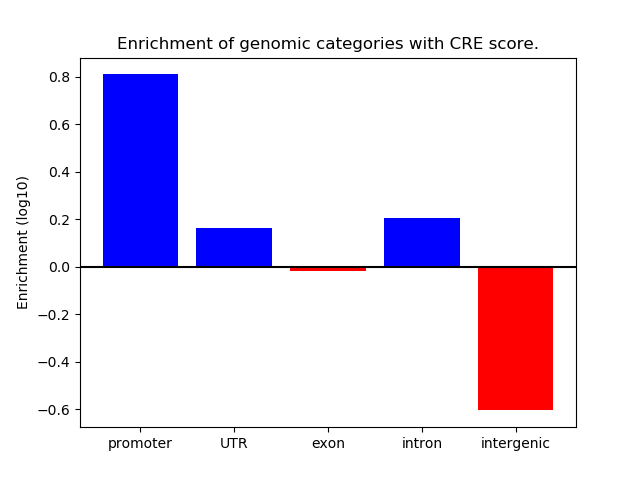
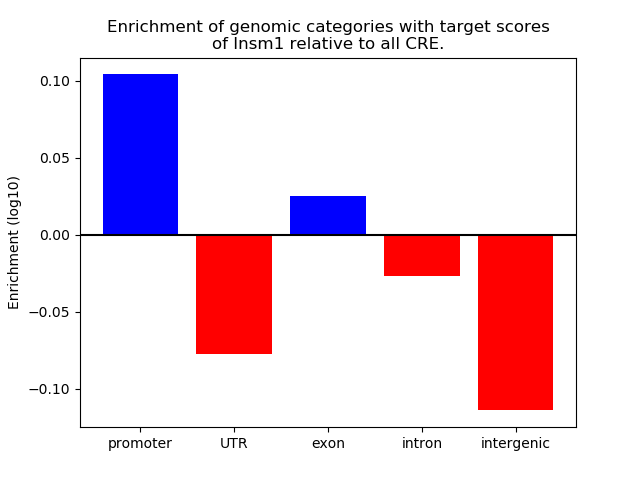
Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
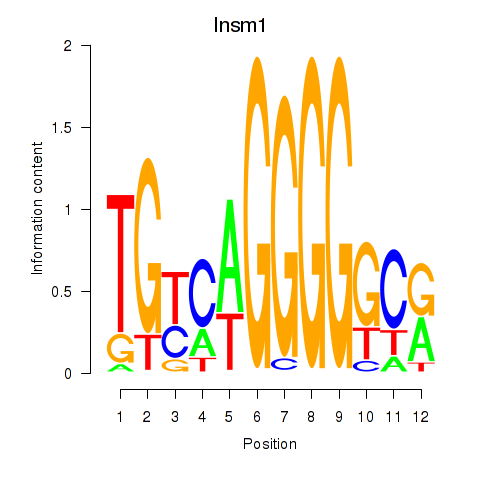
Results for Insm1
Z-value: 1.54
Transcription factors associated with Insm1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Insm1
|
ENSMUSG00000068154.4 | insulinoma-associated 1 |
Activity of the Insm1 motif across conditions
Conditions sorted by the z-value of the Insm1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
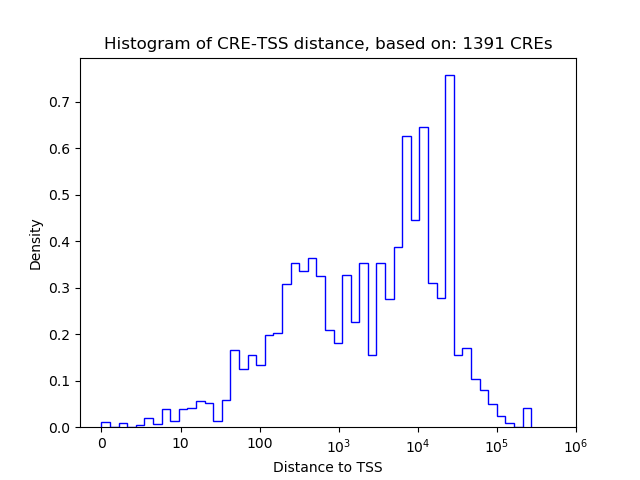
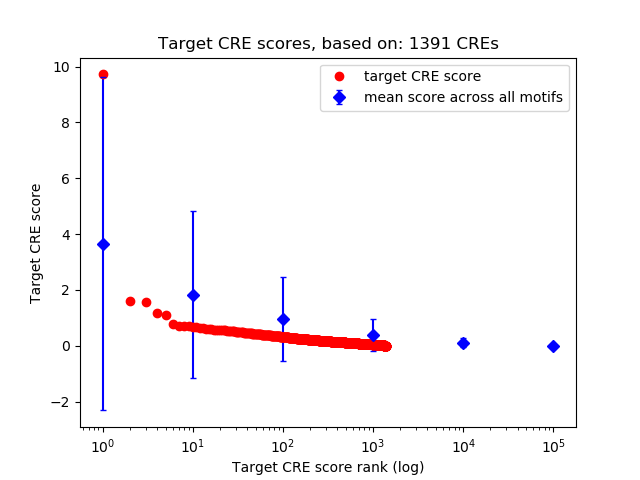
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr9_63555594_63556179 | 9.72 |
Gm16759 |
predicted gene, 16759 |
24534 |
0.17 |
| chr11_95232020_95232325 | 1.59 |
Gm11515 |
predicted gene 11515 |
6540 |
0.15 |
| chr2_31485846_31486386 | 1.56 |
Ass1 |
argininosuccinate synthetase 1 |
11656 |
0.18 |
| chr19_8412638_8412828 | 1.16 |
Slc22a30 |
solute carrier family 22, member 30 |
7622 |
0.21 |
| chr19_8139481_8139632 | 1.09 |
Slc22a28 |
solute carrier family 22, member 28 |
7574 |
0.21 |
| chr1_184101076_184101417 | 0.79 |
Dusp10 |
dual specificity phosphatase 10 |
66865 |
0.11 |
| chr18_59175547_59176237 | 0.71 |
Chsy3 |
chondroitin sulfate synthase 3 |
242 |
0.96 |
| chr7_109566693_109566893 | 0.70 |
Denn2b |
DENN domain containing 2B |
26360 |
0.12 |
| chr17_34024149_34024494 | 0.70 |
Ring1 |
ring finger protein 1 |
331 |
0.42 |
| chr15_39800082_39800233 | 0.67 |
Gm16291 |
predicted gene 16291 |
3112 |
0.26 |
| chr13_57675035_57675197 | 0.65 |
Spock1 |
sparc/osteonectin, cwcv and kazal-like domains proteoglycan 1 |
232471 |
0.02 |
| chr4_141148421_141148589 | 0.64 |
Fbxo42 |
F-box protein 42 |
583 |
0.63 |
| chr11_98962485_98962641 | 0.63 |
Rara |
retinoic acid receptor, alpha |
2151 |
0.18 |
| chr15_79328216_79328601 | 0.61 |
Pla2g6 |
phospholipase A2, group VI |
18 |
0.96 |
| chr16_59600745_59600902 | 0.60 |
Crybg3 |
beta-gamma crystallin domain containing 3 |
156 |
0.96 |
| chr16_4866490_4866641 | 0.59 |
4930562C15Rik |
RIKEN cDNA 4930562C15 gene |
2242 |
0.21 |
| chr5_125074638_125074861 | 0.58 |
Gm42839 |
predicted gene 42839 |
12092 |
0.16 |
| chr4_135745411_135745562 | 0.57 |
Gm12988 |
predicted gene 12988 |
11209 |
0.12 |
| chr9_86721274_86721452 | 0.57 |
Gm37484 |
predicted gene, 37484 |
9359 |
0.11 |
| chr12_29082462_29082658 | 0.56 |
4833405L11Rik |
RIKEN cDNA 4833405L11 gene |
24177 |
0.19 |
| chr14_19808226_19808383 | 0.55 |
Nid2 |
nidogen 2 |
3068 |
0.21 |
| chr15_59040831_59041076 | 0.55 |
Mtss1 |
MTSS I-BAR domain containing 1 |
356 |
0.89 |
| chr3_152763675_152763826 | 0.55 |
Pigk |
phosphatidylinositol glycan anchor biosynthesis, class K |
369 |
0.88 |
| chr7_63899068_63899509 | 0.54 |
Gm27252 |
predicted gene 27252 |
1314 |
0.37 |
| chr8_107311781_107311960 | 0.53 |
Nfat5 |
nuclear factor of activated T cells 5 |
18311 |
0.18 |
| chr14_27298745_27298912 | 0.52 |
Arhgef3 |
Rho guanine nucleotide exchange factor (GEF) 3 |
37238 |
0.15 |
| chr13_51089168_51089510 | 0.52 |
Spin1 |
spindlin 1 |
11541 |
0.24 |
| chr2_58773576_58773851 | 0.52 |
Upp2 |
uridine phosphorylase 2 |
8388 |
0.21 |
| chr15_38299804_38299955 | 0.51 |
Klf10 |
Kruppel-like factor 10 |
84 |
0.96 |
| chr11_100568089_100568249 | 0.51 |
Cnp |
2',3'-cyclic nucleotide 3' phosphodiesterase |
6735 |
0.1 |
| chr3_18186474_18186625 | 0.50 |
Gm23686 |
predicted gene, 23686 |
8924 |
0.24 |
| chr3_142948874_142949040 | 0.50 |
Gm2574 |
predicted pseudogene 2574 |
13654 |
0.17 |
| chr16_37685226_37685393 | 0.50 |
Gm4600 |
predicted gene 4600 |
12031 |
0.18 |
| chr15_79333263_79333463 | 0.50 |
Pla2g6 |
phospholipase A2, group VI |
4973 |
0.12 |
| chr7_98423598_98423760 | 0.49 |
Gm44507 |
predicted gene 44507 |
8055 |
0.13 |
| chr5_125495068_125495293 | 0.49 |
Aacs |
acetoacetyl-CoA synthetase |
10969 |
0.13 |
| chr17_46052256_46052407 | 0.47 |
Vegfa |
vascular endothelial growth factor A |
19962 |
0.13 |
| chr9_44083438_44083791 | 0.47 |
Usp2 |
ubiquitin specific peptidase 2 |
1325 |
0.2 |
| chr3_96172047_96172606 | 0.46 |
Sf3b4 |
splicing factor 3b, subunit 4 |
6 |
0.54 |
| chr10_79684018_79684169 | 0.46 |
Cdc34 |
cell division cycle 34 |
294 |
0.76 |
| chr17_68285429_68285617 | 0.45 |
L3mbtl4 |
L3MBTL4 histone methyl-lysine binding protein |
11726 |
0.26 |
| chr6_47396063_47396225 | 0.45 |
Gm18584 |
predicted gene, 18584 |
36945 |
0.16 |
| chr17_34972548_34972711 | 0.44 |
Hspa1l |
heat shock protein 1-like |
74 |
0.8 |
| chr5_125516996_125517147 | 0.44 |
Aacs |
acetoacetyl-CoA synthetase |
1828 |
0.29 |
| chr10_127188743_127188894 | 0.44 |
Arhgef25 |
Rho guanine nucleotide exchange factor (GEF) 25 |
46 |
0.94 |
| chr6_31261904_31262196 | 0.44 |
2210408F21Rik |
RIKEN cDNA 2210408F21 gene |
15798 |
0.16 |
| chr2_31508672_31508887 | 0.44 |
Ass1 |
argininosuccinate synthetase 1 |
8053 |
0.18 |
| chr12_104338853_104339031 | 0.43 |
Serpina3k |
serine (or cysteine) peptidase inhibitor, clade A, member 3K |
456 |
0.43 |
| chr15_31253435_31253717 | 0.43 |
Dap |
death-associated protein |
14760 |
0.15 |
| chr19_3623839_3624006 | 0.43 |
Lrp5 |
low density lipoprotein receptor-related protein 5 |
8130 |
0.16 |
| chr6_90492906_90493086 | 0.42 |
Aldh1l1 |
aldehyde dehydrogenase 1 family, member L1 |
6569 |
0.11 |
| chr13_58208598_58208750 | 0.42 |
Ubqln1 |
ubiquilin 1 |
6945 |
0.11 |
| chr7_123378143_123378338 | 0.41 |
Lcmt1 |
leucine carboxyl methyltransferase 1 |
184 |
0.94 |
| chr1_66816824_66817235 | 0.41 |
Kansl1l |
KAT8 regulatory NSL complex subunit 1-like |
517 |
0.58 |
| chr8_19784863_19785335 | 0.41 |
Gm10060 |
predicted gene 10060 |
353 |
0.89 |
| chr3_81039705_81039904 | 0.40 |
Gm16000 |
predicted gene 16000 |
633 |
0.66 |
| chr2_154558857_154559033 | 0.40 |
Necab3 |
N-terminal EF-hand calcium binding protein 3 |
55 |
0.95 |
| chr6_146339917_146340100 | 0.40 |
Gm44087 |
predicted gene, 44087 |
36409 |
0.16 |
| chr17_83069993_83070341 | 0.40 |
Gm29052 |
predicted gene 29052 |
1262 |
0.48 |
| chr11_69547199_69547367 | 0.39 |
Dnah2 |
dynein, axonemal, heavy chain 2 |
1825 |
0.17 |
| chr3_138286503_138286658 | 0.39 |
Adh1 |
alcohol dehydrogenase 1 (class I) |
8929 |
0.12 |
| chr14_25488854_25489267 | 0.39 |
Gm47921 |
predicted gene, 47921 |
9006 |
0.14 |
| chr11_3535686_3535900 | 0.39 |
Gm11947 |
predicted gene 11947 |
1555 |
0.2 |
| chr2_164359753_164359904 | 0.39 |
Slpi |
secretory leukocyte peptidase inhibitor |
3321 |
0.13 |
| chr4_126467726_126468262 | 0.39 |
Ago1 |
argonaute RISC catalytic subunit 1 |
427 |
0.74 |
| chr3_52257597_52258053 | 0.38 |
Foxo1 |
forkhead box O1 |
10511 |
0.12 |
| chrX_140458132_140458283 | 0.38 |
Prps1 |
phosphoribosyl pyrophosphate synthetase 1 |
1594 |
0.39 |
| chr6_24527214_24527411 | 0.37 |
Ndufa5 |
NADH:ubiquinone oxidoreductase subunit A5 |
214 |
0.85 |
| chr12_45037426_45037748 | 0.37 |
Stxbp6 |
syntaxin binding protein 6 (amisyn) |
24623 |
0.19 |
| chr9_65968905_65969060 | 0.37 |
Csnk1g1 |
casein kinase 1, gamma 1 |
10566 |
0.16 |
| chr17_25397945_25398101 | 0.37 |
Cacna1h |
calcium channel, voltage-dependent, T type, alpha 1H subunit |
8551 |
0.1 |
| chr6_70709789_70709957 | 0.37 |
Gm30211 |
predicted gene, 30211 |
9201 |
0.13 |
| chr7_97788226_97788577 | 0.37 |
Pak1 |
p21 (RAC1) activated kinase 1 |
140 |
0.96 |
| chr8_20122500_20122829 | 0.37 |
Gm21769 |
predicted gene, 21769 |
280 |
0.92 |
| chr16_90934526_90934702 | 0.37 |
Cfap298 |
cilia and flagella associate protien 298 |
108 |
0.77 |
| chr6_66859498_66859662 | 0.37 |
Gm9794 |
predicted pseudogene 9794 |
16163 |
0.13 |
| chr12_98628007_98628201 | 0.36 |
Spata7 |
spermatogenesis associated 7 |
53 |
0.96 |
| chr11_4186980_4187537 | 0.36 |
Tbc1d10a |
TBC1 domain family, member 10a |
436 |
0.69 |
| chr2_167324607_167324945 | 0.36 |
B4galt5 |
UDP-Gal:betaGlcNAc beta 1,4-galactosyltransferase, polypeptide 5 |
10876 |
0.18 |
| chr5_33253374_33253525 | 0.36 |
Gm43850 |
predicted gene 43850 |
1869 |
0.27 |
| chr8_20297005_20297289 | 0.35 |
6820431F20Rik |
RIKEN cDNA 6820431F20 gene |
285 |
0.93 |
| chr8_93183570_93183871 | 0.35 |
Gm45909 |
predicted gene 45909 |
7638 |
0.14 |
| chr19_4493105_4493260 | 0.35 |
2010003K11Rik |
RIKEN cDNA 2010003K11 gene |
5401 |
0.13 |
| chr11_43927096_43927431 | 0.35 |
Adra1b |
adrenergic receptor, alpha 1b |
26053 |
0.2 |
| chrX_9272438_9272819 | 0.35 |
Xk |
X-linked Kx blood group |
128 |
0.94 |
| chr11_113127851_113128293 | 0.35 |
2610035D17Rik |
RIKEN cDNA 2610035D17 gene |
45005 |
0.18 |
| chr1_177796375_177796892 | 0.35 |
Adss |
adenylosuccinate synthetase, non muscle |
122 |
0.96 |
| chr2_60269617_60269768 | 0.34 |
Rpl10a-ps4 |
ribosomal protein L10A, pseudogene 4 |
5882 |
0.17 |
| chr14_20185433_20185783 | 0.34 |
Kcnk5 |
potassium channel, subfamily K, member 5 |
3799 |
0.16 |
| chr6_17306389_17306751 | 0.34 |
Cav1 |
caveolin 1, caveolae protein |
147 |
0.96 |
| chr11_103697219_103697553 | 0.34 |
Gosr2 |
golgi SNAP receptor complex member 2 |
308 |
0.5 |
| chr9_55253735_55254087 | 0.34 |
Nrg4 |
neuregulin 4 |
11450 |
0.18 |
| chr16_20097870_20098456 | 0.33 |
Klhl24 |
kelch-like 24 |
571 |
0.77 |
| chr19_46328692_46329074 | 0.33 |
Fbxl15 |
F-box and leucine-rich repeat protein 15 |
696 |
0.41 |
| chr11_78422437_78422874 | 0.32 |
Slc13a2 |
solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 2 |
438 |
0.69 |
| chr8_85519627_85519778 | 0.32 |
Gpt2 |
glutamic pyruvate transaminase (alanine aminotransferase) 2 |
293 |
0.89 |
| chr9_106888487_106888638 | 0.32 |
Rbm15b |
RNA binding motif protein 15B |
1134 |
0.3 |
| chr8_40903804_40903955 | 0.32 |
Slc7a2 |
solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 |
5441 |
0.18 |
| chr6_129528344_129528723 | 0.32 |
Gabarapl1 |
gamma-aminobutyric acid (GABA) A receptor-associated protein-like 1 |
4627 |
0.09 |
| chr7_28597835_28598003 | 0.32 |
Pak4 |
p21 (RAC1) activated kinase 4 |
167 |
0.56 |
| chr15_76294294_76294662 | 0.32 |
Smpd5 |
sphingomyelin phosphodiesterase 5 |
130 |
0.88 |
| chr3_130681571_130681744 | 0.32 |
Ostc |
oligosaccharyltransferase complex subunit (non-catalytic) |
14965 |
0.13 |
| chr5_147748468_147748664 | 0.32 |
Gm43156 |
predicted gene 43156 |
3050 |
0.26 |
| chr15_59007009_59007401 | 0.31 |
4930544F09Rik |
RIKEN cDNA 4930544F09 gene |
23069 |
0.16 |
| chr17_46131074_46131225 | 0.31 |
Rsph9 |
radial spoke head 9 homolog (Chlamydomonas) |
5646 |
0.11 |
| chr5_32034405_32034556 | 0.31 |
Babam2 |
BRISC and BRCA1 A complex member 2 |
11498 |
0.19 |
| chr11_117821867_117822183 | 0.31 |
Tk1 |
thymidine kinase 1 |
3792 |
0.09 |
| chr2_163288924_163289099 | 0.31 |
Tox2 |
TOX high mobility group box family member 2 |
31367 |
0.16 |
| chr1_51741438_51741641 | 0.31 |
Gm28055 |
predicted gene 28055 |
5404 |
0.23 |
| chr10_125354052_125354243 | 0.31 |
Slc16a7 |
solute carrier family 16 (monocarboxylic acid transporters), member 7 |
11968 |
0.22 |
| chr10_127167327_127167512 | 0.30 |
B4galnt1 |
beta-1,4-N-acetyl-galactosaminyl transferase 1 |
1560 |
0.19 |
| chr15_59002495_59002896 | 0.30 |
4930544F09Rik |
RIKEN cDNA 4930544F09 gene |
18559 |
0.16 |
| chr3_51159499_51159691 | 0.30 |
Gm38246 |
predicted gene, 38246 |
55935 |
0.09 |
| chr2_34775525_34776380 | 0.30 |
Hspa5 |
heat shock protein 5 |
1105 |
0.4 |
| chr10_123693502_123693679 | 0.30 |
Gm19169 |
predicted gene, 19169 |
110002 |
0.07 |
| chr5_120475950_120476223 | 0.29 |
Sds |
serine dehydratase |
440 |
0.7 |
| chr3_52617818_52618183 | 0.29 |
Gm10293 |
predicted pseudogene 10293 |
5165 |
0.27 |
| chr4_123748931_123749287 | 0.29 |
Akirin1 |
akirin 1 |
1122 |
0.42 |
| chr12_103907475_103907626 | 0.29 |
Serpina1c |
serine (or cysteine) peptidase inhibitor, clade A, member 1C |
2663 |
0.15 |
| chr14_73776184_73776350 | 0.29 |
Gm24540 |
predicted gene, 24540 |
8299 |
0.18 |
| chr18_76119534_76119864 | 0.29 |
2900057B20Rik |
RIKEN cDNA 2900057B20 gene |
21228 |
0.19 |
| chr9_83834067_83834712 | 0.29 |
Ttk |
Ttk protein kinase |
300 |
0.91 |
| chr2_31504934_31505459 | 0.29 |
Ass1 |
argininosuccinate synthetase 1 |
4470 |
0.21 |
| chr3_32396117_32396611 | 0.28 |
Pik3ca |
phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha |
1307 |
0.41 |
| chr14_66103708_66103859 | 0.28 |
Ephx2 |
epoxide hydrolase 2, cytoplasmic |
6877 |
0.17 |
| chr12_85339310_85339673 | 0.28 |
Nek9 |
NIMA (never in mitosis gene a)-related expressed kinase 9 |
129 |
0.93 |
| chr13_119488836_119489180 | 0.28 |
Tmem267 |
transmembrane protein 267 |
148 |
0.85 |
| chr7_44256470_44256632 | 0.28 |
Acp4 |
acid phosphatase 4 |
654 |
0.31 |
| chr16_33684835_33685093 | 0.28 |
Heg1 |
heart development protein with EGF-like domains 1 |
419 |
0.88 |
| chr1_130715772_130716198 | 0.28 |
Pfkfb2 |
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2 |
171 |
0.49 |
| chr4_9616122_9616273 | 0.27 |
Asph |
aspartate-beta-hydroxylase |
20508 |
0.22 |
| chr18_62025539_62025690 | 0.27 |
Sh3tc2 |
SH3 domain and tetratricopeptide repeats 2 |
47655 |
0.14 |
| chr3_36532798_36533054 | 0.27 |
Gm11549 |
predicted gene 11549 |
542 |
0.65 |
| chr3_97226229_97226380 | 0.27 |
Bcl9 |
B cell CLL/lymphoma 9 |
1060 |
0.5 |
| chr4_3939020_3939731 | 0.27 |
Chchd7 |
coiled-coil-helix-coiled-coil-helix domain containing 7 |
47 |
0.94 |
| chr2_4717227_4717378 | 0.27 |
Bend7 |
BEN domain containing 7 |
529 |
0.79 |
| chr19_57363088_57363331 | 0.27 |
Fam160b1 |
family with sequence similarity 160, member B1 |
2179 |
0.23 |
| chr11_115178601_115178800 | 0.26 |
Nat9 |
N-acetyltransferase 9 (GCN5-related, putative) |
8381 |
0.11 |
| chr17_84935947_84936728 | 0.26 |
Ppm1b |
protein phosphatase 1B, magnesium dependent, beta isoform |
20404 |
0.15 |
| chr9_74874885_74875056 | 0.26 |
Onecut1 |
one cut domain, family member 1 |
8486 |
0.16 |
| chr2_165337323_165337478 | 0.26 |
Elmo2 |
engulfment and cell motility 2 |
10921 |
0.14 |
| chr9_43738133_43738287 | 0.26 |
Gm30015 |
predicted gene, 30015 |
1255 |
0.34 |
| chr8_84712053_84712451 | 0.26 |
Nfix |
nuclear factor I/X |
1816 |
0.2 |
| chr2_151494417_151494783 | 0.26 |
Nsfl1c |
NSFL1 (p97) cofactor (p47) |
289 |
0.84 |
| chr15_77823733_77823908 | 0.26 |
Myh9 |
myosin, heavy polypeptide 9, non-muscle |
10589 |
0.14 |
| chr6_139852911_139853089 | 0.26 |
Pik3c2g |
phosphatidylinositol-4-phosphate 3-kinase catalytic subunit type 2 gamma |
9352 |
0.21 |
| chr2_167696514_167696665 | 0.26 |
A530013C23Rik |
RIKEN cDNA A530013C23 gene |
5408 |
0.12 |
| chr7_12927207_12927358 | 0.26 |
Rnf225 |
ring finger protein 225 |
134 |
0.9 |
| chr2_11778396_11778580 | 0.26 |
Ankrd16 |
ankyrin repeat domain 16 |
3 |
0.95 |
| chr17_34859046_34859197 | 0.26 |
Cfb |
complement factor B |
80 |
0.87 |
| chr2_58741116_58741490 | 0.26 |
Gm13559 |
predicted gene 13559 |
664 |
0.72 |
| chr8_122716674_122716851 | 0.26 |
Cbfa2t3 |
CBFA2/RUNX1 translocation partner 3 |
17653 |
0.09 |
| chrX_99975701_99975883 | 0.25 |
Eda |
ectodysplasin-A |
11 |
0.99 |
| chr2_104815840_104816064 | 0.25 |
Qser1 |
glutamine and serine rich 1 |
744 |
0.58 |
| chr9_15306463_15306690 | 0.25 |
4931406C07Rik |
RIKEN cDNA 4931406C07 gene |
128 |
0.61 |
| chr1_59670971_59671144 | 0.25 |
Sumo1 |
small ubiquitin-like modifier 1 |
223 |
0.86 |
| chr2_31509568_31509756 | 0.25 |
Ass1 |
argininosuccinate synthetase 1 |
8828 |
0.18 |
| chr19_46707203_46707544 | 0.25 |
As3mt |
arsenic (+3 oxidation state) methyltransferase |
85 |
0.96 |
| chr8_95146444_95146776 | 0.25 |
Kifc3 |
kinesin family member C3 |
4063 |
0.14 |
| chr4_10459968_10460323 | 0.25 |
Gm11814 |
predicted gene 11814 |
20402 |
0.22 |
| chr15_8131587_8131766 | 0.25 |
Nup155 |
nucleoporin 155 |
1578 |
0.41 |
| chr17_15376604_15376755 | 0.25 |
Dll1 |
delta like canonical Notch ligand 1 |
193 |
0.93 |
| chr4_62697341_62697495 | 0.25 |
Rgs3 |
regulator of G-protein signaling 3 |
2048 |
0.28 |
| chr17_5755888_5756129 | 0.25 |
3300005D01Rik |
RIKEN cDNA 3300005D01 gene |
42649 |
0.13 |
| chr10_44701947_44702137 | 0.25 |
Gm47388 |
predicted gene, 47388 |
4862 |
0.13 |
| chr10_75411570_75411753 | 0.25 |
Upb1 |
ureidopropionase, beta |
1618 |
0.35 |
| chr8_22878603_22878765 | 0.25 |
Gm45555 |
predicted gene 45555 |
5095 |
0.18 |
| chrX_140540637_140540856 | 0.24 |
Tsc22d3 |
TSC22 domain family, member 3 |
1922 |
0.35 |
| chr19_7240751_7240928 | 0.24 |
Naa40 |
N(alpha)-acetyltransferase 40, NatD catalytic subunit |
195 |
0.89 |
| chr14_30913507_30913826 | 0.24 |
Itih3 |
inter-alpha trypsin inhibitor, heavy chain 3 |
463 |
0.7 |
| chr4_24574389_24574545 | 0.24 |
Mms22l |
MMS22-like, DNA repair protein |
4381 |
0.32 |
| chr5_102571751_102571934 | 0.24 |
1700013M08Rik |
RIKEN cDNA 1700013M08 gene |
88553 |
0.09 |
| chr7_99837512_99837672 | 0.24 |
Neu3 |
neuraminidase 3 |
9175 |
0.12 |
| chr4_125122748_125123219 | 0.24 |
Zc3h12a |
zinc finger CCCH type containing 12A |
463 |
0.76 |
| chr17_25839280_25839490 | 0.24 |
Rhot2 |
ras homolog family member T2 |
1214 |
0.15 |
| chr9_77744429_77744594 | 0.24 |
Gclc |
glutamate-cysteine ligase, catalytic subunit |
10024 |
0.13 |
| chr9_63414369_63414559 | 0.24 |
Gm16759 |
predicted gene, 16759 |
15083 |
0.14 |
| chrX_142489982_142490297 | 0.24 |
Gm25915 |
predicted gene, 25915 |
4692 |
0.21 |
| chr6_129233700_129233907 | 0.23 |
2310001H17Rik |
RIKEN cDNA 2310001H17 gene |
171 |
0.92 |
| chr14_21143556_21143728 | 0.23 |
Adk |
adenosine kinase |
67490 |
0.11 |
| chr14_73162192_73162413 | 0.23 |
Rcbtb2 |
regulator of chromosome condensation (RCC1) and BTB (POZ) domain containing protein 2 |
362 |
0.86 |
| chr10_75475274_75475445 | 0.23 |
Gm48158 |
predicted gene, 48158 |
8033 |
0.13 |
| chr11_77884094_77884410 | 0.23 |
Pipox |
pipecolic acid oxidase |
292 |
0.87 |
| chr1_182817032_182817419 | 0.23 |
Susd4 |
sushi domain containing 4 |
52330 |
0.12 |
| chr5_138187143_138187299 | 0.23 |
Taf6 |
TATA-box binding protein associated factor 6 |
0 |
0.75 |
| chr3_89912250_89912528 | 0.23 |
Gm42809 |
predicted gene 42809 |
527 |
0.51 |
| chr4_149426608_149426780 | 0.23 |
Ube4b |
ubiquitination factor E4B |
55 |
0.96 |
| chr10_87920604_87920755 | 0.22 |
Tyms-ps |
thymidylate synthase, pseudogene |
46168 |
0.12 |
| chr14_34317951_34318429 | 0.22 |
Glud1 |
glutamate dehydrogenase 1 |
7463 |
0.1 |
| chr16_92301405_92301603 | 0.22 |
Smim11 |
small integral membrane protein 11 |
208 |
0.9 |
| chr18_38600801_38601184 | 0.22 |
Spry4 |
sprouty RTK signaling antagonist 4 |
423 |
0.54 |
| chr10_84440530_84440894 | 0.22 |
Gm47962 |
predicted gene, 47962 |
114 |
0.49 |
| chr2_128412432_128412601 | 0.22 |
Morrbid |
myeloid RNA regulator of BCL2L11 induced cell death |
9551 |
0.18 |
| chr19_5490661_5490908 | 0.22 |
Cfl1 |
cofilin 1, non-muscle |
219 |
0.76 |
| chr3_27690451_27690616 | 0.22 |
Fndc3b |
fibronectin type III domain containing 3B |
19865 |
0.25 |
| chr1_131828122_131828273 | 0.22 |
Slc41a1 |
solute carrier family 41, member 1 |
220 |
0.91 |
| chr10_87873898_87874083 | 0.22 |
Igf1os |
insulin-like growth factor 1, opposite strand |
10609 |
0.2 |
| chr7_45131841_45131992 | 0.22 |
Flt3l |
FMS-like tyrosine kinase 3 ligand |
159 |
0.78 |
| chr16_55844021_55844404 | 0.22 |
Nfkbiz |
nuclear factor of kappa light polypeptide gene enhancer in B cells inhibitor, zeta |
5313 |
0.18 |
| chr17_78735372_78735770 | 0.22 |
Strn |
striatin, calmodulin binding protein |
1625 |
0.31 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0090154 | positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 0.2 | 0.6 | GO:0010046 | response to mycotoxin(GO:0010046) |
| 0.2 | 0.5 | GO:0097296 | activation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097296) |
| 0.1 | 0.6 | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.1 | 0.4 | GO:0019065 | receptor-mediated endocytosis of virus by host cell(GO:0019065) endocytosis involved in viral entry into host cell(GO:0075509) |
| 0.1 | 0.4 | GO:0006481 | C-terminal protein methylation(GO:0006481) |
| 0.1 | 0.4 | GO:0042851 | L-alanine metabolic process(GO:0042851) |
| 0.1 | 0.4 | GO:0061535 | glutamate secretion, neurotransmission(GO:0061535) |
| 0.1 | 0.5 | GO:0010961 | cellular magnesium ion homeostasis(GO:0010961) |
| 0.1 | 0.3 | GO:0001757 | somite specification(GO:0001757) |
| 0.1 | 0.5 | GO:0006167 | AMP biosynthetic process(GO:0006167) |
| 0.1 | 0.5 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.1 | 0.3 | GO:0061153 | trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 0.1 | 0.3 | GO:0071672 | negative regulation of smooth muscle cell chemotaxis(GO:0071672) |
| 0.1 | 0.2 | GO:0090271 | positive regulation of fibroblast growth factor production(GO:0090271) |
| 0.1 | 0.2 | GO:0070375 | ERK5 cascade(GO:0070375) |
| 0.1 | 0.4 | GO:0090435 | protein localization to nuclear envelope(GO:0090435) |
| 0.1 | 0.4 | GO:1903546 | protein localization to photoreceptor outer segment(GO:1903546) |
| 0.1 | 0.2 | GO:0008626 | granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 0.1 | 0.2 | GO:0090290 | positive regulation of osteoclast proliferation(GO:0090290) |
| 0.1 | 0.2 | GO:1902075 | cellular response to salt(GO:1902075) |
| 0.1 | 0.2 | GO:0019859 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) thymine metabolic process(GO:0019859) |
| 0.1 | 0.4 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.1 | 0.2 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.1 | 0.2 | GO:0002337 | B-1a B cell differentiation(GO:0002337) |
| 0.0 | 0.2 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 0.0 | 0.0 | GO:0010536 | positive regulation of activation of Janus kinase activity(GO:0010536) |
| 0.0 | 0.1 | GO:0060125 | negative regulation of growth hormone secretion(GO:0060125) |
| 0.0 | 0.1 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.0 | 0.1 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.0 | 0.1 | GO:1901078 | negative regulation of relaxation of muscle(GO:1901078) |
| 0.0 | 0.1 | GO:0018197 | peptidyl-aspartic acid modification(GO:0018197) |
| 0.0 | 0.1 | GO:0006059 | hexitol metabolic process(GO:0006059) |
| 0.0 | 0.4 | GO:0042984 | amyloid precursor protein biosynthetic process(GO:0042983) regulation of amyloid precursor protein biosynthetic process(GO:0042984) |
| 0.0 | 0.1 | GO:0060266 | negative regulation of respiratory burst involved in inflammatory response(GO:0060266) |
| 0.0 | 0.1 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 0.0 | 0.2 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.0 | 0.1 | GO:0071727 | response to triacyl bacterial lipopeptide(GO:0071725) cellular response to triacyl bacterial lipopeptide(GO:0071727) |
| 0.0 | 0.1 | GO:0030070 | insulin processing(GO:0030070) |
| 0.0 | 0.1 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.0 | 0.2 | GO:2001184 | positive regulation of interleukin-12 secretion(GO:2001184) |
| 0.0 | 0.2 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.0 | 0.1 | GO:1903223 | positive regulation of oxidative stress-induced neuron death(GO:1903223) |
| 0.0 | 0.1 | GO:2000686 | regulation of rubidium ion transmembrane transporter activity(GO:2000686) |
| 0.0 | 0.1 | GO:1901492 | positive regulation of lymphangiogenesis(GO:1901492) |
| 0.0 | 0.1 | GO:0019482 | beta-alanine metabolic process(GO:0019482) |
| 0.0 | 0.1 | GO:0015819 | lysine transport(GO:0015819) |
| 0.0 | 0.1 | GO:0006742 | NADP catabolic process(GO:0006742) |
| 0.0 | 0.3 | GO:2000288 | positive regulation of myoblast proliferation(GO:2000288) |
| 0.0 | 0.1 | GO:0097680 | double-strand break repair via classical nonhomologous end joining(GO:0097680) |
| 0.0 | 0.2 | GO:0035280 | miRNA loading onto RISC involved in gene silencing by miRNA(GO:0035280) |
| 0.0 | 0.1 | GO:0006987 | activation of signaling protein activity involved in unfolded protein response(GO:0006987) |
| 0.0 | 0.2 | GO:0001842 | neural fold formation(GO:0001842) |
| 0.0 | 0.2 | GO:0006007 | glucose catabolic process(GO:0006007) |
| 0.0 | 0.1 | GO:0048320 | axial mesoderm formation(GO:0048320) |
| 0.0 | 0.1 | GO:0034241 | positive regulation of macrophage fusion(GO:0034241) |
| 0.0 | 0.1 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.0 | 0.3 | GO:0035518 | histone H2A monoubiquitination(GO:0035518) |
| 0.0 | 0.1 | GO:2000259 | positive regulation of complement activation(GO:0045917) positive regulation of protein activation cascade(GO:2000259) |
| 0.0 | 0.1 | GO:0021699 | cerebellar cortex maturation(GO:0021699) |
| 0.0 | 0.3 | GO:0036315 | cellular response to sterol(GO:0036315) |
| 0.0 | 0.1 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.0 | 0.1 | GO:0003099 | positive regulation of the force of heart contraction by epinephrine-norepinephrine(GO:0001997) positive regulation of the force of heart contraction by chemical signal(GO:0003099) |
| 0.0 | 0.1 | GO:0034756 | regulation of iron ion transport(GO:0034756) |
| 0.0 | 0.1 | GO:0003241 | growth involved in heart morphogenesis(GO:0003241) |
| 0.0 | 0.1 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.0 | 0.1 | GO:0060591 | chondroblast differentiation(GO:0060591) |
| 0.0 | 0.1 | GO:0000414 | regulation of histone H3-K36 methylation(GO:0000414) |
| 0.0 | 0.1 | GO:0006361 | transcription initiation from RNA polymerase I promoter(GO:0006361) |
| 0.0 | 0.4 | GO:0042026 | protein refolding(GO:0042026) |
| 0.0 | 0.7 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.1 | GO:0036072 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.0 | 0.1 | GO:0036166 | phenotypic switching(GO:0036166) |
| 0.0 | 0.0 | GO:0097460 | ferrous iron import into cell(GO:0097460) |
| 0.0 | 0.6 | GO:0045672 | positive regulation of osteoclast differentiation(GO:0045672) |
| 0.0 | 0.1 | GO:0019243 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.0 | 0.2 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.0 | 0.1 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.0 | 0.1 | GO:0032819 | positive regulation of natural killer cell proliferation(GO:0032819) |
| 0.0 | 0.1 | GO:0046878 | positive regulation of saliva secretion(GO:0046878) |
| 0.0 | 0.1 | GO:0042780 | tRNA 3'-end processing(GO:0042780) |
| 0.0 | 0.1 | GO:0048294 | negative regulation of isotype switching to IgE isotypes(GO:0048294) |
| 0.0 | 0.1 | GO:0070782 | phosphatidylserine exposure on apoptotic cell surface(GO:0070782) |
| 0.0 | 0.2 | GO:0046051 | UTP metabolic process(GO:0046051) |
| 0.0 | 0.1 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) |
| 0.0 | 0.2 | GO:0034244 | negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.0 | 0.1 | GO:0097477 | lateral motor column neuron migration(GO:0097477) |
| 0.0 | 0.1 | GO:1901475 | pyruvate transmembrane transport(GO:1901475) |
| 0.0 | 0.1 | GO:0008065 | establishment of blood-nerve barrier(GO:0008065) |
| 0.0 | 0.1 | GO:0060633 | negative regulation of transcription initiation from RNA polymerase II promoter(GO:0060633) negative regulation of DNA-templated transcription, initiation(GO:2000143) |
| 0.0 | 0.1 | GO:0010626 | negative regulation of Schwann cell proliferation(GO:0010626) |
| 0.0 | 0.1 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.0 | 0.2 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 0.1 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.0 | 0.2 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 | 0.0 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.0 | 0.0 | GO:0060948 | cardiac vascular smooth muscle cell development(GO:0060948) |
| 0.0 | 0.1 | GO:0060903 | positive regulation of meiosis I(GO:0060903) |
| 0.0 | 0.1 | GO:1900454 | positive regulation of long term synaptic depression(GO:1900454) |
| 0.0 | 0.1 | GO:0036112 | medium-chain fatty-acyl-CoA metabolic process(GO:0036112) |
| 0.0 | 0.1 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.0 | 0.2 | GO:0033147 | negative regulation of intracellular estrogen receptor signaling pathway(GO:0033147) |
| 0.0 | 0.1 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.0 | 0.1 | GO:0046271 | phenylpropanoid catabolic process(GO:0046271) |
| 0.0 | 0.1 | GO:0015884 | folic acid transport(GO:0015884) |
| 0.0 | 0.1 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.0 | 0.1 | GO:0008655 | pyrimidine-containing compound salvage(GO:0008655) pyrimidine nucleoside salvage(GO:0043097) |
| 0.0 | 0.0 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.0 | 0.0 | GO:0071895 | odontoblast differentiation(GO:0071895) |
| 0.0 | 0.0 | GO:0090241 | negative regulation of histone H4 acetylation(GO:0090241) |
| 0.0 | 0.1 | GO:0048861 | leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.0 | 0.1 | GO:2000653 | regulation of genetic imprinting(GO:2000653) |
| 0.0 | 0.0 | GO:0010897 | negative regulation of triglyceride catabolic process(GO:0010897) |
| 0.0 | 0.3 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.1 | GO:1903772 | regulation of viral budding via host ESCRT complex(GO:1903772) |
| 0.0 | 0.0 | GO:1900060 | negative regulation of ceramide biosynthetic process(GO:1900060) |
| 0.0 | 0.0 | GO:0015888 | thiamine transport(GO:0015888) |
| 0.0 | 0.1 | GO:0090383 | phagosome acidification(GO:0090383) |
| 0.0 | 0.0 | GO:0002432 | granuloma formation(GO:0002432) |
| 0.0 | 0.0 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.0 | 0.1 | GO:0046543 | development of secondary female sexual characteristics(GO:0046543) |
| 0.0 | 0.1 | GO:0072015 | glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.0 | 0.0 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.0 | 0.0 | GO:2001269 | positive regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001269) |
| 0.0 | 0.0 | GO:0046104 | thymidine metabolic process(GO:0046104) |
| 0.0 | 0.0 | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 0.0 | 0.1 | GO:1903799 | negative regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903799) |
| 0.0 | 0.1 | GO:1900109 | regulation of histone H3-K9 dimethylation(GO:1900109) |
| 0.0 | 0.2 | GO:0009404 | toxin metabolic process(GO:0009404) |
| 0.0 | 0.0 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.0 | 0.1 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.0 | 0.1 | GO:0042219 | cellular modified amino acid catabolic process(GO:0042219) |
| 0.0 | 0.1 | GO:1900194 | negative regulation of oocyte maturation(GO:1900194) |
| 0.0 | 0.1 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.0 | 0.1 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.0 | 0.1 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.0 | 0.1 | GO:0032960 | regulation of inositol trisphosphate biosynthetic process(GO:0032960) |
| 0.0 | 0.1 | GO:0015705 | iodide transport(GO:0015705) |
| 0.0 | 0.0 | GO:0090365 | regulation of mRNA modification(GO:0090365) |
| 0.0 | 0.0 | GO:1900368 | regulation of RNA interference(GO:1900368) |
| 0.0 | 0.1 | GO:0032825 | positive regulation of natural killer cell differentiation(GO:0032825) |
| 0.0 | 0.0 | GO:1901187 | regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.0 | 0.1 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.0 | GO:0072674 | multinuclear osteoclast differentiation(GO:0072674) osteoclast fusion(GO:0072675) |
| 0.0 | 0.0 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.0 | 0.0 | GO:0042939 | glutathione transport(GO:0034635) tripeptide transport(GO:0042939) |
| 0.0 | 0.0 | GO:0003062 | regulation of heart rate by chemical signal(GO:0003062) |
| 0.0 | 0.2 | GO:0006613 | cotranslational protein targeting to membrane(GO:0006613) |
| 0.0 | 0.0 | GO:1903660 | complement-dependent cytotoxicity(GO:0097278) regulation of complement-dependent cytotoxicity(GO:1903659) negative regulation of complement-dependent cytotoxicity(GO:1903660) |
| 0.0 | 0.1 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.0 | 0.0 | GO:0016103 | diterpenoid catabolic process(GO:0016103) retinoic acid catabolic process(GO:0034653) |
| 0.0 | 0.0 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.0 | 0.0 | GO:0033159 | negative regulation of protein import into nucleus, translocation(GO:0033159) |
| 0.0 | 0.1 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.0 | GO:0006166 | purine ribonucleoside salvage(GO:0006166) |
| 0.0 | 0.0 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.0 | 0.1 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.0 | 0.1 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.0 | 0.0 | GO:0044828 | negative regulation by host of viral genome replication(GO:0044828) |
| 0.0 | 0.0 | GO:1901525 | regulation of macromitophagy(GO:1901524) negative regulation of macromitophagy(GO:1901525) |
| 0.0 | 0.1 | GO:0060294 | cilium movement involved in cell motility(GO:0060294) |
| 0.0 | 0.1 | GO:0035814 | negative regulation of renal sodium excretion(GO:0035814) |
| 0.0 | 0.1 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.0 | 0.0 | GO:1902774 | late endosome to lysosome transport(GO:1902774) |
| 0.0 | 0.0 | GO:0045047 | protein targeting to ER(GO:0045047) |
| 0.0 | 0.1 | GO:0071038 | nuclear polyadenylation-dependent tRNA catabolic process(GO:0071038) |
| 0.0 | 0.0 | GO:0006438 | valyl-tRNA aminoacylation(GO:0006438) |
| 0.0 | 0.0 | GO:0060112 | generation of ovulation cycle rhythm(GO:0060112) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.1 | 0.4 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.1 | 0.5 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.1 | 0.7 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.1 | 0.6 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.3 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.0 | 0.1 | GO:0031074 | nucleocytoplasmic shuttling complex(GO:0031074) |
| 0.0 | 0.2 | GO:0032021 | NELF complex(GO:0032021) |
| 0.0 | 0.2 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 0.2 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.0 | 0.3 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.0 | 0.3 | GO:0071437 | invadopodium(GO:0071437) |
| 0.0 | 0.1 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.0 | 0.2 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.0 | 0.1 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 0.0 | 0.1 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.0 | 0.1 | GO:1990761 | growth cone lamellipodium(GO:1990761) |
| 0.0 | 0.1 | GO:0097651 | phosphatidylinositol 3-kinase complex, class I(GO:0097651) |
| 0.0 | 0.2 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.4 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.0 | 0.1 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.0 | 0.1 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.0 | 0.1 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.1 | GO:0000444 | MIS12/MIND type complex(GO:0000444) |
| 0.0 | 0.2 | GO:0070578 | RISC-loading complex(GO:0070578) |
| 0.0 | 0.1 | GO:0031523 | Myb complex(GO:0031523) |
| 0.0 | 0.1 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.0 | 0.1 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.0 | 0.1 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.2 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.0 | 0.2 | GO:0034464 | BBSome(GO:0034464) |
| 0.0 | 0.1 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.0 | 0.1 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 0.1 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 0.1 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 0.1 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 0.1 | GO:0036454 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) insulin-like growth factor ternary complex(GO:0042567) |
| 0.0 | 0.1 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.0 | 0.2 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.4 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 0.1 | GO:0005845 | mRNA cap binding complex(GO:0005845) RNA cap binding complex(GO:0034518) |
| 0.0 | 0.1 | GO:0097433 | dense body(GO:0097433) |
| 0.0 | 0.2 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 0.1 | GO:1990851 | Wnt-Frizzled-LRP5/6 complex(GO:1990851) |
| 0.0 | 0.1 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.0 | 0.2 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.0 | 0.5 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.1 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.2 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.0 | 0.0 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 0.2 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.0 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 0.0 | 0.1 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.1 | 0.6 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.1 | 0.2 | GO:0017153 | sodium:dicarboxylate symporter activity(GO:0017153) |
| 0.1 | 0.2 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.1 | 0.2 | GO:0003941 | L-serine ammonia-lyase activity(GO:0003941) |
| 0.1 | 0.4 | GO:0005072 | transforming growth factor beta receptor, cytoplasmic mediator activity(GO:0005072) |
| 0.1 | 0.2 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.0 | 0.3 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.3 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 0.4 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.0 | 0.2 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.0 | 0.2 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.0 | 0.5 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.1 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.3 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.1 | GO:0001224 | RNA polymerase II transcription cofactor binding(GO:0001224) |
| 0.0 | 0.1 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.0 | 0.2 | GO:0047760 | butyrate-CoA ligase activity(GO:0047760) |
| 0.0 | 0.2 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.0 | 0.6 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.0 | 0.1 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.1 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.0 | 0.1 | GO:0015189 | arginine transmembrane transporter activity(GO:0015181) L-lysine transmembrane transporter activity(GO:0015189) |
| 0.0 | 0.4 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.1 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.0 | 0.2 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.1 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 0.2 | GO:0043426 | MRF binding(GO:0043426) |
| 0.0 | 0.5 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.1 | GO:0030619 | U1 snRNA binding(GO:0030619) |
| 0.0 | 0.5 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.1 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.0 | 0.1 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.0 | 0.1 | GO:0015111 | iodide transmembrane transporter activity(GO:0015111) |
| 0.0 | 0.3 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.1 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 0.1 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.1 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) |
| 0.0 | 0.1 | GO:0004897 | ciliary neurotrophic factor receptor activity(GO:0004897) |
| 0.0 | 0.3 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.1 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.0 | 0.1 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.1 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.0 | 0.1 | GO:0005168 | neurotrophin TRKA receptor binding(GO:0005168) |
| 0.0 | 0.1 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.0 | 0.1 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 0.1 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.1 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.0 | 0.1 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 0.1 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.0 | 0.1 | GO:0008517 | folic acid transporter activity(GO:0008517) |
| 0.0 | 0.1 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.0 | 0.0 | GO:0031697 | beta-1 adrenergic receptor binding(GO:0031697) |
| 0.0 | 0.1 | GO:0035276 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) ethanol binding(GO:0035276) |
| 0.0 | 0.4 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.0 | 0.1 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.1 | GO:0043184 | vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 0.0 | 0.0 | GO:0003985 | acetyl-CoA C-acetyltransferase activity(GO:0003985) C-acetyltransferase activity(GO:0016453) |
| 0.0 | 0.0 | GO:0015563 | uptake transmembrane transporter activity(GO:0015563) |
| 0.0 | 0.0 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.1 | GO:0008948 | oxaloacetate decarboxylase activity(GO:0008948) |
| 0.0 | 0.1 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.0 | 0.1 | GO:0047105 | aminobutyraldehyde dehydrogenase activity(GO:0019145) 4-trimethylammoniobutyraldehyde dehydrogenase activity(GO:0047105) |
| 0.0 | 0.0 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.0 | 0.1 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.1 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.2 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.1 | GO:0031748 | D1 dopamine receptor binding(GO:0031748) |
| 0.0 | 0.1 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.0 | 0.1 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.1 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.0 | 0.1 | GO:0047391 | alkylglycerophosphoethanolamine phosphodiesterase activity(GO:0047391) |
| 0.0 | 0.0 | GO:0004946 | bombesin receptor activity(GO:0004946) |
| 0.0 | 0.1 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.0 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 0.0 | 0.1 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.0 | 0.1 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.0 | 0.1 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.0 | 0.1 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.0 | GO:0070996 | type 1 melanocortin receptor binding(GO:0070996) |
| 0.0 | 0.1 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.0 | 0.1 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.0 | 0.3 | GO:0052770 | coenzyme F390-A hydrolase activity(GO:0052770) coenzyme F390-G hydrolase activity(GO:0052771) |
| 0.0 | 0.0 | GO:0032551 | UTP binding(GO:0002134) pyrimidine ribonucleoside binding(GO:0032551) |
| 0.0 | 0.1 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.0 | 0.1 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.0 | 0.1 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.0 | 0.2 | GO:0004889 | acetylcholine-activated cation-selective channel activity(GO:0004889) |
| 0.0 | 0.0 | GO:0036435 | K48-linked polyubiquitin binding(GO:0036435) |
| 0.0 | 0.1 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.0 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.1 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) |
| 0.0 | 0.1 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.0 | GO:0004832 | valine-tRNA ligase activity(GO:0004832) |
| 0.0 | 0.2 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.1 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.7 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 0.2 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 0.2 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.4 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 0.4 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.5 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.3 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 0.0 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.0 | 0.5 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 0.2 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 0.4 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.0 | 0.1 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.0 | 0.1 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.0 | 0.3 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.3 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.5 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.4 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 0.6 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.7 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |
| 0.0 | 0.3 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.3 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.3 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 0.3 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.2 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.3 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.3 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.5 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.0 | 0.2 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.1 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.0 | 0.4 | REACTOME RORA ACTIVATES CIRCADIAN EXPRESSION | Genes involved in RORA Activates Circadian Expression |
| 0.0 | 0.3 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.4 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 0.1 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.2 | REACTOME TRAF6 MEDIATED INDUCTION OF TAK1 COMPLEX | Genes involved in TRAF6 mediated induction of TAK1 complex |
| 0.0 | 0.2 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.2 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.0 | 0.2 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.0 | 0.1 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.3 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 0.3 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 0.1 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.0 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.1 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |