Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
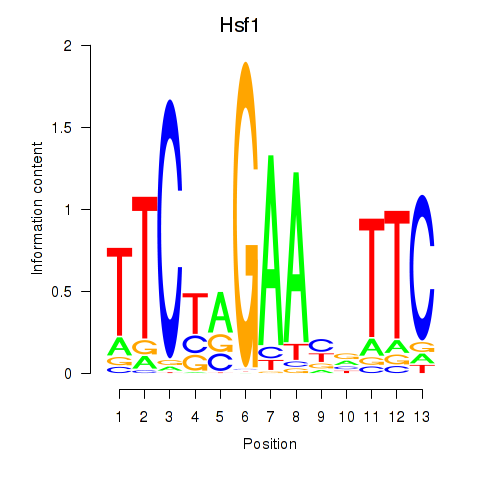
Results for Hsf1
Z-value: 1.99
Transcription factors associated with Hsf1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hsf1
|
ENSMUSG00000022556.9 | heat shock factor 1 |
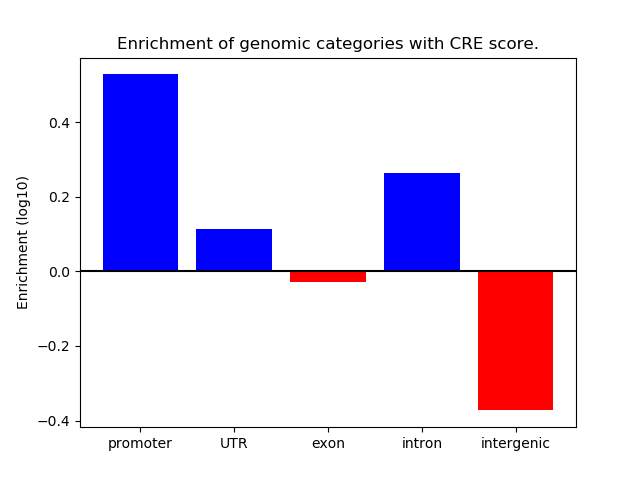
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr15_76477583_76477960 | Hsf1 | 85 | 0.838651 | -0.76 | 7.9e-02 | Click! |
| chr15_76478000_76478246 | Hsf1 | 437 | 0.543659 | -0.75 | 8.8e-02 | Click! |
| chr15_76477284_76477443 | Hsf1 | 59 | 0.516696 | 0.67 | 1.4e-01 | Click! |
| chr15_76480940_76481264 | Hsf1 | 3416 | 0.097576 | -0.38 | 4.6e-01 | Click! |
Activity of the Hsf1 motif across conditions
Conditions sorted by the z-value of the Hsf1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
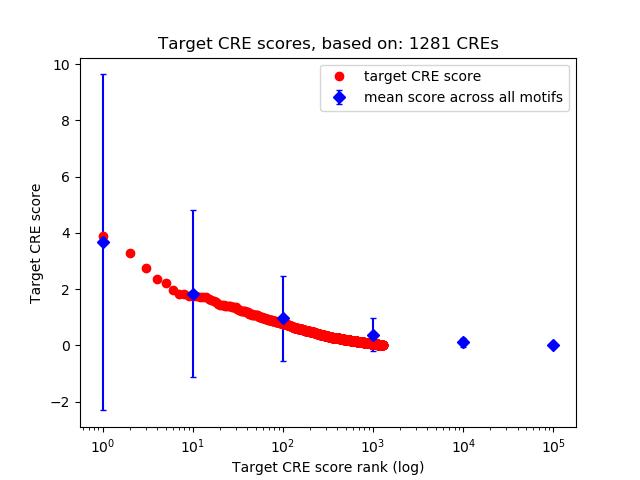
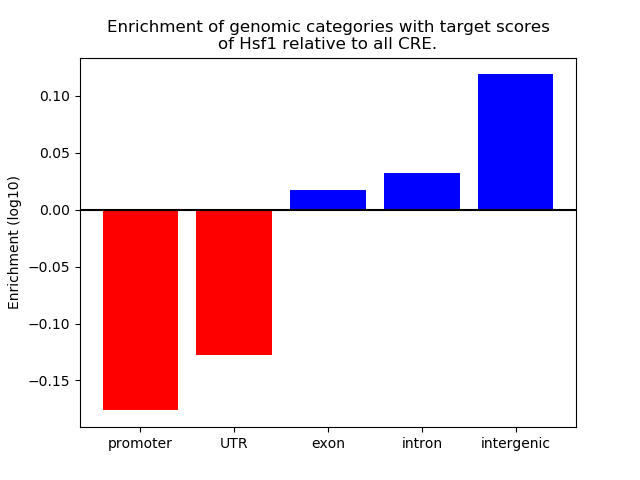
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr11_7199193_7199344 | 3.88 |
Igfbp1 |
insulin-like growth factor binding protein 1 |
1486 |
0.37 |
| chr7_43440689_43440840 | 3.27 |
Cldnd2 |
claudin domain containing 2 |
18 |
0.92 |
| chr10_41992198_41992354 | 2.76 |
Armc2 |
armadillo repeat containing 2 |
890 |
0.62 |
| chr2_144182222_144182616 | 2.35 |
Gm5535 |
predicted gene 5535 |
3333 |
0.2 |
| chr11_82948457_82948616 | 2.21 |
Slfn5 |
schlafen 5 |
2813 |
0.11 |
| chr6_128493697_128493874 | 1.98 |
Pzp |
PZP, alpha-2-macroglobulin like |
1438 |
0.23 |
| chr5_90424675_90424898 | 1.82 |
Gm43363 |
predicted gene 43363 |
27513 |
0.13 |
| chr8_116344914_116345106 | 1.82 |
1700018P08Rik |
RIKEN cDNA 1700018P08 gene |
12051 |
0.29 |
| chr6_136419356_136419519 | 1.76 |
Gm25882 |
predicted gene, 25882 |
1625 |
0.3 |
| chr8_89311386_89311545 | 1.76 |
Gm5356 |
predicted pseudogene 5356 |
123905 |
0.06 |
| chr15_25942169_25942359 | 1.76 |
Retreg1 |
reticulophagy regulator 1 |
415 |
0.85 |
| chr5_135118877_135119300 | 1.73 |
Gm43500 |
predicted gene 43500 |
4205 |
0.12 |
| chr10_61313715_61314040 | 1.72 |
Rpl27a-ps1 |
ribosomal protein L27A, pseudogene 1 |
4047 |
0.16 |
| chr5_52309795_52309946 | 1.72 |
Gm43179 |
predicted gene 43179 |
32490 |
0.12 |
| chr8_70634514_70634904 | 1.64 |
Gdf15 |
growth differentiation factor 15 |
2253 |
0.14 |
| chr12_24615310_24615477 | 1.61 |
Gm6969 |
predicted pseudogene 6969 |
23617 |
0.13 |
| chr19_61057398_61057702 | 1.58 |
Gm22520 |
predicted gene, 22520 |
44005 |
0.12 |
| chr15_63364555_63364706 | 1.53 |
Gm41335 |
predicted gene, 41335 |
7611 |
0.27 |
| chr12_100089427_100089592 | 1.46 |
4930477G07Rik |
RIKEN cDNA 4930477G07 gene |
7709 |
0.15 |
| chr9_102864196_102864634 | 1.44 |
Ryk |
receptor-like tyrosine kinase |
4651 |
0.18 |
| chr7_66196612_66196796 | 1.43 |
Gm45080 |
predicted gene 45080 |
6149 |
0.17 |
| chr12_3862523_3862680 | 1.43 |
Dnmt3aos |
DNA methyltransferase 3A, opposite strand |
357 |
0.82 |
| chr12_83582014_83582393 | 1.42 |
Zfyve1 |
zinc finger, FYVE domain containing 1 |
13484 |
0.14 |
| chr2_133495524_133495962 | 1.41 |
Gm14100 |
predicted gene 14100 |
32625 |
0.16 |
| chr5_112956750_112957247 | 1.41 |
Grk3 |
G protein-coupled receptor kinase 3 |
31200 |
0.15 |
| chr13_18868301_18868479 | 1.39 |
Vps41 |
VPS41 HOPS complex subunit |
23527 |
0.19 |
| chr5_66165832_66165983 | 1.38 |
Gm22929 |
predicted gene, 22929 |
1245 |
0.3 |
| chr3_135605510_135605876 | 1.37 |
Nfkb1 |
nuclear factor of kappa light polypeptide gene enhancer in B cells 1, p105 |
1834 |
0.34 |
| chr6_128485251_128485402 | 1.37 |
Pzp |
PZP, alpha-2-macroglobulin like |
2529 |
0.14 |
| chr8_126832324_126832475 | 1.36 |
A630001O12Rik |
RIKEN cDNA A630001O12 gene |
6834 |
0.22 |
| chr15_73535998_73536192 | 1.28 |
Dennd3 |
DENN/MADD domain containing 3 |
11961 |
0.18 |
| chr17_86779499_86780092 | 1.28 |
Gm50008 |
predicted gene, 50008 |
6525 |
0.19 |
| chr10_68248229_68248529 | 1.25 |
Arid5b |
AT rich interactive domain 5B (MRF1-like) |
30342 |
0.19 |
| chr4_105261100_105261481 | 1.24 |
Plpp3 |
phospholipid phosphatase 3 |
103943 |
0.07 |
| chr12_35418013_35418379 | 1.23 |
Gm26071 |
predicted gene, 26071 |
22994 |
0.21 |
| chr5_135479247_135479465 | 1.23 |
Gm42731 |
predicted gene 42731 |
5937 |
0.16 |
| chr9_77707832_77708042 | 1.22 |
Gm47789 |
predicted gene, 47789 |
55 |
0.97 |
| chr2_35338418_35338609 | 1.21 |
Stom |
stomatin |
1537 |
0.27 |
| chr7_105596543_105596888 | 1.20 |
Hpx |
hemopexin |
3397 |
0.12 |
| chr9_107576082_107576366 | 1.18 |
Hyal1 |
hyaluronoglucosaminidase 1 |
703 |
0.32 |
| chr2_167324318_167324524 | 1.17 |
B4galt5 |
UDP-Gal:betaGlcNAc beta 1,4-galactosyltransferase, polypeptide 5 |
10521 |
0.18 |
| chr13_32492661_32492824 | 1.12 |
Gm48073 |
predicted gene, 48073 |
43673 |
0.17 |
| chr2_101827994_101828363 | 1.12 |
Prr5l |
proline rich 5 like |
10802 |
0.22 |
| chr2_62655069_62655412 | 1.11 |
Ifih1 |
interferon induced with helicase C domain 1 |
8985 |
0.18 |
| chr1_191803404_191803597 | 1.11 |
Gm20203 |
predicted gene, 20203 |
12186 |
0.13 |
| chr2_174858524_174858697 | 1.10 |
Gm14616 |
predicted gene 14616 |
2321 |
0.26 |
| chr2_153694524_153694737 | 1.09 |
Dnmt3c |
DNA methyltransferase 3C |
2022 |
0.27 |
| chr2_172511032_172511203 | 1.09 |
Gm14303 |
predicted gene 14303 |
1426 |
0.37 |
| chr11_79725104_79725267 | 1.08 |
Mir365-2 |
microRNA 365-2 |
1215 |
0.37 |
| chr9_122028437_122028754 | 1.08 |
Gm47117 |
predicted gene, 47117 |
16455 |
0.1 |
| chr16_30258016_30258167 | 1.07 |
Gm49645 |
predicted gene, 49645 |
2939 |
0.19 |
| chr2_80502470_80502665 | 1.06 |
Gm13687 |
predicted gene 13687 |
3337 |
0.2 |
| chr2_35496007_35496180 | 1.06 |
Gm13445 |
predicted gene 13445 |
4574 |
0.15 |
| chr10_8228115_8228580 | 1.05 |
Gm30906 |
predicted gene, 30906 |
52216 |
0.15 |
| chr12_41451963_41452119 | 1.02 |
Lrrn3 |
leucine rich repeat protein 3, neuronal |
34390 |
0.15 |
| chr19_55087908_55088315 | 1.00 |
Gpam |
glycerol-3-phosphate acyltransferase, mitochondrial |
11340 |
0.21 |
| chr13_98956568_98956727 | 1.00 |
Gm35215 |
predicted gene, 35215 |
10731 |
0.12 |
| chr5_74784041_74784346 | 1.00 |
Gm42575 |
predicted gene 42575 |
34311 |
0.15 |
| chr4_137890706_137890863 | 0.98 |
Gm13012 |
predicted gene 13012 |
6847 |
0.21 |
| chr4_155566693_155567032 | 0.98 |
Nadk |
NAD kinase |
2790 |
0.15 |
| chr1_72830155_72830306 | 0.97 |
Igfbp2 |
insulin-like growth factor binding protein 2 |
4908 |
0.25 |
| chr1_136686557_136686994 | 0.95 |
Gm19705 |
predicted gene, 19705 |
3146 |
0.18 |
| chr16_24482781_24482948 | 0.95 |
Lpp |
LIM domain containing preferred translocation partner in lipoma |
34773 |
0.16 |
| chr13_48572250_48572425 | 0.95 |
Gm25232 |
predicted gene, 25232 |
47 |
0.95 |
| chr15_34510659_34511041 | 0.95 |
Pop1 |
processing of precursor 1, ribonuclease P/MRP family, (S. cerevisiae) |
943 |
0.43 |
| chr3_31394556_31394733 | 0.95 |
Gm38025 |
predicted gene, 38025 |
53632 |
0.14 |
| chr5_16470797_16470948 | 0.95 |
Gm8984 |
predicted gene 8984 |
38311 |
0.17 |
| chr8_36609265_36609450 | 0.94 |
Dlc1 |
deleted in liver cancer 1 |
4517 |
0.32 |
| chr2_59726132_59726380 | 0.92 |
Tanc1 |
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 1 |
16546 |
0.26 |
| chr4_150665601_150665815 | 0.90 |
Gm16079 |
predicted gene 16079 |
13084 |
0.18 |
| chr3_57604930_57605098 | 0.89 |
Mir6377 |
microRNA 6377 |
11595 |
0.12 |
| chr14_76179509_76179660 | 0.89 |
Rps2-ps6 |
ribosomal protein S2, pseudogene 6 |
57961 |
0.11 |
| chr11_86257735_86257956 | 0.89 |
Ints2 |
integrator complex subunit 2 |
270 |
0.91 |
| chr1_46908404_46908555 | 0.89 |
Gm28527 |
predicted gene 28527 |
11600 |
0.18 |
| chr15_77410201_77410378 | 0.89 |
Apol9a |
apolipoprotein L 9a |
738 |
0.48 |
| chr11_57981089_57981255 | 0.88 |
Gm12249 |
predicted gene 12249 |
8038 |
0.16 |
| chr8_119824884_119825204 | 0.87 |
Cotl1 |
coactosin-like 1 (Dictyostelium) |
2327 |
0.27 |
| chr15_51888034_51888200 | 0.87 |
Gm34678 |
predicted gene, 34678 |
3834 |
0.19 |
| chr15_7138595_7138765 | 0.87 |
Lifr |
LIF receptor alpha |
1862 |
0.47 |
| chr18_10826881_10827044 | 0.85 |
Mib1 |
mindbomb E3 ubiquitin protein ligase 1 |
28597 |
0.11 |
| chr18_20279203_20279365 | 0.85 |
Dsg1a |
desmoglein 1 alpha |
31527 |
0.17 |
| chr5_112281749_112281914 | 0.85 |
Tpst2 |
protein-tyrosine sulfotransferase 2 |
5124 |
0.13 |
| chr10_39451218_39451393 | 0.84 |
Fyn |
Fyn proto-oncogene |
3500 |
0.26 |
| chr4_136123469_136123639 | 0.83 |
Gm26001 |
predicted gene, 26001 |
13942 |
0.13 |
| chr18_60891316_60891467 | 0.83 |
Arsi |
arylsulfatase i |
20389 |
0.13 |
| chr17_29323723_29323898 | 0.83 |
Gm46603 |
predicted gene, 46603 |
1261 |
0.28 |
| chr7_144038114_144038283 | 0.83 |
Shank2 |
SH3 and multiple ankyrin repeat domains 2 |
6934 |
0.21 |
| chr17_85052584_85052811 | 0.83 |
Slc3a1 |
solute carrier family 3, member 1 |
24321 |
0.15 |
| chr1_13236210_13236392 | 0.82 |
Gm37409 |
predicted gene, 37409 |
133 |
0.95 |
| chr10_80255545_80256082 | 0.82 |
Ndufs7 |
NADH:ubiquinone oxidoreductase core subunit S7 |
2498 |
0.11 |
| chr12_7983690_7984451 | 0.82 |
Apob |
apolipoprotein B |
6291 |
0.23 |
| chr8_109970299_109970623 | 0.81 |
Gm45795 |
predicted gene 45795 |
3891 |
0.15 |
| chr6_121880801_121881009 | 0.81 |
Mug1 |
murinoglobulin 1 |
4672 |
0.22 |
| chr17_86761972_86762144 | 0.81 |
Epas1 |
endothelial PAS domain protein 1 |
8358 |
0.18 |
| chr15_55266394_55266563 | 0.80 |
Gm26296 |
predicted gene, 26296 |
3875 |
0.27 |
| chr7_35131018_35131347 | 0.80 |
Gm35665 |
predicted gene, 35665 |
10267 |
0.1 |
| chr5_146976747_146977100 | 0.79 |
Mtif3 |
mitochondrial translational initiation factor 3 |
13123 |
0.16 |
| chr6_117276510_117276795 | 0.79 |
Rpl28-ps4 |
ribosomal protein L28, pseudogene 4 |
62586 |
0.12 |
| chr7_142622273_142622480 | 0.78 |
Gm33148 |
predicted gene, 33148 |
30632 |
0.08 |
| chr11_113076302_113076554 | 0.77 |
2610035D17Rik |
RIKEN cDNA 2610035D17 gene |
96649 |
0.08 |
| chr15_82422998_82423175 | 0.76 |
Gm19267 |
predicted gene, 19267 |
361 |
0.65 |
| chr8_70531884_70532043 | 0.75 |
Fkbp8 |
FK506 binding protein 8 |
158 |
0.89 |
| chr13_41019879_41020036 | 0.75 |
Tmem14c |
transmembrane protein 14C |
3665 |
0.15 |
| chr10_61337527_61337708 | 0.75 |
Pald1 |
phosphatase domain containing, paladin 1 |
4710 |
0.15 |
| chr15_81399706_81399886 | 0.75 |
St13 |
suppression of tumorigenicity 13 |
202 |
0.64 |
| chr3_30934508_30934659 | 0.75 |
Phc3 |
polyhomeotic 3 |
1472 |
0.36 |
| chr10_87030552_87030730 | 0.74 |
Stab2 |
stabilin 2 |
22616 |
0.14 |
| chr7_28867571_28867753 | 0.74 |
Lgals7 |
lectin, galactose binding, soluble 7 |
3207 |
0.11 |
| chr7_30968614_30968895 | 0.74 |
Lsr |
lipolysis stimulated lipoprotein receptor |
4568 |
0.08 |
| chr18_11915892_11916043 | 0.74 |
Gm49968 |
predicted gene, 49968 |
2836 |
0.3 |
| chr1_55088026_55088196 | 0.74 |
Hspe1 |
heat shock protein 1 (chaperonin 10) |
21 |
0.55 |
| chr9_40801064_40801254 | 0.74 |
Hspa8 |
heat shock protein 8 |
117 |
0.91 |
| chrX_20829007_20829192 | 0.73 |
Gm24824 |
predicted gene, 24824 |
5285 |
0.12 |
| chr5_92674843_92675048 | 0.73 |
Ccdc158 |
coiled-coil domain containing 158 |
180 |
0.94 |
| chr2_31455583_31455753 | 0.73 |
Hmcn2 |
hemicentin 2 |
960 |
0.57 |
| chr1_54006103_54006274 | 0.71 |
Gm24251 |
predicted gene, 24251 |
119247 |
0.05 |
| chr16_30827849_30828032 | 0.71 |
Gm26295 |
predicted gene, 26295 |
13778 |
0.14 |
| chr13_45891782_45891969 | 0.71 |
Atxn1 |
ataxin 1 |
19587 |
0.21 |
| chr1_164215221_164215636 | 0.70 |
Slc19a2 |
solute carrier family 19 (thiamine transporter), member 2 |
33618 |
0.11 |
| chr5_130768145_130768304 | 0.69 |
Gm23761 |
predicted gene, 23761 |
14477 |
0.25 |
| chr8_121583597_121583942 | 0.69 |
Gm17786 |
predicted gene, 17786 |
2004 |
0.19 |
| chr10_75160838_75161062 | 0.68 |
Bcr |
BCR activator of RhoGEF and GTPase |
590 |
0.78 |
| chr5_66579243_66579412 | 0.68 |
C530043K16Rik |
RIKEN cDNA C530043K16 gene |
16243 |
0.15 |
| chr4_149922172_149922323 | 0.67 |
Spsb1 |
splA/ryanodine receptor domain and SOCS box containing 1 |
224 |
0.92 |
| chr5_65351142_65351385 | 0.67 |
Klb |
klotho beta |
2855 |
0.16 |
| chr4_47151808_47151959 | 0.67 |
Gm12423 |
predicted gene 12423 |
19786 |
0.15 |
| chr1_134163496_134163749 | 0.66 |
Chil1 |
chitinase-like 1 |
18554 |
0.12 |
| chr9_66276951_66277383 | 0.64 |
Dapk2 |
death-associated protein kinase 2 |
8793 |
0.23 |
| chr7_141460493_141460686 | 0.64 |
Cracr2b |
calcium release activated channel regulator 2B |
505 |
0.5 |
| chr19_10122021_10122209 | 0.64 |
Fads2 |
fatty acid desaturase 2 |
20369 |
0.12 |
| chr7_130246644_130247139 | 0.64 |
Fgfr2 |
fibroblast growth factor receptor 2 |
14966 |
0.26 |
| chr7_28334687_28334838 | 0.63 |
Supt5 |
suppressor of Ty 5, DSIF elongation factor subunit |
3308 |
0.1 |
| chr16_23293902_23294209 | 0.63 |
St6gal1 |
beta galactoside alpha 2,6 sialyltransferase 1 |
3585 |
0.22 |
| chr11_77371279_77371455 | 0.63 |
Ssh2 |
slingshot protein phosphatase 2 |
20070 |
0.17 |
| chr13_94250295_94250470 | 0.62 |
Scamp1 |
secretory carrier membrane protein 1 |
3824 |
0.21 |
| chr9_73105883_73106055 | 0.62 |
Khdc3 |
KH domain containing 3, subcortical maternal complex member |
3459 |
0.12 |
| chr7_135254550_135254751 | 0.62 |
Nps |
neuropeptide S |
4053 |
0.24 |
| chr1_180402576_180402752 | 0.62 |
Gm36933 |
predicted gene, 36933 |
26220 |
0.12 |
| chr9_48746335_48746564 | 0.62 |
Zbtb16 |
zinc finger and BTB domain containing 16 |
89496 |
0.08 |
| chr15_82547824_82547987 | 0.61 |
Gm28023 |
predicted gene, 28023 |
747 |
0.37 |
| chr12_21183267_21183628 | 0.61 |
AC156032.1 |
|
63876 |
0.08 |
| chr16_22951195_22951399 | 0.61 |
Hrg |
histidine-rich glycoprotein |
197 |
0.91 |
| chr1_156986305_156986506 | 0.61 |
4930439D14Rik |
RIKEN cDNA 4930439D14 gene |
46567 |
0.11 |
| chr17_31401280_31401456 | 0.60 |
Pde9a |
phosphodiesterase 9A |
12788 |
0.13 |
| chr9_31145374_31145534 | 0.60 |
Aplp2 |
amyloid beta (A4) precursor-like protein 2 |
8191 |
0.17 |
| chr10_79838143_79838304 | 0.60 |
Misp |
mitotic spindle positioning |
8937 |
0.06 |
| chrX_136742828_136742985 | 0.59 |
Morf4l2 |
mortality factor 4 like 2 |
12 |
0.54 |
| chr9_54918904_54919072 | 0.59 |
Hykk |
hydroxylysine kinase 1 |
1641 |
0.29 |
| chr13_37649588_37649765 | 0.59 |
Gm40916 |
predicted gene, 40916 |
1440 |
0.29 |
| chr10_20673746_20674078 | 0.59 |
Gm17230 |
predicted gene 17230 |
48277 |
0.14 |
| chr13_23683913_23684244 | 0.59 |
H2bc4 |
H2B clustered histone 4 |
121 |
0.45 |
| chr15_78448062_78448253 | 0.59 |
Tmprss6 |
transmembrane serine protease 6 |
3778 |
0.12 |
| chr2_114775396_114775571 | 0.59 |
Gm13975 |
predicted gene 13975 |
67371 |
0.11 |
| chr11_44156189_44156382 | 0.59 |
Gm12154 |
predicted gene 12154 |
110924 |
0.06 |
| chr11_29808774_29809133 | 0.58 |
Eml6 |
echinoderm microtubule associated protein like 6 |
5854 |
0.17 |
| chr1_58237983_58238146 | 0.58 |
Aox4 |
aldehyde oxidase 4 |
2542 |
0.27 |
| chr8_11323805_11323964 | 0.58 |
Col4a1 |
collagen, type IV, alpha 1 |
11058 |
0.16 |
| chr8_117723783_117723934 | 0.58 |
Hsd17b2 |
hydroxysteroid (17-beta) dehydrogenase 2 |
2922 |
0.19 |
| chr16_34949305_34949469 | 0.58 |
Mylk |
myosin, light polypeptide kinase |
3671 |
0.21 |
| chr2_158734851_158735002 | 0.58 |
Gm26141 |
predicted gene, 26141 |
5212 |
0.19 |
| chr11_110167486_110168045 | 0.57 |
Abca9 |
ATP-binding cassette, sub-family A (ABC1), member 9 |
290 |
0.93 |
| chr2_34977060_34977295 | 0.57 |
Gm13450 |
predicted gene 13450 |
12273 |
0.11 |
| chr9_61810010_61810175 | 0.57 |
Gm19208 |
predicted gene, 19208 |
41612 |
0.15 |
| chr12_28670841_28670992 | 0.57 |
Adi1 |
acireductone dioxygenase 1 |
4315 |
0.15 |
| chr17_73131950_73132101 | 0.56 |
Lclat1 |
lysocardiolipin acyltransferase 1 |
5598 |
0.24 |
| chr8_126903217_126903593 | 0.56 |
Gm31718 |
predicted gene, 31718 |
4810 |
0.2 |
| chr2_167886467_167886618 | 0.56 |
Gm14319 |
predicted gene 14319 |
27957 |
0.14 |
| chr8_126586607_126587402 | 0.55 |
Irf2bp2 |
interferon regulatory factor 2 binding protein 2 |
6982 |
0.23 |
| chr13_107841338_107841500 | 0.55 |
Gm22118 |
predicted gene, 22118 |
12139 |
0.24 |
| chr4_46913567_46913730 | 0.55 |
Gabbr2 |
gamma-aminobutyric acid (GABA) B receptor, 2 |
53746 |
0.13 |
| chr3_153930744_153930906 | 0.54 |
Acadm |
acyl-Coenzyme A dehydrogenase, medium chain |
683 |
0.51 |
| chr10_7142433_7142596 | 0.54 |
Cnksr3 |
Cnksr family member 3 |
7008 |
0.29 |
| chr17_27969528_27969712 | 0.54 |
Gm15597 |
predicted gene 15597 |
4304 |
0.14 |
| chr9_85568984_85569135 | 0.53 |
Gm48834 |
predicted gene, 48834 |
15294 |
0.22 |
| chr6_94729670_94729855 | 0.53 |
Lrig1 |
leucine-rich repeats and immunoglobulin-like domains 1 |
29604 |
0.17 |
| chr3_53372078_53372254 | 0.53 |
Gm43472 |
predicted gene 43472 |
24937 |
0.15 |
| chr3_57428830_57429005 | 0.52 |
Tm4sf4 |
transmembrane 4 superfamily member 4 |
3603 |
0.27 |
| chr5_109557797_109558252 | 0.52 |
Crlf2 |
cytokine receptor-like factor 2 |
912 |
0.5 |
| chr7_145047841_145048023 | 0.52 |
Ccnd1 |
cyclin D1 |
108007 |
0.05 |
| chr6_145360771_145361132 | 0.52 |
Gm23498 |
predicted gene, 23498 |
5907 |
0.15 |
| chr19_7040189_7040358 | 0.52 |
Stip1 |
stress-induced phosphoprotein 1 |
306 |
0.79 |
| chr7_71232911_71233088 | 0.52 |
6030442E23Rik |
RIKEN cDNA 6030442E23 gene |
15950 |
0.2 |
| chr16_38618378_38618532 | 0.52 |
Tmem39a |
transmembrane protein 39a |
30416 |
0.13 |
| chrX_73715619_73715775 | 0.52 |
Bcap31 |
B cell receptor associated protein 31 |
478 |
0.57 |
| chr5_151312471_151312634 | 0.51 |
Gm7041 |
predicted gene 7041 |
31429 |
0.15 |
| chr19_7039938_7040112 | 0.51 |
Stip1 |
stress-induced phosphoprotein 1 |
58 |
0.94 |
| chr18_84854418_84854822 | 0.51 |
Cyb5a |
cytochrome b5 type A (microsomal) |
349 |
0.85 |
| chr18_46636342_46636847 | 0.51 |
Gm3734 |
predicted gene 3734 |
5787 |
0.17 |
| chr6_129097928_129098079 | 0.50 |
Clec2e |
C-type lectin domain family 2, member e |
2910 |
0.16 |
| chr6_72279240_72279429 | 0.50 |
Sftpb |
surfactant associated protein B |
25276 |
0.11 |
| chr7_141327534_141327875 | 0.50 |
Deaf1 |
DEAF1, transcription factor |
14 |
0.5 |
| chr10_80826930_80827098 | 0.50 |
Oaz1 |
ornithine decarboxylase antizyme 1 |
321 |
0.7 |
| chr8_115823982_115824181 | 0.50 |
Maf |
avian musculoaponeurotic fibrosarcoma oncogene homolog |
116287 |
0.06 |
| chr2_6129018_6129180 | 0.50 |
Proser2 |
proline and serine rich 2 |
1040 |
0.45 |
| chr2_36243894_36244091 | 0.50 |
Ptgs1 |
prostaglandin-endoperoxide synthase 1 |
1517 |
0.27 |
| chr7_19793257_19793455 | 0.50 |
Cblc |
Casitas B-lineage lymphoma c |
140 |
0.89 |
| chr7_123192367_123192758 | 0.50 |
Tnrc6a |
trinucleotide repeat containing 6a |
12641 |
0.2 |
| chr19_30207260_30207548 | 0.49 |
Mbl2 |
mannose-binding lectin (protein C) 2 |
25538 |
0.16 |
| chr1_88282912_88283223 | 0.49 |
Trpm8 |
transient receptor potential cation channel, subfamily M, member 8 |
5406 |
0.12 |
| chr9_56418511_56418671 | 0.49 |
Hmg20a |
high mobility group 20A |
18 |
0.9 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.2 | GO:0061083 | regulation of protein refolding(GO:0061083) |
| 0.2 | 0.6 | GO:0034395 | regulation of transcription from RNA polymerase II promoter in response to iron(GO:0034395) |
| 0.2 | 0.5 | GO:1902564 | negative regulation of neutrophil activation(GO:1902564) |
| 0.2 | 0.5 | GO:1903416 | response to glycoside(GO:1903416) |
| 0.1 | 0.5 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.1 | 0.4 | GO:1900104 | hyaluranon cable assembly(GO:0036118) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 0.1 | 0.3 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.1 | 0.5 | GO:1903626 | positive regulation of apoptotic DNA fragmentation(GO:1902512) positive regulation of DNA catabolic process(GO:1903626) |
| 0.1 | 0.3 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.1 | 0.4 | GO:0035799 | ureter maturation(GO:0035799) |
| 0.1 | 0.3 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.1 | 0.3 | GO:1900095 | regulation of dosage compensation by inactivation of X chromosome(GO:1900095) |
| 0.1 | 0.4 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.1 | 0.3 | GO:0019448 | cysteine catabolic process(GO:0009093) L-cysteine catabolic process(GO:0019448) L-cysteine metabolic process(GO:0046439) |
| 0.1 | 0.3 | GO:0044829 | positive regulation by host of viral genome replication(GO:0044829) |
| 0.1 | 0.4 | GO:0071865 | regulation of apoptotic process in bone marrow(GO:0071865) negative regulation of apoptotic process in bone marrow(GO:0071866) |
| 0.1 | 1.8 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.1 | 0.2 | GO:0006578 | amino-acid betaine biosynthetic process(GO:0006578) |
| 0.1 | 0.3 | GO:0003433 | chondrocyte development involved in endochondral bone morphogenesis(GO:0003433) |
| 0.1 | 0.4 | GO:0016078 | tRNA catabolic process(GO:0016078) |
| 0.1 | 0.3 | GO:0090206 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.1 | 0.2 | GO:0032776 | DNA methylation on cytosine within a CG sequence(GO:0010424) DNA methylation on cytosine(GO:0032776) |
| 0.1 | 0.2 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.1 | 0.2 | GO:0000711 | meiotic DNA repair synthesis(GO:0000711) |
| 0.1 | 0.2 | GO:0007161 | calcium-independent cell-matrix adhesion(GO:0007161) |
| 0.1 | 0.2 | GO:0007354 | zygotic determination of anterior/posterior axis, embryo(GO:0007354) |
| 0.1 | 0.3 | GO:0044791 | modulation by host of viral release from host cell(GO:0044789) positive regulation by host of viral release from host cell(GO:0044791) |
| 0.1 | 0.5 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.1 | 0.6 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.1 | 0.3 | GO:0039530 | MDA-5 signaling pathway(GO:0039530) |
| 0.1 | 0.2 | GO:0042524 | negative regulation of tyrosine phosphorylation of Stat5 protein(GO:0042524) |
| 0.1 | 0.3 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.1 | 0.4 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.1 | 0.2 | GO:0061643 | chemorepulsion of axon(GO:0061643) |
| 0.1 | 0.1 | GO:0032672 | regulation of interleukin-3 production(GO:0032672) |
| 0.1 | 0.4 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.1 | 0.2 | GO:2001268 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001268) |
| 0.1 | 0.2 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.1 | 0.3 | GO:1902774 | late endosome to lysosome transport(GO:1902774) |
| 0.1 | 0.3 | GO:0009115 | xanthine catabolic process(GO:0009115) |
| 0.1 | 0.8 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.0 | 0.1 | GO:0015772 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.0 | 0.2 | GO:0065001 | specification of axis polarity(GO:0065001) |
| 0.0 | 0.1 | GO:0010255 | carbohydrate mediated signaling(GO:0009756) hexose mediated signaling(GO:0009757) sugar mediated signaling pathway(GO:0010182) glucose mediated signaling pathway(GO:0010255) |
| 0.0 | 0.3 | GO:0034154 | toll-like receptor 7 signaling pathway(GO:0034154) |
| 0.0 | 0.1 | GO:0060737 | prostate gland morphogenetic growth(GO:0060737) |
| 0.0 | 0.2 | GO:0097278 | complement-dependent cytotoxicity(GO:0097278) |
| 0.0 | 0.1 | GO:0097503 | sialylation(GO:0097503) |
| 0.0 | 0.1 | GO:2000646 | positive regulation of receptor catabolic process(GO:2000646) |
| 0.0 | 0.1 | GO:0030997 | regulation of centriole-centriole cohesion(GO:0030997) |
| 0.0 | 0.3 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) |
| 0.0 | 0.1 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.0 | 0.2 | GO:0044004 | killing by symbiont of host cells(GO:0001907) disruption by symbiont of host cell(GO:0044004) |
| 0.0 | 0.1 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.0 | 0.6 | GO:0050872 | white fat cell differentiation(GO:0050872) |
| 0.0 | 0.2 | GO:0052428 | negative regulation by host of symbiont molecular function(GO:0052405) modification by host of symbiont molecular function(GO:0052428) |
| 0.0 | 0.1 | GO:1901740 | negative regulation of myoblast fusion(GO:1901740) |
| 0.0 | 0.5 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.0 | 0.1 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.0 | 0.1 | GO:0060750 | epithelial cell proliferation involved in mammary gland duct elongation(GO:0060750) |
| 0.0 | 0.2 | GO:0010841 | positive regulation of circadian sleep/wake cycle, wakefulness(GO:0010841) |
| 0.0 | 0.1 | GO:2000504 | positive regulation of blood vessel remodeling(GO:2000504) |
| 0.0 | 0.3 | GO:0035745 | T-helper 2 cell cytokine production(GO:0035745) |
| 0.0 | 0.1 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.0 | 0.1 | GO:0010911 | regulation of isomerase activity(GO:0010911) positive regulation of isomerase activity(GO:0010912) |
| 0.0 | 0.0 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.0 | 0.2 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 0.3 | GO:0055091 | phospholipid homeostasis(GO:0055091) |
| 0.0 | 0.1 | GO:0060332 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.0 | 0.1 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 0.0 | 0.2 | GO:0034374 | low-density lipoprotein particle remodeling(GO:0034374) |
| 0.0 | 0.1 | GO:2000418 | positive regulation of eosinophil migration(GO:2000418) |
| 0.0 | 0.1 | GO:0043134 | hindgut contraction(GO:0043133) regulation of hindgut contraction(GO:0043134) |
| 0.0 | 0.1 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.0 | 0.2 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.0 | 0.1 | GO:0000480 | endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.0 | 0.1 | GO:0015888 | thiamine transport(GO:0015888) |
| 0.0 | 0.1 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.0 | 0.1 | GO:0060672 | epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 0.0 | 0.1 | GO:0003383 | apical constriction(GO:0003383) |
| 0.0 | 0.1 | GO:0021699 | cerebellar cortex maturation(GO:0021699) |
| 0.0 | 0.1 | GO:0000491 | small nucleolar ribonucleoprotein complex assembly(GO:0000491) box C/D snoRNP assembly(GO:0000492) |
| 0.0 | 0.1 | GO:1902075 | cellular response to salt(GO:1902075) |
| 0.0 | 0.2 | GO:0002002 | regulation of angiotensin levels in blood(GO:0002002) |
| 0.0 | 0.3 | GO:0001771 | immunological synapse formation(GO:0001771) |
| 0.0 | 0.1 | GO:0051031 | tRNA transport(GO:0051031) |
| 0.0 | 0.1 | GO:0051665 | membrane raft localization(GO:0051665) |
| 0.0 | 0.1 | GO:1903336 | negative regulation of vacuolar transport(GO:1903336) |
| 0.0 | 0.1 | GO:0009233 | menaquinone metabolic process(GO:0009233) |
| 0.0 | 0.2 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.0 | 0.2 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 | 0.1 | GO:0042758 | long-chain fatty acid catabolic process(GO:0042758) |
| 0.0 | 0.1 | GO:0098902 | regulation of membrane depolarization during action potential(GO:0098902) |
| 0.0 | 0.3 | GO:0042523 | regulation of tyrosine phosphorylation of Stat5 protein(GO:0042522) positive regulation of tyrosine phosphorylation of Stat5 protein(GO:0042523) |
| 0.0 | 0.3 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.0 | 0.7 | GO:0021904 | dorsal/ventral neural tube patterning(GO:0021904) |
| 0.0 | 0.1 | GO:1901164 | negative regulation of trophoblast cell migration(GO:1901164) |
| 0.0 | 0.1 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.0 | 0.1 | GO:0060266 | negative regulation of respiratory burst involved in inflammatory response(GO:0060266) |
| 0.0 | 0.3 | GO:0001553 | luteinization(GO:0001553) |
| 0.0 | 0.1 | GO:0071033 | nuclear retention of pre-mRNA at the site of transcription(GO:0071033) |
| 0.0 | 0.1 | GO:0033031 | positive regulation of neutrophil apoptotic process(GO:0033031) |
| 0.0 | 0.0 | GO:0000430 | regulation of transcription from RNA polymerase II promoter by glucose(GO:0000430) positive regulation of transcription from RNA polymerase II promoter by glucose(GO:0000432) |
| 0.0 | 0.1 | GO:0002017 | regulation of blood volume by renal aldosterone(GO:0002017) |
| 0.0 | 0.2 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.0 | 0.2 | GO:0002934 | desmosome organization(GO:0002934) |
| 0.0 | 0.1 | GO:0006546 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.0 | 0.1 | GO:0016561 | protein import into peroxisome matrix, translocation(GO:0016561) |
| 0.0 | 0.1 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.1 | GO:0070366 | regulation of hepatocyte differentiation(GO:0070366) |
| 0.0 | 0.1 | GO:0034755 | iron ion transmembrane transport(GO:0034755) |
| 0.0 | 0.2 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.0 | 0.1 | GO:1903288 | regulation of potassium ion import(GO:1903286) positive regulation of potassium ion import(GO:1903288) |
| 0.0 | 0.2 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.1 | GO:1902414 | protein localization to cell junction(GO:1902414) |
| 0.0 | 0.1 | GO:0061030 | epithelial cell differentiation involved in mammary gland alveolus development(GO:0061030) |
| 0.0 | 0.1 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.1 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 0.0 | 0.1 | GO:0006768 | biotin metabolic process(GO:0006768) |
| 0.0 | 0.1 | GO:0072675 | osteoclast fusion(GO:0072675) |
| 0.0 | 0.1 | GO:0090080 | positive regulation of MAPKKK cascade by fibroblast growth factor receptor signaling pathway(GO:0090080) |
| 0.0 | 0.3 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.0 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.0 | 0.1 | GO:1902947 | regulation of tau-protein kinase activity(GO:1902947) |
| 0.0 | 0.0 | GO:0030397 | membrane disassembly(GO:0030397) nuclear envelope disassembly(GO:0051081) |
| 0.0 | 0.1 | GO:0021873 | forebrain neuroblast division(GO:0021873) |
| 0.0 | 0.1 | GO:0006548 | histidine catabolic process(GO:0006548) imidazole-containing compound catabolic process(GO:0052805) |
| 0.0 | 0.1 | GO:0008354 | germ cell migration(GO:0008354) |
| 0.0 | 0.1 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.0 | 0.1 | GO:0070669 | response to interleukin-2(GO:0070669) |
| 0.0 | 0.1 | GO:0035814 | negative regulation of renal sodium excretion(GO:0035814) |
| 0.0 | 0.0 | GO:1904180 | negative regulation of membrane depolarization(GO:1904180) |
| 0.0 | 0.2 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.0 | 0.1 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.0 | 0.1 | GO:0043504 | mitochondrial DNA repair(GO:0043504) |
| 0.0 | 0.1 | GO:1902309 | negative regulation of peptidyl-serine dephosphorylation(GO:1902309) |
| 0.0 | 0.2 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.0 | 0.2 | GO:0051895 | negative regulation of focal adhesion assembly(GO:0051895) |
| 0.0 | 0.1 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.0 | 0.1 | GO:0061052 | negative regulation of cell growth involved in cardiac muscle cell development(GO:0061052) |
| 0.0 | 0.0 | GO:0019859 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) thymine metabolic process(GO:0019859) |
| 0.0 | 0.1 | GO:1990086 | lens fiber cell apoptotic process(GO:1990086) |
| 0.0 | 0.1 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.0 | 0.2 | GO:0071625 | vocalization behavior(GO:0071625) |
| 0.0 | 0.1 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.0 | 0.0 | GO:0097029 | mature conventional dendritic cell differentiation(GO:0097029) |
| 0.0 | 0.1 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 0.1 | GO:0002525 | acute inflammatory response to non-antigenic stimulus(GO:0002525) |
| 0.0 | 0.1 | GO:0045658 | regulation of neutrophil differentiation(GO:0045658) |
| 0.0 | 0.1 | GO:0010046 | response to mycotoxin(GO:0010046) |
| 0.0 | 0.1 | GO:0015691 | cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.0 | 0.1 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.0 | 0.1 | GO:0042790 | transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:0042790) |
| 0.0 | 0.0 | GO:0060462 | lung lobe development(GO:0060462) lung lobe morphogenesis(GO:0060463) |
| 0.0 | 0.0 | GO:0055099 | response to high density lipoprotein particle(GO:0055099) |
| 0.0 | 0.1 | GO:0032485 | regulation of Ral protein signal transduction(GO:0032485) |
| 0.0 | 0.3 | GO:0007190 | activation of adenylate cyclase activity(GO:0007190) |
| 0.0 | 0.0 | GO:0086073 | bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 0.0 | 0.1 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.0 | 0.1 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.0 | 0.1 | GO:1904526 | regulation of microtubule binding(GO:1904526) positive regulation of microtubule binding(GO:1904528) |
| 0.0 | 0.4 | GO:0010762 | regulation of fibroblast migration(GO:0010762) |
| 0.0 | 0.0 | GO:0032988 | ribonucleoprotein complex disassembly(GO:0032988) |
| 0.0 | 0.1 | GO:0072307 | metanephric nephron tubule epithelial cell differentiation(GO:0072257) regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072307) |
| 0.0 | 0.0 | GO:2000611 | positive regulation of thyroid hormone generation(GO:2000611) |
| 0.0 | 0.0 | GO:1903275 | positive regulation of sodium ion export(GO:1903275) positive regulation of sodium ion export from cell(GO:1903278) |
| 0.0 | 0.0 | GO:0002432 | granuloma formation(GO:0002432) |
| 0.0 | 0.0 | GO:0035754 | B cell chemotaxis(GO:0035754) |
| 0.0 | 0.2 | GO:0007097 | nuclear migration(GO:0007097) |
| 0.0 | 0.0 | GO:0009629 | response to gravity(GO:0009629) |
| 0.0 | 0.0 | GO:0033029 | regulation of neutrophil apoptotic process(GO:0033029) negative regulation of neutrophil apoptotic process(GO:0033030) |
| 0.0 | 0.0 | GO:0023021 | termination of signal transduction(GO:0023021) |
| 0.0 | 0.0 | GO:0002331 | pre-B cell allelic exclusion(GO:0002331) |
| 0.0 | 0.0 | GO:0070814 | hydrogen sulfide biosynthetic process(GO:0070814) |
| 0.0 | 0.2 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.0 | 0.0 | GO:0009106 | lipoate metabolic process(GO:0009106) |
| 0.0 | 0.2 | GO:0006030 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.0 | 0.0 | GO:1905216 | positive regulation of mRNA binding(GO:1902416) positive regulation of RNA binding(GO:1905216) |
| 0.0 | 0.1 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.0 | 0.0 | GO:0030382 | sperm mitochondrion organization(GO:0030382) |
| 0.0 | 0.0 | GO:0008355 | olfactory learning(GO:0008355) |
| 0.0 | 0.0 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 0.0 | 0.1 | GO:0072386 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.0 | 0.0 | GO:0042427 | serotonin biosynthetic process(GO:0042427) primary amino compound biosynthetic process(GO:1901162) |
| 0.0 | 0.0 | GO:1901843 | positive regulation of high voltage-gated calcium channel activity(GO:1901843) |
| 0.0 | 0.1 | GO:0010992 | ubiquitin homeostasis(GO:0010992) |
| 0.0 | 0.2 | GO:0050832 | defense response to fungus(GO:0050832) |
| 0.0 | 0.2 | GO:0003094 | glomerular filtration(GO:0003094) |
| 0.0 | 0.1 | GO:0046813 | receptor-mediated virion attachment to host cell(GO:0046813) |
| 0.0 | 0.1 | GO:0008054 | negative regulation of cyclin-dependent protein serine/threonine kinase by cyclin degradation(GO:0008054) |
| 0.0 | 0.1 | GO:2000270 | negative regulation of fibroblast apoptotic process(GO:2000270) |
| 0.0 | 0.2 | GO:0044065 | regulation of respiratory system process(GO:0044065) |
| 0.0 | 0.1 | GO:0050955 | thermoception(GO:0050955) |
| 0.0 | 0.0 | GO:0071503 | response to heparin(GO:0071503) cellular response to heparin(GO:0071504) |
| 0.0 | 0.2 | GO:2000651 | positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.0 | 0.1 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.0 | 0.2 | GO:0051818 | disruption of cells of other organism involved in symbiotic interaction(GO:0051818) killing of cells in other organism involved in symbiotic interaction(GO:0051883) |
| 0.0 | 0.0 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.0 | 0.1 | GO:0007176 | regulation of epidermal growth factor-activated receptor activity(GO:0007176) |
| 0.0 | 0.0 | GO:1990791 | dorsal root ganglion development(GO:1990791) |
| 0.0 | 0.0 | GO:0072223 | metanephric glomerular mesangium development(GO:0072223) metanephric glomerular mesangial cell proliferation involved in metanephros development(GO:0072262) |
| 0.0 | 0.1 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.0 | 0.0 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.0 | 0.0 | GO:0070671 | response to interleukin-12(GO:0070671) |
| 0.0 | 0.1 | GO:0034080 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.0 | 0.0 | GO:1900378 | positive regulation of melanin biosynthetic process(GO:0048023) positive regulation of secondary metabolite biosynthetic process(GO:1900378) |
| 0.0 | 0.1 | GO:0033133 | positive regulation of glucokinase activity(GO:0033133) |
| 0.0 | 0.0 | GO:2001180 | negative regulation of interleukin-10 secretion(GO:2001180) |
| 0.0 | 0.1 | GO:0046689 | response to mercury ion(GO:0046689) |
| 0.0 | 0.0 | GO:0009051 | pentose-phosphate shunt, oxidative branch(GO:0009051) |
| 0.0 | 0.0 | GO:1902307 | positive regulation of sodium ion transmembrane transport(GO:1902307) |
| 0.0 | 0.1 | GO:0046501 | protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.0 | 0.2 | GO:0045780 | positive regulation of bone resorption(GO:0045780) positive regulation of bone remodeling(GO:0046852) |
| 0.0 | 0.0 | GO:0060830 | ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) |
| 0.0 | 0.0 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 0.0 | 0.2 | GO:0050732 | negative regulation of peptidyl-tyrosine phosphorylation(GO:0050732) |
| 0.0 | 0.0 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.0 | 0.0 | GO:0006172 | ADP biosynthetic process(GO:0006172) |
| 0.0 | 0.0 | GO:0070314 | G1 to G0 transition(GO:0070314) |
| 0.0 | 0.0 | GO:0014858 | positive regulation of skeletal muscle cell proliferation(GO:0014858) |
| 0.0 | 0.0 | GO:2000653 | regulation of genetic imprinting(GO:2000653) |
| 0.0 | 0.3 | GO:0031440 | regulation of mRNA 3'-end processing(GO:0031440) |
| 0.0 | 0.1 | GO:0033148 | positive regulation of intracellular estrogen receptor signaling pathway(GO:0033148) |
| 0.0 | 0.0 | GO:0051365 | cellular response to potassium ion starvation(GO:0051365) |
| 0.0 | 0.1 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 0.0 | GO:0014717 | regulation of satellite cell activation involved in skeletal muscle regeneration(GO:0014717) satellite cell activation involved in skeletal muscle regeneration(GO:0014901) |
| 0.0 | 0.5 | GO:0007566 | embryo implantation(GO:0007566) |
| 0.0 | 0.1 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.1 | GO:0061050 | regulation of cell growth involved in cardiac muscle cell development(GO:0061050) |
| 0.0 | 0.0 | GO:0018171 | peptidyl-cysteine oxidation(GO:0018171) |
| 0.0 | 0.0 | GO:2000609 | regulation of thyroid hormone generation(GO:2000609) |
| 0.0 | 0.0 | GO:0036265 | RNA (guanine-N7)-methylation(GO:0036265) |
| 0.0 | 0.1 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 0.0 | GO:0010957 | negative regulation of vitamin D biosynthetic process(GO:0010957) |
| 0.0 | 0.1 | GO:1902510 | regulation of apoptotic DNA fragmentation(GO:1902510) |
| 0.0 | 0.1 | GO:2000288 | positive regulation of myoblast proliferation(GO:2000288) |
| 0.0 | 0.0 | GO:0060214 | endocardium formation(GO:0060214) |
| 0.0 | 0.1 | GO:0045974 | negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.0 | 0.1 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.0 | 0.0 | GO:0006203 | dGTP catabolic process(GO:0006203) |
| 0.0 | 0.0 | GO:0051133 | regulation of NK T cell activation(GO:0051133) positive regulation of NK T cell activation(GO:0051135) |
| 0.0 | 0.0 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.0 | 0.0 | GO:0046104 | thymidine metabolic process(GO:0046104) |
| 0.0 | 0.0 | GO:0035188 | blastocyst hatching(GO:0001835) hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.0 | 0.1 | GO:1901727 | positive regulation of histone deacetylase activity(GO:1901727) |
| 0.0 | 0.0 | GO:0051799 | negative regulation of hair follicle development(GO:0051799) |
| 0.0 | 0.0 | GO:0018197 | peptidyl-aspartic acid modification(GO:0018197) |
| 0.0 | 0.0 | GO:0045578 | negative regulation of B cell differentiation(GO:0045578) |
| 0.0 | 0.1 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.0 | 0.3 | GO:0006284 | base-excision repair(GO:0006284) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0032010 | phagolysosome(GO:0032010) |
| 0.1 | 0.4 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.1 | 0.5 | GO:0036454 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) insulin-like growth factor ternary complex(GO:0042567) |
| 0.1 | 0.7 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.1 | 0.3 | GO:0097629 | extrinsic component of omegasome membrane(GO:0097629) |
| 0.1 | 0.3 | GO:0005784 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.1 | 0.8 | GO:0032039 | integrator complex(GO:0032039) |
| 0.1 | 0.3 | GO:0044194 | cytolytic granule(GO:0044194) |
| 0.1 | 0.4 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.1 | 0.2 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.1 | 0.3 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.1 | 0.2 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.0 | 0.2 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.0 | 0.2 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.0 | 0.1 | GO:0036488 | CHOP-C/EBP complex(GO:0036488) |
| 0.0 | 0.2 | GO:0005587 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.0 | 0.1 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.0 | 0.3 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.0 | 0.1 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.0 | 0.2 | GO:0031415 | NatA complex(GO:0031415) |
| 0.0 | 0.2 | GO:0000801 | central element(GO:0000801) |
| 0.0 | 0.2 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.1 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.0 | 0.1 | GO:1990761 | growth cone lamellipodium(GO:1990761) |
| 0.0 | 0.4 | GO:0038201 | TOR complex(GO:0038201) |
| 0.0 | 0.1 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.2 | GO:0005883 | neurofilament(GO:0005883) |
| 0.0 | 0.1 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.1 | GO:0042585 | germinal vesicle(GO:0042585) |
| 0.0 | 0.1 | GO:0071008 | U2-type post-mRNA release spliceosomal complex(GO:0071008) |
| 0.0 | 0.1 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 0.6 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.1 | GO:0038037 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor complex(GO:0097648) |
| 0.0 | 0.1 | GO:0097543 | ciliary inversin compartment(GO:0097543) |
| 0.0 | 0.1 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.0 | 0.1 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 0.1 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.0 | 0.1 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.0 | 0.2 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.3 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) |
| 0.0 | 0.1 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.0 | 0.1 | GO:1990597 | AIP1-IRE1 complex(GO:1990597) |
| 0.0 | 0.2 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.0 | 0.1 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.0 | 0.1 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 0.2 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.1 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.0 | 0.1 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.0 | 0.2 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.1 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.0 | 0.1 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.3 | GO:1902562 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.0 | 0.0 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.0 | 0.7 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 0.1 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.1 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.1 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.0 | 0.1 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 0.1 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.0 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 0.1 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.0 | 0.1 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 0.0 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.0 | 0.0 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.0 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 0.0 | 0.0 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 0.0 | 0.0 | GO:0005767 | secondary lysosome(GO:0005767) |
| 0.0 | 0.1 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.1 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.1 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.1 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.2 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.2 | 0.7 | GO:0031686 | A1 adenosine receptor binding(GO:0031686) |
| 0.2 | 0.5 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.1 | 0.4 | GO:0000171 | ribonuclease MRP activity(GO:0000171) |
| 0.1 | 0.6 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) |
| 0.1 | 0.3 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.1 | 0.4 | GO:0032564 | dATP binding(GO:0032564) |
| 0.1 | 0.4 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.1 | 0.2 | GO:0051718 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) DNA (cytosine-5-)-methyltransferase activity, acting on CpG substrates(GO:0051718) |
| 0.1 | 0.2 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 0.1 | 0.2 | GO:0047276 | N-acetyllactosaminide 3-alpha-galactosyltransferase activity(GO:0047276) |
| 0.1 | 0.4 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.1 | 0.2 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.1 | 0.4 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) |
| 0.1 | 0.2 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 0.1 | 0.3 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.1 | 0.3 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.1 | 0.3 | GO:0050897 | cobalt ion binding(GO:0050897) |
| 0.1 | 0.4 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.1 | 0.3 | GO:0016726 | xanthine dehydrogenase activity(GO:0004854) oxidoreductase activity, acting on CH or CH2 groups, NAD or NADP as acceptor(GO:0016726) |
| 0.0 | 0.1 | GO:0015157 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.0 | 0.2 | GO:0045340 | mercury ion binding(GO:0045340) |
| 0.0 | 0.2 | GO:0009374 | biotin binding(GO:0009374) |
| 0.0 | 0.1 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.2 | GO:0043559 | insulin binding(GO:0043559) |
| 0.0 | 0.3 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.0 | 0.2 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.0 | 0.4 | GO:0070635 | single-strand selective uracil DNA N-glycosylase activity(GO:0017065) nicotinamide riboside hydrolase activity(GO:0070635) nicotinic acid riboside hydrolase activity(GO:0070636) deoxyribonucleoside 5'-monophosphate N-glycosidase activity(GO:0070694) |
| 0.0 | 0.1 | GO:0030943 | mitochondrion targeting sequence binding(GO:0030943) |
| 0.0 | 0.2 | GO:0016401 | palmitoyl-CoA oxidase activity(GO:0016401) |
| 0.0 | 0.2 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 0.2 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 0.2 | GO:0070991 | medium-chain-acyl-CoA dehydrogenase activity(GO:0070991) |
| 0.0 | 0.1 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.0 | 0.2 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.0 | 0.2 | GO:0035473 | lipase binding(GO:0035473) |
| 0.0 | 0.2 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.1 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.0 | 0.0 | GO:0030620 | U2 snRNA binding(GO:0030620) |
| 0.0 | 0.6 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.3 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.0 | 0.1 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 0.1 | GO:0047045 | testosterone 17-beta-dehydrogenase (NADP+) activity(GO:0047045) |
| 0.0 | 0.1 | GO:0015563 | uptake transmembrane transporter activity(GO:0015563) |
| 0.0 | 0.1 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.0 | 0.2 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.0 | 0.1 | GO:0019211 | phosphatase activator activity(GO:0019211) |
| 0.0 | 0.1 | GO:0048045 | 4-hydroxybenzoate octaprenyltransferase activity(GO:0008412) protoheme IX farnesyltransferase activity(GO:0008495) (S)-2,3-di-O-geranylgeranylglyceryl phosphate synthase activity(GO:0043888) cadaverine aminopropyltransferase activity(GO:0043918) agmatine aminopropyltransferase activity(GO:0043919) 1,4-dihydroxy-2-naphthoate octaprenyltransferase activity(GO:0046428) trans-pentaprenyltranstransferase activity(GO:0048045) ATP dimethylallyltransferase activity(GO:0052622) ADP dimethylallyltransferase activity(GO:0052623) |
| 0.0 | 0.1 | GO:0080130 | L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.0 | 0.3 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 0.1 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 0.1 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.0 | 0.1 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.0 | 0.1 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.3 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
| 0.0 | 0.0 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.0 | 0.3 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.1 | GO:0008309 | double-stranded DNA exodeoxyribonuclease activity(GO:0008309) |
| 0.0 | 0.1 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 0.2 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.2 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.0 | 0.2 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.1 | GO:0030350 | iron-responsive element binding(GO:0030350) |
| 0.0 | 0.1 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.0 | 0.1 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.0 | 0.2 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.0 | 0.4 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.4 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.1 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.0 | 0.3 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.0 | 0.1 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.0 | 0.1 | GO:0016728 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.0 | 0.6 | GO:0042805 | actinin binding(GO:0042805) |
| 0.0 | 1.3 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 0.1 | GO:0102345 | 3-hydroxy-behenoyl-CoA dehydratase activity(GO:0102344) 3-hydroxy-lignoceroyl-CoA dehydratase activity(GO:0102345) |
| 0.0 | 0.1 | GO:0034902 | alkyl sulfatase activity(GO:0018741) endosulfan hemisulfate sulfatase activity(GO:0034889) endosulfan sulfate hydrolase activity(GO:0034902) |
| 0.0 | 0.1 | GO:0016892 | endoribonuclease activity, producing 3'-phosphomonoesters(GO:0016892) |
| 0.0 | 0.2 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 0.0 | GO:0030984 | kininogen binding(GO:0030984) |
| 0.0 | 0.1 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.5 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.1 | GO:0070698 | type I activin receptor binding(GO:0070698) |
| 0.0 | 0.2 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.1 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.0 | 0.0 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.0 | 0.2 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 0.3 | GO:0022829 | wide pore channel activity(GO:0022829) |
| 0.0 | 0.1 | GO:0004030 | aldehyde dehydrogenase [NAD(P)+] activity(GO:0004030) |
| 0.0 | 0.1 | GO:0008297 | single-stranded DNA exodeoxyribonuclease activity(GO:0008297) |
| 0.0 | 0.0 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.0 | 0.1 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.0 | 0.4 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.0 | GO:0004946 | bombesin receptor activity(GO:0004946) |
| 0.0 | 0.2 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 0.1 | GO:0001013 | RNA polymerase I regulatory region DNA binding(GO:0001013) |
| 0.0 | 0.1 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.0 | 0.1 | GO:0004792 | thiosulfate sulfurtransferase activity(GO:0004792) |
| 0.0 | 0.1 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.0 | 0.2 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.0 | 0.0 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.0 | 0.0 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.0 | 0.3 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 0.1 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.0 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.0 | 0.1 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.0 | 0.3 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.1 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 0.1 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.2 | GO:0008198 | ferrous iron binding(GO:0008198) |
| 0.0 | 0.0 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.0 | 0.0 | GO:0071208 | histone pre-mRNA DCP binding(GO:0071208) |
| 0.0 | 0.0 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.0 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.0 | GO:0034739 | histone deacetylase activity (H4-K16 specific)(GO:0034739) |
| 0.0 | 0.6 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 0.4 | GO:0004177 | aminopeptidase activity(GO:0004177) |
| 0.0 | 0.0 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.0 | 0.2 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.0 | 0.1 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.0 | 0.1 | GO:0016780 | phosphotransferase activity, for other substituted phosphate groups(GO:0016780) |
| 0.0 | 0.1 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.0 | 0.1 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.1 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.0 | 0.2 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.0 | 0.1 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.1 | GO:0008599 | protein phosphatase type 1 regulator activity(GO:0008599) |
| 0.0 | 0.1 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.0 | 0.2 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.0 | 0.1 | GO:0035174 | histone serine kinase activity(GO:0035174) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.1 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.4 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.4 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.3 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 0.5 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 0.5 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 0.3 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 0.1 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 0.1 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.3 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.1 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.0 | 0.0 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.1 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.0 | 0.7 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.5 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.1 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
| 0.0 | 0.5 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 0.2 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.0 | 0.2 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.3 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.1 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
| 0.0 | 0.3 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.2 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 0.2 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.0 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.3 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 0.6 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.1 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.0 | 0.7 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 0.6 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.7 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.3 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.5 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.3 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 0.4 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.3 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.0 | 0.4 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 0.4 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.0 | 0.2 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 1.1 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.0 | REACTOME CTLA4 INHIBITORY SIGNALING | Genes involved in CTLA4 inhibitory signaling |
| 0.0 | 0.4 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.1 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.1 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.0 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.0 | 0.3 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 0.1 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.1 | REACTOME P75NTR RECRUITS SIGNALLING COMPLEXES | Genes involved in p75NTR recruits signalling complexes |
| 0.0 | 0.5 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 0.1 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.1 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.2 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 0.2 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.1 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.3 | REACTOME FGFR LIGAND BINDING AND ACTIVATION | Genes involved in FGFR ligand binding and activation |
| 0.0 | 0.1 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.1 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |
| 0.0 | 0.9 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |
| 0.0 | 0.3 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 0.3 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 0.1 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.1 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.7 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.1 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.5 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.1 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.1 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.5 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.0 | 0.0 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |