Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
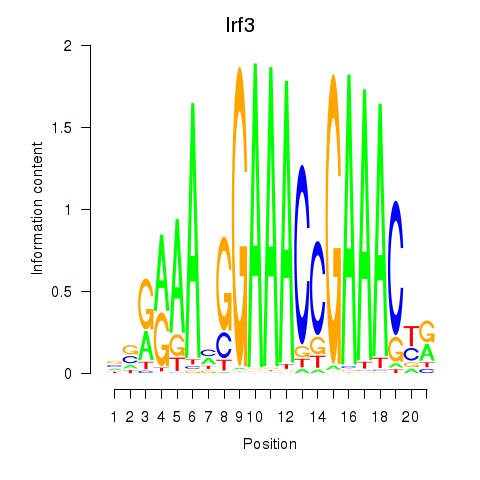
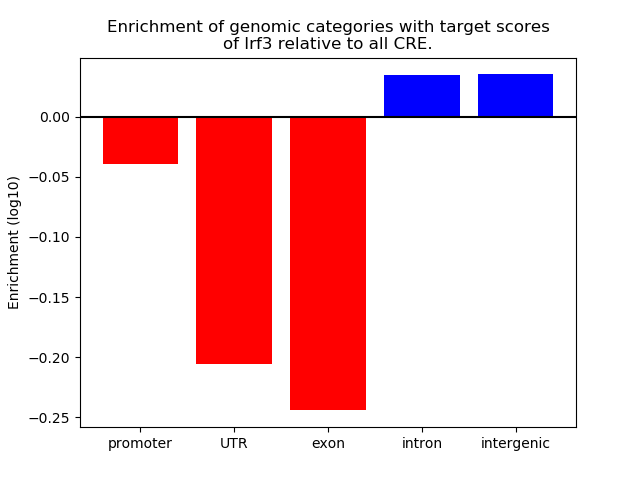
Results for Irf3
Z-value: 2.60
Transcription factors associated with Irf3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Irf3
|
ENSMUSG00000003184.8 | interferon regulatory factor 3 |
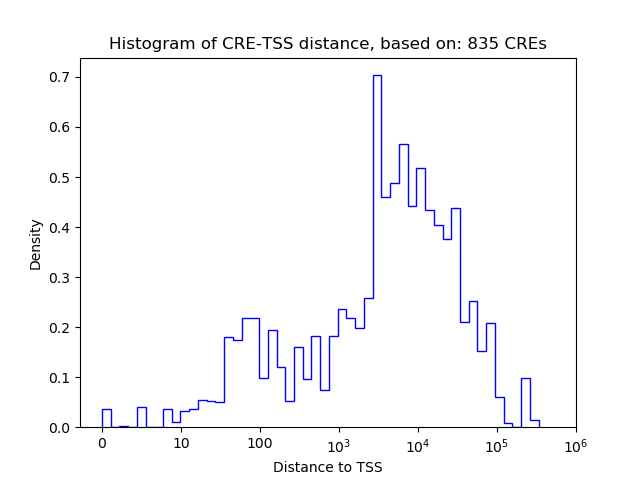
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr7_45000808_45000995 | Irf3 | 153 | 0.842730 | 0.84 | 3.7e-02 | Click! |
| chr7_44997547_44997706 | Irf3 | 22 | 0.496455 | 0.77 | 7.4e-02 | Click! |
| chr7_44999841_45000005 | Irf3 | 244 | 0.731095 | 0.60 | 2.1e-01 | Click! |
| chr7_44997789_44998013 | Irf3 | 48 | 0.774386 | -0.41 | 4.2e-01 | Click! |
| chr7_44998259_44998621 | Irf3 | 114 | 0.665814 | -0.09 | 8.7e-01 | Click! |
Activity of the Irf3 motif across conditions
Conditions sorted by the z-value of the Irf3 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
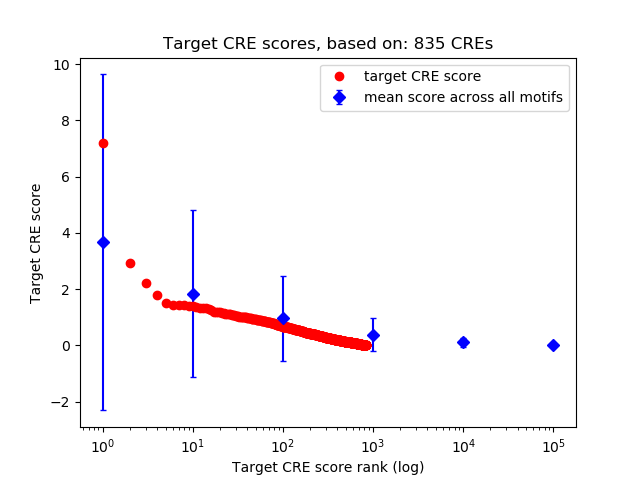
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr5_129701203_129701458 | 7.19 |
Septin14 |
septin 14 |
2929 |
0.16 |
| chr5_103592755_103593053 | 2.91 |
Gm15844 |
predicted gene 15844 |
31567 |
0.12 |
| chr9_32906282_32906466 | 2.22 |
Gm27162 |
predicted gene 27162 |
22592 |
0.19 |
| chr12_79532159_79532332 | 1.79 |
Rad51b |
RAD51 paralog B |
204892 |
0.02 |
| chr14_31903760_31903916 | 1.52 |
D830044D21Rik |
RIKEN cDNA D830044D21 gene |
2690 |
0.29 |
| chr5_36726541_36726703 | 1.45 |
Gm43701 |
predicted gene 43701 |
21996 |
0.11 |
| chr2_72114198_72114385 | 1.43 |
Rapgef4 |
Rap guanine nucleotide exchange factor (GEF) 4 |
26420 |
0.18 |
| chr5_106465177_106465353 | 1.43 |
Gm26872 |
predicted gene, 26872 |
5742 |
0.18 |
| chr4_107285558_107285727 | 1.40 |
Gm12850 |
predicted gene 12850 |
15208 |
0.11 |
| chr11_115199220_115199371 | 1.39 |
Nat9 |
N-acetyltransferase 9 (GCN5-related, putative) |
11436 |
0.11 |
| chr5_86118821_86118972 | 1.36 |
Stap1 |
signal transducing adaptor family member 1 |
32438 |
0.12 |
| chr13_104116230_104116397 | 1.33 |
Sgtb |
small glutamine-rich tetratricopeptide repeat (TPR)-containing, beta |
6484 |
0.18 |
| chr11_53423107_53423311 | 1.33 |
Leap2 |
liver-expressed antimicrobial peptide 2 |
39 |
0.94 |
| chr8_115980743_115980903 | 1.33 |
Gm45733 |
predicted gene 45733 |
8752 |
0.32 |
| chr8_116581400_116581581 | 1.29 |
Dynlrb2 |
dynein light chain roadblock-type 2 |
76475 |
0.1 |
| chr8_46546530_46546687 | 1.24 |
Cenpu |
centromere protein U |
5420 |
0.17 |
| chr11_65776990_65777160 | 1.19 |
Map2k4 |
mitogen-activated protein kinase kinase 4 |
11210 |
0.21 |
| chr1_9876847_9876998 | 1.18 |
Sgk3 |
serum/glucocorticoid regulated kinase 3 |
6545 |
0.15 |
| chr6_142752473_142752975 | 1.17 |
Cmas |
cytidine monophospho-N-acetylneuraminic acid synthetase |
3962 |
0.23 |
| chr2_167062613_167062781 | 1.17 |
Znfx1 |
zinc finger, NFX1-type containing 1 |
19 |
0.72 |
| chr14_45219045_45219298 | 1.14 |
Txndc16 |
thioredoxin domain containing 16 |
193 |
0.78 |
| chr13_39441574_39441735 | 1.13 |
Gm47348 |
predicted gene, 47348 |
1089 |
0.53 |
| chr9_34258652_34258813 | 1.12 |
Gm27161 |
predicted gene 27161 |
58504 |
0.15 |
| chr7_104244343_104244526 | 1.11 |
Trim34a |
tripartite motif-containing 34A |
23 |
0.94 |
| chr4_156200761_156200940 | 1.11 |
Isg15 |
ISG15 ubiquitin-like modifier |
54 |
0.94 |
| chr7_104353298_104353500 | 1.11 |
Trim12c |
tripartite motif-containing 12C |
37 |
0.94 |
| chr9_14717790_14718063 | 1.09 |
Piwil4 |
piwi-like RNA-mediated gene silencing 4 |
547 |
0.6 |
| chr11_48994173_48994324 | 1.09 |
Tgtp1 |
T cell specific GTPase 1 |
76 |
0.93 |
| chr1_164252428_164252629 | 1.06 |
Slc19a2 |
solute carrier family 19 (thiamine transporter), member 2 |
3275 |
0.2 |
| chr4_11147012_11147305 | 1.04 |
Gm11830 |
predicted gene 11830 |
2448 |
0.18 |
| chr11_26413799_26413986 | 1.04 |
Gm12713 |
predicted gene 12713 |
4685 |
0.26 |
| chr6_141333358_141333670 | 1.03 |
Gm10400 |
predicted gene 10400 |
7039 |
0.26 |
| chr12_104230931_104231112 | 1.02 |
Gm8918 |
predicted gene 8918 |
1018 |
0.33 |
| chr2_68434673_68434858 | 1.01 |
Stk39 |
serine/threonine kinase 39 |
37175 |
0.16 |
| chr2_85047886_85048056 | 1.00 |
Tnks1bp1 |
tankyrase 1 binding protein 1 |
51 |
0.96 |
| chr1_85526452_85526618 | 1.00 |
AC147806.2 |
predicted 7592 |
67 |
0.59 |
| chr13_29292838_29292989 | 1.00 |
Gm11364 |
predicted gene 11364 |
56293 |
0.16 |
| chr2_132169135_132169286 | 1.00 |
Slc23a2 |
solute carrier family 23 (nucleobase transporters), member 2 |
24102 |
0.14 |
| chr17_5574750_5574911 | 0.99 |
Zdhhc14 |
zinc finger, DHHC domain containing 14 |
82273 |
0.08 |
| chr16_97535057_97535341 | 0.99 |
Mx2 |
MX dynamin-like GTPase 2 |
109 |
0.96 |
| chr17_31075917_31076069 | 0.98 |
Gm25447 |
predicted gene, 25447 |
16389 |
0.12 |
| chr14_25686626_25686801 | 0.96 |
Ppif |
peptidylprolyl isomerase F (cyclophilin F) |
7441 |
0.14 |
| chr5_100587706_100587862 | 0.96 |
Gm43510 |
predicted gene 43510 |
4972 |
0.16 |
| chr5_21131284_21131470 | 0.96 |
Gm42660 |
predicted gene 42660 |
301 |
0.86 |
| chr10_37145852_37146014 | 0.95 |
5930403N24Rik |
RIKEN cDNA 5930403N24 gene |
4844 |
0.21 |
| chr5_102485247_102485411 | 0.95 |
1700013M08Rik |
RIKEN cDNA 1700013M08 gene |
2040 |
0.35 |
| chr14_17410906_17411057 | 0.94 |
Thrb |
thyroid hormone receptor beta |
249280 |
0.02 |
| chr1_177260954_177261105 | 0.94 |
Akt3 |
thymoma viral proto-oncogene 3 |
2826 |
0.24 |
| chr1_85649860_85650030 | 0.93 |
Sp100 |
nuclear antigen Sp100 |
43 |
0.91 |
| chr3_27267495_27267649 | 0.93 |
Gm37191 |
predicted gene, 37191 |
10488 |
0.19 |
| chr7_83581375_83581526 | 0.92 |
Tmc3 |
transmembrane channel-like gene family 3 |
3477 |
0.19 |
| chr5_136084893_136085068 | 0.91 |
Rasa4 |
RAS p21 protein activator 4 |
90 |
0.93 |
| chr9_79154890_79155051 | 0.91 |
Gm47499 |
predicted gene, 47499 |
47037 |
0.14 |
| chr3_142530130_142530290 | 0.90 |
Gbp7 |
guanylate binding protein 7 |
132 |
0.95 |
| chr3_58719714_58719873 | 0.89 |
4930593A02Rik |
RIKEN cDNA 4930593A02 gene |
13037 |
0.14 |
| chr9_65536208_65536383 | 0.89 |
Gm17749 |
predicted gene, 17749 |
6489 |
0.14 |
| chr4_98923183_98923363 | 0.88 |
Usp1 |
ubiquitin specific peptidase 1 |
537 |
0.77 |
| chr15_58016804_58016970 | 0.88 |
9130401M01Rik |
RIKEN cDNA 9130401M01 gene |
11008 |
0.14 |
| chr17_26753562_26753741 | 0.87 |
Crebrf |
CREB3 regulatory factor |
4051 |
0.18 |
| chr11_21046361_21046512 | 0.86 |
Peli1 |
pellino 1 |
44855 |
0.14 |
| chr4_72122622_72122773 | 0.86 |
Tle1 |
transducin-like enhancer of split 1 |
1963 |
0.41 |
| chr7_104315439_104315612 | 0.85 |
Trim12a |
tripartite motif-containing 12A |
59 |
0.86 |
| chr2_15635875_15636043 | 0.85 |
Gm37595 |
predicted gene, 37595 |
104846 |
0.07 |
| chr6_97464282_97464478 | 0.85 |
Frmd4b |
FERM domain containing 4B |
5094 |
0.23 |
| chr15_58019663_58019825 | 0.84 |
9130401M01Rik |
RIKEN cDNA 9130401M01 gene |
8151 |
0.14 |
| chr17_86470447_86470748 | 0.84 |
Prkce |
protein kinase C, epsilon |
19965 |
0.22 |
| chr4_150784707_150784885 | 0.83 |
Gm13049 |
predicted gene 13049 |
40937 |
0.13 |
| chr1_85327350_85327504 | 0.83 |
Gm2619 |
predicted gene 2619 |
142 |
0.87 |
| chr2_132114570_132114788 | 0.82 |
Slc23a2 |
solute carrier family 23 (nucleobase transporters), member 2 |
3231 |
0.22 |
| chr11_58199417_58199593 | 0.82 |
Igtp |
interferon gamma induced GTPase |
51 |
0.57 |
| chr15_77411058_77411216 | 0.81 |
Apol9a |
apolipoprotein L 9a |
2 |
0.95 |
| chr1_67092581_67092813 | 0.81 |
Cps1 |
carbamoyl-phosphate synthetase 1 |
30329 |
0.17 |
| chr12_111071868_111072033 | 0.81 |
Gm46379 |
predicted gene, 46379 |
3922 |
0.17 |
| chr6_120112510_120112682 | 0.81 |
Ninj2 |
ninjurin 2 |
19246 |
0.17 |
| chr11_100704164_100704495 | 0.80 |
Dhx58 |
DEXH (Asp-Glu-X-His) box polypeptide 58 |
58 |
0.94 |
| chr10_20005611_20005762 | 0.80 |
Map3k5 |
mitogen-activated protein kinase kinase kinase 5 |
54631 |
0.13 |
| chr1_85199424_85199611 | 0.80 |
AC123856.1 |
nuclear antigen Sp100 (Sp100) pseudogene |
96 |
0.49 |
| chr11_60076243_60076400 | 0.79 |
Rai1 |
retinoic acid induced 1 |
28692 |
0.12 |
| chr1_182670647_182670811 | 0.78 |
Gm37516 |
predicted gene, 37516 |
9210 |
0.19 |
| chr17_36042825_36042981 | 0.77 |
Gm6034 |
predicted gene 6034 |
58 |
0.61 |
| chr15_79063149_79063317 | 0.75 |
Ankrd54 |
ankyrin repeat domain 54 |
340 |
0.68 |
| chr7_104288066_104288279 | 0.75 |
Trim5 |
tripartite motif-containing 5 |
78 |
0.93 |
| chr5_21054265_21054635 | 0.75 |
Ptpn12 |
protein tyrosine phosphatase, non-receptor type 12 |
1275 |
0.41 |
| chr3_142496733_142496931 | 0.74 |
Gbp5 |
guanylate binding protein 5 |
68 |
0.97 |
| chr2_84799293_84799459 | 0.73 |
Ube2l6 |
ubiquitin-conjugating enzyme E2L 6 |
542 |
0.62 |
| chr2_48333003_48333182 | 0.73 |
Gm13482 |
predicted gene 13482 |
34184 |
0.18 |
| chr17_35263604_35263786 | 0.73 |
H2-D1 |
histocompatibility 2, D region locus 1 |
599 |
0.4 |
| chr15_77728956_77729123 | 0.73 |
Apol9b |
apolipoprotein L 9b |
0 |
0.95 |
| chr4_86886599_86886793 | 0.72 |
Acer2 |
alkaline ceramidase 2 |
12282 |
0.21 |
| chr5_92093819_92093994 | 0.72 |
G3bp2 |
GTPase activating protein (SH3 domain) binding protein 2 |
10187 |
0.12 |
| chr16_78540459_78540629 | 0.71 |
D16Ertd472e |
DNA segment, Chr 16, ERATO Doi 472, expressed |
19700 |
0.16 |
| chr1_92906015_92906166 | 0.71 |
Dusp28 |
dual specificity phosphatase 28 |
891 |
0.37 |
| chr11_58214796_58214953 | 0.71 |
Irgm2 |
immunity-related GTPase family M member 2 |
134 |
0.93 |
| chr12_79841402_79841571 | 0.69 |
9430078K24Rik |
RIKEN cDNA 9430078K24 gene |
83247 |
0.09 |
| chr1_61374326_61374510 | 0.69 |
9530026F06Rik |
RIKEN cDNA 9530026F06 gene |
4014 |
0.24 |
| chr11_113101446_113101611 | 0.69 |
2610035D17Rik |
RIKEN cDNA 2610035D17 gene |
71549 |
0.12 |
| chr2_131089744_131089895 | 0.69 |
Siglec1 |
sialic acid binding Ig-like lectin 1, sialoadhesin |
3054 |
0.15 |
| chr1_186478678_186478837 | 0.68 |
A730004F24Rik |
RIKEN cDNA A730004F24 gene |
79715 |
0.1 |
| chr3_148998735_148998917 | 0.67 |
Gm43572 |
predicted gene 43572 |
6670 |
0.18 |
| chr2_62645973_62646316 | 0.67 |
Ifih1 |
interferon induced with helicase C domain 1 |
9 |
0.98 |
| chr2_167698008_167698170 | 0.67 |
A530013C23Rik |
RIKEN cDNA A530013C23 gene |
6908 |
0.12 |
| chr9_99491119_99491307 | 0.67 |
Nme9 |
NME/NM23 family member 9 |
20792 |
0.16 |
| chr1_150496695_150496846 | 0.66 |
Prg4 |
proteoglycan 4 (megakaryocyte stimulating factor, articular superficial zone protein) |
30605 |
0.15 |
| chr8_84974794_84975075 | 0.66 |
AC163703.1 |
|
1608 |
0.14 |
| chr2_162976070_162976402 | 0.66 |
Gm11453 |
predicted gene 11453 |
7025 |
0.13 |
| chr16_30959779_30959977 | 0.66 |
Gm15742 |
predicted gene 15742 |
13640 |
0.17 |
| chr10_20674107_20674323 | 0.66 |
Gm17230 |
predicted gene 17230 |
48580 |
0.14 |
| chr9_120721526_120721702 | 0.65 |
Gm47063 |
predicted gene, 47063 |
15377 |
0.13 |
| chr8_107561268_107561433 | 0.65 |
Wwp2 |
WW domain containing E3 ubiquitin protein ligase 2 |
10013 |
0.17 |
| chr1_161951168_161951368 | 0.65 |
4930558K02Rik |
RIKEN cDNA 4930558K02 gene |
17880 |
0.12 |
| chr9_109054788_109055359 | 0.64 |
Shisa5 |
shisa family member 5 |
61 |
0.94 |
| chr17_36020984_36021135 | 0.64 |
H2-T24 |
histocompatibility 2, T region locus 24 |
499 |
0.51 |
| chr4_145640689_145640849 | 0.64 |
Gm13231 |
predicted gene 13231 |
588 |
0.58 |
| chr17_36023597_36023890 | 0.63 |
H2-T24 |
histocompatibility 2, T region locus 24 |
3183 |
0.08 |
| chr7_19563093_19563255 | 0.63 |
Ppp1r37 |
protein phosphatase 1, regulatory subunit 37 |
98 |
0.93 |
| chr7_119527230_119527542 | 0.63 |
Acsm5 |
acyl-CoA synthetase medium-chain family member 5 |
1045 |
0.36 |
| chr2_22623304_22623471 | 0.62 |
Gad2 |
glutamic acid decarboxylase 2 |
83 |
0.96 |
| chr18_84906289_84906638 | 0.62 |
Fbxo15 |
F-box protein 15 |
28319 |
0.13 |
| chr3_135423942_135424111 | 0.62 |
Cisd2 |
CDGSH iron sulfur domain 2 |
101 |
0.94 |
| chr15_74989416_74989586 | 0.62 |
Gm37042 |
predicted gene, 37042 |
751 |
0.35 |
| chr5_9059465_9059641 | 0.61 |
Gm40264 |
predicted gene, 40264 |
24429 |
0.13 |
| chr15_76951965_76952120 | 0.61 |
1110038F14Rik |
RIKEN cDNA 1110038F14 gene |
3293 |
0.13 |
| chr14_66872697_66872883 | 0.61 |
Gm5464 |
predicted gene 5464 |
3940 |
0.18 |
| chr6_22010400_22010560 | 0.61 |
Cped1 |
cadherin-like and PC-esterase domain containing 1 |
6586 |
0.27 |
| chr13_34738568_34738735 | 0.61 |
Fam50b |
family with sequence similarity 50, member B |
961 |
0.45 |
| chr1_156009145_156009296 | 0.59 |
Tor1aip1 |
torsin A interacting protein 1 |
9399 |
0.14 |
| chr9_107975178_107975582 | 0.59 |
Uba7 |
ubiquitin-like modifier activating enzyme 7 |
125 |
0.87 |
| chr18_75378610_75378854 | 0.59 |
Smad7 |
SMAD family member 7 |
3818 |
0.25 |
| chr16_95432244_95432403 | 0.58 |
Erg |
ETS transcription factor |
26922 |
0.22 |
| chr2_156857523_156857697 | 0.58 |
4930518I15Rik |
RIKEN cDNA 4930518I15 gene |
336 |
0.71 |
| chr8_8334587_8334760 | 0.57 |
A630009H07Rik |
RIKEN cDNA A630009H07 gene |
404 |
0.8 |
| chr13_109604491_109604696 | 0.57 |
Pde4d |
phosphodiesterase 4D, cAMP specific |
23188 |
0.29 |
| chr1_152677737_152678058 | 0.57 |
Gm15479 |
predicted gene 15479 |
9604 |
0.19 |
| chr3_127123389_127123738 | 0.57 |
Ank2 |
ankyrin 2, brain |
1299 |
0.38 |
| chr10_81247872_81248124 | 0.56 |
Zfr2 |
zinc finger RNA binding protein 2 |
418 |
0.6 |
| chr19_34553644_34553830 | 0.56 |
Ifit2 |
interferon-induced protein with tetratricopeptide repeats 2 |
3027 |
0.16 |
| chr2_105396213_105396364 | 0.56 |
Rcn1 |
reticulocalbin 1 |
940 |
0.67 |
| chr19_29940298_29940515 | 0.55 |
Il33 |
interleukin 33 |
5384 |
0.21 |
| chr6_121196988_121197205 | 0.55 |
Gm44014 |
predicted gene, 44014 |
1087 |
0.4 |
| chr8_111543847_111544013 | 0.55 |
Znrf1 |
zinc and ring finger 1 |
2671 |
0.26 |
| chr11_48871336_48871517 | 0.55 |
Irgm1 |
immunity-related GTPase family M member 1 |
6 |
0.95 |
| chr10_95297928_95298105 | 0.55 |
Gm48880 |
predicted gene, 48880 |
16837 |
0.14 |
| chr2_122145860_122146082 | 0.55 |
B2m |
beta-2 microglobulin |
1715 |
0.25 |
| chr16_10417919_10418097 | 0.55 |
Nubp1 |
nucleotide binding protein 1 |
3228 |
0.2 |
| chr4_124646730_124646881 | 0.54 |
Pou3f1 |
POU domain, class 3, transcription factor 1 |
10002 |
0.12 |
| chr2_91216142_91216316 | 0.54 |
Acp2 |
acid phosphatase 2, lysosomal |
13301 |
0.09 |
| chr17_36121547_36121705 | 0.54 |
Gm19684 |
predicted gene, 19684 |
52 |
0.61 |
| chr19_47222639_47222810 | 0.54 |
Gm50339 |
predicted gene, 50339 |
815 |
0.48 |
| chr18_68011811_68011962 | 0.54 |
Ldlrad4 |
low density lipoprotein receptor class A domain containing 4 |
78629 |
0.09 |
| chr2_60855128_60855295 | 0.53 |
Rbms1 |
RNA binding motif, single stranded interacting protein 1 |
26227 |
0.23 |
| chr17_71101556_71101726 | 0.53 |
Myom1 |
myomesin 1 |
15245 |
0.15 |
| chr19_11540212_11540374 | 0.53 |
Ms4a4d |
membrane-spanning 4-domains, subfamily A, member 4D |
3492 |
0.12 |
| chr15_76251814_76251971 | 0.53 |
Mir6953 |
microRNA 6953 |
3701 |
0.08 |
| chr6_116107833_116108253 | 0.52 |
Tmcc1 |
transmembrane and coiled coil domains 1 |
184 |
0.72 |
| chr18_32693876_32694200 | 0.52 |
Gm25826 |
predicted gene, 25826 |
71982 |
0.1 |
| chr8_3211565_3211796 | 0.52 |
Insr |
insulin receptor |
18963 |
0.18 |
| chr2_84761940_84762112 | 0.52 |
Serping1 |
serine (or cysteine) peptidase inhibitor, clade G, member 1 |
13102 |
0.08 |
| chr17_27790101_27790294 | 0.52 |
Rps2-ps9 |
ribosomal protein S2, pseudogene 9 |
5773 |
0.14 |
| chr9_78149872_78150023 | 0.51 |
Cilk1 |
ciliogenesis associated kinase 1 |
5376 |
0.13 |
| chr12_99407029_99407580 | 0.51 |
Foxn3 |
forkhead box N3 |
5301 |
0.18 |
| chr11_119393297_119393480 | 0.50 |
Rnf213 |
ring finger protein 213 |
288 |
0.86 |
| chr1_183298173_183298484 | 0.50 |
Aida |
axin interactor, dorsalization associated |
758 |
0.46 |
| chr7_123025255_123025406 | 0.49 |
Gm45846 |
predicted gene 45846 |
6044 |
0.14 |
| chr5_102440458_102440632 | 0.49 |
Gm42932 |
predicted gene 42932 |
24342 |
0.19 |
| chr6_95018081_95018258 | 0.49 |
4930511E03Rik |
RIKEN cDNA 4930511E03 gene |
74335 |
0.09 |
| chr11_88528923_88529091 | 0.49 |
Msi2 |
musashi RNA-binding protein 2 |
61140 |
0.12 |
| chr12_17701834_17702059 | 0.49 |
Hpcal1 |
hippocalcin-like 1 |
11090 |
0.22 |
| chr11_95858752_95858950 | 0.48 |
4833417C18Rik |
RIKEN cDNA 4833417C18 gene |
14 |
0.82 |
| chr4_136213750_136213912 | 0.48 |
Asap3 |
ArfGAP with SH3 domain, ankyrin repeat and PH domain 3 |
7466 |
0.15 |
| chr5_105110364_105110535 | 0.48 |
Gbp9 |
guanylate-binding protein 9 |
172 |
0.95 |
| chr13_23669732_23669907 | 0.47 |
Gm22452 |
predicted gene, 22452 |
6798 |
0.06 |
| chr3_84637962_84638124 | 0.47 |
Tmem154 |
transmembrane protein 154 |
28149 |
0.16 |
| chr15_94948227_94948378 | 0.47 |
Gm23129 |
predicted gene, 23129 |
38836 |
0.22 |
| chr10_128270229_128270557 | 0.47 |
Stat2 |
signal transducer and activator of transcription 2 |
166 |
0.86 |
| chr8_25777362_25777542 | 0.47 |
Bag4 |
BCL2-associated athanogene 4 |
130 |
0.92 |
| chr19_8806965_8807121 | 0.46 |
Zbtb3 |
zinc finger and BTB domain containing 3 |
4488 |
0.05 |
| chr6_87531647_87531807 | 0.46 |
Arhgap25 |
Rho GTPase activating protein 25 |
1508 |
0.29 |
| chr5_105139632_105139812 | 0.46 |
Gbp4 |
guanylate binding protein 4 |
136 |
0.4 |
| chr10_95623611_95623772 | 0.45 |
Gm33336 |
predicted gene, 33336 |
8548 |
0.13 |
| chr7_17090307_17090480 | 0.45 |
Psg16 |
pregnancy specific glycoprotein 16 |
2459 |
0.18 |
| chr9_70761117_70761279 | 0.45 |
Adam10 |
a disintegrin and metallopeptidase domain 10 |
14855 |
0.2 |
| chr3_69264430_69264585 | 0.45 |
Arl14 |
ADP-ribosylation factor-like 14 |
42088 |
0.12 |
| chr18_60211943_60212106 | 0.45 |
Gm4951 |
predicted gene 4951 |
56 |
0.97 |
| chr13_23710505_23710674 | 0.45 |
Hfe |
homeostatic iron regulator |
125 |
0.85 |
| chr10_91183536_91183698 | 0.45 |
Tmpo |
thymopoietin |
2302 |
0.26 |
| chr8_33471202_33471366 | 0.45 |
Hmgb1-rs17 |
high mobility group box 1, related sequence 17 |
12912 |
0.21 |
| chr4_9658072_9658230 | 0.44 |
Asph |
aspartate-beta-hydroxylase |
10935 |
0.2 |
| chr17_86601302_86601488 | 0.44 |
Gm18832 |
predicted gene, 18832 |
84101 |
0.08 |
| chr1_63192366_63192525 | 0.44 |
Gm27512 |
predicted gene, 27512 |
5586 |
0.09 |
| chr4_99185506_99185668 | 0.44 |
Atg4c |
autophagy related 4C, cysteine peptidase |
8347 |
0.18 |
| chr19_43495970_43496152 | 0.43 |
Cnnm1 |
cyclin M1 |
2545 |
0.19 |
| chr15_82643084_82643235 | 0.43 |
AC118710.1 |
cytochrome P450, family 2, subfamily d, polypeptide 13, pseudogene |
1113 |
0.22 |
| chr8_72409462_72409669 | 0.43 |
Eps15l1 |
epidermal growth factor receptor pathway substrate 15-like 1 |
11873 |
0.1 |
| chr11_19016500_19016765 | 0.43 |
Meis1 |
Meis homeobox 1 |
343 |
0.83 |
| chr18_70622479_70622643 | 0.43 |
Mbd2 |
methyl-CpG binding domain protein 2 |
4788 |
0.23 |
| chr7_24884378_24884550 | 0.43 |
Rps19 |
ribosomal protein S19 |
93 |
0.93 |
| chr15_28001392_28001573 | 0.43 |
Trio |
triple functional domain (PTPRF interacting) |
6119 |
0.24 |
| chr3_59228865_59229051 | 0.43 |
P2ry12 |
purinergic receptor P2Y, G-protein coupled 12 |
1377 |
0.34 |
| chr13_107568726_107569034 | 0.43 |
Gm32004 |
predicted gene, 32004 |
3549 |
0.29 |
| chr3_118507933_118508602 | 0.42 |
Gm37773 |
predicted gene, 37773 |
17884 |
0.14 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.0 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.3 | 0.8 | GO:0039534 | negative regulation of MDA-5 signaling pathway(GO:0039534) |
| 0.2 | 0.7 | GO:0006538 | glutamate catabolic process(GO:0006538) |
| 0.2 | 0.6 | GO:0097460 | ferrous iron import into cell(GO:0097460) |
| 0.1 | 0.7 | GO:1900227 | positive regulation of NLRP3 inflammasome complex assembly(GO:1900227) |
| 0.1 | 0.4 | GO:0090365 | regulation of mRNA modification(GO:0090365) |
| 0.1 | 0.4 | GO:0039530 | MDA-5 signaling pathway(GO:0039530) |
| 0.1 | 0.4 | GO:0060266 | negative regulation of respiratory burst involved in inflammatory response(GO:0060266) |
| 0.1 | 0.4 | GO:1901252 | regulation of intracellular transport of viral material(GO:1901252) |
| 0.1 | 0.3 | GO:0010847 | regulation of chromatin assembly(GO:0010847) |
| 0.1 | 0.3 | GO:1904139 | microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) |
| 0.1 | 0.3 | GO:0032687 | negative regulation of interferon-alpha production(GO:0032687) |
| 0.1 | 0.3 | GO:0001692 | histamine metabolic process(GO:0001692) |
| 0.1 | 2.3 | GO:0035458 | cellular response to interferon-beta(GO:0035458) |
| 0.1 | 1.4 | GO:0044406 | adhesion of symbiont to host(GO:0044406) |
| 0.1 | 0.4 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.1 | 0.4 | GO:0090086 | negative regulation of protein deubiquitination(GO:0090086) |
| 0.1 | 0.7 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.1 | 0.2 | GO:0002741 | positive regulation of cytokine secretion involved in immune response(GO:0002741) |
| 0.1 | 0.2 | GO:0002036 | regulation of L-glutamate transport(GO:0002036) |
| 0.1 | 0.2 | GO:0090283 | regulation of protein glycosylation in Golgi(GO:0090283) |
| 0.1 | 0.3 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.1 | 0.2 | GO:1902809 | regulation of skeletal muscle fiber differentiation(GO:1902809) |
| 0.1 | 0.2 | GO:0015888 | thiamine transport(GO:0015888) |
| 0.1 | 0.4 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.1 | 1.3 | GO:0034340 | response to type I interferon(GO:0034340) |
| 0.1 | 0.2 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.1 | 0.2 | GO:0051562 | negative regulation of mitochondrial calcium ion concentration(GO:0051562) |
| 0.1 | 0.1 | GO:0002488 | antigen processing and presentation of endogenous peptide antigen via MHC class Ib via ER pathway(GO:0002488) antigen processing and presentation of endogenous peptide antigen via MHC class Ib via ER pathway, TAP-dependent(GO:0002489) |
| 0.1 | 0.5 | GO:0002485 | antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway(GO:0002484) antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway, TAP-dependent(GO:0002485) |
| 0.1 | 0.2 | GO:0098904 | regulation of AV node cell action potential(GO:0098904) |
| 0.1 | 0.2 | GO:0045006 | DNA deamination(GO:0045006) |
| 0.1 | 0.2 | GO:1901078 | negative regulation of relaxation of muscle(GO:1901078) |
| 0.1 | 0.2 | GO:0055059 | asymmetric neuroblast division(GO:0055059) |
| 0.1 | 0.2 | GO:0002309 | T cell proliferation involved in immune response(GO:0002309) |
| 0.1 | 0.4 | GO:0019852 | L-ascorbic acid metabolic process(GO:0019852) |
| 0.0 | 0.2 | GO:1902093 | positive regulation of sperm motility(GO:1902093) |
| 0.0 | 0.3 | GO:0090188 | negative regulation of pancreatic juice secretion(GO:0090188) |
| 0.0 | 0.2 | GO:2000015 | regulation of determination of dorsal identity(GO:2000015) |
| 0.0 | 0.1 | GO:2000275 | regulation of oxidative phosphorylation uncoupler activity(GO:2000275) |
| 0.0 | 0.2 | GO:2000504 | positive regulation of blood vessel remodeling(GO:2000504) |
| 0.0 | 0.2 | GO:1902459 | positive regulation of stem cell population maintenance(GO:1902459) |
| 0.0 | 0.7 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.0 | 0.1 | GO:0002432 | granuloma formation(GO:0002432) |
| 0.0 | 0.3 | GO:0032464 | positive regulation of protein homooligomerization(GO:0032464) |
| 0.0 | 1.0 | GO:0042832 | defense response to protozoan(GO:0042832) |
| 0.0 | 0.3 | GO:1904417 | regulation of xenophagy(GO:1904415) positive regulation of xenophagy(GO:1904417) |
| 0.0 | 0.1 | GO:0018197 | peptidyl-aspartic acid modification(GO:0018197) |
| 0.0 | 0.4 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 0.1 | GO:0060300 | regulation of cytokine activity(GO:0060300) |
| 0.0 | 0.1 | GO:0042939 | glutathione transport(GO:0034635) tripeptide transport(GO:0042939) |
| 0.0 | 0.2 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.0 | 0.2 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.0 | 0.1 | GO:0034436 | glycoprotein transport(GO:0034436) |
| 0.0 | 0.1 | GO:0043321 | regulation of natural killer cell degranulation(GO:0043321) |
| 0.0 | 0.1 | GO:0051025 | negative regulation of immunoglobulin secretion(GO:0051025) |
| 0.0 | 0.9 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.0 | 0.1 | GO:0051088 | PMA-inducible membrane protein ectodomain proteolysis(GO:0051088) |
| 0.0 | 0.1 | GO:0052055 | modulation by symbiont of host molecular function(GO:0052055) |
| 0.0 | 0.1 | GO:0061366 | behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 0.0 | 0.2 | GO:0035404 | histone-serine phosphorylation(GO:0035404) |
| 0.0 | 0.1 | GO:1902309 | negative regulation of peptidyl-serine dephosphorylation(GO:1902309) |
| 0.0 | 0.2 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.0 | 0.1 | GO:0010528 | regulation of transposition(GO:0010528) negative regulation of transposition(GO:0010529) |
| 0.0 | 0.1 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.0 | 0.2 | GO:0044539 | long-chain fatty acid import(GO:0044539) |
| 0.0 | 0.2 | GO:0000820 | regulation of glutamine family amino acid metabolic process(GO:0000820) |
| 0.0 | 0.1 | GO:0060830 | ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) |
| 0.0 | 0.1 | GO:0042536 | negative regulation of tumor necrosis factor biosynthetic process(GO:0042536) |
| 0.0 | 0.1 | GO:2000418 | positive regulation of eosinophil migration(GO:2000418) |
| 0.0 | 0.1 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.0 | 0.3 | GO:0010801 | negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 0.0 | 0.1 | GO:0021764 | amygdala development(GO:0021764) |
| 0.0 | 0.2 | GO:0085020 | protein K6-linked ubiquitination(GO:0085020) |
| 0.0 | 0.1 | GO:0018101 | protein citrullination(GO:0018101) |
| 0.0 | 0.1 | GO:1904252 | negative regulation of bile acid biosynthetic process(GO:0070858) negative regulation of bile acid metabolic process(GO:1904252) |
| 0.0 | 0.2 | GO:0048102 | autophagic cell death(GO:0048102) |
| 0.0 | 0.1 | GO:0072697 | protein localization to cell cortex(GO:0072697) |
| 0.0 | 0.0 | GO:0097252 | oligodendrocyte apoptotic process(GO:0097252) |
| 0.0 | 0.1 | GO:1900045 | negative regulation of protein K63-linked ubiquitination(GO:1900045) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.0 | 0.1 | GO:0045409 | negative regulation of interleukin-6 biosynthetic process(GO:0045409) |
| 0.0 | 0.1 | GO:1901856 | negative regulation of cellular respiration(GO:1901856) |
| 0.0 | 0.3 | GO:0009226 | nucleotide-sugar biosynthetic process(GO:0009226) |
| 0.0 | 0.1 | GO:0090435 | protein localization to nuclear envelope(GO:0090435) |
| 0.0 | 0.1 | GO:0002913 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.0 | 0.1 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.0 | 0.0 | GO:0002536 | respiratory burst involved in inflammatory response(GO:0002536) |
| 0.0 | 0.1 | GO:0032907 | transforming growth factor beta3 production(GO:0032907) regulation of transforming growth factor beta3 production(GO:0032910) |
| 0.0 | 0.1 | GO:0097211 | response to gonadotropin-releasing hormone(GO:0097210) cellular response to gonadotropin-releasing hormone(GO:0097211) |
| 0.0 | 0.1 | GO:0016344 | meiotic chromosome movement towards spindle pole(GO:0016344) |
| 0.0 | 0.2 | GO:1903405 | protein localization to nuclear body(GO:1903405) protein localization to Cajal body(GO:1904867) regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) protein localization to nucleoplasm(GO:1990173) |
| 0.0 | 0.1 | GO:0050747 | positive regulation of lipoprotein metabolic process(GO:0050747) |
| 0.0 | 0.1 | GO:0034145 | positive regulation of toll-like receptor 4 signaling pathway(GO:0034145) |
| 0.0 | 0.1 | GO:1902990 | mitotic telomere maintenance via semi-conservative replication(GO:1902990) |
| 0.0 | 0.2 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 0.0 | 0.1 | GO:0033058 | directional locomotion(GO:0033058) |
| 0.0 | 0.2 | GO:0032607 | interferon-alpha production(GO:0032607) |
| 0.0 | 0.2 | GO:1902033 | regulation of hematopoietic stem cell proliferation(GO:1902033) |
| 0.0 | 0.0 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.0 | 0.1 | GO:0035457 | cellular response to interferon-alpha(GO:0035457) |
| 0.0 | 0.0 | GO:0030885 | regulation of myeloid dendritic cell activation(GO:0030885) |
| 0.0 | 0.6 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.0 | 0.1 | GO:0006203 | dGTP catabolic process(GO:0006203) |
| 0.0 | 0.1 | GO:0051697 | protein delipidation(GO:0051697) |
| 0.0 | 0.6 | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) |
| 0.0 | 0.1 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.0 | 0.3 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 0.1 | GO:0030167 | proteoglycan catabolic process(GO:0030167) |
| 0.0 | 0.1 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.0 | 0.1 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.0 | 0.1 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.0 | 0.2 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.0 | 0.1 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.0 | 0.3 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.0 | 0.0 | GO:1900044 | regulation of protein K63-linked ubiquitination(GO:1900044) |
| 0.0 | 0.1 | GO:0046341 | CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.0 | 0.1 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 0.0 | GO:0097527 | necroptotic signaling pathway(GO:0097527) |
| 0.0 | 0.1 | GO:0001927 | exocyst assembly(GO:0001927) |
| 0.0 | 0.1 | GO:0019478 | D-amino acid catabolic process(GO:0019478) |
| 0.0 | 0.1 | GO:0031584 | activation of phospholipase D activity(GO:0031584) |
| 0.0 | 0.0 | GO:0034165 | positive regulation of toll-like receptor 9 signaling pathway(GO:0034165) |
| 0.0 | 0.0 | GO:0045357 | interferon-beta biosynthetic process(GO:0045350) regulation of interferon-beta biosynthetic process(GO:0045357) |
| 0.0 | 0.2 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.0 | 0.0 | GO:0046504 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) cellular lipid biosynthetic process(GO:0097384) ether biosynthetic process(GO:1901503) |
| 0.0 | 0.0 | GO:0051891 | positive regulation of cardioblast differentiation(GO:0051891) |
| 0.0 | 0.0 | GO:0046104 | thymidine metabolic process(GO:0046104) |
| 0.0 | 0.1 | GO:0010452 | histone H3-K36 methylation(GO:0010452) |
| 0.0 | 0.0 | GO:1902075 | cellular response to salt(GO:1902075) |
| 0.0 | 0.1 | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.0 | 0.0 | GO:0030578 | PML body organization(GO:0030578) |
| 0.0 | 0.0 | GO:0045343 | MHC class I biosynthetic process(GO:0045341) regulation of MHC class I biosynthetic process(GO:0045343) positive regulation of MHC class I biosynthetic process(GO:0045345) |
| 0.0 | 0.4 | GO:0010259 | multicellular organism aging(GO:0010259) |
| 0.0 | 0.0 | GO:0001794 | type IIa hypersensitivity(GO:0001794) regulation of type IIa hypersensitivity(GO:0001796) positive regulation of type IIa hypersensitivity(GO:0001798) type II hypersensitivity(GO:0002445) regulation of type II hypersensitivity(GO:0002892) positive regulation of type II hypersensitivity(GO:0002894) |
| 0.0 | 0.1 | GO:0051770 | positive regulation of nitric-oxide synthase biosynthetic process(GO:0051770) |
| 0.0 | 0.3 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.0 | 0.0 | GO:1903242 | regulation of cardiac muscle adaptation(GO:0010612) regulation of cardiac muscle hypertrophy in response to stress(GO:1903242) |
| 0.0 | 0.0 | GO:0038171 | cannabinoid signaling pathway(GO:0038171) endocannabinoid signaling pathway(GO:0071926) |
| 0.0 | 0.4 | GO:0000002 | mitochondrial genome maintenance(GO:0000002) |
| 0.0 | 0.1 | GO:2000258 | negative regulation of complement activation(GO:0045916) negative regulation of protein activation cascade(GO:2000258) |
| 0.0 | 0.0 | GO:0002874 | regulation of chronic inflammatory response to antigenic stimulus(GO:0002874) |
| 0.0 | 0.1 | GO:0070234 | positive regulation of T cell apoptotic process(GO:0070234) |
| 0.0 | 0.1 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.0 | 0.0 | GO:0070777 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.0 | 0.1 | GO:0080154 | regulation of fertilization(GO:0080154) |
| 0.0 | 0.0 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) negative regulation of cellular response to vascular endothelial growth factor stimulus(GO:1902548) |
| 0.0 | 0.1 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.0 | 0.1 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.0 | 0.0 | GO:0002337 | B-1a B cell differentiation(GO:0002337) |
| 0.0 | 0.0 | GO:0030423 | targeting of mRNA for destruction involved in RNA interference(GO:0030423) |
| 0.0 | 0.1 | GO:0006977 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) |
| 0.0 | 0.0 | GO:0030070 | insulin processing(GO:0030070) |
| 0.0 | 0.1 | GO:0010991 | regulation of SMAD protein complex assembly(GO:0010990) negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.0 | 0.0 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 | 0.1 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.0 | 0.0 | GO:0036112 | medium-chain fatty-acyl-CoA metabolic process(GO:0036112) |
| 0.0 | 0.1 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) |
| 0.0 | 0.0 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.0 | 0.0 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.0 | 0.1 | GO:0042985 | negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 0.0 | 0.0 | GO:0006499 | N-terminal protein myristoylation(GO:0006499) |
| 0.0 | 0.1 | GO:0060789 | hair follicle placode formation(GO:0060789) |
| 0.0 | 0.1 | GO:1904707 | positive regulation of vascular smooth muscle cell proliferation(GO:1904707) |
| 0.0 | 0.0 | GO:0071677 | positive regulation of mononuclear cell migration(GO:0071677) |
| 0.0 | 0.3 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.0 | GO:0034093 | positive regulation of maintenance of sister chromatid cohesion(GO:0034093) positive regulation of maintenance of mitotic sister chromatid cohesion(GO:0034184) |
| 0.0 | 0.2 | GO:0006474 | N-terminal protein amino acid acetylation(GO:0006474) |
| 0.0 | 0.6 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 0.0 | GO:0043031 | negative regulation of macrophage activation(GO:0043031) |
| 0.0 | 0.0 | GO:0032911 | negative regulation of transforming growth factor beta1 production(GO:0032911) |
| 0.0 | 0.0 | GO:0002322 | B cell proliferation involved in immune response(GO:0002322) |
| 0.0 | 0.0 | GO:2001180 | negative regulation of interleukin-10 secretion(GO:2001180) |
| 0.0 | 0.1 | GO:0014049 | positive regulation of glutamate secretion(GO:0014049) |
| 0.0 | 0.1 | GO:0000042 | protein targeting to Golgi(GO:0000042) |
| 0.0 | 0.0 | GO:2001171 | positive regulation of ATP biosynthetic process(GO:2001171) |
| 0.0 | 0.2 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.0 | 0.2 | GO:0099514 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.0 | 0.0 | GO:0061153 | trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 0.0 | 0.2 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.0 | 0.0 | GO:0099558 | maintenance of synapse structure(GO:0099558) |
| 0.0 | 0.0 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.0 | 0.1 | GO:1903147 | negative regulation of mitophagy(GO:1903147) |
| 0.0 | 0.3 | GO:0010762 | regulation of fibroblast migration(GO:0010762) |
| 0.0 | 0.0 | GO:1901299 | negative regulation of hydrogen peroxide-mediated programmed cell death(GO:1901299) |
| 0.0 | 0.4 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.0 | 0.1 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) |
| 0.0 | 0.1 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.1 | GO:1901407 | regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.0 | 0.1 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.0 | 0.0 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.0 | 0.0 | GO:0051791 | medium-chain fatty acid metabolic process(GO:0051791) |
| 0.0 | 0.0 | GO:2001046 | positive regulation of integrin-mediated signaling pathway(GO:2001046) |
| 0.0 | 0.0 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.0 | 0.5 | GO:0019882 | antigen processing and presentation(GO:0019882) |
| 0.0 | 0.0 | GO:0019230 | proprioception(GO:0019230) |
| 0.0 | 0.0 | GO:0017187 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.8 | GO:1990462 | omegasome(GO:1990462) |
| 0.3 | 2.4 | GO:0020003 | symbiont-containing vacuole(GO:0020003) |
| 0.1 | 0.7 | GO:0071547 | piP-body(GO:0071547) |
| 0.1 | 0.6 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.1 | 0.9 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.1 | 0.3 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.1 | 1.0 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.1 | 0.2 | GO:0042585 | germinal vesicle(GO:0042585) |
| 0.1 | 0.2 | GO:0071256 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.0 | 0.3 | GO:0005767 | secondary lysosome(GO:0005767) |
| 0.0 | 0.1 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.2 | GO:0042825 | TAP complex(GO:0042825) |
| 0.0 | 0.1 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.2 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 0.0 | 0.2 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.6 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 0.3 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.4 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 0.3 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 0.1 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.6 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.1 | GO:0036488 | CHOP-C/EBP complex(GO:0036488) |
| 0.0 | 0.3 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.1 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.0 | 0.1 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 0.1 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.0 | 0.1 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.0 | 0.1 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.0 | 0.2 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 0.1 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.0 | 0.3 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.0 | 0.1 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.6 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.1 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.2 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.7 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.1 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.2 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 0.1 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.0 | 0.1 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.0 | 0.1 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.5 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.0 | GO:0010009 | cytoplasmic side of endosome membrane(GO:0010009) |
| 0.0 | 0.0 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 0.0 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.0 | 0.1 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 0.3 | GO:0022624 | proteasome accessory complex(GO:0022624) |
| 0.0 | 0.2 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.0 | 0.0 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.0 | 0.1 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.0 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.1 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.0 | 0.1 | GO:0031264 | death-inducing signaling complex(GO:0031264) |
| 0.0 | 0.0 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.0 | 0.0 | GO:0032127 | dense core granule membrane(GO:0032127) |
| 0.0 | 0.0 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.0 | 0.4 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.2 | 0.7 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.2 | 0.5 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.2 | 0.6 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.1 | 0.4 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.1 | 1.8 | GO:0008329 | signaling pattern recognition receptor activity(GO:0008329) pattern recognition receptor activity(GO:0038187) |
| 0.1 | 1.0 | GO:0031386 | protein tag(GO:0031386) |
| 0.1 | 0.5 | GO:0090482 | vitamin transmembrane transporter activity(GO:0090482) |
| 0.1 | 0.3 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.1 | 0.2 | GO:0015440 | peptide-transporting ATPase activity(GO:0015440) MHC class Ib protein binding(GO:0023029) |
| 0.1 | 0.1 | GO:0032139 | dinucleotide insertion or deletion binding(GO:0032139) |
| 0.1 | 0.3 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.1 | 0.4 | GO:1990459 | transferrin receptor binding(GO:1990459) |
| 0.1 | 0.2 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.1 | 0.4 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.1 | 0.3 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.1 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.0 | 0.2 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 0.0 | 0.1 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.0 | 0.2 | GO:0044682 | N-cyclopropylmelamine deaminase activity(GO:0034547) N-cyclopropylammeline deaminase activity(GO:0034548) N-cyclopropylammelide alkylamino hydrolase activity(GO:0034549) 2,5-diamino-6-ribitylamino-4(3H)-pyrimidinone 5'-phosphate deaminase activity(GO:0043723) tRNA-specific adenosine-37 deaminase activity(GO:0043829) archaeal-specific GTP cyclohydrolase activity(GO:0044682) tRNA-specific adenosine-34 deaminase activity(GO:0052717) |
| 0.0 | 0.7 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.1 | GO:0034040 | lipid-transporting ATPase activity(GO:0034040) |
| 0.0 | 0.2 | GO:0005072 | transforming growth factor beta receptor, cytoplasmic mediator activity(GO:0005072) |
| 0.0 | 0.1 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.0 | 1.1 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.0 | 0.1 | GO:0016520 | growth hormone-releasing hormone receptor activity(GO:0016520) |
| 0.0 | 0.1 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.0 | 0.1 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.1 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 0.0 | 0.4 | GO:0070567 | cytidylyltransferase activity(GO:0070567) |
| 0.0 | 0.3 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.2 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.4 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.1 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.0 | 0.1 | GO:0033677 | DNA/RNA helicase activity(GO:0033677) |
| 0.0 | 0.2 | GO:0047760 | butyrate-CoA ligase activity(GO:0047760) |
| 0.0 | 0.1 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.0 | 0.2 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.1 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.0 | 0.1 | GO:0080130 | L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.0 | 0.1 | GO:0004305 | ethanolamine kinase activity(GO:0004305) |
| 0.0 | 0.5 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.7 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 4.8 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 0.1 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.0 | 0.1 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 0.3 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.0 | 0.1 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.0 | 0.1 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.2 | GO:0001608 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.1 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 0.2 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.0 | 0.1 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.1 | GO:0002094 | polyprenyltransferase activity(GO:0002094) |
| 0.0 | 0.1 | GO:0052658 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) |
| 0.0 | 0.6 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 0.1 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.0 | 0.1 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.0 | 0.1 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 0.2 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.0 | 0.1 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 1.2 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 0.1 | GO:0035197 | siRNA binding(GO:0035197) |
| 0.0 | 0.1 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.0 | 0.1 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 0.1 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.0 | 0.5 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.2 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.0 | 0.1 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.0 | 0.4 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 0.1 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.4 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.0 | 0.4 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) cGMP-inhibited cyclic-nucleotide phosphodiesterase activity(GO:0004119) photoreceptor cyclic-nucleotide phosphodiesterase activity(GO:0004120) 7,8-dihydro-D-neopterin 2',3'-cyclic phosphate phosphodiesterase activity(GO:0044688) inositol phosphosphingolipid phospholipase activity(GO:0052712) inositol phosphorylceramide phospholipase activity(GO:0052713) mannosyl-inositol phosphorylceramide phospholipase activity(GO:0052714) mannosyl-diinositol phosphorylceramide phospholipase activity(GO:0052715) |
| 0.0 | 0.0 | GO:0017098 | sulfonylurea receptor binding(GO:0017098) |
| 0.0 | 0.2 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.0 | 0.1 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.1 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.0 | 0.0 | GO:0051733 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) |
| 0.0 | 0.1 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 0.7 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 0.1 | GO:0016215 | acyl-CoA desaturase activity(GO:0016215) |
| 0.0 | 0.1 | GO:0032554 | purine deoxyribonucleotide binding(GO:0032554) |
| 0.0 | 0.3 | GO:0015301 | anion:anion antiporter activity(GO:0015301) |
| 0.0 | 0.0 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.0 | 0.1 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 0.1 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.0 | 0.0 | GO:0015038 | glutathione disulfide oxidoreductase activity(GO:0015038) |
| 0.0 | 0.0 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.0 | 0.1 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.0 | 0.2 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.1 | GO:0036122 | BMP binding(GO:0036122) |
| 0.0 | 0.0 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.0 | 0.0 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.1 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.1 | GO:0017108 | 5'-flap endonuclease activity(GO:0017108) |
| 0.0 | 0.0 | GO:0017116 | single-stranded DNA-dependent ATP-dependent DNA helicase activity(GO:0017116) |
| 0.0 | 0.5 | GO:0005080 | protein kinase C binding(GO:0005080) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.0 | 1.1 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.3 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 0.1 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.0 | 0.2 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.9 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 0.6 | ST GA13 PATHWAY | G alpha 13 Pathway |
| 0.0 | 0.3 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.5 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.0 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.1 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 0.2 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 0.6 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.1 | PID IL5 PATHWAY | IL5-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.1 | 2.9 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.1 | 0.2 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.1 | 1.0 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.1 | 1.7 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.1 | 2.2 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.1 | 0.5 | REACTOME RIG I MDA5 MEDIATED INDUCTION OF IFN ALPHA BETA PATHWAYS | Genes involved in RIG-I/MDA5 mediated induction of IFN-alpha/beta pathways |
| 0.0 | 0.1 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.0 | 0.2 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.1 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 0.3 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.3 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.0 | 0.4 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.0 | 0.4 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 0.2 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.2 | REACTOME ER PHAGOSOME PATHWAY | Genes involved in ER-Phagosome pathway |
| 0.0 | 0.2 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.1 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.0 | REACTOME SIGNALING BY ERBB2 | Genes involved in Signaling by ERBB2 |
| 0.0 | 0.3 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.2 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.2 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.1 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.2 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 0.0 | REACTOME TRANSPORT OF MATURE MRNA DERIVED FROM AN INTRONLESS TRANSCRIPT | Genes involved in Transport of Mature mRNA Derived from an Intronless Transcript |
| 0.0 | 0.1 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.0 | REACTOME TRANSCRIPTIONAL ACTIVITY OF SMAD2 SMAD3 SMAD4 HETEROTRIMER | Genes involved in Transcriptional activity of SMAD2/SMAD3:SMAD4 heterotrimer |
| 0.0 | 0.3 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.1 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.2 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 0.1 | REACTOME P53 DEPENDENT G1 DNA DAMAGE RESPONSE | Genes involved in p53-Dependent G1 DNA Damage Response |
| 0.0 | 0.1 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 0.1 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.4 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |