Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
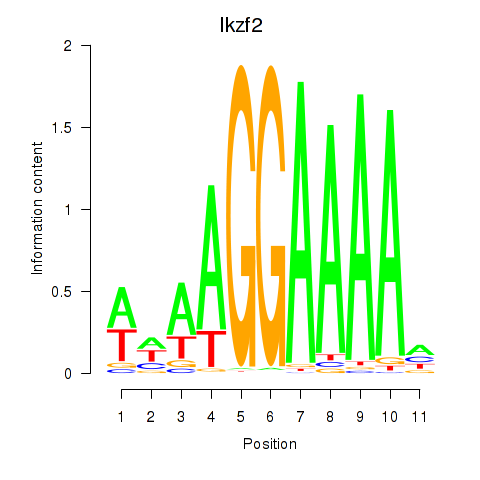
Results for Ikzf2
Z-value: 1.43
Transcription factors associated with Ikzf2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Ikzf2
|
ENSMUSG00000025997.7 | IKAROS family zinc finger 2 |
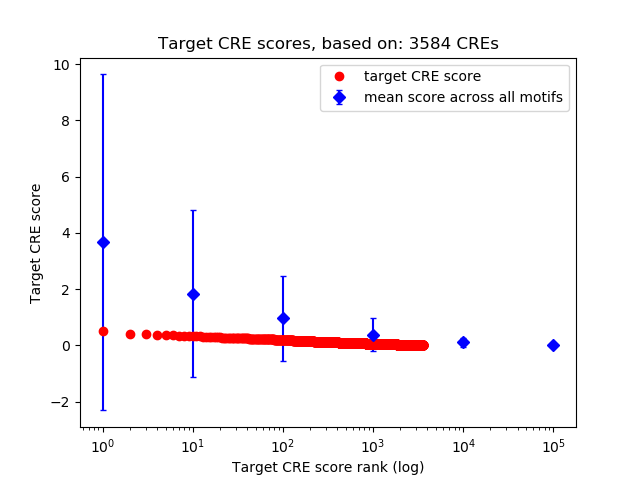
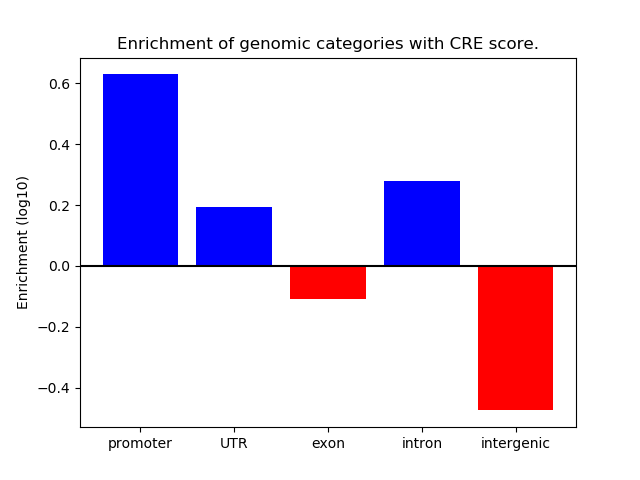
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr1_69687670_69688235 | Ikzf2 | 707 | 0.720596 | 0.67 | 1.4e-01 | Click! |
| chr1_69686936_69687313 | Ikzf2 | 16 | 0.981527 | 0.45 | 3.7e-01 | Click! |
| chr1_69687475_69687652 | Ikzf2 | 318 | 0.909213 | 0.38 | 4.6e-01 | Click! |
| chr1_69688260_69688444 | Ikzf2 | 1107 | 0.553126 | 0.28 | 6.0e-01 | Click! |
| chr1_69688919_69689070 | Ikzf2 | 1749 | 0.398677 | 0.18 | 7.4e-01 | Click! |
Activity of the Ikzf2 motif across conditions
Conditions sorted by the z-value of the Ikzf2 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
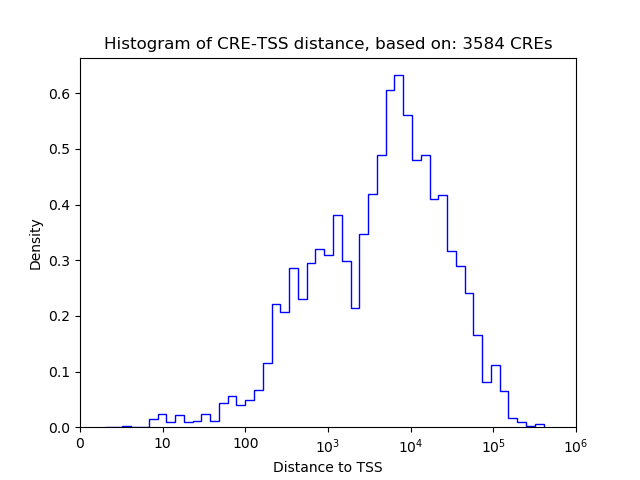
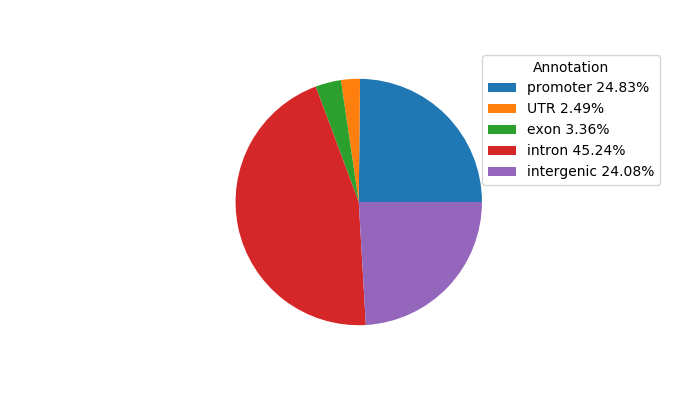
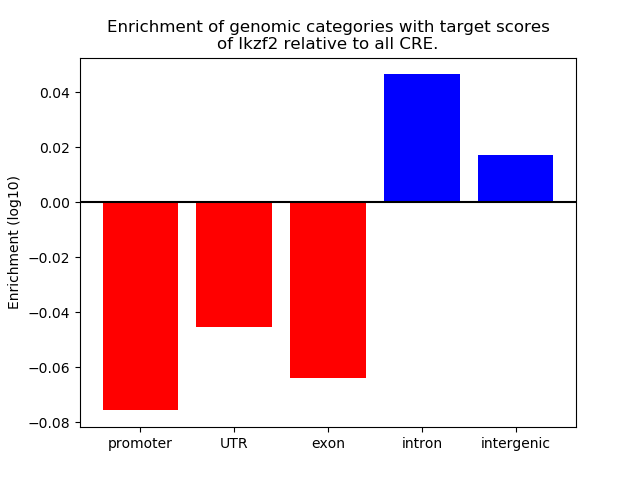
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr17_88625924_88626075 | 0.51 |
Ston1 |
stonin 1 |
556 |
0.73 |
| chr2_94265573_94265724 | 0.41 |
Mir670hg |
MIR670 host gene (non-protein coding) |
730 |
0.59 |
| chr3_121802727_121802878 | 0.40 |
Abcd3 |
ATP-binding cassette, sub-family D (ALD), member 3 |
12107 |
0.12 |
| chr18_39498437_39498588 | 0.35 |
Nr3c1 |
nuclear receptor subfamily 3, group C, member 1 |
7211 |
0.27 |
| chr1_39192284_39192607 | 0.35 |
Npas2 |
neuronal PAS domain protein 2 |
1286 |
0.45 |
| chr18_4967569_4967720 | 0.35 |
Svil |
supervillin |
26169 |
0.25 |
| chr4_58508983_58509134 | 0.33 |
Lpar1 |
lysophosphatidic acid receptor 1 |
9655 |
0.2 |
| chr12_78878863_78879046 | 0.33 |
Eif2s1 |
eukaryotic translation initiation factor 2, subunit 1 alpha |
16824 |
0.15 |
| chr4_155565156_155565307 | 0.33 |
Nadk |
NAD kinase |
1159 |
0.33 |
| chr8_23207373_23207572 | 0.32 |
Gpat4 |
glycerol-3-phosphate acyltransferase 4 |
873 |
0.36 |
| chr4_123419684_123419835 | 0.32 |
Macf1 |
microtubule-actin crosslinking factor 1 |
7531 |
0.17 |
| chr15_75966705_75966865 | 0.32 |
Eif2s3x-ps1 |
eukaryotic translation initiation factor 2, subunit 3, pseudogene 1 |
1550 |
0.17 |
| chr8_25774623_25774814 | 0.31 |
Bag4 |
BCL2-associated athanogene 4 |
2864 |
0.14 |
| chr1_65174138_65174289 | 0.30 |
Idh1 |
isocitrate dehydrogenase 1 (NADP+), soluble |
1351 |
0.35 |
| chr2_103788326_103788477 | 0.29 |
Caprin1 |
cell cycle associated protein 1 |
8143 |
0.1 |
| chr4_129928314_129928470 | 0.29 |
Spocd1 |
SPOC domain containing 1 |
857 |
0.48 |
| chr10_84418574_84418728 | 0.29 |
Nuak1 |
NUAK family, SNF1-like kinase, 1 |
21946 |
0.15 |
| chr3_149090724_149090881 | 0.28 |
Gm25127 |
predicted gene, 25127 |
61870 |
0.11 |
| chr6_31061871_31062042 | 0.28 |
Mir29a |
microRNA 29a |
791 |
0.38 |
| chr14_88347313_88347498 | 0.28 |
Gm9274 |
predicted gene 9274 |
2950 |
0.22 |
| chr14_35174214_35174365 | 0.28 |
Gm49034 |
predicted gene, 49034 |
45149 |
0.2 |
| chr19_3340116_3340320 | 0.28 |
Cpt1a |
carnitine palmitoyltransferase 1a, liver |
1409 |
0.32 |
| chr8_107697000_107697171 | 0.27 |
Gm8940 |
predicted gene 8940 |
6241 |
0.27 |
| chr2_37338925_37339078 | 0.27 |
Olfr368 |
olfactory receptor 368 |
7252 |
0.14 |
| chr11_7184254_7184557 | 0.27 |
Ccdc201 |
coiled coil domain 201 |
4164 |
0.2 |
| chr1_139130984_139131140 | 0.26 |
Gm16304 |
predicted gene 16304 |
3603 |
0.18 |
| chr11_50642967_50643387 | 0.26 |
Gm12198 |
predicted gene 12198 |
40729 |
0.12 |
| chr17_24355260_24355582 | 0.26 |
Abca3 |
ATP-binding cassette, sub-family A (ABC1), member 3 |
3319 |
0.12 |
| chr19_3329840_3329991 | 0.26 |
Cpt1a |
carnitine palmitoyltransferase 1a, liver |
3810 |
0.15 |
| chr17_85057101_85057425 | 0.26 |
Prepl |
prolyl endopeptidase-like |
20211 |
0.16 |
| chr18_20938375_20938560 | 0.26 |
Rnf125 |
ring finger protein 125 |
6158 |
0.22 |
| chr11_77892588_77892765 | 0.26 |
Pipox |
pipecolic acid oxidase |
1420 |
0.34 |
| chr10_122343701_122343852 | 0.26 |
Gm36041 |
predicted gene, 36041 |
43116 |
0.16 |
| chr17_78991834_78992047 | 0.25 |
Prkd3 |
protein kinase D3 |
6492 |
0.16 |
| chr15_82785072_82785230 | 0.25 |
Gm27618 |
predicted gene, 27618 |
3010 |
0.13 |
| chr6_127446671_127447024 | 0.25 |
Parp11 |
poly (ADP-ribose) polymerase family, member 11 |
7 |
0.98 |
| chr9_70270798_70270974 | 0.25 |
Myo1e |
myosin IE |
63518 |
0.11 |
| chr3_83013969_83014357 | 0.25 |
Gm30097 |
predicted gene, 30097 |
5675 |
0.15 |
| chr8_36457473_36457639 | 0.25 |
Trmt9b |
tRNA methyltransferase 9B |
8 |
0.98 |
| chr3_27267495_27267649 | 0.25 |
Gm37191 |
predicted gene, 37191 |
10488 |
0.19 |
| chr4_45393790_45393968 | 0.24 |
Gm25692 |
predicted gene, 25692 |
3239 |
0.18 |
| chr8_106609643_106609823 | 0.24 |
Cdh1 |
cadherin 1 |
5590 |
0.19 |
| chr5_105826898_105827072 | 0.24 |
Lrrc8d |
leucine rich repeat containing 8D |
2474 |
0.21 |
| chr12_99514762_99515114 | 0.24 |
Foxn3 |
forkhead box N3 |
48570 |
0.1 |
| chr2_128746581_128746778 | 0.24 |
Gm14011 |
predicted gene 14011 |
7026 |
0.16 |
| chr10_24958779_24958984 | 0.23 |
Gm36172 |
predicted gene, 36172 |
31262 |
0.12 |
| chr12_103681578_103682027 | 0.23 |
Serpina16 |
serine (or cysteine) peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 16 |
5663 |
0.11 |
| chr6_145759082_145759277 | 0.23 |
Rassf8 |
Ras association (RalGDS/AF-6) domain family (N-terminal) member 8 |
8608 |
0.2 |
| chr13_17696784_17696935 | 0.23 |
Sugct |
succinyl-CoA glutarate-CoA transferase |
1306 |
0.28 |
| chr2_160664743_160664933 | 0.23 |
Top1 |
topoisomerase (DNA) I |
5295 |
0.2 |
| chr9_77942306_77942515 | 0.23 |
Elovl5 |
ELOVL family member 5, elongation of long chain fatty acids (yeast) |
1119 |
0.43 |
| chr6_128818479_128818642 | 0.23 |
Gm44511 |
predicted gene 44511 |
7659 |
0.09 |
| chr5_150772189_150772418 | 0.23 |
Pds5b |
PDS5 cohesin associated factor B |
12939 |
0.15 |
| chr11_102531919_102532070 | 0.23 |
Mdk-ps1 |
midkine pseudogene 1 |
13708 |
0.1 |
| chr18_68226642_68226813 | 0.23 |
Ldlrad4 |
low density lipoprotein receptor class A domain containing 4 |
1340 |
0.44 |
| chr2_85145666_85145817 | 0.23 |
Aplnr |
apelin receptor |
8349 |
0.12 |
| chr13_41226845_41227201 | 0.22 |
Elovl2 |
elongation of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 2 |
6618 |
0.12 |
| chr12_101981443_101981628 | 0.22 |
Ndufb1-ps |
NADH:ubiquinone oxidoreductase subunit B1 |
4467 |
0.18 |
| chr16_30945152_30945303 | 0.22 |
Gm46565 |
predicted gene, 46565 |
9598 |
0.17 |
| chr11_60941556_60941974 | 0.22 |
Map2k3 |
mitogen-activated protein kinase kinase 3 |
115 |
0.95 |
| chr1_59381982_59382225 | 0.22 |
Gm29016 |
predicted gene 29016 |
3173 |
0.26 |
| chr9_86395532_86395683 | 0.22 |
Ube2cbp |
ubiquitin-conjugating enzyme E2C binding protein |
69343 |
0.09 |
| chr12_52097440_52097602 | 0.22 |
Nubpl |
nucleotide binding protein-like |
216 |
0.94 |
| chr1_131473536_131473703 | 0.22 |
Gm24273 |
predicted gene, 24273 |
25183 |
0.12 |
| chr13_41321516_41321722 | 0.22 |
Nedd9 |
neural precursor cell expressed, developmentally down-regulated gene 9 |
37628 |
0.1 |
| chr11_120800693_120800844 | 0.22 |
Dus1l |
dihydrouridine synthase 1-like (S. cerevisiae) |
4365 |
0.09 |
| chr6_24305876_24306056 | 0.22 |
Slc13a1 |
solute carrier family 13 (sodium/sulfate symporters), member 1 |
137874 |
0.04 |
| chr19_55794640_55795127 | 0.22 |
Ppnr |
per-pentamer repeat gene |
46789 |
0.16 |
| chr13_37903137_37903504 | 0.22 |
Rreb1 |
ras responsive element binding protein 1 |
14449 |
0.21 |
| chr12_80827547_80827734 | 0.22 |
Susd6 |
sushi domain containing 6 |
37081 |
0.11 |
| chr5_8055937_8056152 | 0.21 |
Sri |
sorcin |
483 |
0.61 |
| chr11_93988000_93988159 | 0.21 |
Spag9 |
sperm associated antigen 9 |
8012 |
0.15 |
| chr5_145877597_145877748 | 0.21 |
Cyp3a11 |
cytochrome P450, family 3, subfamily a, polypeptide 11 |
981 |
0.52 |
| chr1_179969316_179969494 | 0.21 |
Cdc42bpa |
CDC42 binding protein kinase alpha |
6494 |
0.22 |
| chr11_7201515_7201726 | 0.21 |
Igfbp1 |
insulin-like growth factor binding protein 1 |
3838 |
0.2 |
| chr2_174773866_174774049 | 0.21 |
Edn3 |
endothelin 3 |
4035 |
0.25 |
| chr16_65561790_65561941 | 0.21 |
Chmp2b |
charged multivesicular body protein 2B |
655 |
0.72 |
| chr9_9091904_9092129 | 0.21 |
Gm16485 |
predicted gene 16485 |
120225 |
0.06 |
| chr7_16526304_16526474 | 0.21 |
Gm17981 |
predicted gene, 17981 |
11251 |
0.12 |
| chr16_14157985_14158341 | 0.21 |
Marf1 |
meiosis regulator and mRNA stability 1 |
1104 |
0.35 |
| chr11_58026255_58026440 | 0.20 |
Larp1 |
La ribonucleoprotein domain family, member 1 |
15266 |
0.14 |
| chr11_53361493_53361644 | 0.20 |
Aff4 |
AF4/FMR2 family, member 4 |
6505 |
0.12 |
| chr13_53126019_53126208 | 0.20 |
Gm48336 |
predicted gene, 48336 |
97195 |
0.07 |
| chr7_19320322_19320473 | 0.20 |
Fosb |
FBJ osteosarcoma oncogene B |
10346 |
0.07 |
| chr13_69564655_69564806 | 0.20 |
A530095I07Rik |
RIKEN cDNA A530095I07 gene |
20707 |
0.11 |
| chr2_132585960_132586261 | 0.20 |
Gpcpd1 |
glycerophosphocholine phosphodiesterase 1 |
1619 |
0.33 |
| chr9_44501949_44502105 | 0.20 |
Bcl9l |
B cell CLL/lymphoma 9-like |
2755 |
0.11 |
| chr14_12074077_12074296 | 0.20 |
Ptprg |
protein tyrosine phosphatase, receptor type, G |
16974 |
0.23 |
| chr9_57910217_57910368 | 0.20 |
Ubl7 |
ubiquitin-like 7 (bone marrow stromal cell-derived) |
687 |
0.66 |
| chr10_40103741_40104111 | 0.20 |
Gm25613 |
predicted gene, 25613 |
5536 |
0.16 |
| chr2_31710916_31711068 | 0.20 |
Abl1 |
c-abl oncogene 1, non-receptor tyrosine kinase |
21381 |
0.11 |
| chr1_183388469_183388665 | 0.19 |
Taf1a |
TATA-box binding protein associated factor, RNA polymerase I, A |
414 |
0.77 |
| chr9_43751868_43752079 | 0.19 |
Gm30015 |
predicted gene, 30015 |
5873 |
0.16 |
| chr18_54123642_54124003 | 0.19 |
Gm8594 |
predicted gene 8594 |
92514 |
0.09 |
| chr1_72173165_72173339 | 0.19 |
Mreg |
melanoregulin |
39055 |
0.11 |
| chr11_100917110_100917261 | 0.19 |
Stat3 |
signal transducer and activator of transcription 3 |
8961 |
0.14 |
| chr14_100344624_100344775 | 0.19 |
Gm41231 |
predicted gene, 41231 |
58534 |
0.11 |
| chr4_115496362_115496756 | 0.19 |
Cyp4a14 |
cytochrome P450, family 4, subfamily a, polypeptide 14 |
417 |
0.75 |
| chr14_65783730_65783949 | 0.19 |
Pbk |
PDZ binding kinase |
21998 |
0.18 |
| chr12_34268416_34268623 | 0.19 |
Gm18025 |
predicted gene, 18025 |
22616 |
0.27 |
| chr9_69383086_69383376 | 0.19 |
Ice2 |
interactor of little elongation complex ELL subunit 2 |
14675 |
0.15 |
| chr4_100222085_100222244 | 0.19 |
Gm12701 |
predicted gene 12701 |
102543 |
0.07 |
| chr7_49633783_49633973 | 0.19 |
Dbx1 |
developing brain homeobox 1 |
2971 |
0.33 |
| chr15_96797720_96797879 | 0.19 |
Gm8888 |
predicted gene 8888 |
30721 |
0.19 |
| chr5_92752273_92752453 | 0.19 |
Gm20500 |
predicted gene 20500 |
16158 |
0.18 |
| chr11_60015685_60015836 | 0.19 |
Pemt |
phosphatidylethanolamine N-methyltransferase |
5466 |
0.18 |
| chr7_14425527_14425694 | 0.19 |
Sult2a8 |
sulfotransferase family 2A, dehydroepiandrosterone (DHEA)-preferring, member 8 |
10837 |
0.16 |
| chr6_121849356_121849578 | 0.19 |
Mug1 |
murinoglobulin 1 |
8363 |
0.22 |
| chr14_64652977_64653152 | 0.19 |
Kif13b |
kinesin family member 13B |
462 |
0.83 |
| chr4_43394262_43394749 | 0.19 |
Rusc2 |
RUN and SH3 domain containing 2 |
6733 |
0.14 |
| chr13_50594068_50594219 | 0.19 |
Mir683-2 |
microRNA 683-2 |
6937 |
0.17 |
| chr9_65327351_65327502 | 0.19 |
Gm39363 |
predicted gene, 39363 |
5094 |
0.1 |
| chr11_53703758_53703909 | 0.18 |
Rad50 |
RAD50 double strand break repair protein |
3462 |
0.14 |
| chr7_140767825_140767976 | 0.18 |
Cyp2e1 |
cytochrome P450, family 2, subfamily e, polypeptide 1 |
3419 |
0.12 |
| chr5_119580336_119580487 | 0.18 |
Tbx3os1 |
T-box 3, opposite strand 1 |
5465 |
0.18 |
| chr4_99126806_99126971 | 0.18 |
Dock7 |
dedicator of cytokinesis 7 |
5973 |
0.16 |
| chr19_44044543_44044826 | 0.18 |
Cyp2c23 |
cytochrome P450, family 2, subfamily c, polypeptide 23 |
15476 |
0.13 |
| chrX_169904110_169904261 | 0.18 |
Mid1 |
midline 1 |
22766 |
0.2 |
| chr13_29049878_29050059 | 0.18 |
A330102I10Rik |
RIKEN cDNA A330102I10 gene |
33260 |
0.19 |
| chr2_84386945_84387393 | 0.18 |
Calcrl |
calcitonin receptor-like |
11811 |
0.2 |
| chr5_146255872_146256023 | 0.18 |
Cdk8 |
cyclin-dependent kinase 8 |
1846 |
0.25 |
| chr13_9277842_9278026 | 0.18 |
Dip2c |
disco interacting protein 2 homolog C |
952 |
0.44 |
| chr7_88278905_88279083 | 0.18 |
Ctsc |
cathepsin C |
856 |
0.66 |
| chr11_83577932_83578097 | 0.18 |
Ccl9 |
chemokine (C-C motif) ligand 9 |
622 |
0.56 |
| chr4_83221216_83221400 | 0.18 |
Ttc39b |
tetratricopeptide repeat domain 39B |
11862 |
0.19 |
| chr8_111776845_111777015 | 0.17 |
Bcar1 |
breast cancer anti-estrogen resistance 1 |
33121 |
0.14 |
| chr18_20668429_20668965 | 0.17 |
Ttr |
transthyretin |
3417 |
0.2 |
| chr8_13977825_13978003 | 0.17 |
Tdrp |
testis development related protein |
2882 |
0.2 |
| chr12_102090498_102090674 | 0.17 |
Slc24a4 |
solute carrier family 24 (sodium/potassium/calcium exchanger), member 4 |
38147 |
0.14 |
| chr7_51862626_51863120 | 0.17 |
Gas2 |
growth arrest specific 2 |
446 |
0.55 |
| chr16_4395580_4396011 | 0.17 |
Adcy9 |
adenylate cyclase 9 |
23792 |
0.18 |
| chr5_36681886_36682047 | 0.17 |
Gm25767 |
predicted gene, 25767 |
10308 |
0.13 |
| chr4_145276264_145276637 | 0.17 |
Tnfrsf1b |
tumor necrosis factor receptor superfamily, member 1b |
29580 |
0.15 |
| chr11_89084716_89084878 | 0.17 |
Gm22534 |
predicted gene, 22534 |
2354 |
0.2 |
| chr9_106263078_106263246 | 0.17 |
Alas1 |
aminolevulinic acid synthase 1 |
14508 |
0.1 |
| chr3_95196135_95196310 | 0.17 |
Gabpb2 |
GA repeat binding protein, beta 2 |
4324 |
0.09 |
| chr5_52998745_52999069 | 0.17 |
5033403H07Rik |
RIKEN cDNA 5033403H07 gene |
4471 |
0.18 |
| chr4_82393411_82393562 | 0.17 |
n-R5s188 |
nuclear encoded rRNA 5S 188 |
45924 |
0.17 |
| chr18_20931708_20931938 | 0.16 |
Rnf125 |
ring finger protein 125 |
12802 |
0.2 |
| chr13_34976836_34976987 | 0.16 |
Eci2 |
enoyl-Coenzyme A delta isomerase 2 |
11445 |
0.1 |
| chr3_85819710_85819869 | 0.16 |
Fam160a1 |
family with sequence similarity 160, member A1 |
2498 |
0.27 |
| chr2_115310363_115310574 | 0.16 |
4930528P14Rik |
RIKEN cDNA 4930528P14 gene |
33812 |
0.18 |
| chr11_49986494_49986652 | 0.16 |
Rasgef1c |
RasGEF domain family, member 1C |
22788 |
0.15 |
| chr1_164704417_164705099 | 0.16 |
Gm37853 |
predicted gene, 37853 |
6525 |
0.16 |
| chr18_23496939_23497187 | 0.16 |
Dtna |
dystrobrevin alpha |
228 |
0.96 |
| chr8_21798514_21798691 | 0.16 |
Gm15316 |
predicted gene 15316 |
18782 |
0.08 |
| chr6_47469541_47469695 | 0.16 |
Cul1 |
cullin 1 |
15230 |
0.17 |
| chr7_81572696_81572857 | 0.16 |
Whamm |
WAS protein homolog associated with actin, golgi membranes and microtubules |
832 |
0.45 |
| chr10_23489926_23490106 | 0.16 |
Gm47843 |
predicted gene, 47843 |
119415 |
0.05 |
| chr14_21853942_21854097 | 0.16 |
Comtd1 |
catechol-O-methyltransferase domain containing 1 |
5042 |
0.14 |
| chr14_30946732_30946883 | 0.16 |
Itih1 |
inter-alpha trypsin inhibitor, heavy chain 1 |
3518 |
0.13 |
| chr13_111789281_111789599 | 0.16 |
Map3k1 |
mitogen-activated protein kinase kinase kinase 1 |
18875 |
0.13 |
| chr7_44930245_44930425 | 0.16 |
Ap2a1 |
adaptor-related protein complex 2, alpha 1 subunit |
839 |
0.33 |
| chr9_65289767_65289960 | 0.16 |
Clpx |
caseinolytic mitochondrial matrix peptidase chaperone subunit |
4397 |
0.12 |
| chr8_41130963_41131266 | 0.16 |
Mtus1 |
mitochondrial tumor suppressor 1 |
2604 |
0.3 |
| chr3_108041042_108041216 | 0.16 |
Gstm4 |
glutathione S-transferase, mu 4 |
3730 |
0.09 |
| chr1_9968795_9969006 | 0.16 |
Tcf24 |
transcription factor 24 |
968 |
0.33 |
| chr11_121550191_121550342 | 0.15 |
Tbcd |
tubulin-specific chaperone d |
155 |
0.96 |
| chr8_10935282_10935610 | 0.15 |
Gm45042 |
predicted gene 45042 |
5559 |
0.11 |
| chr2_30203720_30204093 | 0.15 |
Kyat1 |
kynurenine aminotransferase 1 |
1869 |
0.19 |
| chr7_73654293_73654474 | 0.15 |
Gm26176 |
predicted gene, 26176 |
8294 |
0.09 |
| chr6_42254392_42254589 | 0.15 |
Tmem139 |
transmembrane protein 139 |
7469 |
0.11 |
| chr1_23187785_23188477 | 0.15 |
Gm29506 |
predicted gene 29506 |
47542 |
0.11 |
| chr3_95106432_95106780 | 0.15 |
Pip5k1a |
phosphatidylinositol-4-phosphate 5-kinase, type 1 alpha |
173 |
0.86 |
| chr11_20144973_20145150 | 0.15 |
Gm12031 |
predicted gene 12031 |
1283 |
0.38 |
| chr2_23073767_23073918 | 0.15 |
Acbd5 |
acyl-Coenzyme A binding domain containing 5 |
4391 |
0.21 |
| chr7_65357420_65357571 | 0.15 |
Gm44794 |
predicted gene 44794 |
9287 |
0.19 |
| chr13_33085687_33085860 | 0.15 |
Serpinb1b |
serine (or cysteine) peptidase inhibitor, clade B, member 1b |
1648 |
0.3 |
| chr2_30591211_30591362 | 0.15 |
Cstad |
CSA-conditional, T cell activation-dependent protein |
3754 |
0.21 |
| chr12_103629264_103629420 | 0.15 |
Serpina10 |
serine (or cysteine) peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 10 |
2079 |
0.21 |
| chr19_36702491_36702642 | 0.15 |
Hectd2os |
Hectd2, opposite strand |
3075 |
0.29 |
| chr6_17178805_17178999 | 0.15 |
Gm4876 |
predicted gene 4876 |
7069 |
0.21 |
| chr10_24316968_24317125 | 0.15 |
Moxd1 |
monooxygenase, DBH-like 1 |
93501 |
0.06 |
| chrX_7649896_7650047 | 0.15 |
Syp |
synaptophysin |
6062 |
0.08 |
| chr11_68825947_68826339 | 0.15 |
Myh10 |
myosin, heavy polypeptide 10, non-muscle |
11781 |
0.14 |
| chr3_40799638_40799878 | 0.15 |
Plk4 |
polo like kinase 4 |
261 |
0.88 |
| chr2_157095128_157095291 | 0.15 |
Tldc2 |
TBC/LysM associated domain containing 2 |
6756 |
0.15 |
| chr15_8292834_8292985 | 0.15 |
Cplane1 |
ciliogenesis and planar polarity effector 1 |
45806 |
0.15 |
| chr17_33968345_33968496 | 0.15 |
Vps52 |
VPS52 GARP complex subunit |
6672 |
0.05 |
| chr6_108093306_108093738 | 0.15 |
Setmar |
SET domain without mariner transposase fusion |
28460 |
0.17 |
| chr14_100323005_100323183 | 0.15 |
Gm41231 |
predicted gene, 41231 |
36929 |
0.15 |
| chr10_19490879_19491054 | 0.15 |
Gm33104 |
predicted gene, 33104 |
3677 |
0.26 |
| chr12_8780551_8780813 | 0.15 |
Sdc1 |
syndecan 1 |
8879 |
0.18 |
| chr6_112696819_112696998 | 0.15 |
Rad18 |
RAD18 E3 ubiquitin protein ligase |
222 |
0.95 |
| chr7_68120013_68120201 | 0.15 |
Igf1r |
insulin-like growth factor I receptor |
28483 |
0.21 |
| chr15_11407226_11407380 | 0.14 |
Tars |
threonyl-tRNA synthetase |
7638 |
0.27 |
| chr3_121931365_121931695 | 0.14 |
Arhgap29 |
Rho GTPase activating protein 29 |
21011 |
0.17 |
| chr11_50201006_50201196 | 0.14 |
Sqstm1 |
sequestosome 1 |
1853 |
0.21 |
| chr8_119442578_119443025 | 0.14 |
Necab2 |
N-terminal EF-hand calcium binding protein 2 |
3918 |
0.18 |
| chr14_52197609_52197778 | 0.14 |
Gm26590 |
predicted gene, 26590 |
191 |
0.58 |
| chr2_104041847_104042280 | 0.14 |
Gm24644 |
predicted gene, 24644 |
1394 |
0.27 |
| chr8_108731557_108731708 | 0.14 |
Gm38042 |
predicted gene, 38042 |
5961 |
0.25 |
| chr12_54671337_54671527 | 0.14 |
Gm24971 |
predicted gene, 24971 |
4646 |
0.08 |
| chr12_11437653_11437828 | 0.14 |
4930511A02Rik |
RIKEN cDNA 4930511A02 gene |
487 |
0.67 |
| chr19_36627452_36627789 | 0.14 |
Hectd2os |
Hectd2, opposite strand |
1596 |
0.43 |
| chr4_101222796_101222966 | 0.14 |
Gm12785 |
predicted gene 12785 |
12294 |
0.14 |
| chr18_84854418_84854822 | 0.14 |
Cyb5a |
cytochrome b5 type A (microsomal) |
349 |
0.85 |
| chr3_98738112_98738263 | 0.14 |
Hsd3b2 |
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 2 |
13644 |
0.13 |
| chr13_29829021_29829203 | 0.14 |
Cdkal1 |
CDK5 regulatory subunit associated protein 1-like 1 |
26319 |
0.24 |
| chr18_60240885_60241266 | 0.14 |
Gm5970 |
predicted gene 5970 |
20232 |
0.13 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.0 | 0.1 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.0 | 0.1 | GO:0002071 | glandular epithelial cell maturation(GO:0002071) |
| 0.0 | 0.1 | GO:0006553 | lysine metabolic process(GO:0006553) |
| 0.0 | 0.1 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 0.1 | GO:2000320 | negative regulation of T-helper 17 type immune response(GO:2000317) negative regulation of T-helper 17 cell differentiation(GO:2000320) |
| 0.0 | 0.1 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.1 | GO:1901844 | regulation of cell communication by electrical coupling involved in cardiac conduction(GO:1901844) |
| 0.0 | 0.1 | GO:0090365 | regulation of mRNA modification(GO:0090365) |
| 0.0 | 0.1 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.0 | 0.1 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.1 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.0 | 0.2 | GO:0042760 | very long-chain fatty acid catabolic process(GO:0042760) |
| 0.0 | 0.1 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.0 | 0.1 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.0 | 0.1 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.0 | 0.1 | GO:0046487 | glyoxylate metabolic process(GO:0046487) |
| 0.0 | 0.1 | GO:0018199 | peptidyl-glutamine modification(GO:0018199) |
| 0.0 | 0.1 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.0 | 0.1 | GO:0009093 | cysteine catabolic process(GO:0009093) L-cysteine catabolic process(GO:0019448) L-cysteine metabolic process(GO:0046439) |
| 0.0 | 0.1 | GO:2000622 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 | 0.1 | GO:0007403 | glial cell fate determination(GO:0007403) |
| 0.0 | 0.1 | GO:0006435 | threonyl-tRNA aminoacylation(GO:0006435) |
| 0.0 | 0.2 | GO:0032000 | positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.0 | 0.2 | GO:0051775 | response to redox state(GO:0051775) |
| 0.0 | 0.1 | GO:0002019 | regulation of renal output by angiotensin(GO:0002019) |
| 0.0 | 0.0 | GO:1904339 | negative regulation of dopaminergic neuron differentiation(GO:1904339) |
| 0.0 | 0.0 | GO:0035771 | interleukin-4-mediated signaling pathway(GO:0035771) |
| 0.0 | 0.1 | GO:1900126 | negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.0 | 0.1 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.0 | 0.1 | GO:0010835 | regulation of protein ADP-ribosylation(GO:0010835) |
| 0.0 | 0.1 | GO:0007161 | calcium-independent cell-matrix adhesion(GO:0007161) |
| 0.0 | 0.1 | GO:0071673 | positive regulation of smooth muscle cell chemotaxis(GO:0071673) |
| 0.0 | 0.1 | GO:0046501 | protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.0 | 0.1 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.0 | 0.1 | GO:0051890 | regulation of cardioblast differentiation(GO:0051890) |
| 0.0 | 0.0 | GO:1901078 | negative regulation of relaxation of muscle(GO:1901078) |
| 0.0 | 0.0 | GO:0048294 | negative regulation of isotype switching to IgE isotypes(GO:0048294) |
| 0.0 | 0.1 | GO:1902512 | positive regulation of apoptotic DNA fragmentation(GO:1902512) positive regulation of DNA catabolic process(GO:1903626) |
| 0.0 | 0.0 | GO:0055005 | ventricular cardiac myofibril assembly(GO:0055005) |
| 0.0 | 0.1 | GO:0016078 | tRNA catabolic process(GO:0016078) |
| 0.0 | 0.0 | GO:0090219 | negative regulation of lipid kinase activity(GO:0090219) |
| 0.0 | 0.1 | GO:2001286 | regulation of caveolin-mediated endocytosis(GO:2001286) |
| 0.0 | 0.1 | GO:2000676 | positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.0 | 0.1 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.1 | GO:0061084 | negative regulation of protein refolding(GO:0061084) |
| 0.0 | 0.1 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.0 | 0.1 | GO:0006578 | amino-acid betaine biosynthetic process(GO:0006578) |
| 0.0 | 0.1 | GO:1990169 | detoxification of copper ion(GO:0010273) stress response to copper ion(GO:1990169) |
| 0.0 | 0.1 | GO:0060916 | mesenchymal cell proliferation involved in lung development(GO:0060916) |
| 0.0 | 0.1 | GO:1902990 | mitotic telomere maintenance via semi-conservative replication(GO:1902990) |
| 0.0 | 0.0 | GO:1902659 | regulation of glucose mediated signaling pathway(GO:1902659) positive regulation of glucose mediated signaling pathway(GO:1902661) |
| 0.0 | 0.1 | GO:0006528 | asparagine metabolic process(GO:0006528) |
| 0.0 | 0.1 | GO:0042904 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.0 | 0.0 | GO:0002337 | B-1a B cell differentiation(GO:0002337) |
| 0.0 | 0.1 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.0 | 0.1 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.0 | 0.1 | GO:0034239 | regulation of macrophage fusion(GO:0034239) |
| 0.0 | 0.0 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.0 | 0.1 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.0 | 0.1 | GO:0070885 | negative regulation of calcineurin-NFAT signaling cascade(GO:0070885) |
| 0.0 | 0.1 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 0.2 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.0 | 0.0 | GO:0003062 | regulation of heart rate by chemical signal(GO:0003062) |
| 0.0 | 0.1 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.0 | 0.0 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.0 | 0.1 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 0.0 | 0.0 | GO:0006210 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) thymine metabolic process(GO:0019859) |
| 0.0 | 0.1 | GO:2001206 | positive regulation of osteoclast development(GO:2001206) |
| 0.0 | 0.1 | GO:0006824 | cobalt ion transport(GO:0006824) |
| 0.0 | 0.1 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.0 | 0.1 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.0 | 0.2 | GO:0010257 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.1 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.0 | 0.1 | GO:0010992 | ubiquitin homeostasis(GO:0010992) |
| 0.0 | 0.0 | GO:0060161 | positive regulation of dopamine receptor signaling pathway(GO:0060161) |
| 0.0 | 0.0 | GO:0009139 | dUDP biosynthetic process(GO:0006227) pyrimidine nucleoside diphosphate biosynthetic process(GO:0009139) pyrimidine deoxyribonucleoside diphosphate metabolic process(GO:0009196) pyrimidine deoxyribonucleoside diphosphate biosynthetic process(GO:0009197) dUDP metabolic process(GO:0046077) |
| 0.0 | 0.2 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.0 | 0.0 | GO:0007354 | zygotic determination of anterior/posterior axis, embryo(GO:0007354) |
| 0.0 | 0.1 | GO:0042780 | tRNA 3'-end processing(GO:0042780) |
| 0.0 | 0.0 | GO:0010963 | regulation of L-arginine import(GO:0010963) |
| 0.0 | 0.1 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.0 | 0.1 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.0 | 0.0 | GO:0090135 | actin filament branching(GO:0090135) |
| 0.0 | 0.0 | GO:0070669 | response to interleukin-2(GO:0070669) |
| 0.0 | 0.1 | GO:0015684 | ferrous iron transport(GO:0015684) |
| 0.0 | 0.0 | GO:0072310 | glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.0 | 0.0 | GO:0035511 | oxidative DNA demethylation(GO:0035511) |
| 0.0 | 0.0 | GO:0051791 | medium-chain fatty acid metabolic process(GO:0051791) |
| 0.0 | 0.1 | GO:0002322 | B cell proliferation involved in immune response(GO:0002322) |
| 0.0 | 0.1 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.0 | 0.0 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.0 | 0.0 | GO:0003340 | negative regulation of mesenchymal to epithelial transition involved in metanephros morphogenesis(GO:0003340) |
| 0.0 | 0.0 | GO:0060662 | tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.0 | 0.0 | GO:0072675 | multinuclear osteoclast differentiation(GO:0072674) osteoclast fusion(GO:0072675) |
| 0.0 | 0.0 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 0.0 | 0.1 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.0 | 0.1 | GO:0009115 | xanthine catabolic process(GO:0009115) |
| 0.0 | 0.1 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.0 | 0.0 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.0 | 0.0 | GO:0070681 | glutaminyl-tRNAGln biosynthesis via transamidation(GO:0070681) |
| 0.0 | 0.0 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.0 | 0.1 | GO:0007097 | nuclear migration(GO:0007097) |
| 0.0 | 0.1 | GO:0035745 | T-helper 2 cell cytokine production(GO:0035745) |
| 0.0 | 0.1 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.0 | 0.1 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 0.0 | GO:0034773 | histone H4-K20 trimethylation(GO:0034773) |
| 0.0 | 0.1 | GO:0061418 | regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061418) |
| 0.0 | 0.0 | GO:0061470 | T follicular helper cell differentiation(GO:0061470) |
| 0.0 | 0.0 | GO:0032849 | positive regulation of cellular pH reduction(GO:0032849) |
| 0.0 | 0.1 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.0 | 0.0 | GO:1902065 | response to L-glutamate(GO:1902065) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.0 | 0.2 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.0 | 0.2 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.0 | 0.1 | GO:0097648 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor complex(GO:0097648) |
| 0.0 | 0.1 | GO:0098536 | deuterosome(GO:0098536) |
| 0.0 | 0.1 | GO:1990597 | AIP1-IRE1 complex(GO:1990597) |
| 0.0 | 0.1 | GO:0097425 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.0 | 0.1 | GO:0034679 | integrin alpha9-beta1 complex(GO:0034679) |
| 0.0 | 0.1 | GO:0034715 | pICln-Sm protein complex(GO:0034715) |
| 0.0 | 0.2 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.1 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 0.2 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 0.1 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.0 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.0 | 0.1 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.0 | 0.1 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 0.1 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 0.0 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.0 | 0.1 | GO:0031467 | Cul7-RING ubiquitin ligase complex(GO:0031467) |
| 0.0 | 0.1 | GO:0044308 | axonal spine(GO:0044308) |
| 0.0 | 0.0 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.0 | 0.1 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.1 | GO:0043219 | lateral loop(GO:0043219) |
| 0.0 | 0.0 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.1 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 0.0 | GO:0030956 | glutamyl-tRNA(Gln) amidotransferase complex(GO:0030956) |
| 0.0 | 0.1 | GO:0005767 | secondary lysosome(GO:0005767) |
| 0.0 | 0.0 | GO:1990357 | terminal web(GO:1990357) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0018685 | alkane 1-monooxygenase activity(GO:0018685) |
| 0.1 | 0.2 | GO:0000171 | ribonuclease MRP activity(GO:0000171) |
| 0.0 | 0.1 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.2 | GO:0000403 | Y-form DNA binding(GO:0000403) |
| 0.0 | 0.1 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.0 | 0.1 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.0 | 0.1 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 0.2 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.1 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.0 | 0.2 | GO:1990239 | steroid hormone binding(GO:1990239) |
| 0.0 | 0.1 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) |
| 0.0 | 0.1 | GO:0004829 | threonine-tRNA ligase activity(GO:0004829) |
| 0.0 | 0.1 | GO:0030350 | iron-responsive element binding(GO:0030350) |
| 0.0 | 0.1 | GO:0031711 | bradykinin receptor binding(GO:0031711) |
| 0.0 | 0.1 | GO:0004165 | dodecenoyl-CoA delta-isomerase activity(GO:0004165) |
| 0.0 | 0.1 | GO:0017153 | sodium:dicarboxylate symporter activity(GO:0017153) |
| 0.0 | 0.1 | GO:0016635 | oxidoreductase activity, acting on the CH-CH group of donors, quinone or related compound as acceptor(GO:0016635) |
| 0.0 | 0.2 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.0 | 0.1 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.0 | 0.1 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 0.0 | 0.1 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 0.1 | GO:0102337 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.2 | GO:0050649 | testosterone 6-beta-hydroxylase activity(GO:0050649) |
| 0.0 | 0.1 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.0 | 0.1 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) |
| 0.0 | 0.1 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.0 | 0.1 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.0 | 0.1 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.0 | 0.1 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.0 | 0.0 | GO:2001070 | starch binding(GO:2001070) |
| 0.0 | 0.1 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.0 | 0.0 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.0 | 0.1 | GO:0015321 | sodium-dependent phosphate transmembrane transporter activity(GO:0015321) |
| 0.0 | 0.1 | GO:0017099 | very-long-chain-acyl-CoA dehydrogenase activity(GO:0017099) |
| 0.0 | 0.3 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 0.1 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 0.1 | GO:0018642 | 3-(3-hydroxyphenyl)propionate hydroxylase activity(GO:0008688) 4-chlorobenzaldehyde oxidase activity(GO:0018471) 3,5-xylenol methylhydroxylase activity(GO:0018630) phenylacetate hydroxylase activity(GO:0018631) 4-nitrophenol 4-monooxygenase activity(GO:0018632) dimethyl sulfide monooxygenase activity(GO:0018633) alpha-pinene monooxygenase [NADH] activity(GO:0018634) 1-hydroxy-2-naphthoate hydroxylase activity(GO:0018637) toluene 4-monooxygenase activity(GO:0018638) xylene monooxygenase activity(GO:0018639) dibenzothiophene monooxygenase activity(GO:0018640) 6-hydroxy-3-methyl-2-oxo-1,2-dihydroquinoline 6-monooxygenase activity(GO:0018641) chlorophenol 4-monooxygenase activity(GO:0018642) carbon disulfide oxygenase activity(GO:0018643) toluene 2-monooxygenase activity(GO:0018644) 1-hydroxy-2-oxolimonene 1,2-monooxygenase activity(GO:0018646) phenanthrene 1,2-monooxygenase activity(GO:0018647) tetrahydrofuran hydroxylase activity(GO:0018649) styrene monooxygenase activity(GO:0018650) toluene-4-sulfonate monooxygenase activity(GO:0018651) toluene-sulfonate methyl-monooxygenase activity(GO:0018652) 3-methyl-2-oxo-1,2-dihydroquinoline 6-monooxygenase activity(GO:0018653) 2-hydroxy-phenylacetate hydroxylase activity(GO:0018654) 2-oxo-delta3-4,5,5-trimethylcyclopentenylacetyl-CoA 1,2-monooxygenase activity(GO:0018655) phenanthrene 3,4-monooxygenase activity(GO:0018656) toluene 3-monooxygenase activity(GO:0018657) 4-hydroxyphenylacetate,NADH:oxygen oxidoreductase (3-hydroxylating) activity(GO:0018660) limonene monooxygenase activity(GO:0019113) 2-methylnaphthalene hydroxylase activity(GO:0034526) 1-methylnaphthalene hydroxylase activity(GO:0034534) bisphenol A hydroxylase A activity(GO:0034560) salicylate 5-hydroxylase activity(GO:0034785) isobutylamine N-hydroxylase activity(GO:0034791) branched-chain dodecylbenzene sulfonate monooxygenase activity(GO:0034802) 3-HSA hydroxylase activity(GO:0034819) 4-hydroxypyridine-3-hydroxylase activity(GO:0034894) 2-octaprenyl-3-methyl-6-methoxy-1,4-benzoquinol hydroxylase activity(GO:0043719) 6-hydroxynicotinate 3-monooxygenase activity(GO:0043731) tocotrienol omega-hydroxylase activity(GO:0052872) thalianol hydroxylase activity(GO:0080014) |
| 0.0 | 0.1 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.0 | 0.1 | GO:0008410 | CoA-transferase activity(GO:0008410) |
| 0.0 | 0.1 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.0 | 0.0 | GO:0008309 | double-stranded DNA exodeoxyribonuclease activity(GO:0008309) |
| 0.0 | 0.0 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.0 | 0.3 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.1 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.0 | 0.1 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.0 | 0.0 | GO:0047045 | testosterone 17-beta-dehydrogenase (NADP+) activity(GO:0047045) |
| 0.0 | 0.0 | GO:0003865 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) cholestenone 5-alpha-reductase activity(GO:0047751) |
| 0.0 | 0.2 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.0 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.1 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.0 | 0.1 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.0 | 0.0 | GO:0035514 | DNA demethylase activity(GO:0035514) |
| 0.0 | 0.1 | GO:0004030 | aldehyde dehydrogenase [NAD(P)+] activity(GO:0004030) |
| 0.0 | 0.0 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.0 | 0.1 | GO:0016151 | nickel cation binding(GO:0016151) |
| 0.0 | 0.2 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.0 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.0 | 0.0 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.1 | GO:0034875 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 0.0 | 0.0 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.0 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.0 | 0.0 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.1 | GO:0016726 | xanthine dehydrogenase activity(GO:0004854) oxidoreductase activity, acting on CH or CH2 groups, NAD or NADP as acceptor(GO:0016726) |
| 0.0 | 0.0 | GO:0050567 | glutaminyl-tRNA synthase (glutamine-hydrolyzing) activity(GO:0050567) |
| 0.0 | 0.1 | GO:0097642 | calcitonin family receptor activity(GO:0097642) |
| 0.0 | 0.1 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.1 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.0 | 0.0 | GO:0044020 | histone methyltransferase activity (H4-R3 specific)(GO:0044020) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.0 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.0 | 0.1 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 0.1 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.1 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.0 | 0.1 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.0 | 0.3 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.4 | REACTOME RORA ACTIVATES CIRCADIAN EXPRESSION | Genes involved in RORA Activates Circadian Expression |
| 0.0 | 0.2 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.0 | REACTOME PKB MEDIATED EVENTS | Genes involved in PKB-mediated events |
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.1 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.1 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.1 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.2 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.0 | 0.2 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.0 | 0.0 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.0 | 0.2 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |