Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
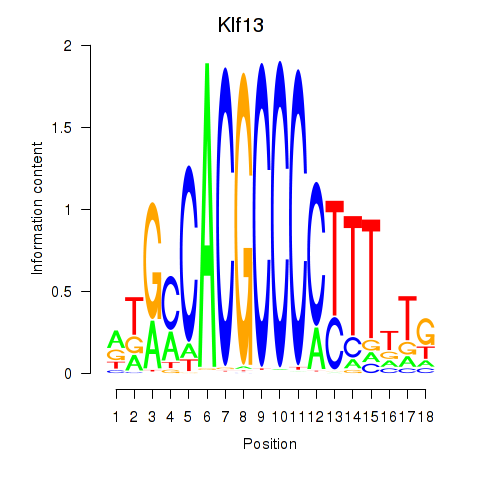
Results for Klf13
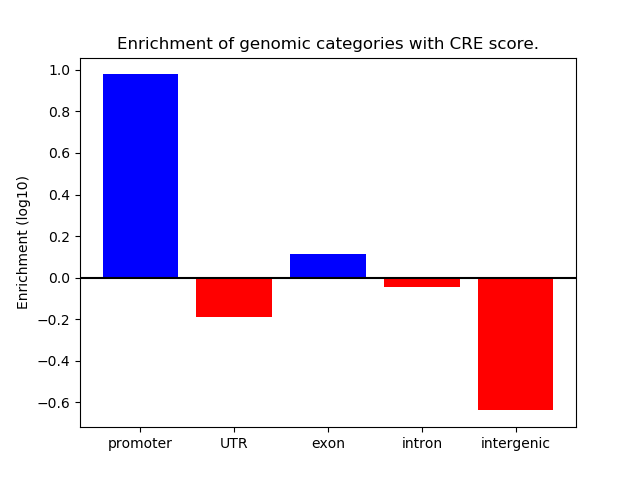
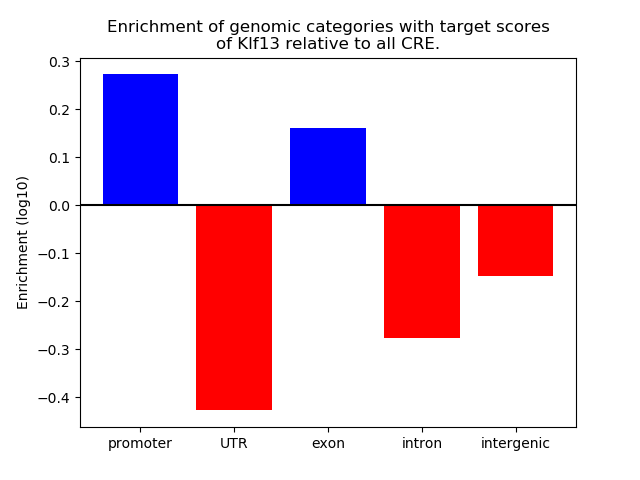
Z-value: 0.97
Transcription factors associated with Klf13
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Klf13
|
ENSMUSG00000052040.9 | Kruppel-like factor 13 |
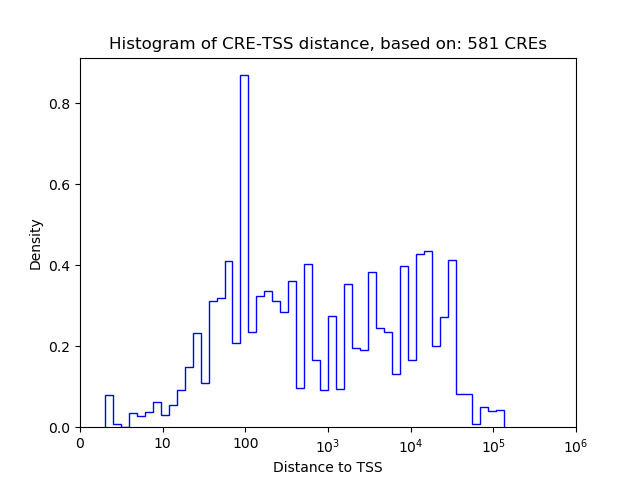
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr7_63934853_63935097 | Klf13 | 1326 | 0.339936 | -0.83 | 3.9e-02 | Click! |
| chr7_63930850_63931034 | Klf13 | 5359 | 0.144137 | -0.80 | 5.5e-02 | Click! |
| chr7_63928069_63928220 | Klf13 | 3274 | 0.172344 | -0.73 | 9.8e-02 | Click! |
| chr7_63924758_63924944 | Klf13 | 19 | 0.966664 | -0.72 | 1.0e-01 | Click! |
| chr7_63936513_63936685 | Klf13 | 267 | 0.876987 | -0.67 | 1.5e-01 | Click! |
Activity of the Klf13 motif across conditions
Conditions sorted by the z-value of the Klf13 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
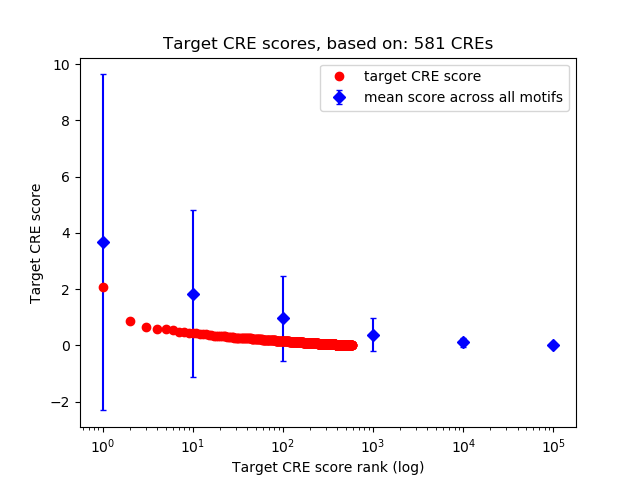
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr7_139020930_139021245 | 2.06 |
Gm45613 |
predicted gene 45613 |
88 |
0.97 |
| chr10_95720533_95720688 | 0.88 |
1110019B22Rik |
RIKEN cDNA 1110019B22 gene |
1879 |
0.2 |
| chr7_66005228_66005393 | 0.66 |
Pcsk6 |
proprotein convertase subtilisin/kexin type 6 |
33108 |
0.12 |
| chrY_90739212_90739561 | 0.60 |
Mid1-ps1 |
midline 1, pseudogene 1 |
13671 |
0.18 |
| chr4_35134107_35134262 | 0.57 |
Ifnk |
interferon kappa |
17872 |
0.17 |
| chr17_14175138_14175307 | 0.54 |
Gm34510 |
predicted gene, 34510 |
28506 |
0.14 |
| chr11_67081331_67081503 | 0.48 |
Myh3 |
myosin, heavy polypeptide 3, skeletal muscle, embryonic |
3117 |
0.17 |
| chr11_97428221_97428386 | 0.46 |
Arhgap23 |
Rho GTPase activating protein 23 |
7982 |
0.16 |
| chr8_71559035_71559219 | 0.45 |
Tmem221 |
transmembrane protein 221 |
256 |
0.79 |
| chr6_90486122_90486491 | 0.44 |
Gm45901 |
predicted gene 45901 |
87 |
0.54 |
| chr6_148212064_148212226 | 0.44 |
Ergic2 |
ERGIC and golgi 2 |
38 |
0.58 |
| chr5_38560875_38561037 | 0.40 |
Wdr1 |
WD repeat domain 1 |
587 |
0.71 |
| chr4_150874506_150874750 | 0.39 |
Errfi1 |
ERBB receptor feedback inhibitor 1 |
19555 |
0.12 |
| chr17_29841749_29841925 | 0.39 |
Mdga1 |
MAM domain containing glycosylphosphatidylinositol anchor 1 |
584 |
0.73 |
| chr10_117418076_117418301 | 0.37 |
Gm40770 |
predicted gene, 40770 |
4284 |
0.16 |
| chr16_22919975_22920126 | 0.36 |
Fetub |
fetuin beta |
187 |
0.91 |
| chr18_60603591_60603778 | 0.35 |
Synpo |
synaptopodin |
3187 |
0.2 |
| chr6_116195979_116196139 | 0.34 |
Gm8213 |
predicted pseudogene 8213 |
201 |
0.89 |
| chr3_105893960_105894139 | 0.34 |
Adora3 |
adenosine A3 receptor |
10372 |
0.12 |
| chr2_158176890_158177041 | 0.34 |
1700060C20Rik |
RIKEN cDNA 1700060C20 gene |
15043 |
0.14 |
| chr1_191186557_191186730 | 0.33 |
Atf3 |
activating transcription factor 3 |
3303 |
0.18 |
| chr7_80156693_80156942 | 0.33 |
Gm45202 |
predicted gene 45202 |
1648 |
0.25 |
| chr4_133804499_133804683 | 0.32 |
Gm25270 |
predicted gene, 25270 |
6872 |
0.14 |
| chr1_182585565_182585739 | 0.31 |
Capn8 |
calpain 8 |
20614 |
0.16 |
| chr15_31338782_31338933 | 0.31 |
Ankrd33b |
ankyrin repeat domain 33B |
13608 |
0.15 |
| chr2_85047886_85048056 | 0.30 |
Tnks1bp1 |
tankyrase 1 binding protein 1 |
51 |
0.96 |
| chr4_47245499_47245708 | 0.30 |
Col15a1 |
collagen, type XV, alpha 1 |
12336 |
0.2 |
| chr19_44997151_44997302 | 0.28 |
Sema4g |
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4G |
5090 |
0.11 |
| chr17_83503811_83503975 | 0.28 |
Cox7a2l |
cytochrome c oxidase subunit 7A2 like |
613 |
0.8 |
| chr7_27178957_27179117 | 0.27 |
Rab4b |
RAB4B, member RAS oncogene family |
141 |
0.86 |
| chr9_31211792_31211960 | 0.27 |
Aplp2 |
amyloid beta (A4) precursor-like protein 2 |
61 |
0.98 |
| chr12_14152257_14152408 | 0.27 |
Lratd1 |
LRAT domain containing 1 |
278 |
0.93 |
| chr10_80168459_80168625 | 0.26 |
Cirbp |
cold inducible RNA binding protein |
656 |
0.47 |
| chr2_167014815_167015114 | 0.26 |
Ddx27 |
DEAD (Asp-Glu-Ala-Asp) box polypeptide 27 |
229 |
0.87 |
| chr13_98689660_98689822 | 0.26 |
Tmem171 |
transmembrane protein 171 |
5027 |
0.15 |
| chr11_69326130_69326304 | 0.26 |
Kcnab3 |
potassium voltage-gated channel, shaker-related subfamily, beta member 3 |
41 |
0.93 |
| chr15_99113382_99113533 | 0.26 |
Spats2 |
spermatogenesis associated, serine-rich 2 |
12459 |
0.09 |
| chrX_8252648_8252921 | 0.26 |
Ftsj1 |
FtsJ RNA methyltransferase homolog 1 (E. coli) |
378 |
0.8 |
| chr2_170155421_170155732 | 0.25 |
Zfp217 |
zinc finger protein 217 |
7473 |
0.29 |
| chr1_39192284_39192607 | 0.25 |
Npas2 |
neuronal PAS domain protein 2 |
1286 |
0.45 |
| chr8_94012059_94012219 | 0.25 |
Amfr |
autocrine motility factor receptor |
12 |
0.97 |
| chr5_129895249_129895421 | 0.25 |
Zbed5 |
zinc finger, BED type containing 5 |
388 |
0.71 |
| chr11_117836449_117836615 | 0.24 |
Afmid |
arylformamidase |
4259 |
0.1 |
| chr5_115732580_115732733 | 0.24 |
Bicdl1 |
BICD family like cargo adaptor 1 |
1035 |
0.52 |
| chr9_94155767_94155925 | 0.24 |
Gm5369 |
predicted gene 5369 |
16384 |
0.29 |
| chr3_142709083_142709286 | 0.24 |
Kyat3 |
kynurenine aminotransferase 3 |
8090 |
0.14 |
| chr7_19665320_19665478 | 0.23 |
Clptm1 |
cleft lip and palate associated transmembrane protein 1 |
366 |
0.7 |
| chr10_79908555_79908720 | 0.23 |
Med16 |
mediator complex subunit 16 |
181 |
0.71 |
| chr15_78340778_78340947 | 0.22 |
Mir7676-2 |
microRNA 7676-2 |
7737 |
0.12 |
| chr7_16874839_16875265 | 0.22 |
9330104G04Rik |
RIKEN cDNA 9330104G04 gene |
166 |
0.6 |
| chr1_35931118_35931293 | 0.22 |
Gm8022 |
predicted gene 8022 |
27714 |
0.14 |
| chr5_74704572_74704742 | 0.22 |
Gm15984 |
predicted gene 15984 |
1637 |
0.29 |
| chr9_107289789_107289967 | 0.22 |
Mapkapk3 |
mitogen-activated protein kinase-activated protein kinase 3 |
1 |
0.95 |
| chr5_114826323_114826479 | 0.22 |
Ywhaq-ps2 |
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein theta, pseudogene 2 |
2814 |
0.12 |
| chr17_31519992_31520147 | 0.21 |
Ndufv3 |
NADH:ubiquinone oxidoreductase core subunit V3 |
46 |
0.53 |
| chr3_39220041_39220211 | 0.21 |
Gm43008 |
predicted gene 43008 |
2280 |
0.42 |
| chr7_27447795_27447954 | 0.21 |
Blvrb |
biliverdin reductase B (flavin reductase (NADPH)) |
104 |
0.61 |
| chr17_84879283_84879445 | 0.21 |
Gm49982 |
predicted gene, 49982 |
23205 |
0.14 |
| chr12_21145146_21145377 | 0.20 |
Asap2 |
ArfGAP with SH3 domain, ankyrin repeat and PH domain 2 |
33307 |
0.16 |
| chr6_4086907_4087076 | 0.20 |
Bet1 |
Bet1 golgi vesicular membrane trafficking protein |
19 |
0.98 |
| chr2_38519062_38519254 | 0.20 |
Nek6 |
NIMA (never in mitosis gene a)-related expressed kinase 6 |
4532 |
0.14 |
| chr2_152707734_152707946 | 0.20 |
H13 |
histocompatibility 13 |
16001 |
0.09 |
| chr19_4300270_4300606 | 0.20 |
Grk2 |
G protein-coupled receptor kinase 2 |
4770 |
0.1 |
| chr15_67039473_67039636 | 0.20 |
Gm31342 |
predicted gene, 31342 |
504 |
0.83 |
| chr1_188858745_188858917 | 0.19 |
Ush2a |
usherin |
94913 |
0.08 |
| chr9_120582294_120582487 | 0.19 |
Gm47062 |
predicted gene, 47062 |
2255 |
0.19 |
| chr2_73892795_73892960 | 0.19 |
Atf2 |
activating transcription factor 2 |
238 |
0.93 |
| chr6_140874934_140875096 | 0.19 |
Gm30524 |
predicted gene, 30524 |
80594 |
0.09 |
| chr1_118627791_118627969 | 0.19 |
Tfcp2l1 |
transcription factor CP2-like 1 |
65 |
0.97 |
| chr10_95193833_95194034 | 0.18 |
Gm48874 |
predicted gene, 48874 |
33572 |
0.14 |
| chr6_82652519_82652678 | 0.18 |
Pole4 |
polymerase (DNA-directed), epsilon 4 (p12 subunit) |
78 |
0.97 |
| chr8_71272393_71272552 | 0.18 |
Haus8 |
4HAUS augmin-like complex, subunit 8 |
6 |
0.77 |
| chr17_35909704_35909977 | 0.18 |
Atat1 |
alpha tubulin acetyltransferase 1 |
101 |
0.88 |
| chrY_90740044_90740606 | 0.18 |
Mid1-ps1 |
midline 1, pseudogene 1 |
12732 |
0.18 |
| chr16_96127966_96128133 | 0.18 |
Hmgn1 |
high mobility group nucleosomal binding domain 1 |
320 |
0.83 |
| chr4_119223756_119224101 | 0.18 |
Gm12898 |
predicted gene 12898 |
754 |
0.42 |
| chr1_165763620_165763778 | 0.18 |
Creg1 |
cellular repressor of E1A-stimulated genes 1 |
47 |
0.95 |
| chr18_12941778_12941952 | 0.17 |
Osbpl1a |
oxysterol binding protein-like 1A |
24 |
0.98 |
| chr14_51884664_51884952 | 0.17 |
Mettl17 |
methyltransferase like 17 |
34 |
0.94 |
| chr17_55956278_55956439 | 0.17 |
Yju2 |
YJU2 splicing factor |
2741 |
0.11 |
| chr2_34897940_34898106 | 0.17 |
Phf19 |
PHD finger protein 19 |
6129 |
0.11 |
| chr3_9038418_9038576 | 0.17 |
Gm37616 |
predicted gene, 37616 |
8995 |
0.17 |
| chr4_123686928_123687079 | 0.17 |
Macf1 |
microtubule-actin crosslinking factor 1 |
2643 |
0.2 |
| chr4_43442956_43443142 | 0.17 |
Tesk1 |
testis specific protein kinase 1 |
272 |
0.82 |
| chr15_88837187_88837359 | 0.17 |
Gm23144 |
predicted gene, 23144 |
1973 |
0.24 |
| chr15_12248377_12248536 | 0.17 |
Mtmr12 |
myotubularin related protein 12 |
25102 |
0.12 |
| chr5_113219993_113220303 | 0.17 |
2900026A02Rik |
RIKEN cDNA 2900026A02 gene |
1088 |
0.42 |
| chr1_136008248_136008409 | 0.17 |
Tmem9 |
transmembrane protein 9 |
178 |
0.92 |
| chr1_42474261_42474413 | 0.17 |
Gm37047 |
predicted gene, 37047 |
17476 |
0.25 |
| chr9_44327277_44327612 | 0.16 |
Dpagt1 |
dolichyl-phosphate (UDP-N-acetylglucosamine) acetylglucosaminephosphotransferase 1 (GlcNAc-1-P transferase) |
539 |
0.46 |
| chr15_67110643_67110805 | 0.16 |
St3gal1 |
ST3 beta-galactoside alpha-2,3-sialyltransferase 1 |
3268 |
0.32 |
| chr4_134796652_134796803 | 0.16 |
Maco1 |
macoilin 1 |
14486 |
0.18 |
| chr2_163308491_163308647 | 0.16 |
Tox2 |
TOX high mobility group box family member 2 |
11809 |
0.2 |
| chr8_84707765_84707923 | 0.16 |
Nfix |
nuclear factor I/X |
128 |
0.92 |
| chr4_43040334_43040495 | 0.16 |
Fam214b |
family with sequence similarity 214, member B |
119 |
0.93 |
| chr7_45714696_45714867 | 0.16 |
Sphk2 |
sphingosine kinase 2 |
426 |
0.47 |
| chr16_30512503_30512806 | 0.16 |
Tmem44 |
transmembrane protein 44 |
28255 |
0.16 |
| chr12_100958810_100958969 | 0.15 |
Ccdc88c |
coiled-coil domain containing 88C |
13716 |
0.12 |
| chr19_5118250_5118577 | 0.15 |
Klc2 |
kinesin light chain 2 |
113 |
0.76 |
| chr11_59787617_59787784 | 0.15 |
Pld6 |
phospholipase D family, member 6 |
55 |
0.95 |
| chr1_184732600_184732772 | 0.15 |
Hlx |
H2.0-like homeobox |
67 |
0.96 |
| chr12_52314917_52315090 | 0.15 |
Gm47431 |
predicted gene, 47431 |
133122 |
0.04 |
| chr14_47189631_47189815 | 0.15 |
Gch1 |
GTP cyclohydrolase 1 |
310 |
0.8 |
| chr15_62203372_62203523 | 0.15 |
Pvt1 |
Pvt1 oncogene |
14776 |
0.23 |
| chrX_150813018_150813184 | 0.14 |
Maged2 |
melanoma antigen, family D, 2 |
23 |
0.97 |
| chr9_21265122_21265283 | 0.14 |
Atg4d |
autophagy related 4D, cysteine peptidase |
91 |
0.93 |
| chr10_24830554_24830712 | 0.14 |
Enpp3 |
ectonucleotide pyrophosphatase/phosphodiesterase 3 |
1173 |
0.49 |
| chr13_49216001_49216172 | 0.14 |
Card19 |
caspase recruitment domain family, member 19 |
49 |
0.97 |
| chr7_19811188_19811394 | 0.14 |
Bcl3 |
B cell leukemia/lymphoma 3 |
598 |
0.46 |
| chr11_119238465_119238619 | 0.14 |
Ccdc40 |
coiled-coil domain containing 40 |
176 |
0.92 |
| chr2_167538011_167538171 | 0.14 |
Snai1 |
snail family zinc finger 1 |
104 |
0.94 |
| chr17_47438043_47438256 | 0.14 |
1700001C19Rik |
RIKEN cDNA 1700001C19 gene |
773 |
0.44 |
| chr17_87282754_87282914 | 0.14 |
4833418N02Rik |
RIKEN cDNA 4833418N02 gene |
20 |
0.52 |
| chr5_138280001_138280359 | 0.14 |
Stag3 |
stromal antigen 3 |
60 |
0.52 |
| chr7_25619112_25619443 | 0.14 |
Dmac2 |
distal membrane arm assembly complex 2 |
137 |
0.9 |
| chr4_156197972_156198140 | 0.14 |
Agrn |
agrin |
568 |
0.56 |
| chr10_61492177_61492335 | 0.14 |
Gm47595 |
predicted gene, 47595 |
4034 |
0.12 |
| chr8_72239914_72240076 | 0.13 |
Ap1m1 |
adaptor-related protein complex AP-1, mu subunit 1 |
23 |
0.95 |
| chr9_119100530_119100792 | 0.13 |
Dlec1 |
deleted in lung and esophageal cancer 1 |
1817 |
0.25 |
| chr11_5445001_5445157 | 0.13 |
Znrf3 |
zinc and ring finger 3 |
232 |
0.93 |
| chr8_125199868_125200019 | 0.13 |
Gm16237 |
predicted gene 16237 |
27380 |
0.22 |
| chr7_16313428_16313735 | 0.13 |
Bbc3 |
BCL2 binding component 3 |
64 |
0.96 |
| chr11_67025097_67025255 | 0.13 |
Tmem220 |
transmembrane protein 220 |
22 |
0.97 |
| chr14_30916422_30916581 | 0.13 |
Itih3 |
inter-alpha trypsin inhibitor, heavy chain 3 |
1114 |
0.35 |
| chr10_40257637_40257797 | 0.13 |
Gtf3c6 |
general transcription factor IIIC, polypeptide 6, alpha |
41 |
0.92 |
| chr12_4874314_4874496 | 0.12 |
Mfsd2b |
major facilitator superfamily domain containing 2B |
46 |
0.96 |
| chr4_152108046_152108214 | 0.12 |
Plekhg5 |
pleckstrin homology domain containing, family G (with RhoGef domain) member 5 |
401 |
0.76 |
| chr2_172826555_172826706 | 0.12 |
Gm22773 |
predicted gene, 22773 |
37711 |
0.18 |
| chr8_57488604_57488764 | 0.12 |
2500002B13Rik |
RIKEN cDNA 2500002B13 gene |
630 |
0.45 |
| chr8_94876193_94876353 | 0.12 |
Dok4 |
docking protein 4 |
57 |
0.95 |
| chr11_75247267_75247439 | 0.12 |
Rtn4rl1 |
reticulon 4 receptor-like 1 |
53570 |
0.07 |
| chr19_29325385_29325550 | 0.12 |
Insl6 |
insulin-like 6 |
111 |
0.95 |
| chr7_19228690_19228852 | 0.12 |
Opa3 |
optic atrophy 3 |
388 |
0.68 |
| chr7_19118353_19118670 | 0.12 |
Gm4969 |
predicted gene 4969 |
143 |
0.87 |
| chr11_23254748_23254899 | 0.12 |
Xpo1 |
exportin 1 |
1218 |
0.51 |
| chr10_76531437_76531608 | 0.12 |
Lss |
lanosterol synthase |
66 |
0.96 |
| chr8_94183220_94183416 | 0.12 |
Gm39228 |
predicted gene, 39228 |
29 |
0.95 |
| chr12_8921530_8921697 | 0.12 |
Laptm4a |
lysosomal-associated protein transmembrane 4A |
51 |
0.97 |
| chr7_44858030_44858203 | 0.12 |
Pnkp |
polynucleotide kinase 3'- phosphatase |
380 |
0.64 |
| chr9_44515546_44515772 | 0.11 |
Cxcr5 |
chemokine (C-X-C motif) receptor 5 |
10762 |
0.07 |
| chr15_9072830_9073005 | 0.11 |
Nadk2 |
NAD kinase 2, mitochondrial |
1230 |
0.52 |
| chr3_145556330_145556492 | 0.11 |
Col24a1 |
collagen, type XXIV, alpha 1 |
11438 |
0.2 |
| chr8_36377048_36377257 | 0.11 |
Gm5787 |
predicted gene 5787 |
3519 |
0.25 |
| chr17_65951168_65951335 | 0.11 |
Twsg1 |
twisted gastrulation BMP signaling modulator 1 |
25 |
0.97 |
| chr13_94177289_94177442 | 0.11 |
Lhfpl2 |
lipoma HMGIC fusion partner-like 2 |
3373 |
0.25 |
| chr2_32751402_32751562 | 0.11 |
Tor2a |
torsin family 2, member A |
5752 |
0.08 |
| chr11_75047480_75047670 | 0.11 |
Gm12333 |
predicted gene 12333 |
8227 |
0.13 |
| chr11_113684528_113684688 | 0.11 |
D11Wsu47e |
DNA segment, Chr 11, Wayne State University 47, expressed |
161 |
0.77 |
| chr13_13954579_13954776 | 0.11 |
B3galnt2 |
UDP-GalNAc:betaGlcNAc beta 1,3-galactosaminyltransferase, polypeptide 2 |
3 |
0.96 |
| chr8_64946983_64947186 | 0.11 |
Tmem192 |
transmembrane protein 192 |
101 |
0.88 |
| chr2_35336999_35337164 | 0.11 |
Stom |
stomatin |
105 |
0.95 |
| chr14_46832037_46832198 | 0.11 |
Cgrrf1 |
cell growth regulator with ring finger domain 1 |
8 |
0.56 |
| chr17_56627213_56627375 | 0.11 |
Lonp1 |
lon peptidase 1, mitochondrial |
407 |
0.62 |
| chr15_10675955_10676106 | 0.11 |
Rai14 |
retinoic acid induced 14 |
37510 |
0.14 |
| chr9_18473377_18473557 | 0.11 |
Zfp558 |
zinc finger protein 558 |
45 |
0.96 |
| chr8_95293260_95293422 | 0.11 |
Cngb1 |
cyclic nucleotide gated channel beta 1 |
328 |
0.85 |
| chr9_21918187_21918359 | 0.11 |
Rab3d |
RAB3D, member RAS oncogene family |
81 |
0.93 |
| chrX_73967268_73967441 | 0.11 |
Hcfc1 |
host cell factor C1 |
997 |
0.39 |
| chr10_21420912_21421063 | 0.11 |
1700021A07Rik |
RIKEN cDNA 1700021A07 gene |
1510 |
0.3 |
| chr10_59951771_59951950 | 0.10 |
Ddit4 |
DNA-damage-inducible transcript 4 |
26 |
0.98 |
| chr14_122105762_122105950 | 0.10 |
A330035P11Rik |
RIKEN cDNA A330035P11 gene |
1097 |
0.31 |
| chr4_155469018_155469192 | 0.10 |
Tmem52 |
transmembrane protein 52 |
9 |
0.96 |
| chr14_63164949_63165115 | 0.10 |
Fdft1 |
farnesyl diphosphate farnesyl transferase 1 |
107 |
0.96 |
| chr16_22899718_22899927 | 0.10 |
Ahsg |
alpha-2-HS-glycoprotein |
4958 |
0.13 |
| chr10_80577406_80577585 | 0.10 |
Klf16 |
Kruppel-like factor 16 |
174 |
0.87 |
| chr11_51856076_51856454 | 0.10 |
Jade2 |
jade family PHD finger 2 |
860 |
0.59 |
| chr3_90509502_90509661 | 0.10 |
Chtop |
chromatin target of PRMT1 |
83 |
0.92 |
| chr13_95894500_95894710 | 0.10 |
Iqgap2 |
IQ motif containing GTPase activating protein 2 |
2122 |
0.3 |
| chr15_79774430_79774597 | 0.10 |
Dnal4 |
dynein, axonemal, light chain 4 |
46 |
0.95 |
| chr16_57167289_57167444 | 0.10 |
Nit2 |
nitrilase family, member 2 |
25 |
0.5 |
| chr1_74721665_74721828 | 0.10 |
Cyp27a1 |
cytochrome P450, family 27, subfamily a, polypeptide 1 |
8172 |
0.13 |
| chr7_115948051_115948232 | 0.10 |
Sox6 |
SRY (sex determining region Y)-box 6 |
46945 |
0.13 |
| chr17_56079134_56079310 | 0.09 |
Hdgfl2 |
HDGF like 2 |
412 |
0.69 |
| chr4_132329477_132329639 | 0.09 |
Trnau1ap |
tRNA selenocysteine 1 associated protein 1 |
20 |
0.92 |
| chr8_83608021_83608180 | 0.09 |
Dnajb1 |
DnaJ heat shock protein family (Hsp40) member B1 |
93 |
0.54 |
| chr9_57295766_57295926 | 0.09 |
1700017B05Rik |
RIKEN cDNA 1700017B05 gene |
33234 |
0.11 |
| chr12_21286198_21286361 | 0.09 |
Itgb1bp1 |
integrin beta 1 binding protein 1 |
5 |
0.55 |
| chr1_105990177_105990357 | 0.09 |
Gm7160 |
predicted gene 7160 |
15 |
0.64 |
| chr4_104975117_104975442 | 0.09 |
Gm12721 |
predicted gene 12721 |
5150 |
0.27 |
| chr11_59191846_59192317 | 0.09 |
Guk1 |
guanylate kinase 1 |
129 |
0.91 |
| chr2_11535513_11535687 | 0.09 |
Gm37975 |
predicted gene, 37975 |
906 |
0.49 |
| chr10_119614856_119615049 | 0.09 |
Gm47027 |
predicted gene, 47027 |
2768 |
0.28 |
| chr10_81320325_81320502 | 0.09 |
Cactin |
cactin, spliceosome C complex subunit |
690 |
0.37 |
| chr7_29170009_29170256 | 0.09 |
Ggn |
gametogenetin |
78 |
0.73 |
| chr3_108467871_108468033 | 0.09 |
5330417C22Rik |
RIKEN cDNA 5330417C22 gene |
2129 |
0.16 |
| chr5_144358410_144358608 | 0.09 |
Dmrt1i |
Dmrt1 interacting ncRNA |
16 |
0.87 |
| chr8_125133982_125134146 | 0.09 |
Disc1 |
disrupted in schizophrenia 1 |
46077 |
0.14 |
| chr8_126516634_126516823 | 0.09 |
Gm6091 |
predicted pseudogene 6091 |
40316 |
0.14 |
| chr11_16803990_16804532 | 0.09 |
Egfros |
epidermal growth factor receptor, opposite strand |
26441 |
0.18 |
| chr6_90395849_90396012 | 0.09 |
C030015A19Rik |
RIKEN cDNA C030015A19 gene |
9123 |
0.12 |
| chr12_86825129_86825313 | 0.09 |
Gm10095 |
predicted gene 10095 |
21246 |
0.17 |
| chr17_25474786_25474993 | 0.09 |
Tekt4 |
tektin 4 |
585 |
0.6 |
| chr7_30626129_30626287 | 0.09 |
Cox6b1 |
cytochrome c oxidase, subunit 6B1 |
57 |
0.91 |
| chr17_84875170_84875325 | 0.09 |
Gm9400 |
predicted gene 9400 |
22709 |
0.14 |
| chr17_80004938_80005095 | 0.09 |
Gm41625 |
predicted gene, 41625 |
23354 |
0.13 |
| chr1_195092267_195092425 | 0.09 |
Cd46 |
CD46 antigen, complement regulatory protein |
97 |
0.95 |
| chr7_99828286_99828448 | 0.09 |
Neu3 |
neuraminidase 3 |
50 |
0.96 |
| chr4_133085038_133085232 | 0.09 |
Ahdc1 |
AT hook, DNA binding motif, containing 1 |
18886 |
0.17 |
| chr14_31641238_31641408 | 0.09 |
Hacl1 |
2-hydroxyacyl-CoA lyase 1 |
37 |
0.72 |
| chr9_26923072_26923237 | 0.08 |
Gm1110 |
predicted gene 1110 |
43 |
0.97 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 0.0 | 0.1 | GO:0000720 | pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) |
| 0.0 | 0.1 | GO:0042167 | heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.0 | 0.1 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.0 | 0.1 | GO:1900739 | regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900739) positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900740) |
| 0.0 | 0.2 | GO:0009256 | 10-formyltetrahydrofolate metabolic process(GO:0009256) |
| 0.0 | 0.2 | GO:0044351 | macropinocytosis(GO:0044351) |
| 0.0 | 0.2 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.0 | 0.1 | GO:0051697 | protein delipidation(GO:0051697) |
| 0.0 | 0.1 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.0 | 0.2 | GO:1900227 | positive regulation of NLRP3 inflammasome complex assembly(GO:1900227) |
| 0.0 | 0.1 | GO:0046654 | tetrahydrofolate biosynthetic process(GO:0046654) |
| 0.0 | 0.3 | GO:0030836 | positive regulation of actin filament depolymerization(GO:0030836) |
| 0.0 | 0.1 | GO:0034436 | glycoprotein transport(GO:0034436) |
| 0.0 | 0.1 | GO:0007354 | zygotic determination of anterior/posterior axis, embryo(GO:0007354) |
| 0.0 | 0.1 | GO:0043382 | positive regulation of memory T cell differentiation(GO:0043382) |
| 0.0 | 0.1 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.0 | 0.2 | GO:0032049 | cardiolipin biosynthetic process(GO:0032049) |
| 0.0 | 0.1 | GO:0072061 | inner medullary collecting duct development(GO:0072061) |
| 0.0 | 0.1 | GO:1903121 | regulation of TRAIL-activated apoptotic signaling pathway(GO:1903121) |
| 0.0 | 0.1 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.0 | 0.2 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 | 0.1 | GO:0036265 | RNA (guanine-N7)-methylation(GO:0036265) |
| 0.0 | 0.2 | GO:0032906 | transforming growth factor beta2 production(GO:0032906) regulation of transforming growth factor beta2 production(GO:0032909) |
| 0.0 | 0.1 | GO:0042091 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 0.0 | 0.1 | GO:0002071 | glandular epithelial cell maturation(GO:0002071) |
| 0.0 | 0.3 | GO:0002082 | regulation of oxidative phosphorylation(GO:0002082) |
| 0.0 | 0.1 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 0.1 | GO:0018199 | peptidyl-glutamine modification(GO:0018199) |
| 0.0 | 0.3 | GO:0006878 | cellular copper ion homeostasis(GO:0006878) |
| 0.0 | 0.0 | GO:0042726 | flavin-containing compound metabolic process(GO:0042726) |
| 0.0 | 0.2 | GO:0035646 | endosome to melanosome transport(GO:0035646) endosome to pigment granule transport(GO:0043485) pigment granule maturation(GO:0048757) |
| 0.0 | 0.1 | GO:0021684 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.0 | 0.1 | GO:0007066 | female meiosis sister chromatid cohesion(GO:0007066) |
| 0.0 | 0.1 | GO:1903273 | regulation of sodium ion export(GO:1903273) regulation of sodium ion export from cell(GO:1903276) |
| 0.0 | 0.1 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.0 | 0.1 | GO:2000017 | positive regulation of determination of dorsal identity(GO:2000017) |
| 0.0 | 0.0 | GO:2000409 | positive regulation of T cell extravasation(GO:2000409) |
| 0.0 | 0.2 | GO:0002070 | epithelial cell maturation(GO:0002070) |
| 0.0 | 0.1 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.0 | 0.1 | GO:0006933 | negative regulation of cell adhesion involved in substrate-bound cell migration(GO:0006933) |
| 0.0 | 0.0 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.0 | 0.1 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 0.1 | GO:0006451 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.0 | 0.0 | GO:0019042 | viral latency(GO:0019042) |
| 0.0 | 0.1 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.0 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.0 | 0.1 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.0 | 0.0 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.0 | 0.1 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.0 | 0.0 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.0 | 0.1 | GO:0044829 | positive regulation by host of viral genome replication(GO:0044829) |
| 0.0 | 0.2 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.0 | GO:0006848 | pyruvate transport(GO:0006848) pyruvate transmembrane transport(GO:1901475) |
| 0.0 | 0.0 | GO:1900194 | negative regulation of oocyte maturation(GO:1900194) |
| 0.0 | 0.1 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.0 | 0.1 | GO:0045820 | negative regulation of glycolytic process(GO:0045820) |
| 0.0 | 0.1 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.0 | 0.0 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.1 | GO:0002326 | B cell lineage commitment(GO:0002326) |
| 0.0 | 0.1 | GO:0072015 | glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.0 | 0.4 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.0 | 0.1 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 | 0.1 | GO:0044539 | long-chain fatty acid import(GO:0044539) |
| 0.0 | 0.1 | GO:0009437 | carnitine metabolic process(GO:0009437) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0000836 | ER ubiquitin ligase complex(GO:0000835) Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.1 | 0.2 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.0 | 0.2 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.0 | 0.1 | GO:1990393 | 3M complex(GO:1990393) |
| 0.0 | 0.1 | GO:0000802 | transverse filament(GO:0000802) |
| 0.0 | 0.1 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 0.1 | GO:0002079 | inner acrosomal membrane(GO:0002079) |
| 0.0 | 0.1 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 0.1 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.2 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 0.1 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.0 | 0.0 | GO:0033647 | host intracellular organelle(GO:0033647) host intracellular membrane-bounded organelle(GO:0033648) |
| 0.0 | 0.0 | GO:0097543 | ciliary inversin compartment(GO:0097543) |
| 0.0 | 0.0 | GO:0071008 | U2-type post-mRNA release spliceosomal complex(GO:0071008) |
| 0.0 | 0.0 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 0.0 | 0.1 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 0.0 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0019145 | aminobutyraldehyde dehydrogenase activity(GO:0019145) 4-trimethylammoniobutyraldehyde dehydrogenase activity(GO:0047105) |
| 0.1 | 0.2 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.0 | 0.1 | GO:0008808 | cardiolipin synthase activity(GO:0008808) phosphatidyltransferase activity(GO:0030572) |
| 0.0 | 0.2 | GO:0030375 | thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 0.3 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.1 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.0 | 0.1 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 0.1 | GO:0003933 | GTP cyclohydrolase activity(GO:0003933) |
| 0.0 | 0.1 | GO:0046404 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) |
| 0.0 | 0.2 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.1 | GO:0052796 | exo-alpha-sialidase activity(GO:0004308) alpha-sialidase activity(GO:0016997) exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.0 | 0.2 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.1 | GO:0030620 | U2 snRNA binding(GO:0030620) |
| 0.0 | 0.1 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.0 | 0.1 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.0 | 0.1 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.1 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.0 | 0.3 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.0 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.0 | 0.1 | GO:0051880 | G-quadruplex DNA binding(GO:0051880) |
| 0.0 | 0.0 | GO:0030621 | U4 snRNA binding(GO:0030621) |
| 0.0 | 0.1 | GO:0005223 | intracellular cGMP activated cation channel activity(GO:0005223) |
| 0.0 | 0.2 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.0 | 0.1 | GO:0031559 | oxidosqualene cyclase activity(GO:0031559) |
| 0.0 | 0.1 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 0.0 | 0.0 | GO:0016532 | superoxide dismutase copper chaperone activity(GO:0016532) |
| 0.0 | 0.1 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.0 | 0.0 | GO:0089720 | death effector domain binding(GO:0035877) caspase binding(GO:0089720) |
| 0.0 | 0.0 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.0 | 0.1 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.0 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.0 | 0.1 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.0 | 0.0 | GO:0016018 | cyclosporin A binding(GO:0016018) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 0.3 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.2 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.0 | 0.3 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.1 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.0 | REACTOME FRS2 MEDIATED CASCADE | Genes involved in FRS2-mediated cascade |
| 0.0 | 0.1 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.2 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.1 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |