Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
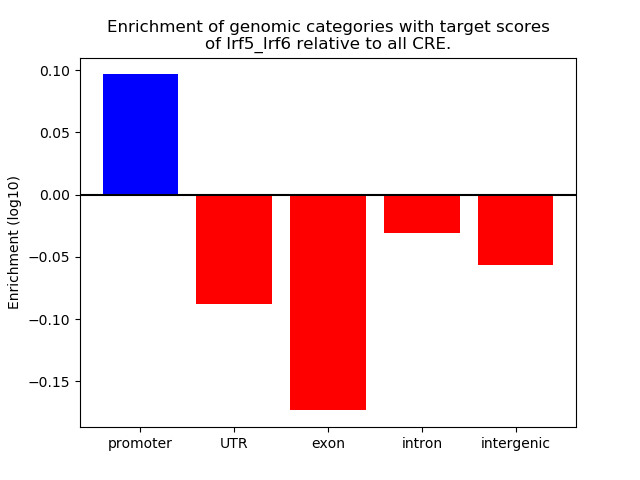
Results for Irf5_Irf6
Z-value: 0.93
Transcription factors associated with Irf5_Irf6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Irf5
|
ENSMUSG00000029771.6 | interferon regulatory factor 5 |
|
Irf6
|
ENSMUSG00000026638.9 | interferon regulatory factor 6 |
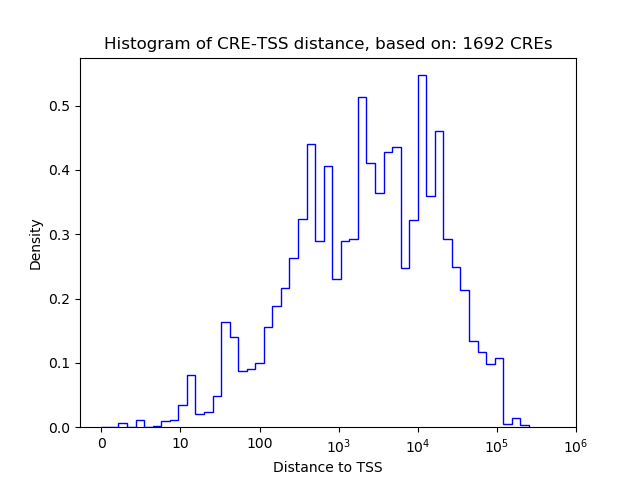
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr6_29523942_29524117 | Irf5 | 2596 | 0.161909 | -0.95 | 3.8e-03 | Click! |
| chr6_29538799_29539193 | Irf5 | 2150 | 0.196327 | -0.89 | 1.8e-02 | Click! |
| chr6_29522778_29522945 | Irf5 | 3764 | 0.131636 | -0.76 | 7.8e-02 | Click! |
| chr6_29522974_29523209 | Irf5 | 3534 | 0.135510 | -0.74 | 9.1e-02 | Click! |
| chr6_29526985_29527412 | Irf5 | 490 | 0.680948 | 0.70 | 1.2e-01 | Click! |
| chr1_193170032_193170189 | Irf6 | 1607 | 0.218081 | -0.90 | 1.5e-02 | Click! |
| chr1_193153538_193153923 | Irf6 | 576 | 0.632924 | 0.89 | 1.7e-02 | Click! |
| chr1_193169761_193169948 | Irf6 | 1351 | 0.254104 | 0.65 | 1.6e-01 | Click! |
| chr1_193153972_193154171 | Irf6 | 917 | 0.438303 | 0.60 | 2.1e-01 | Click! |
| chr1_193159245_193159427 | Irf6 | 2667 | 0.165053 | -0.54 | 2.7e-01 | Click! |
Activity of the Irf5_Irf6 motif across conditions
Conditions sorted by the z-value of the Irf5_Irf6 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
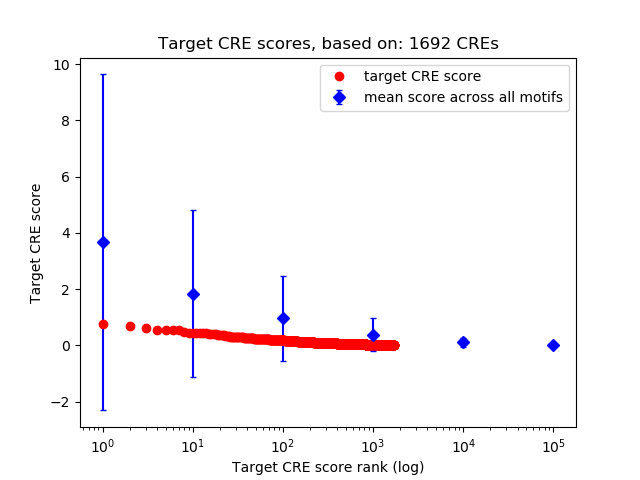
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr6_6206187_6206411 | 0.75 |
Slc25a13 |
solute carrier family 25 (mitochondrial carrier, adenine nucleotide translocator), member 13 |
10819 |
0.23 |
| chr6_88641424_88641714 | 0.69 |
Kbtbd12 |
kelch repeat and BTB (POZ) domain containing 12 |
3619 |
0.23 |
| chr19_46645853_46646011 | 0.62 |
Gm36602 |
predicted gene, 36602 |
20194 |
0.11 |
| chr9_65675680_65676009 | 0.56 |
Oaz2 |
ornithine decarboxylase antizyme 2 |
704 |
0.64 |
| chr13_31808871_31809301 | 0.55 |
Foxc1 |
forkhead box C1 |
2453 |
0.26 |
| chr6_134538835_134539031 | 0.54 |
Lrp6 |
low density lipoprotein receptor-related protein 6 |
2781 |
0.28 |
| chr13_101811863_101812015 | 0.53 |
Gm19108 |
predicted gene, 19108 |
30367 |
0.15 |
| chr9_56929068_56929260 | 0.48 |
Snx33 |
sorting nexin 33 |
793 |
0.44 |
| chr13_13994363_13994818 | 0.45 |
B3galnt2 |
UDP-GalNAc:betaGlcNAc beta 1,3-galactosaminyltransferase, polypeptide 2 |
170 |
0.92 |
| chr2_147961290_147961525 | 0.44 |
9030622O22Rik |
RIKEN cDNA 9030622O22 gene |
14778 |
0.22 |
| chr11_101070404_101070591 | 0.44 |
Naglu |
alpha-N-acetylglucosaminidase (Sanfilippo disease IIIB) |
485 |
0.62 |
| chr9_14381410_14381579 | 0.44 |
Endod1 |
endonuclease domain containing 1 |
13 |
0.96 |
| chr1_138617784_138617983 | 0.42 |
Nek7 |
NIMA (never in mitosis gene a)-related expressed kinase 7 |
1791 |
0.35 |
| chr9_9230047_9230379 | 0.42 |
Gm16833 |
predicted gene, 16833 |
6075 |
0.24 |
| chr11_88614641_88614847 | 0.40 |
Msi2 |
musashi RNA-binding protein 2 |
24597 |
0.21 |
| chr1_130761129_130761587 | 0.39 |
Gm28856 |
predicted gene 28856 |
5026 |
0.11 |
| chr2_128377797_128377975 | 0.39 |
Morrbid |
myeloid RNA regulator of BCL2L11 induced cell death |
25079 |
0.18 |
| chr6_108506487_108506825 | 0.39 |
Itpr1 |
inositol 1,4,5-trisphosphate receptor 1 |
1657 |
0.32 |
| chr1_10039266_10039475 | 0.37 |
Cspp1 |
centrosome and spindle pole associated protein 1 |
397 |
0.7 |
| chr11_119109577_119109728 | 0.37 |
Gm11754 |
predicted gene 11754 |
1773 |
0.27 |
| chr1_84051880_84052058 | 0.37 |
Pid1 |
phosphotyrosine interaction domain containing 1 |
1284 |
0.56 |
| chr14_46277334_46277485 | 0.35 |
Gm15217 |
predicted gene 15217 |
102005 |
0.06 |
| chr9_111138334_111138716 | 0.32 |
Lrrfip2 |
leucine rich repeat (in FLII) interacting protein 2 |
18850 |
0.15 |
| chr16_24061978_24062260 | 0.32 |
Gm46545 |
predicted gene, 46545 |
20571 |
0.15 |
| chr4_59546423_59546594 | 0.32 |
Ptbp3 |
polypyrimidine tract binding protein 3 |
2229 |
0.23 |
| chr8_23207373_23207572 | 0.31 |
Gpat4 |
glycerol-3-phosphate acyltransferase 4 |
873 |
0.36 |
| chr13_119966643_119967735 | 0.31 |
Gm20784 |
predicted gene, 20784 |
4945 |
0.11 |
| chr16_6901477_6901805 | 0.31 |
Rbfox1 |
RNA binding protein, fox-1 homolog (C. elegans) 1 |
92419 |
0.1 |
| chr13_51101532_51101693 | 0.30 |
Spin1 |
spindlin 1 |
732 |
0.73 |
| chr5_125189474_125189632 | 0.30 |
Ncor2 |
nuclear receptor co-repressor 2 |
10334 |
0.21 |
| chr13_60508473_60508632 | 0.30 |
A530001N23Rik |
RIKEN cDNA A530001N23 gene |
19744 |
0.15 |
| chr19_47311513_47311735 | 0.29 |
Sh3pxd2a |
SH3 and PX domains 2A |
3127 |
0.23 |
| chr19_29367023_29367182 | 0.29 |
Plgrkt |
plasminogen receptor, C-terminal lysine transmembrane protein |
206 |
0.65 |
| chr4_123645434_123645623 | 0.28 |
Gm12926 |
predicted gene 12926 |
15233 |
0.13 |
| chr1_105356184_105356368 | 0.28 |
Gm17634 |
predicted gene, 17634 |
269 |
0.62 |
| chr4_59452546_59452697 | 0.28 |
Susd1 |
sushi domain containing 1 |
13988 |
0.19 |
| chr19_5529673_5529945 | 0.28 |
Gm31166 |
predicted gene, 31166 |
1821 |
0.14 |
| chr2_38225178_38225332 | 0.27 |
Gm44455 |
predicted gene, 44455 |
133 |
0.96 |
| chr14_64154802_64154953 | 0.26 |
9630015K15Rik |
RIKEN cDNA 9630015K15 gene |
38563 |
0.11 |
| chr10_111490434_111490604 | 0.26 |
Nap1l1 |
nucleosome assembly protein 1-like 1 |
326 |
0.84 |
| chr5_130204701_130204852 | 0.26 |
Rabgef1 |
RAB guanine nucleotide exchange factor (GEF) 1 |
2277 |
0.16 |
| chr5_129715571_129715984 | 0.25 |
Mrps17 |
mitochondrial ribosomal protein S17 |
80 |
0.95 |
| chr10_7589267_7589435 | 0.25 |
Lrp11 |
low density lipoprotein receptor-related protein 11 |
449 |
0.78 |
| chr9_27106238_27106393 | 0.25 |
Gm48796 |
predicted gene, 48796 |
7764 |
0.17 |
| chr19_46140028_46140457 | 0.25 |
Pitx3 |
paired-like homeodomain transcription factor 3 |
741 |
0.53 |
| chr4_15066667_15066833 | 0.25 |
Gm11844 |
predicted gene 11844 |
44168 |
0.16 |
| chr18_44661527_44661720 | 0.25 |
A930012L18Rik |
RIKEN cDNA A930012L18 gene |
42 |
0.93 |
| chr4_102583077_102583228 | 0.24 |
Pde4b |
phosphodiesterase 4B, cAMP specific |
4406 |
0.34 |
| chr5_99978647_99978803 | 0.24 |
Hnrnpd |
heterogeneous nuclear ribonucleoprotein D |
101 |
0.78 |
| chr16_17838275_17838426 | 0.24 |
Car15 |
carbonic anhydrase 15 |
14 |
0.95 |
| chr5_99269909_99270098 | 0.24 |
Gm35394 |
predicted gene, 35394 |
4092 |
0.27 |
| chr15_74967628_74968021 | 0.24 |
Ly6e |
lymphocyte antigen 6 complex, locus E |
11269 |
0.08 |
| chr13_119966017_119966201 | 0.23 |
Gm20784 |
predicted gene, 20784 |
3865 |
0.12 |
| chr5_87573106_87573296 | 0.23 |
Sult1d1 |
sulfotransferase family 1D, member 1 |
4174 |
0.13 |
| chr8_61927981_61928360 | 0.23 |
Ddx60 |
DEAD (Asp-Glu-Ala-Asp) box polypeptide 60 |
81 |
0.58 |
| chr6_13870954_13871322 | 0.22 |
2610001J05Rik |
RIKEN cDNA 2610001J05 gene |
327 |
0.54 |
| chr10_20150520_20150679 | 0.22 |
Map7 |
microtubule-associated protein 7 |
1601 |
0.37 |
| chr9_55541381_55541949 | 0.22 |
Isl2 |
insulin related protein 2 (islet 2) |
458 |
0.61 |
| chr7_13005790_13006506 | 0.22 |
Zbtb45 |
zinc finger and BTB domain containing 45 |
3652 |
0.09 |
| chr3_121531769_121531936 | 0.22 |
A530020G20Rik |
RIKEN cDNA A530020G20 gene |
173 |
0.82 |
| chr7_30955354_30955627 | 0.22 |
Usf2 |
upstream transcription factor 2 |
40 |
0.79 |
| chr3_131552705_131552871 | 0.22 |
Papss1 |
3'-phosphoadenosine 5'-phosphosulfate synthase 1 |
11980 |
0.23 |
| chr19_16029303_16029488 | 0.22 |
Rpl37-ps1 |
ribosomal protein 37, pseudogene 1 |
3944 |
0.23 |
| chr10_20170266_20170434 | 0.22 |
Map7 |
microtubule-associated protein 7 |
430 |
0.82 |
| chr14_11699956_11700511 | 0.22 |
Gm48602 |
predicted gene, 48602 |
67710 |
0.12 |
| chr8_111299402_111299697 | 0.21 |
Rfwd3 |
ring finger and WD repeat domain 3 |
642 |
0.64 |
| chr14_62557485_62557724 | 0.21 |
Fam124a |
family with sequence similarity 124, member A |
1829 |
0.24 |
| chr10_127355157_127355314 | 0.21 |
Inhbe |
inhibin beta-E |
824 |
0.38 |
| chr11_58100366_58100593 | 0.21 |
Gm12247 |
predicted gene 12247 |
2706 |
0.16 |
| chr4_115057217_115057403 | 0.21 |
Tal1 |
T cell acute lymphocytic leukemia 1 |
128 |
0.95 |
| chr17_6782237_6782457 | 0.21 |
Ezr |
ezrin |
437 |
0.8 |
| chr1_131620226_131620377 | 0.21 |
Gm8532 |
predicted gene 8532 |
12668 |
0.14 |
| chr7_139401907_139402067 | 0.21 |
Inpp5a |
inositol polyphosphate-5-phosphatase A |
1498 |
0.49 |
| chr13_8651016_8651177 | 0.20 |
Gm48262 |
predicted gene, 48262 |
98076 |
0.06 |
| chr4_33033624_33033916 | 0.20 |
Ube2j1 |
ubiquitin-conjugating enzyme E2J 1 |
2242 |
0.18 |
| chr12_100158931_100159295 | 0.20 |
Nrde2 |
nrde-2 necessary for RNA interference, domain containing |
535 |
0.7 |
| chr13_60479839_60480430 | 0.20 |
Gm48500 |
predicted gene, 48500 |
1164 |
0.45 |
| chr16_96363004_96363159 | 0.20 |
Igsf5 |
immunoglobulin superfamily, member 5 |
1287 |
0.33 |
| chr15_98869205_98869356 | 0.20 |
Kmt2d |
lysine (K)-specific methyltransferase 2D |
1903 |
0.14 |
| chr11_43767190_43767354 | 0.20 |
Ttc1 |
tetratricopeptide repeat domain 1 |
19264 |
0.19 |
| chr2_27725053_27725318 | 0.20 |
Rxra |
retinoid X receptor alpha |
34 |
0.99 |
| chr15_74955213_74955440 | 0.20 |
Ly6e |
lymphocyte antigen 6 complex, locus E |
66 |
0.93 |
| chr3_145912337_145912507 | 0.20 |
Bcl10 |
B cell leukemia/lymphoma 10 |
10382 |
0.17 |
| chr6_116240202_116240376 | 0.20 |
Washc2 |
WASH complex subunit 2` |
14317 |
0.11 |
| chr10_75187490_75187671 | 0.19 |
Bcr |
BCR activator of RhoGEF and GTPase |
11204 |
0.2 |
| chr16_91043514_91043963 | 0.19 |
Paxbp1 |
PAX3 and PAX7 binding protein 1 |
426 |
0.63 |
| chr5_145871647_145871798 | 0.19 |
Cyp3a11 |
cytochrome P450, family 3, subfamily a, polypeptide 11 |
4969 |
0.18 |
| chr3_27183172_27183371 | 0.19 |
Nceh1 |
neutral cholesterol ester hydrolase 1 |
267 |
0.91 |
| chr8_35826443_35826604 | 0.19 |
Cldn23 |
claudin 23 |
36 |
0.97 |
| chr18_75370151_75370534 | 0.19 |
Smad7 |
SMAD family member 7 |
366 |
0.87 |
| chr11_69080646_69080973 | 0.19 |
Gm25371 |
predicted gene, 25371 |
5794 |
0.07 |
| chr14_63247987_63248412 | 0.19 |
Gata4 |
GATA binding protein 4 |
2928 |
0.24 |
| chr17_71485826_71486209 | 0.19 |
Gm18738 |
predicted gene, 18738 |
2625 |
0.16 |
| chr5_72410672_72410832 | 0.18 |
Gm19560 |
predicted gene, 19560 |
1308 |
0.39 |
| chr13_59556477_59556954 | 0.18 |
Agtpbp1 |
ATP/GTP binding protein 1 |
204 |
0.85 |
| chr15_60823871_60824050 | 0.18 |
9930014A18Rik |
RIKEN cDNA 9930014A18 gene |
381 |
0.77 |
| chr2_91084552_91084841 | 0.18 |
Spi1 |
spleen focus forming virus (SFFV) proviral integration oncogene |
2288 |
0.17 |
| chr2_93470457_93470622 | 0.18 |
Gm10804 |
predicted gene 10804 |
2100 |
0.3 |
| chr4_98322017_98322210 | 0.18 |
0610025J13Rik |
RIKEN cDNA 0610025J13 gene |
665 |
0.68 |
| chr10_12996840_12997182 | 0.18 |
Sf3b5 |
splicing factor 3b, subunit 5 |
8332 |
0.19 |
| chr16_8736038_8736588 | 0.18 |
Usp7 |
ubiquitin specific peptidase 7 |
2029 |
0.28 |
| chr15_65787239_65787398 | 0.18 |
Efr3a |
EFR3 homolog A |
270 |
0.94 |
| chr19_25383146_25383377 | 0.18 |
Kank1 |
KN motif and ankyrin repeat domains 1 |
12042 |
0.21 |
| chr6_126848563_126848769 | 0.17 |
Ndufa9 |
NADH:ubiquinone oxidoreductase subunit A9 |
459 |
0.74 |
| chr18_26015437_26015603 | 0.17 |
Gm33228 |
predicted gene, 33228 |
3217 |
0.39 |
| chr16_52081908_52082365 | 0.17 |
Gm23513 |
predicted gene, 23513 |
23011 |
0.21 |
| chr2_31501565_31501761 | 0.17 |
Ass1 |
argininosuccinate synthetase 1 |
937 |
0.56 |
| chr9_21031078_21031624 | 0.17 |
Icam5 |
intercellular adhesion molecule 5, telencephalin |
722 |
0.27 |
| chr10_9901181_9901520 | 0.17 |
Gm46210 |
predicted gene, 46210 |
37 |
0.79 |
| chr6_3620652_3620816 | 0.17 |
Vps50 |
VPS50 EARP/GARPII complex subunit |
26716 |
0.17 |
| chr13_38122267_38122440 | 0.17 |
Dsp |
desmoplakin |
28941 |
0.13 |
| chr3_85273227_85273668 | 0.16 |
1700036G14Rik |
RIKEN cDNA 1700036G14 gene |
44072 |
0.16 |
| chr7_16738552_16738717 | 0.16 |
Ap2s1 |
adaptor-related protein complex 2, sigma 1 subunit |
96 |
0.94 |
| chr18_46280937_46281101 | 0.16 |
Pggt1b |
protein geranylgeranyltransferase type I, beta subunit |
26 |
0.97 |
| chr4_101448515_101448685 | 0.16 |
Ak4 |
adenylate kinase 4 |
1377 |
0.4 |
| chr3_60965282_60965682 | 0.16 |
P2ry1 |
purinergic receptor P2Y, G-protein coupled 1 |
37313 |
0.16 |
| chr5_32469434_32469600 | 0.16 |
Gm43313 |
predicted gene 43313 |
2322 |
0.19 |
| chr3_108114477_108114651 | 0.16 |
Gnat2 |
guanine nucleotide binding protein, alpha transducing 2 |
17321 |
0.08 |
| chr8_85555887_85556054 | 0.15 |
Dnaja2 |
DnaJ heat shock protein family (Hsp40) member A2 |
626 |
0.69 |
| chr8_12720450_12720739 | 0.15 |
Gm15348 |
predicted gene 15348 |
1467 |
0.36 |
| chr3_28026863_28027216 | 0.15 |
Pld1 |
phospholipase D1 |
4223 |
0.33 |
| chr11_5520050_5520258 | 0.15 |
Xbp1 |
X-box binding protein 1 |
505 |
0.71 |
| chr12_55490259_55490936 | 0.15 |
Nfkbia |
nuclear factor of kappa light polypeptide gene enhancer in B cells inhibitor, alpha |
1213 |
0.45 |
| chr6_134543337_134543527 | 0.15 |
Lrp6 |
low density lipoprotein receptor-related protein 6 |
1718 |
0.37 |
| chr11_49064151_49064330 | 0.15 |
Tgtp2 |
T cell specific GTPase 2 |
34 |
0.95 |
| chr1_151501177_151501494 | 0.15 |
Rnf2 |
ring finger protein 2 |
380 |
0.75 |
| chr8_48733650_48734100 | 0.15 |
Tenm3 |
teneurin transmembrane protein 3 |
59185 |
0.14 |
| chr10_75415055_75415209 | 0.15 |
Upb1 |
ureidopropionase, beta |
5089 |
0.19 |
| chr1_37890066_37890236 | 0.14 |
Mitd1 |
MIT, microtubule interacting and transport, domain containing 1 |
125 |
0.73 |
| chr4_135269728_135270123 | 0.14 |
Clic4 |
chloride intracellular channel 4 (mitochondrial) |
2889 |
0.19 |
| chr8_93333775_93334133 | 0.14 |
Ces1g |
carboxylesterase 1G |
3354 |
0.19 |
| chr1_134962882_134963043 | 0.14 |
Ube2t |
ubiquitin-conjugating enzyme E2T |
344 |
0.87 |
| chr6_122819451_122819712 | 0.14 |
Foxj2 |
forkhead box J2 |
333 |
0.71 |
| chr18_64338075_64338233 | 0.14 |
Onecut2 |
one cut domain, family member 2 |
1866 |
0.3 |
| chr9_65193311_65193501 | 0.14 |
Parp16 |
poly (ADP-ribose) polymerase family, member 16 |
1947 |
0.21 |
| chr7_39841847_39842021 | 0.14 |
4930558N11Rik |
RIKEN cDNA 4930558N11 gene |
44192 |
0.11 |
| chr9_42262292_42262705 | 0.14 |
Sc5d |
sterol-C5-desaturase |
1702 |
0.34 |
| chr8_110629373_110629540 | 0.14 |
Vac14 |
Vac14 homolog (S. cerevisiae) |
10844 |
0.21 |
| chr3_122274519_122274867 | 0.14 |
Dnttip2 |
deoxynucleotidyltransferase, terminal, interacting protein 2 |
275 |
0.82 |
| chr2_72286436_72286609 | 0.14 |
Map3k20 |
mitogen-activated protein kinase kinase kinase 20 |
728 |
0.67 |
| chr7_110894004_110894327 | 0.14 |
Gm16336 |
predicted gene 16336 |
10820 |
0.16 |
| chr9_8043039_8043256 | 0.14 |
Cfap300 |
cilia and flagella associated protein 300 |
324 |
0.88 |
| chr16_22259188_22259423 | 0.14 |
Tra2b |
transformer 2 beta |
317 |
0.83 |
| chr3_65527969_65528149 | 0.14 |
4931440P22Rik |
RIKEN cDNA 4931440P22 gene |
125 |
0.75 |
| chr4_150371202_150371353 | 0.14 |
Rere |
arginine glutamic acid dipeptide (RE) repeats |
35117 |
0.15 |
| chr16_93816432_93816740 | 0.14 |
Dop1b |
DOP1 leucine zipper like protein B |
7152 |
0.14 |
| chr7_92681661_92681812 | 0.14 |
Pcf11 |
PCF11 cleavage and polyadenylation factor subunit |
11802 |
0.12 |
| chr13_120096376_120096558 | 0.14 |
Gm21378 |
predicted gene, 21378 |
3865 |
0.15 |
| chr5_100636918_100637077 | 0.14 |
Coq2 |
coenzyme Q2 4-hydroxybenzoate polyprenyltransferase |
27422 |
0.12 |
| chr7_97736977_97737173 | 0.14 |
Aqp11 |
aquaporin 11 |
863 |
0.54 |
| chr1_51774175_51774474 | 0.13 |
Myo1b |
myosin IB |
2112 |
0.33 |
| chr11_72690308_72690642 | 0.13 |
Ankfy1 |
ankyrin repeat and FYVE domain containing 1 |
469 |
0.8 |
| chr4_56222157_56222524 | 0.13 |
2310081O03Rik |
RIKEN cDNA 2310081O03 gene |
2181 |
0.42 |
| chr3_108022402_108022739 | 0.13 |
Gm12498 |
predicted gene 12498 |
2679 |
0.1 |
| chr15_58415328_58415520 | 0.13 |
Fam91a1 |
family with sequence similarity 91, member A1 |
44 |
0.97 |
| chr16_26505107_26505285 | 0.13 |
Tmem207 |
transmembrane protein 207 |
21566 |
0.21 |
| chr5_136506363_136506567 | 0.13 |
Gm38082 |
predicted gene, 38082 |
30012 |
0.18 |
| chr6_108091683_108091857 | 0.13 |
Setmar |
SET domain without mariner transposase fusion |
26708 |
0.18 |
| chr10_84885783_84886161 | 0.13 |
Ric8b |
RIC8 guanine nucleotide exchange factor B |
31644 |
0.17 |
| chr14_66279141_66279338 | 0.13 |
Ptk2b |
PTK2 protein tyrosine kinase 2 beta |
1743 |
0.32 |
| chr2_68947159_68947379 | 0.13 |
Cers6 |
ceramide synthase 6 |
9 |
0.97 |
| chr6_50037597_50037914 | 0.13 |
Gm3455 |
predicted gene 3455 |
13431 |
0.26 |
| chr13_99092586_99092968 | 0.13 |
2310020H05Rik |
RIKEN cDNA 2310020H05 gene |
4856 |
0.18 |
| chr11_102432106_102432323 | 0.13 |
Grn |
granulin |
571 |
0.58 |
| chr11_79747289_79747692 | 0.13 |
Mir365-2 |
microRNA 365-2 |
21090 |
0.12 |
| chr6_91703317_91703496 | 0.13 |
Slc6a6 |
solute carrier family 6 (neurotransmitter transporter, taurine), member 6 |
1749 |
0.25 |
| chr12_72490399_72490568 | 0.13 |
Lrrc9 |
leucine rich repeat containing 9 |
5184 |
0.21 |
| chr17_81064683_81064872 | 0.13 |
Thumpd2 |
THUMP domain containing 2 |
290 |
0.94 |
| chr6_139852445_139852613 | 0.13 |
Pik3c2g |
phosphatidylinositol-4-phosphate 3-kinase catalytic subunit type 2 gamma |
8881 |
0.22 |
| chr11_58020856_58021032 | 0.13 |
Larp1 |
La ribonucleoprotein domain family, member 1 |
11880 |
0.15 |
| chr8_111737464_111737633 | 0.13 |
Bcar1 |
breast cancer anti-estrogen resistance 1 |
5412 |
0.2 |
| chr16_23609739_23610190 | 0.12 |
Rtp4 |
receptor transporter protein 4 |
39 |
0.98 |
| chr9_20868532_20868915 | 0.12 |
Shfl |
shiftless antiviral inhibitor of ribosomal frameshifting |
43 |
0.94 |
| chr8_121083645_121083858 | 0.12 |
Foxf1 |
forkhead box F1 |
635 |
0.34 |
| chr1_92179149_92179712 | 0.12 |
Hdac4 |
histone deacetylase 4 |
867 |
0.7 |
| chr12_69790259_69790597 | 0.12 |
4930512B01Rik |
RIKEN cDNA 4930512B01 gene |
73 |
0.76 |
| chr12_82265513_82265664 | 0.12 |
Sipa1l1 |
signal-induced proliferation-associated 1 like 1 |
9003 |
0.27 |
| chr13_67683658_67684247 | 0.12 |
Zfp738 |
zinc finger protein 738 |
401 |
0.71 |
| chr13_75225448_75225621 | 0.12 |
Gm19095 |
predicted gene, 19095 |
26320 |
0.24 |
| chr5_89341929_89342080 | 0.12 |
Gc |
vitamin D binding protein |
93624 |
0.09 |
| chr6_97310793_97311248 | 0.12 |
Frmd4b |
FERM domain containing 4B |
1360 |
0.49 |
| chr2_146429524_146429960 | 0.12 |
Ralgapa2 |
Ral GTPase activating protein, alpha subunit 2 (catalytic) |
16866 |
0.26 |
| chr5_115565672_115565823 | 0.12 |
Gcn1 |
GCN1 activator of EIF2AK4 |
444 |
0.69 |
| chr5_44244398_44244549 | 0.12 |
Tapt1 |
transmembrane anterior posterior transformation 1 |
17847 |
0.11 |
| chr2_94010991_94011165 | 0.12 |
Alkbh3 |
alkB homolog 3, alpha-ketoglutarate-dependent dioxygenase |
271 |
0.89 |
| chrX_49624123_49624288 | 0.12 |
Gm14719 |
predicted gene 14719 |
36755 |
0.15 |
| chr14_21357195_21357346 | 0.12 |
Adk |
adenosine kinase |
11800 |
0.25 |
| chr4_53826592_53826930 | 0.12 |
Tmem38b |
transmembrane protein 38B |
704 |
0.65 |
| chr11_22871923_22872271 | 0.11 |
Gm24917 |
predicted gene, 24917 |
3196 |
0.16 |
| chr7_114778861_114779234 | 0.11 |
Insc |
INSC spindle orientation adaptor protein |
7726 |
0.19 |
| chrX_38218134_38218423 | 0.11 |
Mir3110 |
microRNA 3110 |
7837 |
0.15 |
| chr18_39486774_39486949 | 0.11 |
Nr3c1 |
nuclear receptor subfamily 3, group C, member 1 |
371 |
0.91 |
| chr1_67192772_67192960 | 0.11 |
Gm15668 |
predicted gene 15668 |
56334 |
0.12 |
| chr13_40971914_40972082 | 0.11 |
Gcnt2 |
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme |
18583 |
0.1 |
| chr6_121302024_121302175 | 0.11 |
Slc6a13 |
solute carrier family 6 (neurotransmitter transporter, GABA), member 13 |
1317 |
0.36 |
| chr11_109705848_109706019 | 0.11 |
Fam20a |
family with sequence similarity 20, member A |
16323 |
0.17 |
| chr19_14596433_14596591 | 0.11 |
Tle4 |
transducin-like enhancer of split 4 |
973 |
0.68 |
| chr18_58659158_58659487 | 0.11 |
Isoc1 |
isochorismatase domain containing 1 |
144 |
0.97 |
| chr13_29985974_29986454 | 0.11 |
E2f3 |
E2F transcription factor 3 |
151 |
0.96 |
| chr13_94398571_94398749 | 0.11 |
Ap3b1 |
adaptor-related protein complex 3, beta 1 subunit |
39700 |
0.13 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.1 | 0.4 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.1 | 0.2 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.1 | 0.2 | GO:0070346 | positive regulation of fat cell proliferation(GO:0070346) |
| 0.1 | 0.2 | GO:0061341 | non-canonical Wnt signaling pathway involved in heart development(GO:0061341) planar cell polarity pathway involved in heart morphogenesis(GO:0061346) |
| 0.1 | 0.2 | GO:0060375 | regulation of mast cell differentiation(GO:0060375) |
| 0.1 | 0.2 | GO:0097167 | circadian regulation of translation(GO:0097167) |
| 0.1 | 0.2 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.1 | 0.2 | GO:0035609 | C-terminal protein deglutamylation(GO:0035609) |
| 0.0 | 0.2 | GO:0009414 | response to water deprivation(GO:0009414) |
| 0.0 | 0.1 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.0 | 0.1 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.0 | 0.1 | GO:0021524 | visceral motor neuron differentiation(GO:0021524) |
| 0.0 | 0.2 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.0 | 0.2 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 0.0 | 0.1 | GO:0071332 | cellular response to fructose stimulus(GO:0071332) |
| 0.0 | 0.1 | GO:0032439 | endosome localization(GO:0032439) |
| 0.0 | 0.1 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.0 | 0.1 | GO:0002071 | glandular epithelial cell maturation(GO:0002071) |
| 0.0 | 0.1 | GO:0003278 | apoptotic process involved in heart morphogenesis(GO:0003278) |
| 0.0 | 0.2 | GO:0044351 | macropinocytosis(GO:0044351) |
| 0.0 | 0.1 | GO:0032911 | negative regulation of transforming growth factor beta1 production(GO:0032911) |
| 0.0 | 0.1 | GO:0035553 | oxidative RNA demethylation(GO:0035513) oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.0 | 0.3 | GO:0048242 | epinephrine secretion(GO:0048242) |
| 0.0 | 0.1 | GO:1901078 | negative regulation of relaxation of muscle(GO:1901078) |
| 0.0 | 0.1 | GO:0000430 | regulation of transcription from RNA polymerase II promoter by glucose(GO:0000430) positive regulation of transcription from RNA polymerase II promoter by glucose(GO:0000432) |
| 0.0 | 0.1 | GO:0035519 | protein K29-linked ubiquitination(GO:0035519) |
| 0.0 | 0.1 | GO:0010046 | response to mycotoxin(GO:0010046) |
| 0.0 | 0.2 | GO:0051771 | negative regulation of nitric-oxide synthase biosynthetic process(GO:0051771) |
| 0.0 | 0.1 | GO:0042045 | epithelial fluid transport(GO:0042045) |
| 0.0 | 0.1 | GO:0048369 | lateral mesoderm morphogenesis(GO:0048369) lateral mesoderm formation(GO:0048370) lateral mesodermal cell differentiation(GO:0048371) |
| 0.0 | 0.1 | GO:0030050 | vesicle transport along actin filament(GO:0030050) |
| 0.0 | 0.3 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.0 | 0.2 | GO:0010944 | negative regulation of transcription by competitive promoter binding(GO:0010944) |
| 0.0 | 0.1 | GO:0070668 | mast cell proliferation(GO:0070662) regulation of mast cell proliferation(GO:0070666) positive regulation of mast cell proliferation(GO:0070668) |
| 0.0 | 0.1 | GO:0010961 | cellular magnesium ion homeostasis(GO:0010961) |
| 0.0 | 0.1 | GO:1903011 | negative regulation of bone development(GO:1903011) |
| 0.0 | 0.1 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.0 | 0.3 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.0 | 0.1 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.0 | 0.1 | GO:1902564 | negative regulation of neutrophil activation(GO:1902564) |
| 0.0 | 0.1 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.1 | GO:0052055 | modulation by symbiont of host molecular function(GO:0052055) |
| 0.0 | 0.1 | GO:2000074 | regulation of type B pancreatic cell development(GO:2000074) |
| 0.0 | 0.1 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 0.0 | 0.1 | GO:0002756 | MyD88-independent toll-like receptor signaling pathway(GO:0002756) |
| 0.0 | 0.1 | GO:0032485 | regulation of Ral protein signal transduction(GO:0032485) |
| 0.0 | 0.2 | GO:0014842 | regulation of skeletal muscle satellite cell proliferation(GO:0014842) |
| 0.0 | 0.1 | GO:0014870 | response to inactivity(GO:0014854) response to muscle inactivity(GO:0014870) response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.0 | 0.1 | GO:0070344 | fat cell proliferation(GO:0070341) regulation of fat cell proliferation(GO:0070344) negative regulation of fat cell proliferation(GO:0070345) |
| 0.0 | 0.1 | GO:0072674 | multinuclear osteoclast differentiation(GO:0072674) osteoclast fusion(GO:0072675) |
| 0.0 | 0.1 | GO:2001181 | positive regulation of interleukin-10 secretion(GO:2001181) |
| 0.0 | 0.1 | GO:0001514 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.0 | 0.1 | GO:0070427 | nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070427) |
| 0.0 | 0.0 | GO:0031587 | positive regulation of inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0031587) |
| 0.0 | 0.1 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.0 | 0.0 | GO:0050427 | 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) |
| 0.0 | 0.0 | GO:0002125 | maternal aggressive behavior(GO:0002125) |
| 0.0 | 0.1 | GO:0034239 | regulation of macrophage fusion(GO:0034239) |
| 0.0 | 0.0 | GO:0090191 | negative regulation of branching involved in ureteric bud morphogenesis(GO:0090191) |
| 0.0 | 0.0 | GO:1903894 | regulation of IRE1-mediated unfolded protein response(GO:1903894) |
| 0.0 | 0.1 | GO:1901727 | positive regulation of histone deacetylase activity(GO:1901727) |
| 0.0 | 0.1 | GO:0071225 | cellular response to muramyl dipeptide(GO:0071225) |
| 0.0 | 0.0 | GO:0035523 | protein K29-linked deubiquitination(GO:0035523) protein K33-linked deubiquitination(GO:1990168) |
| 0.0 | 0.1 | GO:1903799 | negative regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903799) |
| 0.0 | 0.1 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.0 | 0.0 | GO:0006562 | proline catabolic process(GO:0006562) |
| 0.0 | 0.1 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) |
| 0.0 | 0.0 | GO:0014022 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.0 | 0.0 | GO:0032484 | Ral protein signal transduction(GO:0032484) |
| 0.0 | 0.0 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.0 | 0.0 | GO:0006987 | activation of signaling protein activity involved in unfolded protein response(GO:0006987) |
| 0.0 | 0.1 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.0 | 0.1 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.0 | 0.1 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.0 | 0.1 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.0 | 0.0 | GO:0072367 | regulation of lipid transport by regulation of transcription from RNA polymerase II promoter(GO:0072367) |
| 0.0 | 0.0 | GO:0038171 | cannabinoid signaling pathway(GO:0038171) endocannabinoid signaling pathway(GO:0071926) |
| 0.0 | 0.0 | GO:0051562 | negative regulation of mitochondrial calcium ion concentration(GO:0051562) |
| 0.0 | 0.0 | GO:0060762 | regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 0.0 | 0.0 | GO:0071873 | response to norepinephrine(GO:0071873) |
| 0.0 | 0.1 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.0 | 0.3 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.1 | GO:1900103 | positive regulation of endoplasmic reticulum unfolded protein response(GO:1900103) |
| 0.0 | 0.1 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 | 0.1 | GO:0009202 | deoxyribonucleoside triphosphate biosynthetic process(GO:0009202) |
| 0.0 | 0.2 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.0 | 0.0 | GO:0035754 | B cell chemotaxis(GO:0035754) |
| 0.0 | 0.0 | GO:0002669 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0044393 | microspike(GO:0044393) |
| 0.1 | 0.2 | GO:1990851 | Wnt-Frizzled-LRP5/6 complex(GO:1990851) |
| 0.0 | 0.1 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.0 | 0.1 | GO:0044354 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.0 | 0.1 | GO:0034666 | integrin alpha2-beta1 complex(GO:0034666) |
| 0.0 | 0.2 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.0 | 0.2 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.1 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.1 | GO:0035355 | Toll-like receptor 2-Toll-like receptor 6 protein complex(GO:0035355) |
| 0.0 | 0.1 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.0 | 0.1 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.0 | 0.1 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 0.0 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.0 | 0.0 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 0.0 | 0.1 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.0 | GO:1990761 | growth cone lamellipodium(GO:1990761) |
| 0.0 | 0.2 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.1 | 0.2 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.1 | 0.3 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.0 | 0.1 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.2 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.0 | 0.1 | GO:0071535 | RING-like zinc finger domain binding(GO:0071535) |
| 0.0 | 0.1 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.1 | GO:0035515 | oxidative RNA demethylase activity(GO:0035515) |
| 0.0 | 0.2 | GO:0004661 | protein geranylgeranyltransferase activity(GO:0004661) |
| 0.0 | 0.1 | GO:0046404 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) |
| 0.0 | 0.2 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.1 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.0 | 0.1 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.0 | 0.1 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 0.0 | 0.1 | GO:0001025 | RNA polymerase III transcription factor binding(GO:0001025) |
| 0.0 | 0.1 | GO:0043125 | ErbB-3 class receptor binding(GO:0043125) |
| 0.0 | 0.1 | GO:0005072 | transforming growth factor beta receptor, cytoplasmic mediator activity(GO:0005072) |
| 0.0 | 0.2 | GO:0019534 | toxin transporter activity(GO:0019534) |
| 0.0 | 0.1 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 0.0 | 0.3 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.1 | GO:1990239 | steroid hormone binding(GO:1990239) |
| 0.0 | 0.1 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.1 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.1 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.0 | 0.1 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.0 | 0.1 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.0 | 0.1 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.0 | GO:0031893 | vasopressin receptor binding(GO:0031893) |
| 0.0 | 0.1 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.0 | 0.0 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.0 | 0.3 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.2 | GO:0042301 | phosphate ion binding(GO:0042301) |
| 0.0 | 0.0 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 0.0 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.0 | 0.2 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.0 | 0.0 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.0 | 0.1 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.0 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.0 | 0.0 | GO:0034928 | mono-butyltin dioxygenase activity(GO:0018586) tri-n-butyltin dioxygenase activity(GO:0018588) di-n-butyltin dioxygenase activity(GO:0018589) methylsilanetriol hydroxylase activity(GO:0018590) methyl tertiary butyl ether 3-monooxygenase activity(GO:0018591) 4-nitrocatechol 4-monooxygenase activity(GO:0018592) 4-chlorophenoxyacetate monooxygenase activity(GO:0018593) tert-butanol 2-monooxygenase activity(GO:0018594) alpha-pinene monooxygenase activity(GO:0018595) dimethylsilanediol hydroxylase activity(GO:0018596) ammonia monooxygenase activity(GO:0018597) hydroxymethylsilanetriol oxidase activity(GO:0018598) 2-hydroxyisobutyrate 3-monooxygenase activity(GO:0018599) alpha-pinene dehydrogenase activity(GO:0018600) bisphenol A hydroxylase B activity(GO:0034559) 2,2-bis(4-hydroxyphenyl)-1-propanol hydroxylase activity(GO:0034562) 9-fluorenone-3,4-dioxygenase activity(GO:0034786) anthracene 9,10-dioxygenase activity(GO:0034816) 2-(methylthio)benzothiazole monooxygenase activity(GO:0034857) 2-hydroxybenzothiazole monooxygenase activity(GO:0034858) benzothiazole monooxygenase activity(GO:0034859) 2,6-dihydroxybenzothiazole monooxygenase activity(GO:0034862) pinacolone 5-monooxygenase activity(GO:0034870) thioacetamide S-oxygenase activity(GO:0034873) thioacetamide S-oxide S-oxygenase activity(GO:0034874) endosulfan monooxygenase I activity(GO:0034888) N-nitrodimethylamine hydroxylase activity(GO:0034893) 4-(1-ethyl-1,4-dimethyl-pentyl)phenol monoxygenase activity(GO:0034897) endosulfan ether monooxygenase activity(GO:0034903) pyrene 4,5-monooxygenase activity(GO:0034925) pyrene 1,2-monooxygenase activity(GO:0034927) 1-hydroxypyrene 6,7-monooxygenase activity(GO:0034928) 1-hydroxypyrene 7,8-monooxygenase activity(GO:0034929) phenylboronic acid monooxygenase activity(GO:0034950) spheroidene monooxygenase activity(GO:0043823) |
| 0.0 | 0.1 | GO:0071723 | lipopeptide binding(GO:0071723) |
| 0.0 | 0.1 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.0 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 0.1 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.0 | 0.0 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.0 | 0.1 | GO:0050700 | CARD domain binding(GO:0050700) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.0 | 0.2 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 0.2 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.3 | ST GAQ PATHWAY | G alpha q Pathway |
| 0.0 | 0.3 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.0 | 0.2 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.0 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.1 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.1 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.0 | 0.3 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.0 | 0.0 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.0 | 0.2 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |