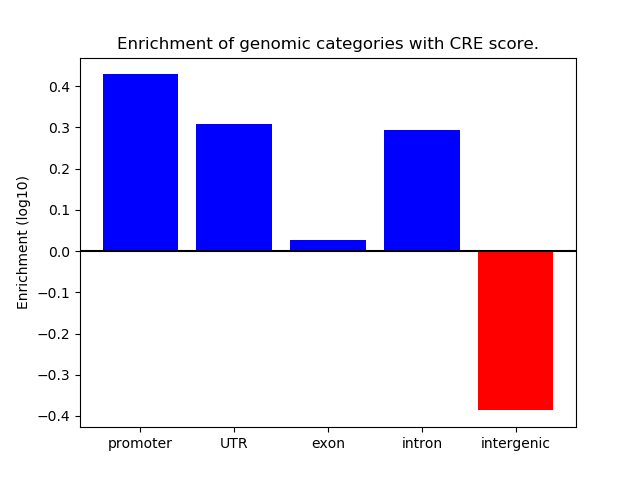
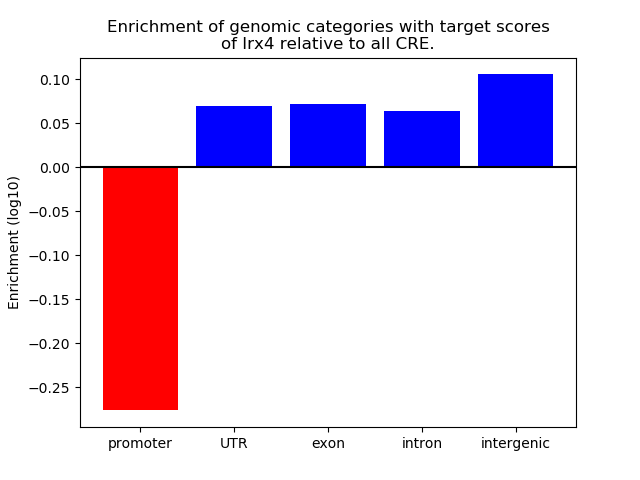
Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
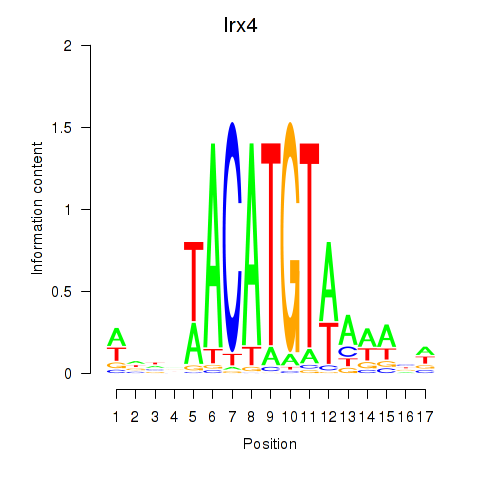
Results for Irx4
Z-value: 1.76
Transcription factors associated with Irx4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Irx4
|
ENSMUSG00000021604.9 | Iroquois homeobox 4 |
Activity of the Irx4 motif across conditions
Conditions sorted by the z-value of the Irx4 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
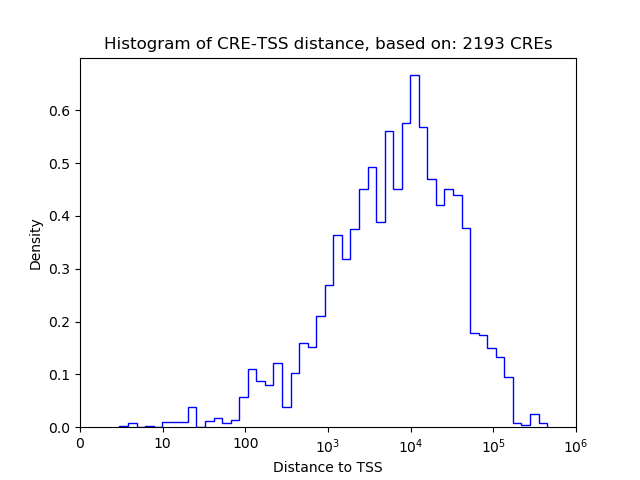
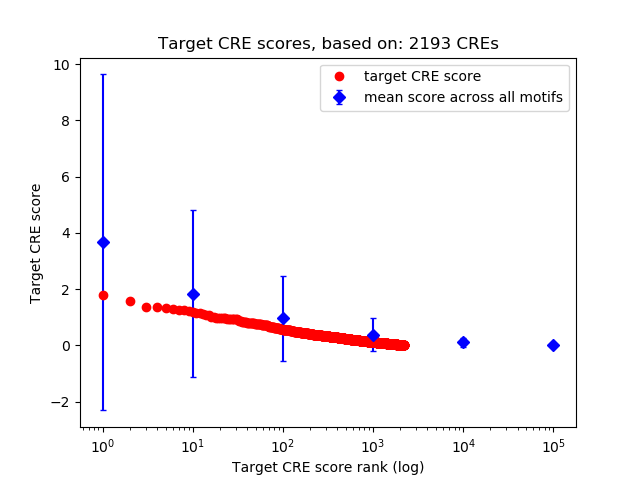
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr16_80273340_80273491 | 1.80 |
Gm23083 |
predicted gene, 23083 |
136063 |
0.05 |
| chr6_128515921_128516189 | 1.56 |
Pzp |
PZP, alpha-2-macroglobulin like |
10648 |
0.09 |
| chr11_87462456_87462620 | 1.37 |
Rnu3b4 |
U3B small nuclear RNA 4 |
252 |
0.83 |
| chr10_19443595_19443775 | 1.36 |
Gm33104 |
predicted gene, 33104 |
43030 |
0.15 |
| chr18_10251145_10251313 | 1.34 |
Gm5686 |
predicted gene 5686 |
10978 |
0.14 |
| chr3_27909385_27909539 | 1.29 |
Tmem212 |
transmembrane protein 212 |
13094 |
0.21 |
| chr8_128687947_128688299 | 1.27 |
Itgb1 |
integrin beta 1 (fibronectin receptor beta) |
2253 |
0.29 |
| chr1_45920584_45920780 | 1.25 |
Slc40a1 |
solute carrier family 40 (iron-regulated transporter), member 1 |
2627 |
0.2 |
| chr6_35874026_35874908 | 1.21 |
Gm43442 |
predicted gene 43442 |
52244 |
0.17 |
| chr7_72345699_72346292 | 1.20 |
Mctp2 |
multiple C2 domains, transmembrane 2 |
39387 |
0.21 |
| chr14_78850784_78850948 | 1.14 |
Vwa8 |
von Willebrand factor A domain containing 8 |
1612 |
0.36 |
| chr9_83575574_83575734 | 1.14 |
Sh3bgrl2 |
SH3 domain binding glutamic acid-rich protein like 2 |
1177 |
0.44 |
| chr1_178330385_178330898 | 1.11 |
Hnrnpu |
heterogeneous nuclear ribonucleoprotein U |
20 |
0.96 |
| chr19_4588592_4588754 | 1.09 |
Pcx |
pyruvate carboxylase |
5673 |
0.13 |
| chr13_100544476_100544653 | 1.08 |
Ocln |
occludin |
7896 |
0.12 |
| chr18_20146742_20147068 | 1.01 |
Dsc1 |
desmocollin 1 |
32034 |
0.2 |
| chr6_101351206_101351526 | 1.00 |
Gm43946 |
predicted gene, 43946 |
2536 |
0.21 |
| chr1_88975960_88976113 | 0.97 |
1700067G17Rik |
RIKEN cDNA 1700067G17 gene |
40077 |
0.14 |
| chr14_114418952_114419116 | 0.97 |
Gm19829 |
predicted gene, 19829 |
108231 |
0.08 |
| chr5_146079224_146079555 | 0.96 |
Cyp3a59 |
cytochrome P450, family 3, subfamily a, polypeptide 59 |
122 |
0.95 |
| chr11_80069895_80070062 | 0.96 |
Crlf3 |
cytokine receptor-like factor 3 |
10883 |
0.16 |
| chr13_81314862_81315034 | 0.96 |
Adgrv1 |
adhesion G protein-coupled receptor V1 |
27888 |
0.21 |
| chr13_73720273_73720547 | 0.96 |
Slc12a7 |
solute carrier family 12, member 7 |
12684 |
0.14 |
| chr6_54438444_54438595 | 0.95 |
9130019P16Rik |
RIKEN cDNA 9130019P16 gene |
8298 |
0.17 |
| chr4_81591457_81591882 | 0.95 |
Gm11411 |
predicted gene 11411 |
104772 |
0.07 |
| chr15_79741187_79741771 | 0.95 |
Sun2 |
Sad1 and UNC84 domain containing 2 |
613 |
0.45 |
| chr1_170607667_170608002 | 0.93 |
Nos1ap |
nitric oxide synthase 1 (neuronal) adaptor protein |
17973 |
0.17 |
| chr8_48944740_48944906 | 0.93 |
Gm25830 |
predicted gene, 25830 |
13658 |
0.25 |
| chrX_50956978_50957135 | 0.93 |
Frmd7 |
FERM domain containing 7 |
14346 |
0.23 |
| chr4_107899351_107899556 | 0.93 |
Czib |
CXXC motif containing zinc binding protein |
6083 |
0.13 |
| chr12_79630375_79630621 | 0.92 |
9430078K24Rik |
RIKEN cDNA 9430078K24 gene |
294235 |
0.01 |
| chr6_15929821_15930118 | 0.90 |
Gm43990 |
predicted gene, 43990 |
93880 |
0.08 |
| chr19_38558391_38558564 | 0.87 |
Plce1 |
phospholipase C, epsilon 1 |
24891 |
0.22 |
| chr14_64441838_64442032 | 0.86 |
Msra |
methionine sulfoxide reductase A |
1018 |
0.64 |
| chr11_49996483_49996658 | 0.84 |
Rnf130 |
ring finger protein 130 |
28776 |
0.14 |
| chr5_122769447_122769598 | 0.84 |
Camkk2 |
calcium/calmodulin-dependent protein kinase kinase 2, beta |
5657 |
0.14 |
| chr17_30640985_30641153 | 0.83 |
Dnah8 |
dynein, axonemal, heavy chain 8 |
5313 |
0.13 |
| chr14_60796696_60796848 | 0.83 |
Mipep |
mitochondrial intermediate peptidase |
12134 |
0.17 |
| chr17_31569080_31569361 | 0.83 |
Gm50107 |
predicted gene, 50107 |
2057 |
0.13 |
| chr1_154149125_154149300 | 0.81 |
A830008E24Rik |
RIKEN cDNA A830008E24 gene |
31261 |
0.15 |
| chr7_119783443_119783771 | 0.81 |
Acsm3 |
acyl-CoA synthetase medium-chain family member 3 |
2871 |
0.15 |
| chr10_38439349_38439537 | 0.80 |
Gm48198 |
predicted gene, 48198 |
5308 |
0.32 |
| chr5_27251468_27251639 | 0.80 |
Dpp6 |
dipeptidylpeptidase 6 |
10422 |
0.23 |
| chr2_170208965_170209511 | 0.79 |
Zfp217 |
zinc finger protein 217 |
61135 |
0.12 |
| chr14_116642069_116642356 | 0.79 |
Gm38045 |
predicted gene, 38045 |
108003 |
0.08 |
| chr7_140771376_140771879 | 0.79 |
Cyp2e1 |
cytochrome P450, family 2, subfamily e, polypeptide 1 |
7146 |
0.09 |
| chr10_43902698_43902849 | 0.78 |
Rtn4ip1 |
reticulon 4 interacting protein 1 |
966 |
0.39 |
| chr16_87690672_87690831 | 0.78 |
Bach1 |
BTB and CNC homology 1, basic leucine zipper transcription factor 1 |
8194 |
0.22 |
| chrX_38422575_38423072 | 0.77 |
Lamp2 |
lysosomal-associated membrane protein 2 |
20327 |
0.15 |
| chr6_31221861_31222046 | 0.77 |
Lncpint |
long non-protein coding RNA, Trp53 induced transcript |
1465 |
0.3 |
| chr7_105596936_105597272 | 0.77 |
Hpx |
hemopexin |
3008 |
0.13 |
| chr10_29821551_29821768 | 0.77 |
Gm6390 |
predicted gene 6390 |
40248 |
0.18 |
| chr16_91045477_91045695 | 0.76 |
4931406G06Rik |
RIKEN cDNA 4931406G06 gene |
930 |
0.33 |
| chr3_152208280_152208431 | 0.76 |
Dnajb4 |
DnaJ heat shock protein family (Hsp40) member B4 |
1664 |
0.24 |
| chr15_67159040_67159221 | 0.76 |
St3gal1 |
ST3 beta-galactoside alpha-2,3-sialyltransferase 1 |
17582 |
0.24 |
| chr8_13161522_13161685 | 0.75 |
Lamp1 |
lysosomal-associated membrane protein 1 |
2442 |
0.16 |
| chr15_62302358_62302526 | 0.74 |
Pvt1 |
Pvt1 oncogene |
79839 |
0.1 |
| chr13_38962916_38963078 | 0.74 |
Slc35b3 |
solute carrier family 35, member B3 |
2122 |
0.28 |
| chr16_52267502_52267712 | 0.74 |
Alcam |
activated leukocyte cell adhesion molecule |
1454 |
0.57 |
| chr6_146765296_146765601 | 0.74 |
Gm44270 |
predicted gene, 44270 |
2867 |
0.2 |
| chr2_77155688_77155851 | 0.73 |
Ccdc141 |
coiled-coil domain containing 141 |
14807 |
0.21 |
| chr1_58116945_58117096 | 0.73 |
Aox3 |
aldehyde oxidase 3 |
3834 |
0.24 |
| chr7_140905824_140905984 | 0.72 |
BC024386 |
cDNA sequence BC023486 |
110 |
0.89 |
| chr11_87471532_87471686 | 0.72 |
Rnu3b2 |
U3B small nuclear RNA 2 |
241 |
0.85 |
| chr13_41263624_41263896 | 0.71 |
Gm17364 |
predicted gene, 17364 |
10217 |
0.13 |
| chr6_9066317_9066493 | 0.71 |
Gm35736 |
predicted gene, 35736 |
80361 |
0.11 |
| chr5_142783226_142783546 | 0.71 |
Tnrc18 |
trinucleotide repeat containing 18 |
5137 |
0.22 |
| chr10_128566469_128566764 | 0.70 |
Pa2g4 |
proliferation-associated 2G4 |
629 |
0.43 |
| chr8_64716760_64716977 | 0.70 |
Msmo1 |
methylsterol monoxygenase 1 |
5063 |
0.17 |
| chr11_102138520_102138679 | 0.68 |
Nags |
N-acetylglutamate synthase |
6914 |
0.08 |
| chr7_130564136_130564299 | 0.65 |
Nsmce4a |
NSE4 homolog A, SMC5-SMC6 complex component |
8901 |
0.15 |
| chr19_44985292_44985474 | 0.65 |
4930414N06Rik |
RIKEN cDNA 4930414N06 gene |
833 |
0.43 |
| chr11_16888258_16888409 | 0.65 |
Egfr |
epidermal growth factor receptor |
10183 |
0.2 |
| chr9_94692147_94692428 | 0.65 |
Gm16262 |
predicted gene 16262 |
10032 |
0.17 |
| chr16_15984456_15984622 | 0.65 |
n-R5s31 |
nuclear encoded rRNA 5S 31 |
26470 |
0.2 |
| chr16_52249852_52250207 | 0.65 |
Alcam |
activated leukocyte cell adhesion molecule |
19032 |
0.27 |
| chr1_74059706_74059994 | 0.65 |
Tns1 |
tensin 1 |
22693 |
0.18 |
| chr3_28722548_28722883 | 0.64 |
Slc2a2 |
solute carrier family 2 (facilitated glucose transporter), member 2 |
1198 |
0.42 |
| chr5_102599017_102599403 | 0.64 |
1700013M08Rik |
RIKEN cDNA 1700013M08 gene |
115921 |
0.06 |
| chr7_19695332_19695494 | 0.63 |
Apoe |
apolipoprotein E |
2218 |
0.12 |
| chr3_22078655_22078806 | 0.63 |
Tbl1xr1 |
transducin (beta)-like 1X-linked receptor 1 |
1446 |
0.33 |
| chrX_42065003_42065280 | 0.63 |
Xiap |
X-linked inhibitor of apoptosis |
2695 |
0.31 |
| chr6_141515419_141515600 | 0.62 |
Slco1c1 |
solute carrier organic anion transporter family, member 1c1 |
8859 |
0.27 |
| chr16_43547072_43547244 | 0.62 |
Zbtb20 |
zinc finger and BTB domain containing 20 |
36850 |
0.17 |
| chr7_27252126_27252620 | 0.61 |
Gm44641 |
predicted gene 44641 |
197 |
0.84 |
| chr2_84650666_84651104 | 0.61 |
Ctnnd1 |
catenin (cadherin associated protein), delta 1 |
120 |
0.92 |
| chr1_187476098_187476254 | 0.61 |
Gm37896 |
predicted gene, 37896 |
28299 |
0.21 |
| chr19_55087908_55088315 | 0.60 |
Gpam |
glycerol-3-phosphate acyltransferase, mitochondrial |
11340 |
0.21 |
| chr2_152817536_152817750 | 0.60 |
Bcl2l1 |
BCL2-like 1 |
10892 |
0.12 |
| chr15_3483921_3484108 | 0.60 |
Ghr |
growth hormone receptor |
12370 |
0.28 |
| chr2_134548299_134548473 | 0.59 |
Hao1 |
hydroxyacid oxidase 1, liver |
5921 |
0.31 |
| chr3_24782914_24783090 | 0.59 |
Naaladl2 |
N-acetylated alpha-linked acidic dipeptidase-like 2 |
620 |
0.85 |
| chr8_73226959_73227110 | 0.59 |
Gm22532 |
predicted gene, 22532 |
51776 |
0.17 |
| chr10_108449802_108449991 | 0.58 |
Gm36283 |
predicted gene, 36283 |
3294 |
0.25 |
| chr18_34899168_34899345 | 0.58 |
Etf1 |
eukaryotic translation termination factor 1 |
32751 |
0.09 |
| chr15_89396778_89397117 | 0.58 |
Klhdc7b |
kelch domain containing 7B |
10056 |
0.07 |
| chr10_10408279_10408449 | 0.58 |
Adgb |
androglobin |
3380 |
0.27 |
| chr7_102224048_102224209 | 0.58 |
Pgap2 |
post-GPI attachment to proteins 2 |
846 |
0.48 |
| chr6_138153579_138153750 | 0.57 |
Mgst1 |
microsomal glutathione S-transferase 1 |
10810 |
0.28 |
| chr16_24487981_24488183 | 0.56 |
Lpp |
LIM domain containing preferred translocation partner in lipoma |
39991 |
0.15 |
| chr4_48315210_48315516 | 0.56 |
Gm12435 |
predicted gene 12435 |
29867 |
0.16 |
| chr2_65172186_65172346 | 0.56 |
Cobll1 |
Cobl-like 1 |
63729 |
0.11 |
| chr4_60659172_60659323 | 0.56 |
Mup11 |
major urinary protein 11 |
491 |
0.78 |
| chr5_145986709_145986916 | 0.56 |
Cyp3a25 |
cytochrome P450, family 3, subfamily a, polypeptide 25 |
4831 |
0.14 |
| chr9_25329643_25329942 | 0.55 |
Gm18891 |
predicted gene, 18891 |
19706 |
0.17 |
| chr5_8902050_8902207 | 0.55 |
Abcb4 |
ATP-binding cassette, sub-family B (MDR/TAP), member 4 |
8179 |
0.15 |
| chr9_60478237_60478388 | 0.55 |
Thsd4 |
thrombospondin, type I, domain containing 4 |
18814 |
0.15 |
| chr3_138291566_138291979 | 0.55 |
Adh1 |
alcohol dehydrogenase 1 (class I) |
14121 |
0.11 |
| chr5_121663580_121663743 | 0.55 |
Brap |
BRCA1 associated protein |
1581 |
0.25 |
| chr5_66095606_66095870 | 0.55 |
Rbm47 |
RNA binding motif protein 47 |
2453 |
0.2 |
| chr13_100579015_100579281 | 0.55 |
Gm8847 |
predicted gene 8847 |
19116 |
0.1 |
| chr5_32441692_32441843 | 0.54 |
Ppp1cb |
protein phosphatase 1 catalytic subunit beta |
17076 |
0.11 |
| chr14_27475610_27475797 | 0.54 |
Tasor |
transcription activation suppressor |
1467 |
0.36 |
| chr1_82974456_82974623 | 0.54 |
Gm47955 |
predicted gene, 47955 |
14253 |
0.08 |
| chr3_65991098_65991265 | 0.54 |
Gm19024 |
predicted gene, 19024 |
1428 |
0.29 |
| chr2_51512722_51513109 | 0.54 |
Gm23505 |
predicted gene, 23505 |
19133 |
0.19 |
| chr4_32538108_32538303 | 0.54 |
Bach2os |
BTB and CNC homology 2, opposite strand |
33412 |
0.14 |
| chr11_70472894_70473060 | 0.53 |
Zmynd15 |
zinc finger, MYND-type containing 15 |
12412 |
0.07 |
| chr15_55265818_55265991 | 0.53 |
Gm26296 |
predicted gene, 26296 |
3301 |
0.29 |
| chr1_172827733_172827924 | 0.53 |
Gm24817 |
predicted gene, 24817 |
15810 |
0.16 |
| chr13_36681541_36681847 | 0.53 |
Gm26395 |
predicted gene, 26395 |
27289 |
0.13 |
| chr4_147406377_147406541 | 0.52 |
Gm8178 |
predicted gene 8178 |
27530 |
0.11 |
| chr5_130139764_130139934 | 0.52 |
Kctd7 |
potassium channel tetramerisation domain containing 7 |
5012 |
0.13 |
| chr10_89518213_89518404 | 0.52 |
Nr1h4 |
nuclear receptor subfamily 1, group H, member 4 |
11650 |
0.21 |
| chr1_190059603_190059918 | 0.51 |
Gm28172 |
predicted gene 28172 |
108910 |
0.06 |
| chr4_128969208_128969368 | 0.51 |
Azin2 |
antizyme inhibitor 2 |
6846 |
0.16 |
| chr11_104392570_104392977 | 0.50 |
Gm47315 |
predicted gene, 47315 |
32527 |
0.14 |
| chr15_61517445_61517596 | 0.50 |
Gm49498 |
predicted gene, 49498 |
26663 |
0.25 |
| chr10_30810298_30810917 | 0.50 |
Ncoa7 |
nuclear receptor coactivator 7 |
7281 |
0.16 |
| chr6_5223673_5223834 | 0.50 |
Gm44250 |
predicted gene, 44250 |
3267 |
0.22 |
| chr7_49751262_49751438 | 0.50 |
Htatip2 |
HIV-1 Tat interactive protein 2 |
7765 |
0.22 |
| chr13_112674167_112674325 | 0.49 |
Slc38a9 |
solute carrier family 38, member 9 |
5875 |
0.15 |
| chr5_114581477_114581628 | 0.49 |
Fam222a |
family with sequence similarity 222, member A |
13535 |
0.15 |
| chr17_86593218_86593401 | 0.49 |
Gm10309 |
predicted gene 10309 |
88077 |
0.07 |
| chr13_36152416_36152579 | 0.49 |
Fars2 |
phenylalanine-tRNA synthetase 2 (mitochondrial) |
22597 |
0.14 |
| chr5_137481515_137481682 | 0.49 |
Epo |
erythropoietin |
4218 |
0.09 |
| chr3_104279351_104279517 | 0.48 |
Magi3 |
membrane associated guanylate kinase, WW and PDZ domain containing 3 |
59060 |
0.08 |
| chr5_67025062_67025433 | 0.48 |
Gm42713 |
predicted gene 42713 |
14129 |
0.12 |
| chr1_184811348_184811540 | 0.48 |
Mtarc1 |
mitochondrial amidoxime reducing component 1 |
131 |
0.95 |
| chr14_46017259_46017572 | 0.47 |
Gm6580 |
predicted gene 6580 |
10013 |
0.17 |
| chr6_90851681_90851855 | 0.47 |
Gm25185 |
predicted gene, 25185 |
5769 |
0.17 |
| chr1_174921018_174921617 | 0.47 |
Grem2 |
gremlin 2, DAN family BMP antagonist |
502 |
0.88 |
| chr16_86826664_86826815 | 0.47 |
Gm32624 |
predicted gene, 32624 |
9865 |
0.19 |
| chr4_33262151_33262376 | 0.47 |
Pnrc1 |
proline-rich nuclear receptor coactivator 1 |
13753 |
0.16 |
| chr5_77340141_77340310 | 0.47 |
Polr2b |
polymerase (RNA) II (DNA directed) polypeptide B |
143 |
0.94 |
| chr13_5845761_5845936 | 0.47 |
1700016G22Rik |
RIKEN cDNA 1700016G22 gene |
11713 |
0.16 |
| chr4_60418791_60418999 | 0.47 |
Mup9 |
major urinary protein 9 |
1689 |
0.29 |
| chr3_129867430_129867724 | 0.47 |
Pla2g12a |
phospholipase A2, group XIIA |
11029 |
0.13 |
| chr13_73730823_73730977 | 0.47 |
Slc12a7 |
solute carrier family 12, member 7 |
2194 |
0.26 |
| chr16_48767631_48767807 | 0.47 |
Trat1 |
T cell receptor associated transmembrane adaptor 1 |
4237 |
0.22 |
| chr3_146631735_146632069 | 0.46 |
Gm16325 |
predicted gene 16325 |
10030 |
0.12 |
| chr3_146604513_146604664 | 0.46 |
Uox |
urate oxidase |
5397 |
0.14 |
| chr14_76797951_76798124 | 0.46 |
Gm30246 |
predicted gene, 30246 |
17210 |
0.17 |
| chr13_30346992_30347295 | 0.46 |
Agtr1a |
angiotensin II receptor, type 1a |
1602 |
0.41 |
| chr16_71730517_71730728 | 0.46 |
Gm22797 |
predicted gene, 22797 |
66834 |
0.13 |
| chr15_8251636_8251790 | 0.46 |
Cplane1 |
ciliogenesis and planar polarity effector 1 |
4610 |
0.26 |
| chr1_118458813_118459291 | 0.46 |
Gm22710 |
predicted gene, 22710 |
213 |
0.44 |
| chr11_45877769_45877933 | 0.45 |
Clint1 |
clathrin interactor 1 |
4546 |
0.19 |
| chr19_12011828_12011982 | 0.45 |
Olfr1421-ps1 |
olfactory receptor 1421, pseudogene 1 |
8270 |
0.08 |
| chr3_133530261_133530426 | 0.45 |
Tet2 |
tet methylcytosine dioxygenase 2 |
13943 |
0.17 |
| chr11_94637499_94637823 | 0.45 |
Lrrc59 |
leucine rich repeat containing 59 |
7847 |
0.1 |
| chr10_94882831_94882985 | 0.44 |
Gm48718 |
predicted gene, 48718 |
884 |
0.59 |
| chr13_24363340_24363497 | 0.44 |
Gm11342 |
predicted gene 11342 |
12532 |
0.12 |
| chr4_138342269_138342429 | 0.44 |
Cda |
cytidine deaminase |
1300 |
0.3 |
| chr15_77773367_77773546 | 0.44 |
Myh9 |
myosin, heavy polypeptide 9, non-muscle |
3677 |
0.15 |
| chr5_117275916_117276216 | 0.44 |
Pebp1 |
phosphatidylethanolamine binding protein 1 |
11459 |
0.11 |
| chr3_27152202_27152442 | 0.44 |
Ect2 |
ect2 oncogene |
1460 |
0.31 |
| chr12_58323949_58324220 | 0.44 |
Clec14a |
C-type lectin domain family 14, member a |
54794 |
0.16 |
| chr13_64364897_64365048 | 0.44 |
Ctsl |
cathepsin L |
2083 |
0.18 |
| chr3_50279626_50279824 | 0.43 |
Gm2345 |
predicted gene 2345 |
24870 |
0.22 |
| chr19_32689880_32690046 | 0.43 |
Atad1 |
ATPase family, AAA domain containing 1 |
4907 |
0.27 |
| chrX_139113469_139113635 | 0.43 |
4930513O06Rik |
RIKEN cDNA 4930513O06 gene |
27309 |
0.16 |
| chr7_145044142_145044344 | 0.43 |
Ccnd1 |
cyclin D1 |
104318 |
0.05 |
| chr12_71912308_71912620 | 0.43 |
Daam1 |
dishevelled associated activator of morphogenesis 1 |
22734 |
0.2 |
| chr15_10308697_10308848 | 0.43 |
Prlr |
prolactin receptor |
5330 |
0.22 |
| chr15_67044966_67045152 | 0.43 |
Gm31342 |
predicted gene, 31342 |
5001 |
0.25 |
| chr2_126950948_126951317 | 0.43 |
Sppl2a |
signal peptide peptidase like 2A |
17897 |
0.16 |
| chr15_39813920_39814243 | 0.43 |
Gm16291 |
predicted gene 16291 |
17036 |
0.19 |
| chr17_44943176_44943431 | 0.43 |
4930564C03Rik |
RIKEN cDNA 4930564C03 gene |
63008 |
0.13 |
| chr7_26139383_26139621 | 0.43 |
Cyp2b26-ps |
cytochrome P450, family 2, subfamily b, polypeptide 26, pseudogene |
9090 |
0.15 |
| chr4_101176228_101176401 | 0.43 |
Jak1 |
Janus kinase 1 |
9111 |
0.15 |
| chr6_137744823_137744982 | 0.42 |
Strap |
serine/threonine kinase receptor associated protein |
138 |
0.97 |
| chr4_57525649_57525867 | 0.42 |
Pakap |
paralemmin A kinase anchor protein |
42401 |
0.16 |
| chr4_35223300_35223499 | 0.42 |
C9orf72 |
C9orf72, member of C9orf72-SMCR8 complex |
2435 |
0.24 |
| chr14_29672536_29672709 | 0.42 |
Cacna2d3 |
calcium channel, voltage-dependent, alpha2/delta subunit 3 |
49173 |
0.13 |
| chr12_101080892_101081278 | 0.42 |
D130020L05Rik |
RIKEN cDNA D130020L05 gene |
1366 |
0.29 |
| chr13_41112373_41112548 | 0.42 |
Gcm2 |
glial cells missing homolog 2 |
1425 |
0.28 |
| chr15_89210520_89210683 | 0.42 |
Ppp6r2 |
protein phosphatase 6, regulatory subunit 2 |
952 |
0.39 |
| chr18_20939203_20939371 | 0.42 |
Rnf125 |
ring finger protein 125 |
5338 |
0.23 |
| chr1_140406769_140406932 | 0.42 |
Kcnt2 |
potassium channel, subfamily T, member 2 |
36086 |
0.22 |
| chr19_5433656_5433971 | 0.42 |
Gm50111 |
predicted gene, 50111 |
2087 |
0.09 |
| chr8_126838292_126838477 | 0.41 |
A630001O12Rik |
RIKEN cDNA A630001O12 gene |
849 |
0.65 |
| chr17_55513425_55513584 | 0.41 |
St6gal2 |
beta galactoside alpha 2,6 sialyltransferase 2 |
67260 |
0.1 |
| chr1_164215221_164215636 | 0.41 |
Slc19a2 |
solute carrier family 19 (thiamine transporter), member 2 |
33618 |
0.11 |
| chr13_80882069_80882492 | 0.41 |
Arrdc3 |
arrestin domain containing 3 |
1104 |
0.47 |
| chr3_102670542_102670863 | 0.41 |
Gm19202 |
predicted gene, 19202 |
16015 |
0.14 |
| chr1_161269196_161269347 | 0.41 |
Prdx6 |
peroxiredoxin 6 |
18052 |
0.16 |
| chr13_28874054_28874215 | 0.41 |
2610307P16Rik |
RIKEN cDNA 2610307P16 gene |
9322 |
0.18 |
| chr15_55224563_55224882 | 0.41 |
Gm26296 |
predicted gene, 26296 |
37881 |
0.16 |
| chr2_154820458_154820609 | 0.40 |
Raly |
hnRNP-associated with lethal yellow |
28818 |
0.13 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0010912 | regulation of isomerase activity(GO:0010911) positive regulation of isomerase activity(GO:0010912) |
| 0.1 | 0.4 | GO:0007161 | calcium-independent cell-matrix adhesion(GO:0007161) |
| 0.1 | 0.4 | GO:0060300 | regulation of cytokine activity(GO:0060300) |
| 0.1 | 0.4 | GO:0043578 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.1 | 0.4 | GO:0071879 | positive regulation of adrenergic receptor signaling pathway(GO:0071879) |
| 0.1 | 0.5 | GO:0030091 | protein repair(GO:0030091) |
| 0.1 | 0.6 | GO:1900086 | positive regulation of peptidyl-tyrosine autophosphorylation(GO:1900086) |
| 0.1 | 0.3 | GO:0070681 | glutaminyl-tRNAGln biosynthesis via transamidation(GO:0070681) |
| 0.1 | 0.4 | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.1 | 0.3 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.1 | 0.2 | GO:0070537 | histone H2A K63-linked deubiquitination(GO:0070537) |
| 0.1 | 0.2 | GO:0002019 | regulation of renal output by angiotensin(GO:0002019) |
| 0.1 | 0.3 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.1 | 0.2 | GO:0002669 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.1 | 0.2 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.1 | 0.3 | GO:0016078 | tRNA catabolic process(GO:0016078) |
| 0.1 | 0.2 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.1 | 0.4 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.1 | 0.2 | GO:0055118 | negative regulation of cardiac muscle contraction(GO:0055118) |
| 0.1 | 0.3 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.1 | 0.2 | GO:0061084 | negative regulation of protein refolding(GO:0061084) |
| 0.1 | 0.3 | GO:0019243 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.1 | 0.5 | GO:0090520 | sphingolipid mediated signaling pathway(GO:0090520) |
| 0.1 | 0.2 | GO:0061091 | regulation of phospholipid translocation(GO:0061091) positive regulation of phospholipid translocation(GO:0061092) |
| 0.0 | 0.1 | GO:0048294 | negative regulation of isotype switching to IgE isotypes(GO:0048294) |
| 0.0 | 0.2 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.0 | 0.1 | GO:0033031 | positive regulation of neutrophil apoptotic process(GO:0033031) |
| 0.0 | 0.1 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.0 | 0.3 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.0 | 0.2 | GO:0006538 | glutamate catabolic process(GO:0006538) |
| 0.0 | 0.1 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.0 | 0.1 | GO:0006982 | response to lipid hydroperoxide(GO:0006982) |
| 0.0 | 0.1 | GO:0006216 | cytidine catabolic process(GO:0006216) cytidine deamination(GO:0009972) cytidine metabolic process(GO:0046087) |
| 0.0 | 0.1 | GO:0014016 | neuroblast differentiation(GO:0014016) |
| 0.0 | 0.2 | GO:0071681 | response to indole-3-methanol(GO:0071680) cellular response to indole-3-methanol(GO:0071681) |
| 0.0 | 0.4 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 | 0.1 | GO:0043321 | regulation of natural killer cell degranulation(GO:0043321) |
| 0.0 | 0.1 | GO:2000304 | positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 0.0 | 0.2 | GO:0060971 | embryonic heart tube left/right pattern formation(GO:0060971) |
| 0.0 | 0.2 | GO:2001280 | positive regulation of prostaglandin biosynthetic process(GO:0031394) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.0 | 0.2 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.0 | 0.3 | GO:0090232 | positive regulation of spindle checkpoint(GO:0090232) |
| 0.0 | 0.1 | GO:0034395 | regulation of transcription from RNA polymerase II promoter in response to iron(GO:0034395) |
| 0.0 | 0.1 | GO:0045897 | regulation of transcription during mitosis(GO:0045896) positive regulation of transcription during mitosis(GO:0045897) |
| 0.0 | 0.1 | GO:0033668 | negative regulation by symbiont of host apoptotic process(GO:0033668) negative regulation by symbiont of host programmed cell death(GO:0052041) negative regulation by organism of programmed cell death in other organism involved in symbiotic interaction(GO:0052490) |
| 0.0 | 0.1 | GO:0006172 | ADP biosynthetic process(GO:0006172) |
| 0.0 | 0.1 | GO:0015888 | thiamine transport(GO:0015888) |
| 0.0 | 0.1 | GO:0060336 | negative regulation of response to interferon-gamma(GO:0060331) negative regulation of interferon-gamma-mediated signaling pathway(GO:0060336) |
| 0.0 | 0.2 | GO:0086073 | bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 0.0 | 0.1 | GO:0061144 | alveolar secondary septum development(GO:0061144) |
| 0.0 | 0.2 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.0 | 0.1 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.0 | 0.1 | GO:2000169 | regulation of peptidyl-cysteine S-nitrosylation(GO:2000169) |
| 0.0 | 0.1 | GO:0035910 | ascending aorta development(GO:0035905) ascending aorta morphogenesis(GO:0035910) |
| 0.0 | 0.3 | GO:0007097 | nuclear migration(GO:0007097) |
| 0.0 | 0.2 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.0 | 0.1 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.0 | 0.1 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.0 | 0.1 | GO:0044789 | modulation by host of viral release from host cell(GO:0044789) positive regulation by host of viral release from host cell(GO:0044791) |
| 0.0 | 0.1 | GO:0097475 | motor neuron migration(GO:0097475) spinal cord motor neuron migration(GO:0097476) |
| 0.0 | 0.1 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 0.1 | GO:0048143 | astrocyte activation(GO:0048143) |
| 0.0 | 0.1 | GO:0007089 | traversing start control point of mitotic cell cycle(GO:0007089) |
| 0.0 | 0.1 | GO:1990791 | dorsal root ganglion development(GO:1990791) |
| 0.0 | 0.1 | GO:2000173 | negative regulation of branching morphogenesis of a nerve(GO:2000173) |
| 0.0 | 0.1 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.3 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.0 | 0.1 | GO:0021800 | cerebral cortex tangential migration(GO:0021800) |
| 0.0 | 0.1 | GO:1902996 | neurofibrillary tangle assembly(GO:1902988) regulation of neurofibrillary tangle assembly(GO:1902996) |
| 0.0 | 0.1 | GO:0060060 | post-embryonic retina morphogenesis in camera-type eye(GO:0060060) |
| 0.0 | 0.1 | GO:0014045 | establishment of endothelial blood-brain barrier(GO:0014045) |
| 0.0 | 0.1 | GO:0061316 | canonical Wnt signaling pathway involved in heart development(GO:0061316) |
| 0.0 | 0.1 | GO:0006041 | glucosamine metabolic process(GO:0006041) |
| 0.0 | 0.1 | GO:0010871 | negative regulation of receptor biosynthetic process(GO:0010871) |
| 0.0 | 0.1 | GO:1902218 | intrinsic apoptotic signaling pathway in response to osmotic stress(GO:0008627) regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902218) negative regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902219) |
| 0.0 | 0.3 | GO:0061418 | regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061418) |
| 0.0 | 0.1 | GO:0002606 | positive regulation of dendritic cell antigen processing and presentation(GO:0002606) |
| 0.0 | 0.4 | GO:0006309 | apoptotic DNA fragmentation(GO:0006309) |
| 0.0 | 0.1 | GO:0003289 | atrial septum primum morphogenesis(GO:0003289) |
| 0.0 | 0.1 | GO:0060671 | epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 0.0 | 0.1 | GO:0021914 | negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021914) |
| 0.0 | 0.2 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.0 | 0.1 | GO:0002894 | type IIa hypersensitivity(GO:0001794) regulation of type IIa hypersensitivity(GO:0001796) positive regulation of type IIa hypersensitivity(GO:0001798) type II hypersensitivity(GO:0002445) regulation of type II hypersensitivity(GO:0002892) positive regulation of type II hypersensitivity(GO:0002894) |
| 0.0 | 0.1 | GO:0015886 | heme transport(GO:0015886) |
| 0.0 | 0.1 | GO:0061643 | chemorepulsion of axon(GO:0061643) |
| 0.0 | 0.1 | GO:0051890 | regulation of cardioblast differentiation(GO:0051890) |
| 0.0 | 0.7 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 0.1 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.0 | 0.1 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.0 | 0.1 | GO:0006578 | amino-acid betaine biosynthetic process(GO:0006578) |
| 0.0 | 0.1 | GO:1901844 | regulation of cell communication by electrical coupling involved in cardiac conduction(GO:1901844) |
| 0.0 | 0.2 | GO:0000183 | chromatin silencing at rDNA(GO:0000183) |
| 0.0 | 0.1 | GO:0009157 | deoxyribonucleoside monophosphate biosynthetic process(GO:0009157) pyrimidine deoxyribonucleoside monophosphate biosynthetic process(GO:0009177) |
| 0.0 | 0.1 | GO:0038026 | reelin-mediated signaling pathway(GO:0038026) |
| 0.0 | 0.1 | GO:1902455 | negative regulation of stem cell population maintenance(GO:1902455) |
| 0.0 | 0.1 | GO:0051791 | medium-chain fatty acid metabolic process(GO:0051791) |
| 0.0 | 0.0 | GO:0043622 | cortical microtubule organization(GO:0043622) |
| 0.0 | 0.4 | GO:1902230 | negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902230) |
| 0.0 | 0.1 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.1 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.0 | 0.1 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 0.0 | 0.1 | GO:0006524 | alanine catabolic process(GO:0006524) pyruvate family amino acid catabolic process(GO:0009080) |
| 0.0 | 0.1 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.0 | 0.1 | GO:0042518 | negative regulation of tyrosine phosphorylation of Stat3 protein(GO:0042518) |
| 0.0 | 0.2 | GO:0060546 | negative regulation of necroptotic process(GO:0060546) |
| 0.0 | 0.1 | GO:1903750 | regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903750) negative regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903751) |
| 0.0 | 0.0 | GO:0021593 | rhombomere morphogenesis(GO:0021593) |
| 0.0 | 0.2 | GO:0002329 | pre-B cell differentiation(GO:0002329) |
| 0.0 | 0.1 | GO:0030953 | astral microtubule organization(GO:0030953) |
| 0.0 | 0.2 | GO:0042635 | positive regulation of hair cycle(GO:0042635) |
| 0.0 | 0.1 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 0.0 | 0.1 | GO:0042117 | monocyte activation(GO:0042117) |
| 0.0 | 0.1 | GO:0046726 | positive regulation by virus of viral protein levels in host cell(GO:0046726) |
| 0.0 | 0.0 | GO:1901166 | neural crest cell migration involved in autonomic nervous system development(GO:1901166) |
| 0.0 | 0.1 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 0.5 | GO:0070830 | bicellular tight junction assembly(GO:0070830) |
| 0.0 | 0.1 | GO:0019859 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) thymine metabolic process(GO:0019859) |
| 0.0 | 0.0 | GO:1903423 | positive regulation of synaptic vesicle recycling(GO:1903423) |
| 0.0 | 0.1 | GO:0098902 | regulation of membrane depolarization during action potential(GO:0098902) |
| 0.0 | 0.1 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.0 | 0.0 | GO:0019448 | cysteine catabolic process(GO:0009093) L-cysteine catabolic process(GO:0019448) L-cysteine metabolic process(GO:0046439) |
| 0.0 | 0.1 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.0 | 0.0 | GO:0003164 | His-Purkinje system development(GO:0003164) bundle of His development(GO:0003166) |
| 0.0 | 0.1 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 0.1 | GO:0090214 | spongiotrophoblast layer developmental growth(GO:0090214) |
| 0.0 | 0.1 | GO:0000492 | small nucleolar ribonucleoprotein complex assembly(GO:0000491) box C/D snoRNP assembly(GO:0000492) |
| 0.0 | 0.0 | GO:1905208 | negative regulation of cardiocyte differentiation(GO:1905208) negative regulation of cardiac muscle cell differentiation(GO:2000726) |
| 0.0 | 0.1 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.0 | 0.1 | GO:0043116 | negative regulation of vascular permeability(GO:0043116) |
| 0.0 | 0.1 | GO:0072429 | response to intra-S DNA damage checkpoint signaling(GO:0072429) |
| 0.0 | 0.2 | GO:0006517 | protein deglycosylation(GO:0006517) |
| 0.0 | 0.1 | GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 0.0 | 0.0 | GO:0060128 | corticotropin hormone secreting cell differentiation(GO:0060128) |
| 0.0 | 0.1 | GO:0030210 | heparin biosynthetic process(GO:0030210) |
| 0.0 | 0.1 | GO:0006551 | leucine metabolic process(GO:0006551) |
| 0.0 | 0.1 | GO:0060696 | regulation of phospholipid catabolic process(GO:0060696) |
| 0.0 | 0.2 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.1 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 0.1 | GO:0009838 | abscission(GO:0009838) |
| 0.0 | 0.2 | GO:0019886 | antigen processing and presentation of exogenous peptide antigen via MHC class II(GO:0019886) |
| 0.0 | 0.1 | GO:0042637 | catagen(GO:0042637) |
| 0.0 | 0.1 | GO:0007343 | egg activation(GO:0007343) |
| 0.0 | 0.4 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.0 | GO:0003062 | regulation of heart rate by chemical signal(GO:0003062) |
| 0.0 | 0.0 | GO:0002018 | renin-angiotensin regulation of aldosterone production(GO:0002018) |
| 0.0 | 0.0 | GO:0021986 | epithalamus development(GO:0021538) habenula development(GO:0021986) |
| 0.0 | 0.1 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 0.1 | GO:0072502 | cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.0 | 0.2 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.0 | 0.1 | GO:0070126 | mitochondrial translational termination(GO:0070126) |
| 0.0 | 0.0 | GO:0007418 | ventral midline development(GO:0007418) |
| 0.0 | 0.1 | GO:0010694 | positive regulation of alkaline phosphatase activity(GO:0010694) |
| 0.0 | 0.0 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 0.1 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.0 | 0.0 | GO:0050713 | negative regulation of interleukin-1 beta secretion(GO:0050713) |
| 0.0 | 0.0 | GO:0015817 | histidine transport(GO:0015817) |
| 0.0 | 0.1 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.0 | 0.0 | GO:0002525 | acute inflammatory response to non-antigenic stimulus(GO:0002525) |
| 0.0 | 0.1 | GO:0006449 | regulation of translational termination(GO:0006449) |
| 0.0 | 0.1 | GO:0006415 | translational termination(GO:0006415) |
| 0.0 | 0.1 | GO:2000418 | positive regulation of eosinophil migration(GO:2000418) |
| 0.0 | 0.0 | GO:0023021 | termination of signal transduction(GO:0023021) |
| 0.0 | 0.0 | GO:0015755 | fructose transport(GO:0015755) |
| 0.0 | 0.0 | GO:0061687 | detoxification of inorganic compound(GO:0061687) |
| 0.0 | 0.1 | GO:0051292 | nuclear pore complex assembly(GO:0051292) |
| 0.0 | 0.0 | GO:0007403 | glial cell fate determination(GO:0007403) |
| 0.0 | 0.1 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.0 | 0.4 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 0.1 | GO:0002031 | G-protein coupled receptor internalization(GO:0002031) |
| 0.0 | 0.1 | GO:0061050 | regulation of cell growth involved in cardiac muscle cell development(GO:0061050) |
| 0.0 | 0.1 | GO:0046051 | UTP metabolic process(GO:0046051) |
| 0.0 | 0.0 | GO:0033029 | regulation of neutrophil apoptotic process(GO:0033029) |
| 0.0 | 0.1 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.0 | 0.0 | GO:2000359 | regulation of binding of sperm to zona pellucida(GO:2000359) |
| 0.0 | 0.0 | GO:0036112 | medium-chain fatty-acyl-CoA metabolic process(GO:0036112) |
| 0.0 | 0.0 | GO:0060748 | tertiary branching involved in mammary gland duct morphogenesis(GO:0060748) |
| 0.0 | 0.2 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.0 | 0.2 | GO:0014850 | response to muscle activity(GO:0014850) |
| 0.0 | 0.0 | GO:0035262 | gonad morphogenesis(GO:0035262) |
| 0.0 | 0.0 | GO:0033686 | positive regulation of luteinizing hormone secretion(GO:0033686) |
| 0.0 | 0.0 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.0 | GO:0033483 | gas homeostasis(GO:0033483) |
| 0.0 | 0.1 | GO:1902373 | negative regulation of mRNA catabolic process(GO:1902373) |
| 0.0 | 0.1 | GO:0035590 | purinergic nucleotide receptor signaling pathway(GO:0035590) |
| 0.0 | 0.1 | GO:0015867 | ATP transport(GO:0015867) |
| 0.0 | 0.2 | GO:0032516 | positive regulation of phosphoprotein phosphatase activity(GO:0032516) |
| 0.0 | 0.0 | GO:2000370 | positive regulation of clathrin-mediated endocytosis(GO:2000370) |
| 0.0 | 0.0 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.0 | 0.1 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.0 | 0.0 | GO:0060710 | chorio-allantoic fusion(GO:0060710) |
| 0.0 | 0.0 | GO:0002337 | B-1a B cell differentiation(GO:0002337) |
| 0.0 | 0.0 | GO:2000659 | regulation of interleukin-1-mediated signaling pathway(GO:2000659) |
| 0.0 | 0.0 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.0 | 0.1 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.0 | 0.0 | GO:2001137 | positive regulation of endocytic recycling(GO:2001137) |
| 0.0 | 0.1 | GO:0010572 | positive regulation of platelet activation(GO:0010572) |
| 0.0 | 0.1 | GO:0043545 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) molybdopterin cofactor biosynthetic process(GO:0032324) molybdopterin cofactor metabolic process(GO:0043545) prosthetic group metabolic process(GO:0051189) |
| 0.0 | 0.0 | GO:0019249 | lactate biosynthetic process from pyruvate(GO:0019244) lactate biosynthetic process(GO:0019249) |
| 0.0 | 0.0 | GO:1900242 | regulation of synaptic vesicle endocytosis(GO:1900242) |
| 0.0 | 0.0 | GO:1902071 | regulation of hypoxia-inducible factor-1alpha signaling pathway(GO:1902071) |
| 0.0 | 0.2 | GO:0010592 | positive regulation of lamellipodium assembly(GO:0010592) |
| 0.0 | 0.0 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.0 | 0.0 | GO:0071895 | odontoblast differentiation(GO:0071895) |
| 0.0 | 0.0 | GO:0044004 | killing by symbiont of host cells(GO:0001907) disruption by symbiont of host cell(GO:0044004) |
| 0.0 | 0.2 | GO:0002021 | response to dietary excess(GO:0002021) |
| 0.0 | 0.1 | GO:0046040 | IMP metabolic process(GO:0046040) |
| 0.0 | 0.0 | GO:0044800 | fusion of virus membrane with host plasma membrane(GO:0019064) membrane fusion involved in viral entry into host cell(GO:0039663) multi-organism membrane fusion(GO:0044800) |
| 0.0 | 0.0 | GO:0040009 | regulation of growth rate(GO:0040009) |
| 0.0 | 0.0 | GO:0050883 | medulla oblongata development(GO:0021550) musculoskeletal movement, spinal reflex action(GO:0050883) |
| 0.0 | 0.0 | GO:0002386 | immune response in mucosal-associated lymphoid tissue(GO:0002386) |
| 0.0 | 0.0 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.0 | 0.0 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.1 | GO:1900619 | acetylcholine metabolic process(GO:0008291) acetate ester metabolic process(GO:1900619) |
| 0.0 | 0.0 | GO:0035022 | positive regulation of Rac protein signal transduction(GO:0035022) |
| 0.0 | 0.0 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 0.0 | 0.0 | GO:2000302 | positive regulation of synaptic vesicle exocytosis(GO:2000302) |
| 0.0 | 0.0 | GO:0034241 | positive regulation of macrophage fusion(GO:0034241) |
| 0.0 | 0.0 | GO:0099515 | actin filament-based transport(GO:0099515) |
| 0.0 | 0.0 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.0 | 0.1 | GO:0009115 | xanthine catabolic process(GO:0009115) xanthine metabolic process(GO:0046110) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.0 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.1 | 0.4 | GO:0034666 | integrin alpha2-beta1 complex(GO:0034666) |
| 0.1 | 0.4 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.1 | 0.3 | GO:0030956 | glutamyl-tRNA(Gln) amidotransferase complex(GO:0030956) |
| 0.1 | 0.6 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.1 | 0.3 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.0 | 0.4 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.1 | GO:0097543 | ciliary inversin compartment(GO:0097543) |
| 0.0 | 0.3 | GO:0031983 | vesicle lumen(GO:0031983) |
| 0.0 | 0.1 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 0.0 | 0.2 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 0.1 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.0 | 0.2 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.0 | 0.2 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 0.3 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.4 | GO:0032797 | SMN complex(GO:0032797) |
| 0.0 | 0.2 | GO:1990696 | USH2 complex(GO:1990696) |
| 0.0 | 0.2 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.0 | 0.2 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.0 | 0.2 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.0 | 0.2 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 0.1 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.6 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 0.2 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 0.1 | GO:0010009 | cytoplasmic side of endosome membrane(GO:0010009) |
| 0.0 | 0.1 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 0.1 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.0 | 0.1 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.0 | 0.1 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.0 | 0.2 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.3 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.0 | 0.5 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.2 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.2 | GO:0042627 | chylomicron(GO:0042627) |
| 0.0 | 0.1 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 0.0 | 0.2 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.1 | GO:0034361 | very-low-density lipoprotein particle(GO:0034361) triglyceride-rich lipoprotein particle(GO:0034385) |
| 0.0 | 0.0 | GO:1990812 | growth cone filopodium(GO:1990812) |
| 0.0 | 0.3 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.3 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
| 0.0 | 0.1 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.1 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.0 | 0.1 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 0.0 | 0.0 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 0.1 | GO:0043219 | lateral loop(GO:0043219) |
| 0.0 | 0.1 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.0 | 0.1 | GO:0005818 | aster(GO:0005818) |
| 0.0 | 0.1 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.1 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 0.1 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.0 | 0.1 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.1 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.0 | 0.0 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.0 | 0.0 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.0 | 0.1 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.0 | 0.1 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.1 | GO:1990851 | Wnt-Frizzled-LRP5/6 complex(GO:1990851) |
| 0.0 | 0.1 | GO:0000801 | central element(GO:0000801) |
| 0.0 | 0.1 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.0 | 0.2 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.1 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.0 | 0.1 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.1 | GO:1990745 | EARP complex(GO:1990745) |
| 0.0 | 0.0 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.0 | 0.0 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.1 | GO:0000805 | X chromosome(GO:0000805) |
| 0.0 | 0.0 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.0 | 0.2 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.0 | 0.0 | GO:0031429 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 0.0 | 0.0 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.0 | 0.5 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.1 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.0 | GO:0032127 | dense core granule membrane(GO:0032127) |
| 0.0 | 0.0 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.0 | 0.0 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.0 | 0.0 | GO:0002142 | stereocilia ankle link complex(GO:0002142) |
| 0.0 | 0.0 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.0 | 0.3 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 0.0 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.0 | 0.0 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.0 | 0.1 | GO:0060091 | kinocilium(GO:0060091) |
| 0.0 | 0.0 | GO:0000811 | GINS complex(GO:0000811) |
| 0.0 | 0.0 | GO:0036488 | CHOP-C/EBP complex(GO:0036488) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.1 | GO:0008390 | testosterone 16-alpha-hydroxylase activity(GO:0008390) |
| 0.1 | 1.0 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.1 | 0.4 | GO:0000171 | ribonuclease MRP activity(GO:0000171) |
| 0.1 | 0.3 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.1 | 0.3 | GO:0050567 | glutaminyl-tRNA synthase (glutamine-hydrolyzing) activity(GO:0050567) |
| 0.1 | 0.4 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.1 | 0.5 | GO:0018647 | 3-(3-hydroxyphenyl)propionate hydroxylase activity(GO:0008688) 4-chlorobenzaldehyde oxidase activity(GO:0018471) 3,5-xylenol methylhydroxylase activity(GO:0018630) phenylacetate hydroxylase activity(GO:0018631) 4-nitrophenol 4-monooxygenase activity(GO:0018632) dimethyl sulfide monooxygenase activity(GO:0018633) alpha-pinene monooxygenase [NADH] activity(GO:0018634) 1-hydroxy-2-naphthoate hydroxylase activity(GO:0018637) toluene 4-monooxygenase activity(GO:0018638) xylene monooxygenase activity(GO:0018639) dibenzothiophene monooxygenase activity(GO:0018640) 6-hydroxy-3-methyl-2-oxo-1,2-dihydroquinoline 6-monooxygenase activity(GO:0018641) chlorophenol 4-monooxygenase activity(GO:0018642) carbon disulfide oxygenase activity(GO:0018643) toluene 2-monooxygenase activity(GO:0018644) 1-hydroxy-2-oxolimonene 1,2-monooxygenase activity(GO:0018646) phenanthrene 1,2-monooxygenase activity(GO:0018647) tetrahydrofuran hydroxylase activity(GO:0018649) styrene monooxygenase activity(GO:0018650) toluene-4-sulfonate monooxygenase activity(GO:0018651) toluene-sulfonate methyl-monooxygenase activity(GO:0018652) 3-methyl-2-oxo-1,2-dihydroquinoline 6-monooxygenase activity(GO:0018653) 2-hydroxy-phenylacetate hydroxylase activity(GO:0018654) 2-oxo-delta3-4,5,5-trimethylcyclopentenylacetyl-CoA 1,2-monooxygenase activity(GO:0018655) phenanthrene 3,4-monooxygenase activity(GO:0018656) toluene 3-monooxygenase activity(GO:0018657) 4-hydroxyphenylacetate,NADH:oxygen oxidoreductase (3-hydroxylating) activity(GO:0018660) limonene monooxygenase activity(GO:0019113) 2-methylnaphthalene hydroxylase activity(GO:0034526) 1-methylnaphthalene hydroxylase activity(GO:0034534) bisphenol A hydroxylase A activity(GO:0034560) salicylate 5-hydroxylase activity(GO:0034785) isobutylamine N-hydroxylase activity(GO:0034791) branched-chain dodecylbenzene sulfonate monooxygenase activity(GO:0034802) 3-HSA hydroxylase activity(GO:0034819) 4-hydroxypyridine-3-hydroxylase activity(GO:0034894) 2-octaprenyl-3-methyl-6-methoxy-1,4-benzoquinol hydroxylase activity(GO:0043719) 6-hydroxynicotinate 3-monooxygenase activity(GO:0043731) thalianol hydroxylase activity(GO:0080014) |
| 0.1 | 0.2 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.1 | 0.2 | GO:0031711 | bradykinin receptor binding(GO:0031711) |
| 0.1 | 0.3 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.1 | 0.2 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.1 | 0.5 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.1 | 0.2 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.1 | 0.4 | GO:0016889 | endodeoxyribonuclease activity, producing 3'-phosphomonoesters(GO:0016889) |
| 0.0 | 0.2 | GO:0005346 | adenine nucleotide transmembrane transporter activity(GO:0000295) purine ribonucleotide transmembrane transporter activity(GO:0005346) purine nucleotide transmembrane transporter activity(GO:0015216) |
| 0.0 | 0.3 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) xenobiotic transporter activity(GO:0042910) |
| 0.0 | 0.2 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.0 | 0.4 | GO:0036122 | BMP binding(GO:0036122) |
| 0.0 | 0.2 | GO:0016899 | oxidoreductase activity, acting on the CH-OH group of donors, oxygen as acceptor(GO:0016899) |
| 0.0 | 0.2 | GO:0022820 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.0 | 0.2 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 0.4 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.0 | 0.1 | GO:0035877 | death effector domain binding(GO:0035877) |
| 0.0 | 0.1 | GO:0004104 | cholinesterase activity(GO:0004104) |
| 0.0 | 0.2 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.1 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.2 | GO:0090482 | vitamin transmembrane transporter activity(GO:0090482) |
| 0.0 | 0.2 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.0 | 0.2 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 0.2 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 0.1 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.0 | 0.2 | GO:0018685 | alkane 1-monooxygenase activity(GO:0018685) |
| 0.0 | 0.1 | GO:0032564 | dATP binding(GO:0032564) |
| 0.0 | 0.2 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.0 | 0.1 | GO:0102344 | 3-hydroxy-behenoyl-CoA dehydratase activity(GO:0102344) 3-hydroxy-lignoceroyl-CoA dehydratase activity(GO:0102345) |
| 0.0 | 0.1 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) |
| 0.0 | 0.1 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.0 | 0.1 | GO:0034618 | arginine binding(GO:0034618) |
| 0.0 | 0.1 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.0 | 0.2 | GO:0047760 | butyrate-CoA ligase activity(GO:0047760) |
| 0.0 | 0.1 | GO:0015198 | oligopeptide transporter activity(GO:0015198) |
| 0.0 | 0.2 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.0 | 0.1 | GO:0015252 | hydrogen ion channel activity(GO:0015252) |
| 0.0 | 0.1 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.0 | 0.1 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) |
| 0.0 | 0.0 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 0.0 | 0.4 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.0 | 0.1 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.1 | GO:0060228 | phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
| 0.0 | 0.1 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.0 | 0.1 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.0 | 0.1 | GO:0030984 | kininogen binding(GO:0030984) |
| 0.0 | 0.0 | GO:0019959 | interleukin-8 binding(GO:0019959) |
| 0.0 | 0.1 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.1 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.0 | 0.1 | GO:0031893 | vasopressin receptor binding(GO:0031893) |
| 0.0 | 0.1 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.0 | 0.1 | GO:0034190 | apolipoprotein receptor binding(GO:0034190) |
| 0.0 | 0.1 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.0 | 0.2 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.2 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.1 | GO:0009374 | biotin binding(GO:0009374) |
| 0.0 | 0.1 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 0.1 | GO:0004305 | ethanolamine kinase activity(GO:0004305) |
| 0.0 | 0.2 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.0 | 0.7 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.1 | GO:0070905 | serine binding(GO:0070905) |
| 0.0 | 0.1 | GO:0004346 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.0 | 0.2 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.0 | 0.1 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.0 | 0.3 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.1 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.0 | 0.1 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.0 | 0.1 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.0 | 0.3 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.1 | GO:0016149 | translation release factor activity, codon specific(GO:0016149) |
| 0.0 | 0.3 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.0 | 0.2 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.1 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.1 | GO:0042979 | ornithine decarboxylase regulator activity(GO:0042979) |
| 0.0 | 0.1 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.0 | 0.4 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.3 | GO:0031690 | adrenergic receptor binding(GO:0031690) |
| 0.0 | 0.0 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.0 | 0.1 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 0.0 | 0.1 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.1 | GO:0045340 | mercury ion binding(GO:0045340) |
| 0.0 | 0.3 | GO:0016780 | phosphotransferase activity, for other substituted phosphate groups(GO:0016780) |
| 0.0 | 0.1 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.0 | 0.2 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.1 | GO:0050897 | cobalt ion binding(GO:0050897) |
| 0.0 | 0.1 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.0 | 0.1 | GO:0008035 | high-density lipoprotein particle binding(GO:0008035) |
| 0.0 | 0.0 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.1 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.0 | 0.0 | GO:0015182 | L-asparagine transmembrane transporter activity(GO:0015182) L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.0 | 0.0 | GO:0030158 | protein xylosyltransferase activity(GO:0030158) |
| 0.0 | 0.2 | GO:0048185 | activin binding(GO:0048185) |
| 0.0 | 0.1 | GO:0052759 | 4-methyloctanoyl-CoA dehydrogenase activity(GO:0034580) naphthyl-2-methyl-succinyl-CoA dehydrogenase activity(GO:0034845) 2-methylhexanoyl-CoA dehydrogenase activity(GO:0034916) propionyl-CoA dehydrogenase activity(GO:0043820) thiol-driven fumarate reductase activity(GO:0043830) coenzyme F420-dependent 2,4,6-trinitrophenol reductase activity(GO:0052758) coenzyme F420-dependent 2,4,6-trinitrophenol hydride reductase activity(GO:0052759) coenzyme F420-dependent 2,4-dinitrophenol reductase activity(GO:0052760) |
| 0.0 | 0.2 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.1 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.0 | GO:0004471 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) |
| 0.0 | 0.3 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.0 | 0.1 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 0.0 | GO:0032767 | copper-dependent protein binding(GO:0032767) |
| 0.0 | 0.3 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 0.1 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.1 | GO:0031782 | melanocortin receptor binding(GO:0031779) type 3 melanocortin receptor binding(GO:0031781) type 4 melanocortin receptor binding(GO:0031782) |
| 0.0 | 0.1 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.0 | 0.1 | GO:0004031 | aldehyde oxidase activity(GO:0004031) oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) |
| 0.0 | 0.0 | GO:0008948 | oxaloacetate decarboxylase activity(GO:0008948) |
| 0.0 | 0.0 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.0 | 0.0 | GO:0004165 | dodecenoyl-CoA delta-isomerase activity(GO:0004165) |
| 0.0 | 0.0 | GO:0015111 | iodide transmembrane transporter activity(GO:0015111) |
| 0.0 | 0.1 | GO:0034863 | pinocarveol dehydrogenase activity(GO:0018446) chloral hydrate dehydrogenase activity(GO:0018447) hydroxymethylmethylsilanediol oxidase activity(GO:0018448) 1-phenylethanol dehydrogenase activity(GO:0018449) myrtenol dehydrogenase activity(GO:0018450) cis-1,2-dihydroxy-1,2-dihydro-8-carboxynaphthalene dehydrogenase activity(GO:0034522) 3-hydroxy-4-methyloctanoyl-CoA dehydrogenase activity(GO:0034582) 2-hydroxy-4-isopropenylcyclohexane-1-carboxyl-CoA dehydrogenase activity(GO:0034778) cis-9,10-dihydroanthracene-9,10-diol dehydrogenase activity(GO:0034817) citronellol dehydrogenase activity(GO:0034821) naphthyl-2-hydroxymethyl-succinyl-CoA dehydrogenase activity(GO:0034847) 2,4,4-trimethyl-1-pentanol dehydrogenase activity(GO:0034863) 2,4,4-trimethyl-3-hydroxypentanoyl-CoA dehydrogenase activity(GO:0034868) 1-hydroxy-4,4-dimethylpentan-3-one dehydrogenase activity(GO:0034871) endosulfan diol dehydrogenase activity(GO:0034891) endosulfan hydroxyether dehydrogenase activity(GO:0034901) 3-hydroxy-2-methylhexanoyl-CoA dehydrogenase activity(GO:0034918) 3-hydroxy-2,6-dimethyl-5-methylene-heptanoyl-CoA dehydrogenase activity(GO:0034944) versicolorin reductase activity(GO:0042469) ketoreductase activity(GO:0045703) |
| 0.0 | 0.0 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.0 | 0.1 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.0 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.1 | GO:0003918 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.0 | 0.0 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.1 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.0 | 0.1 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 0.0 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.0 | 0.2 | GO:1900750 | glutathione binding(GO:0043295) oligopeptide binding(GO:1900750) |
| 0.0 | 0.1 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 0.0 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.2 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.1 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 0.0 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 0.0 | 0.1 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) |
| 0.0 | 0.0 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 0.2 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 0.0 | GO:0008240 | tripeptidyl-peptidase activity(GO:0008240) |
| 0.0 | 0.3 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.7 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.0 | 0.3 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.1 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 0.0 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 0.1 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 0.3 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.0 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 0.2 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 0.1 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.4 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.1 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.0 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 0.3 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 0.2 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.0 | 0.6 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 0.3 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.4 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.3 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.4 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 0.5 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.3 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.2 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 0.6 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.4 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 0.4 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.2 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.5 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.2 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.1 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.2 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.3 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.2 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 0.2 | REACTOME MRNA CAPPING | Genes involved in mRNA Capping |
| 0.0 | 0.2 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.2 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 0.0 | REACTOME CTLA4 INHIBITORY SIGNALING | Genes involved in CTLA4 inhibitory signaling |
| 0.0 | 0.0 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS | Genes involved in Synthesis of bile acids and bile salts |
| 0.0 | 1.1 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.2 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.0 | 0.2 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.0 | REACTOME SIGNALING BY EGFR IN CANCER | Genes involved in Signaling by EGFR in Cancer |
| 0.0 | 0.0 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 0.0 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 0.2 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.0 | REACTOME DNA STRAND ELONGATION | Genes involved in DNA strand elongation |