.31 |
C230014O12Rik |
RIKEN cDNA C230014O12 gene |
58180 |
0.11 |
| chr12_54439488_54439663 | 0.31 |
Gm7557 |
predicted gene 7557 |
9177 |
0.15 |
| chr18_46655480_46655793 | 0.31 |
Gm3734 |
predicted gene 3734 |
24829 |
0.13 |
| chr11_4832480_4832698 | 0.31 |
Nf2 |
neurofibromin 2 |
544 |
0.74 |
| chr1_62469805_62469956 | 0.31 |
Pard3bos3 |
par-3 family cell polarity regulator beta, opposite strand 3 |
29381 |
0.21 |
| chr2_107628721_107628872 | 0.31 |
Gm9864 |
predicted gene 9864 |
28257 |
0.26 |
| chr5_30286285_30286785 | 0.31 |
Drc1 |
dynein regulatory complex subunit 1 |
5147 |
0.17 |
| chr1_62899396_62899571 | 0.30 |
Gm4208 |
predicted gene 4208 |
67113 |
0.09 |
| chr8_93277306_93277457 | 0.30 |
Ces1f |
carboxylesterase 1F |
2035 |
0.26 |
| chr18_38370754_38370932 | 0.30 |
Gm4949 |
predicted gene 4949 |
10305 |
0.12 |
| chr9_46479855_46480133 | 0.30 |
Gm47144 |
predicted gene, 47144 |
25079 |
0.15 |
| chr16_25221904_25222082 | 0.30 |
Tprg |
transformation related protein 63 regulated |
64824 |
0.14 |
| chr10_61444315_61444902 | 0.30 |
Gm48086 |
predicted gene, 48086 |
6109 |
0.11 |
| chr1_67182326_67182537 | 0.30 |
Cps1 |
carbamoyl-phosphate synthetase 1 |
59405 |
0.11 |
| chr3_149234408_149234559 | 0.30 |
Gm10287 |
predicted gene 10287 |
8738 |
0.21 |
| chr17_10187170_10187336 | 0.30 |
Qk |
quaking |
26321 |
0.25 |
| chr1_51751367_51751637 | 0.30 |
Gm28055 |
predicted gene 28055 |
4559 |
0.23 |
| chr4_109104769_109104920 | 0.30 |
Osbpl9 |
oxysterol binding protein-like 9 |
3187 |
0.27 |
| chr1_118448989_118449484 | 0.29 |
Gm26080 |
predicted gene, 26080 |
4249 |
0.15 |
| chr6_124880400_124880682 | 0.29 |
Cd4 |
CD4 antigen |
7658 |
0.08 |
| chr1_88134414_88134588 | 0.29 |
Ugt1a6a |
UDP glucuronosyltransferase 1 family, polypeptide A6A |
308 |
0.71 |
| chr2_52470397_52470548 | 0.29 |
A430018G15Rik |
RIKEN cDNA A430018G15 gene |
45460 |
0.12 |
| chr13_9306293_9306452 | 0.29 |
Gm48887 |
predicted gene, 48887 |
15055 |
0.11 |
| chr16_65665507_65665824 | 0.29 |
Gm49633 |
predicted gene, 49633 |
52246 |
0.14 |
| chr1_67195986_67196354 | 0.29 |
Gm15668 |
predicted gene 15668 |
53030 |
0.13 |
| chr16_56714341_56714503 | 0.29 |
Tfg |
Trk-fused gene |
1446 |
0.47 |
| chr10_111149650_111150090 | 0.29 |
Gm8873 |
predicted gene 8873 |
6346 |
0.14 |
| chr2_32606655_32606830 | 0.29 |
St6galnac6 |
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 6 |
219 |
0.84 |
| chr1_66816824_66817235 | 0.29 |
Kansl1l |
KAT8 regulatory NSL complex subunit 1-like |
517 |
0.58 |
| chr9_84497400_84497569 | 0.29 |
Gm8226 |
predicted gene 8226 |
73500 |
0.1 |
| chr15_54596223_54596485 | 0.29 |
Mal2 |
mal, T cell differentiation protein 2 |
25162 |
0.22 |
| chr19_32603829_32604061 | 0.29 |
Papss2 |
3'-phosphoadenosine 5'-phosphosulfate synthase 2 |
8155 |
0.23 |
| chr15_59316490_59316736 | 0.29 |
Sqle |
squalene epoxidase |
1506 |
0.35 |
| chr2_88007915_88008090 | 0.29 |
Olfr1160 |
olfactory receptor 1160 |
266 |
0.8 |
| chr16_84807428_84807749 | 0.29 |
Jam2 |
junction adhesion molecule 2 |
1819 |
0.26 |
| chr13_117745874_117746186 | 0.29 |
4933413L06Rik |
RIKEN cDNA 4933413L06 gene |
26019 |
0.26 |
| chr17_32934878_32935069 | 0.29 |
Cyp4f13 |
cytochrome P450, family 4, subfamily f, polypeptide 13 |
3965 |
0.12 |
| chr13_8526113_8526309 | 0.28 |
Gm48262 |
predicted gene, 48262 |
26809 |
0.23 |
| chr15_102752976_102753138 | 0.28 |
Calcoco1 |
calcium binding and coiled coil domain 1 |
30879 |
0.11 |
| chr10_93196295_93196461 | 0.28 |
Cdk17 |
cyclin-dependent kinase 17 |
10609 |
0.2 |
| chr11_5524693_5525093 | 0.28 |
Xbp1 |
X-box binding protein 1 |
2970 |
0.18 |
| chr14_20164716_20164945 | 0.28 |
Kcnk5 |
potassium channel, subfamily K, member 5 |
16979 |
0.13 |
| chr15_41832966_41833117 | 0.28 |
Oxr1 |
oxidation resistance 1 |
2048 |
0.35 |
| chr11_60178593_60178767 | 0.28 |
Rai1 |
retinoic acid induced 1 |
2801 |
0.18 |
| chr16_26727363_26727514 | 0.28 |
Il1rap |
interleukin 1 receptor accessory protein |
396 |
0.9 |
| chr1_163817675_163817827 | 0.28 |
Kifap3 |
kinesin-associated protein 3 |
38168 |
0.16 |
| chr18_11317026_11317247 | 0.28 |
Gata6 |
GATA binding protein 6 |
258089 |
0.02 |
| chr7_49764294_49764503 | 0.28 |
Htatip2 |
HIV-1 Tat interactive protein 2 |
5245 |
0.23 |
| chr17_71213183_71213334 | 0.28 |
Lpin2 |
lipin 2 |
8582 |
0.17 |
| chr3_106497089_106497388 | 0.28 |
Dennd2d |
DENN/MADD domain containing 2D |
4063 |
0.16 |
| chr17_89653592_89653743 | 0.28 |
Gm50061 |
predicted gene, 50061 |
98859 |
0.09 |
| chr3_100683705_100684061 | 0.28 |
Man1a2 |
mannosidase, alpha, class 1A, member 2 |
730 |
0.55 |
| chr9_112785054_112785229 | 0.28 |
Gm24957 |
predicted gene, 24957 |
262612 |
0.02 |
| chr1_119661376_119661532 | 0.27 |
Epb41l5 |
erythrocyte membrane protein band 4.1 like 5 |
12454 |
0.16 |
| chr10_125967716_125967898 | 0.27 |
Lrig3 |
leucine-rich repeats and immunoglobulin-like domains 3 |
1554 |
0.53 |
| chr2_43592382_43592533 | 0.27 |
Kynu |
kynureninase |
7158 |
0.3 |
| chr8_68094755_68094906 | 0.27 |
Psd3 |
pleckstrin and Sec7 domain containing 3 |
26483 |
0.22 |
| chr15_62746422_62746596 | 0.27 |
Gm22521 |
predicted gene, 22521 |
46692 |
0.18 |
| chr1_23261263_23261446 | 0.27 |
Gm6420 |
predicted gene 6420 |
4947 |
0.13 |
| chr11_109027604_109027760 | 0.27 |
Gm11668 |
predicted gene 11668 |
20031 |
0.18 |
| chr6_144895356_144895507 | 0.27 |
Gm22792 |
predicted gene, 22792 |
91799 |
0.07 |
| chr18_5617566_5617751 | 0.27 |
Gm22709 |
predicted gene, 22709 |
3185 |
0.18 |
| chr11_106889132_106889465 | 0.27 |
Smurf2 |
SMAD specific E3 ubiquitin protein ligase 2 |
30977 |
0.11 |
| chr13_6515622_6515787 | 0.27 |
2900042G08Rik |
RIKEN cDNA 2900042G08 gene |
14891 |
0.21 |
| chr5_106140040_106140240 | 0.27 |
Gm32736 |
predicted gene, 32736 |
45049 |
0.13 |
| chr14_79629212_79629374 | 0.27 |
Sugt1 |
SGT1, suppressor of G2 allele of SKP1 (S. cerevisiae) |
4654 |
0.19 |
| chr13_19394098_19394409 | 0.27 |
Gm42683 |
predicted gene 42683 |
1200 |
0.38 |
| chr2_75605802_75605972 | 0.27 |
Gm13655 |
predicted gene 13655 |
27495 |
0.15 |
| chr8_40564203_40564371 | 0.27 |
Vps37a |
vacuolar protein sorting 37A |
20863 |
0.15 |
| chr16_94606252_94606403 | 0.27 |
Gm15971 |
predicted gene 15971 |
14050 |
0.19 |
| chr11_75495112_75495263 | 0.27 |
Prpf8 |
pre-mRNA processing factor 8 |
2941 |
0.12 |
| chr11_102741952_102742137 | 0.26 |
Gm16342 |
predicted gene 16342 |
9639 |
0.11 |
| chr2_31499430_31499808 | 0.26 |
Ass1 |
argininosuccinate synthetase 1 |
1107 |
0.5 |
| chr2_48404312_48404494 | 0.26 |
Gm13472 |
predicted gene 13472 |
16366 |
0.2 |
| chr10_86690446_86690597 | 0.26 |
Gm15344 |
predicted gene 15344 |
2574 |
0.11 |
| chr4_124809297_124809476 | 0.26 |
Mtf1 |
metal response element binding transcription factor 1 |
6708 |
0.1 |
| chr11_117845425_117845576 | 0.26 |
Birc5 |
baculoviral IAP repeat-containing 5 |
3751 |
0.1 |
| chr2_31519719_31520357 | 0.26 |
Ass1 |
argininosuccinate synthetase 1 |
1548 |
0.36 |
| chr1_51760527_51760678 | 0.26 |
Myo1b |
myosin IB |
3868 |
0.24 |
| chr11_60877088_60877239 | 0.26 |
Tmem11 |
transmembrane protein 11 |
1624 |
0.26 |
| chr12_101954170_101954556 | 0.26 |
Atxn3 |
ataxin 3 |
3785 |
0.17 |
| chr2_77155937_77156088 | 0.26 |
Ccdc141 |
coiled-coil domain containing 141 |
14564 |
0.21 |
| chr6_108834740_108834891 | 0.26 |
Edem1 |
ER degradation enhancer, mannosidase alpha-like 1 |
1595 |
0.43 |
| chr17_31155739_31155923 | 0.26 |
Gm15318 |
predicted gene 15318 |
3035 |
0.15 |
| chr19_39739857_39740024 | 0.26 |
Cyp2c68 |
cytochrome P450, family 2, subfamily c, polypeptide 68 |
1114 |
0.6 |
| chr2_128972020_128972207 | 0.26 |
Gm10762 |
predicted gene 10762 |
4069 |
0.12 |
| chr3_51437862_51438013 | 0.26 |
Naa15 |
N(alpha)-acetyltransferase 15, NatA auxiliary subunit |
2980 |
0.14 |
| chr16_17340724_17340946 | 0.26 |
Gm24927 |
predicted gene, 24927 |
1177 |
0.36 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0019676 | ammonia assimilation cycle(GO:0019676) |
| 0.1 | 0.1 | GO:0034310 | primary alcohol catabolic process(GO:0034310) |
| 0.1 | 0.3 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.1 | 0.3 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.1 | 0.6 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.1 | 0.2 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.1 | 0.2 | GO:0033031 | positive regulation of neutrophil apoptotic process(GO:0033031) |
| 0.1 | 0.2 | GO:0046271 | phenylpropanoid catabolic process(GO:0046271) |
| 0.1 | 0.2 | GO:0003331 | regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) |
| 0.1 | 0.2 | GO:0030327 | prenylated protein catabolic process(GO:0030327) |
| 0.1 | 0.2 | GO:0072361 | regulation of glycolytic process by regulation of transcription from RNA polymerase II promoter(GO:0072361) |
| 0.1 | 0.2 | GO:0060741 | prostate gland stromal morphogenesis(GO:0060741) |
| 0.1 | 0.3 | GO:1901525 | negative regulation of macromitophagy(GO:1901525) |
| 0.1 | 0.2 | GO:1903984 | positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) |
| 0.1 | 0.2 | GO:0036112 | medium-chain fatty-acyl-CoA metabolic process(GO:0036112) |
| 0.1 | 0.2 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.1 | 0.2 | GO:0060523 | prostate epithelial cord elongation(GO:0060523) |
| 0.1 | 0.2 | GO:0061724 | lipophagy(GO:0061724) |
| 0.1 | 0.5 | GO:2000980 | regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.0 | 0.1 | GO:0010963 | regulation of L-arginine import(GO:0010963) |
| 0.0 | 0.5 | GO:0009404 | toxin metabolic process(GO:0009404) |
| 0.0 | 0.1 | GO:0015747 | urate transport(GO:0015747) |
| 0.0 | 0.2 | GO:0060839 | endothelial cell fate commitment(GO:0060839) |
| 0.0 | 0.2 | GO:0042524 | negative regulation of tyrosine phosphorylation of Stat5 protein(GO:0042524) |
| 0.0 | 0.0 | GO:0034140 | negative regulation of toll-like receptor 3 signaling pathway(GO:0034140) |
| 0.0 | 0.2 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.0 | 0.2 | GO:0072602 | interleukin-4 secretion(GO:0072602) |
| 0.0 | 0.2 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.0 | 0.1 | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 0.0 | 0.1 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.0 | 0.2 | GO:0009202 | deoxyribonucleoside triphosphate biosynthetic process(GO:0009202) |
| 0.0 | 0.2 | GO:0030647 | polyketide metabolic process(GO:0030638) aminoglycoside antibiotic metabolic process(GO:0030647) daunorubicin metabolic process(GO:0044597) doxorubicin metabolic process(GO:0044598) |
| 0.0 | 0.3 | GO:0052697 | xenobiotic glucuronidation(GO:0052697) |
| 0.0 | 0.5 | GO:0048742 | regulation of skeletal muscle fiber development(GO:0048742) |
| 0.0 | 0.1 | GO:1904154 | positive regulation of retrograde protein transport, ER to cytosol(GO:1904154) |
| 0.0 | 0.1 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.0 | 0.2 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.0 | 0.1 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.0 | 0.2 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.0 | 0.1 | GO:0009826 | unidimensional cell growth(GO:0009826) |
| 0.0 | 0.1 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.0 | 0.1 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.1 | GO:0006987 | activation of signaling protein activity involved in unfolded protein response(GO:0006987) |
| 0.0 | 0.1 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.0 | 0.3 | GO:0048671 | negative regulation of collateral sprouting(GO:0048671) |
| 0.0 | 0.1 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.0 | 0.2 | GO:0015871 | choline transport(GO:0015871) |
| 0.0 | 0.1 | GO:0061419 | positive regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061419) |
| 0.0 | 0.1 | GO:2000302 | positive regulation of synaptic vesicle exocytosis(GO:2000302) |
| 0.0 | 0.0 | GO:0006714 | sesquiterpenoid metabolic process(GO:0006714) |
| 0.0 | 0.1 | GO:1902564 | negative regulation of neutrophil activation(GO:1902564) |
| 0.0 | 0.2 | GO:0044829 | positive regulation by host of viral genome replication(GO:0044829) |
| 0.0 | 0.1 | GO:0006562 | proline catabolic process(GO:0006562) |
| 0.0 | 0.2 | GO:0071447 | cellular response to hydroperoxide(GO:0071447) |
| 0.0 | 0.1 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 0.0 | 0.3 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 0.1 | GO:0019086 | late viral transcription(GO:0019086) |
| 0.0 | 0.1 | GO:0050427 | 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) |
| 0.0 | 0.1 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.0 | 0.1 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.0 | 0.1 | GO:0042939 | glutathione transport(GO:0034635) tripeptide transport(GO:0042939) |
| 0.0 | 0.2 | GO:0086013 | membrane repolarization during cardiac muscle cell action potential(GO:0086013) |
| 0.0 | 0.1 | GO:0044337 | canonical Wnt signaling pathway involved in positive regulation of apoptotic process(GO:0044337) |
| 0.0 | 0.1 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 0.0 | 0.1 | GO:0002282 | microglial cell activation involved in immune response(GO:0002282) |
| 0.0 | 0.1 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.0 | 0.1 | GO:0042737 | drug catabolic process(GO:0042737) |
| 0.0 | 0.1 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.0 | 0.2 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.0 | 0.2 | GO:0019530 | taurine metabolic process(GO:0019530) |
| 0.0 | 0.1 | GO:0018879 | biphenyl metabolic process(GO:0018879) |
| 0.0 | 0.1 | GO:0002036 | regulation of L-glutamate transport(GO:0002036) |
| 0.0 | 0.1 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.0 | 0.2 | GO:0006707 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.0 | 0.2 | GO:0006677 | glycosylceramide metabolic process(GO:0006677) |
| 0.0 | 0.0 | GO:2000409 | positive regulation of T cell extravasation(GO:2000409) |
| 0.0 | 0.0 | GO:0032741 | positive regulation of interleukin-18 production(GO:0032741) |
| 0.0 | 0.1 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.0 | 0.1 | GO:0097105 | presynaptic membrane assembly(GO:0097105) |
| 0.0 | 0.1 | GO:0032488 | Cdc42 protein signal transduction(GO:0032488) |
| 0.0 | 0.1 | GO:0021553 | olfactory nerve development(GO:0021553) |
| 0.0 | 0.1 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.0 | 0.0 | GO:2000437 | monocyte extravasation(GO:0035696) regulation of monocyte extravasation(GO:2000437) |
| 0.0 | 0.1 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 0.0 | 0.2 | GO:0006655 | phosphatidylglycerol biosynthetic process(GO:0006655) |
| 0.0 | 0.1 | GO:0097112 | gamma-aminobutyric acid receptor clustering(GO:0097112) |
| 0.0 | 0.2 | GO:0002072 | optic cup morphogenesis involved in camera-type eye development(GO:0002072) |
| 0.0 | 0.1 | GO:0061511 | centriole elongation(GO:0061511) |
| 0.0 | 0.0 | GO:0003213 | cardiac right atrium morphogenesis(GO:0003213) |
| 0.0 | 0.0 | GO:1904742 | regulation of telomeric DNA binding(GO:1904742) |
| 0.0 | 0.1 | GO:0001842 | neural fold formation(GO:0001842) |
| 0.0 | 0.1 | GO:1900103 | positive regulation of endoplasmic reticulum unfolded protein response(GO:1900103) |
| 0.0 | 0.1 | GO:0070948 | regulation of neutrophil mediated cytotoxicity(GO:0070948) regulation of neutrophil mediated killing of symbiont cell(GO:0070949) |
| 0.0 | 0.0 | GO:1901078 | negative regulation of relaxation of muscle(GO:1901078) |
| 0.0 | 0.1 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.0 | 0.1 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.0 | 0.1 | GO:2000483 | negative regulation of interleukin-8 secretion(GO:2000483) |
| 0.0 | 0.2 | GO:0060546 | negative regulation of necroptotic process(GO:0060546) |
| 0.0 | 0.0 | GO:0048550 | negative regulation of pinocytosis(GO:0048550) |
| 0.0 | 0.1 | GO:0034436 | glycoprotein transport(GO:0034436) |
| 0.0 | 0.1 | GO:0071681 | response to indole-3-methanol(GO:0071680) cellular response to indole-3-methanol(GO:0071681) |
| 0.0 | 0.2 | GO:0009143 | nucleoside triphosphate catabolic process(GO:0009143) |
| 0.0 | 0.1 | GO:0001905 | activation of membrane attack complex(GO:0001905) regulation of activation of membrane attack complex(GO:0001969) |
| 0.0 | 0.2 | GO:0033147 | negative regulation of intracellular estrogen receptor signaling pathway(GO:0033147) |
| 0.0 | 0.2 | GO:0006517 | protein deglycosylation(GO:0006517) |
| 0.0 | 0.0 | GO:0030397 | membrane disassembly(GO:0030397) nuclear envelope disassembly(GO:0051081) |
| 0.0 | 0.1 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.0 | 0.1 | GO:2000370 | positive regulation of clathrin-mediated endocytosis(GO:2000370) |
| 0.0 | 0.1 | GO:0007567 | parturition(GO:0007567) |
| 0.0 | 0.1 | GO:0048318 | axial mesoderm development(GO:0048318) |
| 0.0 | 0.1 | GO:0046104 | thymidine metabolic process(GO:0046104) |
| 0.0 | 0.1 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.0 | 0.1 | GO:0090210 | regulation of establishment of blood-brain barrier(GO:0090210) |
| 0.0 | 0.1 | GO:0042447 | hormone catabolic process(GO:0042447) |
| 0.0 | 0.1 | GO:0046218 | tryptophan catabolic process(GO:0006569) tryptophan catabolic process to kynurenine(GO:0019441) indole-containing compound catabolic process(GO:0042436) indolalkylamine catabolic process(GO:0046218) |
| 0.0 | 0.2 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.0 | 0.1 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.1 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.0 | 0.1 | GO:1900084 | regulation of peptidyl-tyrosine autophosphorylation(GO:1900084) |
| 0.0 | 0.1 | GO:0061158 | 3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.0 | 0.1 | GO:0070375 | ERK5 cascade(GO:0070375) |
| 0.0 | 0.1 | GO:2000467 | positive regulation of glycogen (starch) synthase activity(GO:2000467) |
| 0.0 | 0.1 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.0 | 0.1 | GO:0046618 | drug export(GO:0046618) |
| 0.0 | 0.1 | GO:0006768 | biotin metabolic process(GO:0006768) |
| 0.0 | 0.1 | GO:0055091 | phospholipid homeostasis(GO:0055091) |
| 0.0 | 0.0 | GO:0060559 | positive regulation of vitamin metabolic process(GO:0046136) positive regulation of vitamin D biosynthetic process(GO:0060557) positive regulation of calcidiol 1-monooxygenase activity(GO:0060559) |
| 0.0 | 0.1 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.3 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.0 | 0.1 | GO:1903966 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.0 | 0.1 | GO:0032789 | saturated monocarboxylic acid metabolic process(GO:0032788) unsaturated monocarboxylic acid metabolic process(GO:0032789) |
| 0.0 | 0.1 | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.0 | 0.1 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.1 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.0 | 0.1 | GO:0070989 | oxidative demethylation(GO:0070989) |
| 0.0 | 0.1 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.0 | 0.1 | GO:0070777 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.0 | 0.1 | GO:0045085 | negative regulation of interleukin-2 biosynthetic process(GO:0045085) |
| 0.0 | 0.1 | GO:0000960 | mitochondrial RNA catabolic process(GO:0000957) regulation of mitochondrial RNA catabolic process(GO:0000960) |
| 0.0 | 0.0 | GO:2000646 | positive regulation of receptor catabolic process(GO:2000646) |
| 0.0 | 0.1 | GO:0032202 | telomere assembly(GO:0032202) |
| 0.0 | 0.1 | GO:0042494 | detection of bacterial lipoprotein(GO:0042494) detection of bacterial lipopeptide(GO:0070340) |
| 0.0 | 0.1 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.0 | 0.0 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 0.0 | GO:0046149 | heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.0 | 0.1 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.0 | 0.1 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.0 | 0.0 | GO:1901187 | regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.0 | 0.0 | GO:0003062 | regulation of heart rate by chemical signal(GO:0003062) |
| 0.0 | 0.1 | GO:0050859 | negative regulation of B cell receptor signaling pathway(GO:0050859) |
| 0.0 | 0.1 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.0 | 0.1 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.0 | 0.0 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.0 | 0.0 | GO:0050904 | diapedesis(GO:0050904) |
| 0.0 | 0.1 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.0 | 0.2 | GO:0009812 | flavonoid metabolic process(GO:0009812) |
| 0.0 | 0.1 | GO:1902177 | positive regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902177) |
| 0.0 | 0.1 | GO:0051657 | maintenance of organelle location(GO:0051657) |
| 0.0 | 0.1 | GO:0008228 | opsonization(GO:0008228) |
| 0.0 | 0.0 | GO:0010248 | establishment or maintenance of transmembrane electrochemical gradient(GO:0010248) |
| 0.0 | 0.0 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.0 | 0.4 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.0 | GO:0006481 | C-terminal protein methylation(GO:0006481) |
| 0.0 | 0.0 | GO:1902630 | regulation of membrane hyperpolarization(GO:1902630) |
| 0.0 | 0.4 | GO:0019432 | triglyceride biosynthetic process(GO:0019432) |
| 0.0 | 0.0 | GO:0006591 | ornithine metabolic process(GO:0006591) |
| 0.0 | 0.0 | GO:2000562 | negative regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000562) |
| 0.0 | 0.0 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 0.0 | 0.1 | GO:0021670 | lateral ventricle development(GO:0021670) |
| 0.0 | 0.1 | GO:0015074 | DNA integration(GO:0015074) |
| 0.0 | 0.1 | GO:0006537 | glutamate biosynthetic process(GO:0006537) |
| 0.0 | 0.0 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 0.0 | 0.1 | GO:1902969 | mitotic DNA replication(GO:1902969) |
| 0.0 | 0.0 | GO:2000599 | regulation of cyclin catabolic process(GO:2000598) negative regulation of cyclin catabolic process(GO:2000599) |
| 0.0 | 0.0 | GO:1903977 | positive regulation of glial cell migration(GO:1903977) |
| 0.0 | 0.1 | GO:2001184 | positive regulation of interleukin-12 secretion(GO:2001184) |
| 0.0 | 0.0 | GO:0018199 | peptidyl-glutamine modification(GO:0018199) |
| 0.0 | 0.0 | GO:0006578 | amino-acid betaine biosynthetic process(GO:0006578) |
| 0.0 | 0.0 | GO:1903799 | negative regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903799) |
| 0.0 | 0.0 | GO:0014732 | skeletal muscle atrophy(GO:0014732) |
| 0.0 | 0.1 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.0 | 0.1 | GO:0006528 | asparagine metabolic process(GO:0006528) |
| 0.0 | 0.0 | GO:0090219 | negative regulation of lipid kinase activity(GO:0090219) |
| 0.0 | 0.1 | GO:0021684 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.0 | 0.1 | GO:1900165 | negative regulation of interleukin-6 secretion(GO:1900165) |
| 0.0 | 0.0 | GO:1900060 | negative regulation of ceramide biosynthetic process(GO:1900060) |
| 0.0 | 0.1 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.0 | GO:0030908 | intein-mediated protein splicing(GO:0016539) protein splicing(GO:0030908) |
| 0.0 | 0.1 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.0 | 0.0 | GO:2000828 | regulation of parathyroid hormone secretion(GO:2000828) |
| 0.0 | 0.1 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 0.0 | 0.2 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.1 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 | 0.1 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.0 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.0 | 0.0 | GO:1904684 | negative regulation of metalloendopeptidase activity(GO:1904684) negative regulation of metallopeptidase activity(GO:1905049) |
| 0.0 | 0.0 | GO:2000224 | regulation of testosterone biosynthetic process(GO:2000224) |
| 0.0 | 0.0 | GO:0046813 | receptor-mediated virion attachment to host cell(GO:0046813) |
| 0.0 | 0.0 | GO:0072025 | distal convoluted tubule development(GO:0072025) DCT cell differentiation(GO:0072069) metanephric distal convoluted tubule development(GO:0072221) metanephric distal tubule development(GO:0072235) metanephric DCT cell differentiation(GO:0072240) |
| 0.0 | 0.1 | GO:0010898 | positive regulation of triglyceride catabolic process(GO:0010898) |
| 0.0 | 0.1 | GO:0045653 | negative regulation of megakaryocyte differentiation(GO:0045653) |
| 0.0 | 0.1 | GO:0000467 | exonucleolytic trimming involved in rRNA processing(GO:0000459) exonucleolytic trimming to generate mature 3'-end of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000467) |
| 0.0 | 0.1 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 | 0.0 | GO:1904469 | positive regulation of tumor necrosis factor secretion(GO:1904469) |
| 0.0 | 0.0 | GO:0035672 | oligopeptide transmembrane transport(GO:0035672) |
| 0.0 | 0.0 | GO:1903975 | regulation of glial cell migration(GO:1903975) |
| 0.0 | 0.1 | GO:0015867 | ATP transport(GO:0015867) |
| 0.0 | 0.2 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 0.1 | GO:0071267 | amino acid salvage(GO:0043102) L-methionine biosynthetic process(GO:0071265) L-methionine salvage(GO:0071267) |
| 0.0 | 0.0 | GO:0051594 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 0.0 | 0.0 | GO:1903347 | negative regulation of bicellular tight junction assembly(GO:1903347) |
| 0.0 | 0.0 | GO:0035247 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) peptidyl-arginine omega-N-methylation(GO:0035247) |
| 0.0 | 0.0 | GO:0097211 | response to gonadotropin-releasing hormone(GO:0097210) cellular response to gonadotropin-releasing hormone(GO:0097211) |
| 0.0 | 0.2 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 0.0 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.0 | 0.0 | GO:0044359 | modulation of molecular function in other organism(GO:0044359) negative regulation of molecular function in other organism(GO:0044362) negative regulation of molecular function in other organism involved in symbiotic interaction(GO:0052204) modulation of molecular function in other organism involved in symbiotic interaction(GO:0052205) |
| 0.0 | 0.0 | GO:0097368 | establishment of Sertoli cell barrier(GO:0097368) |
| 0.0 | 0.1 | GO:0002176 | male germ cell proliferation(GO:0002176) |
| 0.0 | 0.1 | GO:1900016 | negative regulation of cytokine production involved in inflammatory response(GO:1900016) |
| 0.0 | 0.0 | GO:0061370 | testosterone biosynthetic process(GO:0061370) |
| 0.0 | 0.0 | GO:0032077 | positive regulation of deoxyribonuclease activity(GO:0032077) |
| 0.0 | 0.0 | GO:0070126 | mitochondrial translational termination(GO:0070126) |
| 0.0 | 0.0 | GO:1901491 | negative regulation of lymphangiogenesis(GO:1901491) |
| 0.0 | 0.0 | GO:0010958 | regulation of amino acid import(GO:0010958) |
| 0.0 | 0.1 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.0 | 0.0 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.0 | 0.1 | GO:0009052 | pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 0.0 | 0.0 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.1 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.0 | 0.0 | GO:1903774 | positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 0.0 | 0.1 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 0.0 | GO:0006701 | progesterone biosynthetic process(GO:0006701) |
| 0.0 | 0.0 | GO:1902897 | regulation of postsynaptic density protein 95 clustering(GO:1902897) |
| 0.0 | 0.0 | GO:0006824 | cobalt ion transport(GO:0006824) |
| 0.0 | 0.0 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.0 | 0.2 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.1 | GO:2000310 | regulation of N-methyl-D-aspartate selective glutamate receptor activity(GO:2000310) |
| 0.0 | 0.1 | GO:0051987 | positive regulation of attachment of spindle microtubules to kinetochore(GO:0051987) |
| 0.0 | 0.0 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.0 | 0.1 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.0 | 0.1 | GO:0010961 | cellular magnesium ion homeostasis(GO:0010961) |
| 0.0 | 0.0 | GO:0060051 | negative regulation of protein glycosylation(GO:0060051) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0045293 | mRNA editing complex(GO:0045293) |
| 0.0 | 0.2 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.0 | 0.1 | GO:0034715 | pICln-Sm protein complex(GO:0034715) |
| 0.0 | 0.2 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.1 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.1 | GO:0000811 | GINS complex(GO:0000811) |
| 0.0 | 0.1 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.0 | 0.1 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.0 | 0.1 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.0 | 0.1 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.2 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.0 | GO:0097418 | neurofibrillary tangle(GO:0097418) |
| 0.0 | 0.0 | GO:0034666 | integrin alpha2-beta1 complex(GO:0034666) |
| 0.0 | 0.1 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 0.1 | GO:0005784 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.0 | 0.1 | GO:0030313 | cell outer membrane(GO:0009279) cell envelope(GO:0030313) external encapsulating structure part(GO:0044462) |
| 0.0 | 0.1 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 0.1 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 0.0 | 0.2 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.1 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 0.0 | 0.1 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.0 | 0.1 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.0 | 0.1 | GO:0016939 | kinesin II complex(GO:0016939) |
| 0.0 | 0.1 | GO:0005593 | FACIT collagen trimer(GO:0005593) |
| 0.0 | 0.1 | GO:0035355 | Toll-like receptor 2-Toll-like receptor 6 protein complex(GO:0035355) |
| 0.0 | 0.1 | GO:0000306 | extrinsic component of vacuolar membrane(GO:0000306) |
| 0.0 | 0.3 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.2 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 0.1 | GO:0035749 | myelin sheath adaxonal region(GO:0035749) |
| 0.0 | 0.1 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.2 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.3 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.0 | 0.2 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.4 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.0 | 0.0 | GO:0070110 | ciliary neurotrophic factor receptor complex(GO:0070110) |
| 0.0 | 0.2 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.0 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 0.0 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 0.1 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 0.1 | GO:0071556 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.0 | 0.1 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.0 | 0.1 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.0 | 0.1 | GO:0001527 | microfibril(GO:0001527) |
| 0.0 | 0.1 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.2 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.2 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.0 | 0.1 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.0 | 0.0 | GO:0097651 | phosphatidylinositol 3-kinase complex, class I(GO:0097651) |
| 0.0 | 0.0 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.0 | GO:1990851 | Wnt-Frizzled-LRP5/6 complex(GO:1990851) |
| 0.0 | 0.3 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 0.0 | GO:1990131 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.0 | 0.1 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.0 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.0 | 0.1 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.1 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 0.1 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.0 | 0.1 | GO:0031211 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0016743 | carboxyl- or carbamoyltransferase activity(GO:0016743) |
| 0.1 | 0.3 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.1 | 0.2 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.1 | 0.3 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.1 | 0.2 | GO:0046573 | lactonohydrolase activity(GO:0046573) acyl-L-homoserine-lactone lactonohydrolase activity(GO:0102007) |
| 0.1 | 0.2 | GO:0038181 | bile acid receptor activity(GO:0038181) |
| 0.1 | 0.6 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.1 | 0.2 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.1 | 0.2 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.1 | 0.2 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.1 | 0.2 | GO:0047045 | testosterone 17-beta-dehydrogenase (NADP+) activity(GO:0047045) |
| 0.0 | 0.1 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.0 | 0.2 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.0 | 0.2 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.0 | 0.1 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.0 | 0.3 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.0 | 0.2 | GO:0043426 | MRF binding(GO:0043426) |
| 0.0 | 0.1 | GO:0030158 | protein xylosyltransferase activity(GO:0030158) |
| 0.0 | 0.2 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.0 | 0.2 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.1 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.0 | 0.2 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.2 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.0 | 0.1 | GO:0031697 | beta-1 adrenergic receptor binding(GO:0031697) |
| 0.0 | 0.3 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.1 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.2 | GO:0035671 | enone reductase activity(GO:0035671) |
| 0.0 | 0.1 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.1 | GO:0016899 | oxidoreductase activity, acting on the CH-OH group of donors, oxygen as acceptor(GO:0016899) |
| 0.0 | 0.1 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 0.3 | GO:0005167 | neurotrophin TRK receptor binding(GO:0005167) |
| 0.0 | 0.2 | GO:0047760 | butyrate-CoA ligase activity(GO:0047760) |
| 0.0 | 0.2 | GO:0018685 | alkane 1-monooxygenase activity(GO:0018685) |
| 0.0 | 0.4 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.2 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.3 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.1 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.3 | GO:0070990 | snRNP binding(GO:0070990) |
| 0.0 | 0.1 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.0 | 0.1 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.0 | 0.2 | GO:0086008 | voltage-gated potassium channel activity involved in cardiac muscle cell action potential repolarization(GO:0086008) |
| 0.0 | 0.1 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.1 | GO:0030572 | cardiolipin synthase activity(GO:0008808) phosphatidyltransferase activity(GO:0030572) |
| 0.0 | 0.0 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.0 | 0.1 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.1 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.0 | GO:0050051 | leukotriene-B4 20-monooxygenase activity(GO:0050051) |
| 0.0 | 0.1 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.0 | 0.1 | GO:0016823 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.0 | 0.4 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.5 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.2 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 0.4 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.0 | 0.2 | GO:0004779 | sulfate adenylyltransferase activity(GO:0004779) |
| 0.0 | 0.1 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.0 | 0.1 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.0 | 0.1 | GO:0015038 | glutathione disulfide oxidoreductase activity(GO:0015038) |
| 0.0 | 0.0 | GO:0001226 | RNA polymerase II transcription corepressor binding(GO:0001226) |
| 0.0 | 0.1 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.0 | 0.2 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.1 | GO:0035663 | Toll-like receptor 2 binding(GO:0035663) |
| 0.0 | 0.1 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) |
| 0.0 | 0.1 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) |
| 0.0 | 0.1 | GO:0052723 | inositol hexakisphosphate kinase activity(GO:0000828) inositol hexakisphosphate 5-kinase activity(GO:0000832) inositol hexakisphosphate 1-kinase activity(GO:0052723) inositol hexakisphosphate 3-kinase activity(GO:0052724) |
| 0.0 | 0.1 | GO:0019797 | procollagen-proline 3-dioxygenase activity(GO:0019797) |
| 0.0 | 0.1 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.0 | 0.1 | GO:0033592 | RNA strand annealing activity(GO:0033592) |
| 0.0 | 0.1 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.0 | 0.1 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.0 | 0.1 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 0.1 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.0 | 0.1 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.0 | 0.1 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.1 | GO:0051880 | G-quadruplex DNA binding(GO:0051880) |
| 0.0 | 0.0 | GO:0008948 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) oxaloacetate decarboxylase activity(GO:0008948) |
| 0.0 | 0.1 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.0 | 0.0 | GO:0000702 | oxidized base lesion DNA N-glycosylase activity(GO:0000702) |
| 0.0 | 0.2 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.2 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.0 | 0.0 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.0 | 0.1 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.1 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.0 | 0.1 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.0 | 0.1 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.0 | 0.1 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 0.2 | GO:0052672 | cobinamide kinase activity(GO:0008819) phytol kinase activity(GO:0010276) phenol kinase activity(GO:0018720) cyclin-dependent protein kinase activating kinase regulator activity(GO:0019914) inositol tetrakisphosphate 2-kinase activity(GO:0032942) heptose 7-phosphate kinase activity(GO:0033785) aminoglycoside phosphotransferase activity(GO:0034071) eukaryotic elongation factor-2 kinase regulator activity(GO:0042556) eukaryotic elongation factor-2 kinase activator activity(GO:0042557) LPPG:FO 2-phospho-L-lactate transferase activity(GO:0043743) cytidine kinase activity(GO:0043771) glycerate 2-kinase activity(GO:0043798) (S)-lactate 2-kinase activity(GO:0043841) phosphoserine:homoserine phosphotransferase activity(GO:0043899) L-seryl-tRNA(Sec) kinase activity(GO:0043915) phosphocholine transferase activity(GO:0044605) GTP-dependent polynucleotide kinase activity(GO:0051735) farnesol kinase activity(GO:0052668) CTP:2-trans,-6-trans-farnesol kinase activity(GO:0052669) geraniol kinase activity(GO:0052670) geranylgeraniol kinase activity(GO:0052671) CTP:geranylgeraniol kinase activity(GO:0052672) prenol kinase activity(GO:0052673) 1-phosphatidylinositol-5-kinase activity(GO:0052810) 1-phosphatidylinositol-3-phosphate 4-kinase activity(GO:0052811) phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) inositol-3,4,6-trisphosphate 1-kinase activity(GO:0052835) inositol 5-diphosphate pentakisphosphate 5-kinase activity(GO:0052836) inositol diphosphate tetrakisphosphate kinase activity(GO:0052839) |
| 0.0 | 0.1 | GO:0042979 | ornithine decarboxylase regulator activity(GO:0042979) |
| 0.0 | 0.0 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.1 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 0.1 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.0 | 0.1 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.1 | GO:0070569 | uridylyltransferase activity(GO:0070569) |
| 0.0 | 0.1 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.1 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 0.0 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.0 | 0.0 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) |
| 0.0 | 0.1 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.1 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.0 | GO:0046870 | cadmium ion binding(GO:0046870) |
| 0.0 | 0.2 | GO:0016502 | purinergic nucleotide receptor activity(GO:0001614) nucleotide receptor activity(GO:0016502) |
| 0.0 | 0.0 | GO:0005280 | hydrogen:amino acid symporter activity(GO:0005280) |
| 0.0 | 0.4 | GO:0051723 | protein methylesterase activity(GO:0051723) |
| 0.0 | 0.1 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.0 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.0 | 0.0 | GO:0016661 | oxidoreductase activity, acting on other nitrogenous compounds as donors(GO:0016661) |
| 0.0 | 0.2 | GO:0008601 | protein phosphatase type 2A regulator activity(GO:0008601) |
| 0.0 | 0.1 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 0.0 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.0 | 0.0 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 0.1 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.0 | 0.1 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 0.1 | GO:0005313 | L-glutamate transmembrane transporter activity(GO:0005313) |
| 0.0 | 0.0 | GO:0051033 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.0 | 0.0 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.0 | 0.0 | GO:0038064 | collagen receptor activity(GO:0038064) |
| 0.0 | 0.0 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.0 | 0.1 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 0.0 | 0.1 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 0.2 | GO:0015924 | mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.0 | 0.0 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 0.0 | 0.3 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.0 | 0.0 | GO:0015252 | hydrogen ion channel activity(GO:0015252) |
| 0.0 | 0.1 | GO:0048273 | mitogen-activated protein kinase p38 binding(GO:0048273) |
| 0.0 | 0.0 | GO:0016635 | oxidoreductase activity, acting on the CH-CH group of donors, quinone or related compound as acceptor(GO:0016635) |
| 0.0 | 0.0 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.1 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.0 | 0.0 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.0 | 0.1 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.0 | 0.0 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.1 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 0.0 | GO:0008898 | S-adenosylmethionine-homocysteine S-methyltransferase activity(GO:0008898) |
| 0.0 | 0.1 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.0 | 0.0 | GO:0016615 | malate dehydrogenase activity(GO:0016615) |
| 0.0 | 0.1 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 0.1 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.0 | 0.0 | GO:0030619 | U1 snRNA binding(GO:0030619) |
| 0.0 | 0.1 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.0 | 0.1 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.0 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 0.0 | 0.1 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 0.1 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 0.0 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 0.3 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.1 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 0.3 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.1 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 0.0 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.3 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 0.2 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.2 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 0.7 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.2 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.4 | REACTOME BILE ACID AND BILE SALT METABOLISM | Genes involved in Bile acid and bile salt metabolism |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.3 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.0 | 0.3 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.0 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.1 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 0.2 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.0 | 0.1 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.1 | REACTOME DESTABILIZATION OF MRNA BY TRISTETRAPROLIN TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 0.0 | 0.4 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 0.1 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.0 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.0 | 0.0 | REACTOME SHC RELATED EVENTS | Genes involved in SHC-related events |
| 0.0 | 0.1 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.0 | REACTOME VIF MEDIATED DEGRADATION OF APOBEC3G | Genes involved in Vif-mediated degradation of APOBEC3G |
| 0.0 | 0.1 | REACTOME HIGHLY CALCIUM PERMEABLE POSTSYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Highly calcium permeable postsynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.1 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.1 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.1 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.1 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.0 | 0.2 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.3 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.0 | 0.1 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |