Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
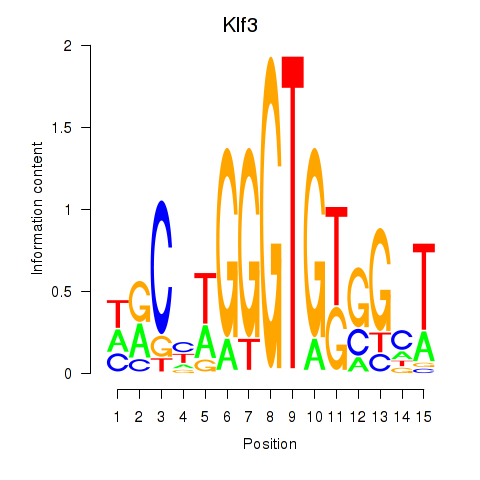
Results for Klf3
Z-value: 1.32
Transcription factors associated with Klf3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Klf3
|
ENSMUSG00000029178.8 | Kruppel-like factor 3 (basic) |
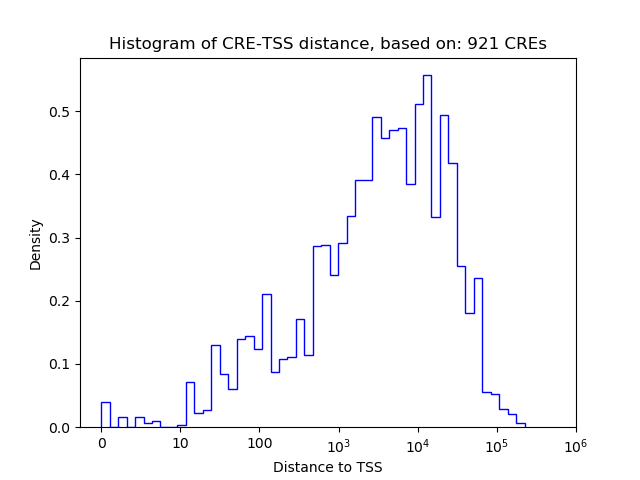
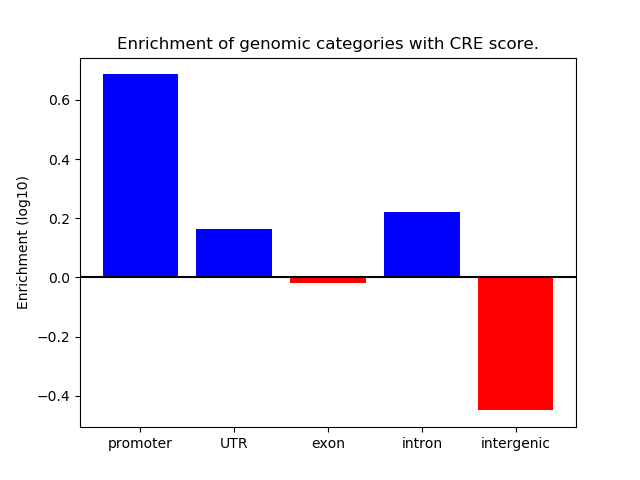
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr5_64811771_64811965 | Klf3 | 471 | 0.766893 | -0.98 | 4.1e-04 | Click! |
| chr5_64840100_64840327 | Klf3 | 13404 | 0.151270 | 0.97 | 1.2e-03 | Click! |
| chr5_64795807_64795970 | Klf3 | 7500 | 0.153458 | 0.86 | 2.7e-02 | Click! |
| chr5_64812184_64812344 | Klf3 | 75 | 0.964728 | 0.86 | 3.0e-02 | Click! |
| chr5_64809339_64809781 | Klf3 | 2779 | 0.206898 | 0.83 | 4.2e-02 | Click! |
Activity of the Klf3 motif across conditions
Conditions sorted by the z-value of the Klf3 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
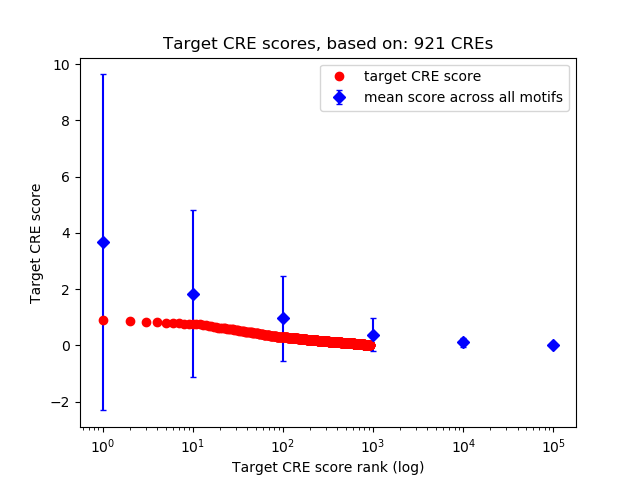
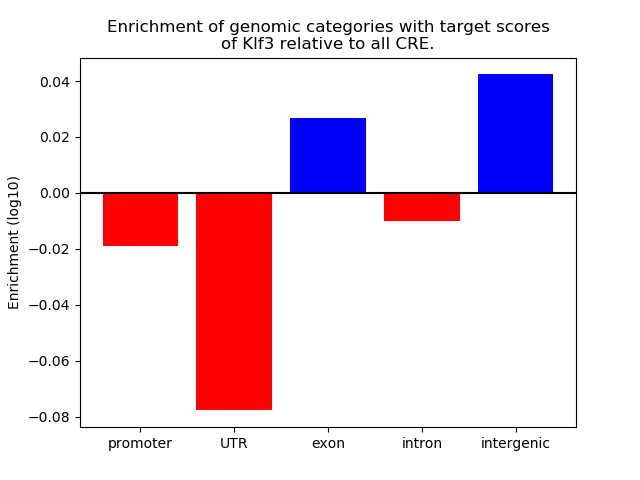
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr12_8778168_8778353 | 0.90 |
Sdc1 |
syndecan 1 |
6457 |
0.19 |
| chr13_100577098_100577269 | 0.87 |
Gm8847 |
predicted gene 8847 |
17151 |
0.1 |
| chr8_104101509_104101690 | 0.82 |
Cdh5 |
cadherin 5 |
26 |
0.97 |
| chr8_121591389_121591573 | 0.82 |
Map1lc3b |
microtubule-associated protein 1 light chain 3 beta |
769 |
0.5 |
| chr13_9431614_9431799 | 0.81 |
Gm48871 |
predicted gene, 48871 |
14194 |
0.17 |
| chr18_56437222_56437386 | 0.79 |
Gramd3 |
GRAM domain containing 3 |
5172 |
0.22 |
| chr12_69299371_69299530 | 0.78 |
Klhdc2 |
kelch domain containing 2 |
2695 |
0.15 |
| chr6_48595408_48595586 | 0.77 |
Repin1 |
replication initiator 1 |
1178 |
0.2 |
| chr15_31344240_31344565 | 0.77 |
Ankrd33b |
ankyrin repeat domain 33B |
19153 |
0.14 |
| chr10_82623685_82624011 | 0.76 |
1190007I07Rik |
RIKEN cDNA 1190007I07 gene |
620 |
0.65 |
| chr10_68355501_68355861 | 0.75 |
4930545H06Rik |
RIKEN cDNA 4930545H06 gene |
34539 |
0.17 |
| chr11_98770256_98770457 | 0.75 |
Nr1d1 |
nuclear receptor subfamily 1, group D, member 1 |
70 |
0.94 |
| chr16_20674169_20674320 | 0.73 |
Eif4g1 |
eukaryotic translation initiation factor 4, gamma 1 |
133 |
0.89 |
| chr3_105893960_105894139 | 0.72 |
Adora3 |
adenosine A3 receptor |
10372 |
0.12 |
| chr12_21227287_21227746 | 0.70 |
AC156032.1 |
|
19807 |
0.14 |
| chr1_165617606_165617906 | 0.67 |
Mpzl1 |
myelin protein zero-like 1 |
3777 |
0.14 |
| chr12_12320785_12320940 | 0.66 |
Fam49a |
family with sequence similarity 49, member A |
58673 |
0.14 |
| chr12_73300604_73300762 | 0.66 |
Slc38a6 |
solute carrier family 38, member 6 |
13623 |
0.17 |
| chr1_133296542_133296715 | 0.63 |
Plekha6 |
pleckstrin homology domain containing, family A member 6 |
1323 |
0.33 |
| chr17_78249106_78249271 | 0.62 |
Crim1 |
cysteine rich transmembrane BMP regulator 1 (chordin like) |
11416 |
0.18 |
| chr3_22076000_22076172 | 0.60 |
Gm43672 |
predicted gene 43672 |
56 |
0.91 |
| chr10_80647071_80647222 | 0.60 |
Btbd2 |
BTB (POZ) domain containing 2 |
564 |
0.54 |
| chr9_44398704_44398895 | 0.60 |
Slc37a4 |
solute carrier family 37 (glucose-6-phosphate transporter), member 4 |
325 |
0.63 |
| chr3_81102286_81102462 | 0.59 |
Gm16000 |
predicted gene 16000 |
61937 |
0.11 |
| chr4_128804978_128805273 | 0.57 |
Zfp362 |
zinc finger protein 362 |
920 |
0.55 |
| chr1_180279676_180280012 | 0.57 |
Psen2 |
presenilin 2 |
16406 |
0.13 |
| chr4_140787577_140787739 | 0.57 |
Padi4 |
peptidyl arginine deiminase, type IV |
13422 |
0.12 |
| chr9_44551273_44551431 | 0.57 |
Cxcr5 |
chemokine (C-X-C motif) receptor 5 |
4934 |
0.09 |
| chr11_69073830_69074123 | 0.55 |
Gm12306 |
predicted gene 12306 |
514 |
0.33 |
| chr10_51455905_51456056 | 0.55 |
Gm46189 |
predicted gene, 46189 |
0 |
0.97 |
| chr13_111957222_111957419 | 0.54 |
Gm15322 |
predicted gene 15322 |
34146 |
0.14 |
| chr1_164788357_164788561 | 0.54 |
Dpt |
dermatopontin |
8185 |
0.18 |
| chr6_3346058_3346227 | 0.52 |
Gm20559 |
predicted gene, 20559 |
14 |
0.96 |
| chr17_36019112_36019264 | 0.52 |
H2-T24 |
histocompatibility 2, T region locus 24 |
1337 |
0.17 |
| chr3_121276958_121277317 | 0.51 |
Gm30517 |
predicted gene, 30517 |
387 |
0.81 |
| chr6_3399444_3399625 | 0.51 |
Samd9l |
sterile alpha motif domain containing 9-like |
38 |
0.97 |
| chr3_89887959_89888297 | 0.49 |
Gm42809 |
predicted gene 42809 |
24788 |
0.1 |
| chr4_35098657_35098808 | 0.49 |
Ifnk |
interferon kappa |
53324 |
0.12 |
| chr8_116993108_116993270 | 0.47 |
Gcsh |
glycine cleavage system protein H (aminomethyl carrier) |
348 |
0.84 |
| chr13_3260533_3260700 | 0.47 |
Gm40653 |
predicted gene, 40653 |
12619 |
0.17 |
| chr1_134406120_134406612 | 0.47 |
Cyb5r1 |
cytochrome b5 reductase 1 |
53 |
0.95 |
| chr7_46839028_46839384 | 0.47 |
Ldha |
lactate dehydrogenase A |
2269 |
0.15 |
| chr5_139382284_139382462 | 0.47 |
Gpr146 |
G protein-coupled receptor 146 |
1792 |
0.22 |
| chr19_23150997_23151148 | 0.46 |
Mir1192 |
microRNA 1192 |
1641 |
0.33 |
| chr5_105698740_105698909 | 0.46 |
Lrrc8d |
leucine rich repeat containing 8D |
1145 |
0.57 |
| chr11_78550155_78550328 | 0.45 |
Tmem97 |
transmembrane protein 97 |
536 |
0.54 |
| chr1_66895610_66896410 | 0.45 |
Gm25832 |
predicted gene, 25832 |
29213 |
0.09 |
| chr7_35119039_35119269 | 0.44 |
Cebpa |
CCAAT/enhancer binding protein (C/EBP), alpha |
139 |
0.78 |
| chr4_139978986_139979192 | 0.43 |
Klhdc7a |
kelch domain containing 7A |
11063 |
0.16 |
| chr17_24443544_24443715 | 0.43 |
Dnase1l2 |
deoxyribonuclease 1-like 2 |
524 |
0.46 |
| chr7_44819003_44819183 | 0.43 |
Nup62 |
nucleoporin 62 |
656 |
0.42 |
| chr10_116297940_116298100 | 0.42 |
Ptprb |
protein tyrosine phosphatase, receptor type, B |
3352 |
0.23 |
| chr4_155162532_155162683 | 0.42 |
Ski |
ski sarcoma viral oncogene homolog (avian) |
25689 |
0.14 |
| chr13_111861024_111861186 | 0.41 |
Gm15324 |
predicted gene 15324 |
4109 |
0.15 |
| chr16_94546158_94546496 | 0.41 |
Vps26c |
VPS26 endosomal protein sorting factor C |
19497 |
0.16 |
| chr6_124694855_124695031 | 0.40 |
Lpcat3 |
lysophosphatidylcholine acyltransferase 3 |
6476 |
0.07 |
| chr4_98508428_98508827 | 0.40 |
Patj |
PATJ, crumbs cell polarity complex component |
10344 |
0.22 |
| chr9_43231970_43232132 | 0.40 |
Oaf |
out at first homolog |
6959 |
0.15 |
| chr2_29426777_29426974 | 0.40 |
Gm13402 |
predicted gene 13402 |
30416 |
0.13 |
| chr1_88765371_88765522 | 0.39 |
Platr5 |
pluripotency associated transcript 5 |
10492 |
0.2 |
| chr15_75892571_75892918 | 0.38 |
Naprt |
nicotinate phosphoribosyltransferase |
176 |
0.87 |
| chr17_34722831_34722982 | 0.38 |
Cyp21a2-ps |
cytochrome P450, family 21, subfamily a, polypeptide 2 pseudogene |
654 |
0.44 |
| chr7_105770059_105770232 | 0.38 |
Gm15645 |
predicted gene 15645 |
7824 |
0.09 |
| chr3_32495485_32495636 | 0.38 |
Kcnmb3 |
potassium large conductance calcium-activated channel, subfamily M, beta member 3 |
3329 |
0.17 |
| chr7_144014599_144014780 | 0.38 |
Mir3105 |
microRNA 3105 |
5435 |
0.21 |
| chr18_60619666_60619817 | 0.37 |
Synpo |
synaptopodin |
3584 |
0.21 |
| chr3_9947781_9947953 | 0.37 |
Gm17877 |
predicted gene, 17877 |
24134 |
0.16 |
| chr3_106545726_106545888 | 0.36 |
Cept1 |
choline/ethanolaminephosphotransferase 1 |
1775 |
0.22 |
| chr9_106452941_106453115 | 0.35 |
Gm28959 |
predicted gene 28959 |
110 |
0.68 |
| chr11_20396557_20396740 | 0.35 |
Gm12034 |
predicted gene 12034 |
49764 |
0.12 |
| chr2_84650666_84651104 | 0.35 |
Ctnnd1 |
catenin (cadherin associated protein), delta 1 |
120 |
0.92 |
| chr10_93488942_93489116 | 0.35 |
Hal |
histidine ammonia lyase |
226 |
0.91 |
| chr13_34742265_34742434 | 0.34 |
Fam50b |
family with sequence similarity 50, member B |
2737 |
0.19 |
| chr7_45132163_45132314 | 0.33 |
Flt3l |
FMS-like tyrosine kinase 3 ligand |
369 |
0.53 |
| chr12_100527851_100528010 | 0.33 |
Ttc7b |
tetratricopeptide repeat domain 7B |
7104 |
0.18 |
| chr5_142440544_142440746 | 0.33 |
Ap5z1 |
adaptor-related protein complex 5, zeta 1 subunit |
23299 |
0.17 |
| chr15_67102477_67102648 | 0.33 |
St3gal1 |
ST3 beta-galactoside alpha-2,3-sialyltransferase 1 |
11430 |
0.24 |
| chr15_75270297_75270448 | 0.33 |
Ly6f |
lymphocyte antigen 6 complex, locus F |
1670 |
0.23 |
| chr7_141096744_141096936 | 0.32 |
Sigirr |
single immunoglobulin and toll-interleukin 1 receptor (TIR) domain |
101 |
0.92 |
| chr13_112751580_112751773 | 0.32 |
Gm47850 |
predicted gene, 47850 |
13172 |
0.14 |
| chr5_124007295_124007675 | 0.32 |
Gm37939 |
predicted gene, 37939 |
2837 |
0.13 |
| chr5_64806228_64806550 | 0.32 |
Gm20033 |
predicted gene, 20033 |
2272 |
0.21 |
| chr1_181228977_181229128 | 0.32 |
A430110L20Rik |
RIKEN cDNA A430110L20 gene |
2976 |
0.19 |
| chr7_80232137_80232317 | 0.32 |
Cib1 |
calcium and integrin binding 1 (calmyrin) |
268 |
0.69 |
| chr12_21136203_21136399 | 0.31 |
Asap2 |
ArfGAP with SH3 domain, ankyrin repeat and PH domain 2 |
24347 |
0.18 |
| chr18_64265864_64266047 | 0.31 |
St8sia3 |
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 3 |
31 |
0.97 |
| chr15_102177432_102177780 | 0.31 |
Csad |
cysteine sulfinic acid decarboxylase |
1413 |
0.26 |
| chr8_84984897_84985063 | 0.31 |
Hook2 |
hook microtubule tethering protein 2 |
5623 |
0.06 |
| chr11_120005928_120006087 | 0.31 |
Aatk |
apoptosis-associated tyrosine kinase |
6401 |
0.11 |
| chr15_100674641_100674912 | 0.31 |
Cela1 |
chymotrypsin-like elastase family, member 1 |
4086 |
0.11 |
| chr10_80033033_80033205 | 0.31 |
Polr2e |
polymerase (RNA) II (DNA directed) polypeptide E |
4066 |
0.1 |
| chr10_61328072_61328223 | 0.30 |
Pald1 |
phosphatase domain containing, paladin 1 |
14180 |
0.12 |
| chr8_106138653_106138804 | 0.30 |
1810019D21Rik |
RIKEN cDNA 1810019D21 gene |
1663 |
0.21 |
| chr16_80273340_80273491 | 0.30 |
Gm23083 |
predicted gene, 23083 |
136063 |
0.05 |
| chr8_104375548_104375738 | 0.30 |
Gm45877 |
predicted gene 45877 |
11975 |
0.09 |
| chr13_41253417_41253699 | 0.30 |
Smim13 |
small integral membrane protein 13 |
3714 |
0.15 |
| chr17_73826207_73826394 | 0.30 |
Ehd3 |
EH-domain containing 3 |
21459 |
0.18 |
| chr2_158121562_158121714 | 0.29 |
Gm20412 |
predicted gene 20412 |
4484 |
0.17 |
| chr1_133863573_133863878 | 0.29 |
Gm28441 |
predicted gene 28441 |
28536 |
0.11 |
| chr11_55126402_55126555 | 0.29 |
Mup-ps22 |
major urinary protein, pseudogene 22 |
2783 |
0.16 |
| chr6_35494062_35494219 | 0.29 |
Gm23053 |
predicted gene, 23053 |
30498 |
0.19 |
| chr11_86985069_86985237 | 0.29 |
Ypel2 |
yippee like 2 |
8554 |
0.17 |
| chr16_94537265_94537456 | 0.29 |
Vps26c |
VPS26 endosomal protein sorting factor C |
10530 |
0.18 |
| chr1_160791931_160792321 | 0.29 |
Rabgap1l |
RAB GTPase activating protein 1-like |
812 |
0.5 |
| chr12_82915523_82915797 | 0.29 |
1700085C21Rik |
RIKEN cDNA 1700085C21 gene |
23495 |
0.21 |
| chr11_100935486_100935649 | 0.29 |
Stat3 |
signal transducer and activator of transcription 3 |
3813 |
0.17 |
| chr11_100623827_100624107 | 0.28 |
Nkiras2 |
NFKB inhibitor interacting Ras-like protein 2 |
956 |
0.37 |
| chr7_98500649_98500827 | 0.28 |
Gm23040 |
predicted gene, 23040 |
6216 |
0.13 |
| chr1_88954676_88954892 | 0.28 |
Gm4753 |
predicted gene 4753 |
22465 |
0.19 |
| chr7_28969946_28970152 | 0.28 |
Eif3k |
eukaryotic translation initiation factor 3, subunit K |
4787 |
0.11 |
| chr15_85675723_85675883 | 0.28 |
Lncppara |
long noncoding RNA near Ppara |
22187 |
0.12 |
| chr13_64153328_64153492 | 0.28 |
Zfp367 |
zinc finger protein 367 |
208 |
0.83 |
| chr8_34442687_34442913 | 0.28 |
Gm45349 |
predicted gene 45349 |
30195 |
0.14 |
| chr1_87764802_87764962 | 0.28 |
Atg16l1 |
autophagy related 16-like 1 (S. cerevisiae) |
2019 |
0.23 |
| chr1_105354986_105355137 | 0.28 |
Gm17634 |
predicted gene, 17634 |
946 |
0.48 |
| chr17_46004571_46004735 | 0.27 |
Vegfa |
vascular endothelial growth factor A |
16719 |
0.16 |
| chr3_88146112_88146278 | 0.27 |
Mef2d |
myocyte enhancer factor 2D |
3621 |
0.13 |
| chr8_23192641_23192801 | 0.27 |
Gpat4 |
glycerol-3-phosphate acyltransferase 4 |
1783 |
0.23 |
| chr1_88654880_88655031 | 0.27 |
Gm29336 |
predicted gene 29336 |
1353 |
0.35 |
| chr11_120507725_120507906 | 0.27 |
Slc25a10 |
solute carrier family 25 (mitochondrial carrier, dicarboxylate transporter), member 10 |
10090 |
0.06 |
| chr11_98750923_98751087 | 0.27 |
Thra |
thyroid hormone receptor alpha |
2581 |
0.15 |
| chr9_70933007_70933160 | 0.27 |
Lipc |
lipase, hepatic |
1530 |
0.39 |
| chr4_102505805_102505983 | 0.27 |
Pde4b |
phosphodiesterase 4B, cAMP specific |
35497 |
0.23 |
| chr2_28525746_28525944 | 0.26 |
Ralgds |
ral guanine nucleotide dissociation stimulator |
2005 |
0.19 |
| chr5_145873496_145873886 | 0.26 |
Cyp3a11 |
cytochrome P450, family 3, subfamily a, polypeptide 11 |
3000 |
0.22 |
| chr2_179292597_179292961 | 0.26 |
Gm14293 |
predicted gene 14293 |
52289 |
0.14 |
| chr18_46631846_46631997 | 0.26 |
Gm3734 |
predicted gene 3734 |
1114 |
0.44 |
| chr17_29050362_29051086 | 0.26 |
Gm41556 |
predicted gene, 41556 |
2612 |
0.13 |
| chr7_143501066_143501235 | 0.26 |
Phlda2 |
pleckstrin homology like domain, family A, member 2 |
1391 |
0.29 |
| chr14_66636173_66636515 | 0.26 |
Adra1a |
adrenergic receptor, alpha 1a |
788 |
0.71 |
| chr1_120075577_120075750 | 0.26 |
Tmem37 |
transmembrane protein 37 |
1589 |
0.37 |
| chr19_5100696_5100847 | 0.26 |
Cnih2 |
cornichon family AMPA receptor auxiliary protein 2 |
2250 |
0.11 |
| chr1_132384418_132384569 | 0.26 |
Gm15849 |
predicted gene 15849 |
3364 |
0.17 |
| chr9_32833494_32833645 | 0.26 |
Gm37167 |
predicted gene, 37167 |
6353 |
0.22 |
| chr5_99994836_99995114 | 0.26 |
Gm17092 |
predicted gene 17092 |
15914 |
0.13 |
| chr6_128650063_128650348 | 0.25 |
Gm44420 |
predicted gene, 44420 |
1140 |
0.26 |
| chr10_93519889_93520065 | 0.25 |
Hal |
histidine ammonia lyase |
18852 |
0.12 |
| chrX_120050578_120050737 | 0.25 |
Gm16420 |
predicted gene 16420 |
64317 |
0.12 |
| chr4_144981227_144981590 | 0.25 |
Vps13d |
vacuolar protein sorting 13D |
2553 |
0.3 |
| chr7_4150769_4150950 | 0.25 |
Leng9 |
leukocyte receptor cluster (LRC) member 9 |
485 |
0.61 |
| chr18_38521952_38522103 | 0.25 |
Gm50349 |
predicted gene, 50349 |
40724 |
0.12 |
| chr1_4496427_4496626 | 0.25 |
Gm37587 |
predicted gene, 37587 |
25 |
0.59 |
| chr4_144901641_144901842 | 0.25 |
Dhrs3 |
dehydrogenase/reductase (SDR family) member 3 |
8522 |
0.21 |
| chr2_163468579_163468765 | 0.25 |
2310001K24Rik |
RIKEN cDNA 2310001K24 gene |
3285 |
0.14 |
| chr1_39869838_39870141 | 0.25 |
1700066B17Rik |
RIKEN cDNA 1700066B17 gene |
22655 |
0.18 |
| chr10_85193452_85193650 | 0.24 |
Cry1 |
cryptochrome 1 (photolyase-like) |
8487 |
0.2 |
| chr1_89467714_89468048 | 0.24 |
Agap1 |
ArfGAP with GTPase domain, ankyrin repeat and PH domain 1 |
12625 |
0.19 |
| chr2_152338389_152338546 | 0.24 |
Trib3 |
tribbles pseudokinase 3 |
5503 |
0.1 |
| chr14_54431415_54431610 | 0.24 |
Mmp14 |
matrix metallopeptidase 14 (membrane-inserted) |
100 |
0.92 |
| chr9_33101315_33101483 | 0.24 |
Gm32171 |
predicted gene, 32171 |
49893 |
0.13 |
| chr14_7990964_7991352 | 0.24 |
Dnase1l3 |
deoxyribonuclease 1-like 3 |
3415 |
0.22 |
| chr12_52376859_52377022 | 0.24 |
Gm47431 |
predicted gene, 47431 |
71185 |
0.11 |
| chr6_28876568_28876734 | 0.24 |
Snd1 |
staphylococcal nuclease and tudor domain containing 1 |
3392 |
0.27 |
| chr3_89843106_89843448 | 0.24 |
She |
src homology 2 domain-containing transforming protein E |
5388 |
0.14 |
| chr1_36636614_36636765 | 0.23 |
Gm24133 |
predicted gene, 24133 |
27276 |
0.09 |
| chr7_141266376_141266537 | 0.23 |
Irf7 |
interferon regulatory factor 7 |
25 |
0.94 |
| chr5_121189338_121189507 | 0.23 |
Ptpn11 |
protein tyrosine phosphatase, non-receptor type 11 |
1907 |
0.24 |
| chr8_11340394_11340565 | 0.23 |
Col4a1 |
collagen, type IV, alpha 1 |
27653 |
0.13 |
| chr1_151346486_151346783 | 0.23 |
Gm10138 |
predicted gene 10138 |
1403 |
0.31 |
| chr12_113141120_113141303 | 0.23 |
Crip2 |
cysteine rich protein 2 |
639 |
0.55 |
| chr12_54313077_54313237 | 0.22 |
Gm47552 |
predicted gene, 47552 |
3858 |
0.22 |
| chr1_119290301_119290473 | 0.22 |
Gm29456 |
predicted gene 29456 |
36446 |
0.14 |
| chr15_81896525_81896724 | 0.22 |
Aco2 |
aconitase 2, mitochondrial |
1318 |
0.26 |
| chr19_36629854_36630038 | 0.22 |
Hectd2os |
Hectd2, opposite strand |
3922 |
0.26 |
| chr14_121129398_121129725 | 0.22 |
Farp1 |
FERM, RhoGEF (Arhgef) and pleckstrin domain protein 1 (chondrocyte-derived) |
27482 |
0.23 |
| chr8_89015607_89015979 | 0.22 |
Mir8110 |
microRNA 8110 |
8942 |
0.24 |
| chr6_91659872_91660034 | 0.22 |
Gm45217 |
predicted gene 45217 |
19270 |
0.12 |
| chr7_112187893_112188109 | 0.22 |
Dkk3 |
dickkopf WNT signaling pathway inhibitor 3 |
28944 |
0.2 |
| chr19_24017522_24017784 | 0.22 |
Gm50305 |
predicted gene, 50305 |
12873 |
0.17 |
| chr5_73103744_73103903 | 0.22 |
Fryl |
FRY like transcription coactivator |
5444 |
0.18 |
| chr3_38155310_38155488 | 0.22 |
Gm43821 |
predicted gene 43821 |
25208 |
0.16 |
| chr5_92822938_92823090 | 0.22 |
Shroom3 |
shroom family member 3 |
13544 |
0.2 |
| chr19_21106934_21107106 | 0.22 |
4930554I06Rik |
RIKEN cDNA 4930554I06 gene |
2310 |
0.37 |
| chr2_25095172_25095346 | 0.22 |
Noxa1 |
NADPH oxidase activator 1 |
110 |
0.92 |
| chr5_73647707_73647869 | 0.21 |
Sgcb |
sarcoglycan, beta (dystrophin-associated glycoprotein) |
2 |
0.72 |
| chr14_73374673_73374886 | 0.21 |
Itm2b |
integral membrane protein 2B |
1166 |
0.51 |
| chr10_77126560_77126749 | 0.21 |
Gm7775 |
predicted gene 7775 |
10446 |
0.17 |
| chr8_80880650_80880819 | 0.21 |
Gab1 |
growth factor receptor bound protein 2-associated protein 1 |
215 |
0.85 |
| chr9_108306051_108306209 | 0.21 |
Rhoa |
ras homolog family member A |
1 |
0.5 |
| chr11_50159467_50159635 | 0.21 |
Tbc1d9b |
TBC1 domain family, member 9B |
2112 |
0.2 |
| chr14_20082742_20082925 | 0.21 |
Saysd1 |
SAYSVFN motif domain containing 1 |
350 |
0.87 |
| chr1_170639090_170639313 | 0.21 |
Olfml2b |
olfactomedin-like 2B |
5331 |
0.2 |
| chrX_48343770_48343921 | 0.21 |
Bcorl1 |
BCL6 co-repressor-like 1 |
75 |
0.97 |
| chr7_27159930_27160235 | 0.20 |
Egln2 |
egl-9 family hypoxia-inducible factor 2 |
789 |
0.36 |
| chr18_38269268_38269419 | 0.20 |
Pcdh12 |
protocadherin 12 |
8616 |
0.1 |
| chr4_139577114_139577275 | 0.20 |
Iffo2 |
intermediate filament family orphan 2 |
2474 |
0.23 |
| chr12_21143230_21143533 | 0.20 |
Asap2 |
ArfGAP with SH3 domain, ankyrin repeat and PH domain 2 |
31427 |
0.16 |
| chr11_60845341_60845511 | 0.20 |
Dhrs7b |
dehydrogenase/reductase (SDR family) member 7B |
14747 |
0.09 |
| chr4_150065927_150066095 | 0.20 |
Mir34a |
microRNA 34a |
2443 |
0.16 |
| chr8_35377910_35378565 | 0.20 |
Ppp1r3b |
protein phosphatase 1, regulatory subunit 3B |
1577 |
0.3 |
| chr4_144905236_144905418 | 0.20 |
Dhrs3 |
dehydrogenase/reductase (SDR family) member 3 |
12108 |
0.2 |
| chr2_118547575_118547732 | 0.20 |
Bmf |
BCL2 modifying factor |
54 |
0.97 |
| chr1_130661373_130661552 | 0.20 |
C4bp |
complement component 4 binding protein |
148 |
0.93 |
| chr17_36023597_36023890 | 0.19 |
H2-T24 |
histocompatibility 2, T region locus 24 |
3183 |
0.08 |
| chr7_127841086_127841237 | 0.19 |
Stx4a |
syntaxin 4A (placental) |
603 |
0.48 |
| chr6_72624193_72624362 | 0.19 |
Gm15401 |
predicted gene 15401 |
5011 |
0.09 |
| chr1_132329567_132329732 | 0.19 |
Nuak2 |
NUAK family, SNF1-like kinase, 2 |
2053 |
0.21 |
| chr19_3363639_3363796 | 0.19 |
Cpt1a |
carnitine palmitoyltransferase 1a, liver |
2613 |
0.2 |
| chr9_56949772_56949935 | 0.19 |
Snupn |
snurportin 1 |
1025 |
0.37 |
| chr11_100523646_100523797 | 0.19 |
Acly |
ATP citrate lyase |
123 |
0.93 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | GO:1901552 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.1 | 0.4 | GO:0019464 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.1 | 0.8 | GO:0034144 | negative regulation of toll-like receptor 4 signaling pathway(GO:0034144) |
| 0.1 | 0.3 | GO:0097503 | sialylation(GO:0097503) |
| 0.1 | 0.3 | GO:0045079 | negative regulation of chemokine biosynthetic process(GO:0045079) |
| 0.1 | 0.4 | GO:0015760 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.1 | 0.4 | GO:0052805 | histidine catabolic process(GO:0006548) imidazole-containing compound catabolic process(GO:0052805) |
| 0.1 | 0.4 | GO:1900086 | positive regulation of peptidyl-tyrosine autophosphorylation(GO:1900086) |
| 0.1 | 0.2 | GO:0097195 | pilomotor reflex(GO:0097195) |
| 0.1 | 0.2 | GO:0090289 | regulation of osteoclast proliferation(GO:0090289) positive regulation of osteoclast proliferation(GO:0090290) |
| 0.1 | 0.5 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.1 | 0.2 | GO:0010728 | regulation of hydrogen peroxide biosynthetic process(GO:0010728) |
| 0.1 | 0.4 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.1 | 0.2 | GO:0021699 | cerebellar cortex maturation(GO:0021699) |
| 0.0 | 0.1 | GO:2000847 | negative regulation of steroid hormone secretion(GO:2000832) negative regulation of corticosteroid hormone secretion(GO:2000847) negative regulation of glucocorticoid secretion(GO:2000850) |
| 0.0 | 0.1 | GO:0030167 | proteoglycan catabolic process(GO:0030167) |
| 0.0 | 0.3 | GO:0043173 | nucleotide salvage(GO:0043173) |
| 0.0 | 0.1 | GO:0070368 | positive regulation of hepatocyte differentiation(GO:0070368) |
| 0.0 | 0.2 | GO:0018101 | protein citrullination(GO:0018101) |
| 0.0 | 0.2 | GO:1903553 | positive regulation of extracellular exosome assembly(GO:1903553) |
| 0.0 | 0.2 | GO:0045653 | negative regulation of megakaryocyte differentiation(GO:0045653) |
| 0.0 | 0.1 | GO:0043366 | beta selection(GO:0043366) |
| 0.0 | 0.1 | GO:0019448 | cysteine catabolic process(GO:0009093) L-cysteine catabolic process(GO:0019448) L-cysteine metabolic process(GO:0046439) |
| 0.0 | 0.2 | GO:0002756 | MyD88-independent toll-like receptor signaling pathway(GO:0002756) |
| 0.0 | 0.1 | GO:1903238 | positive regulation of leukocyte tethering or rolling(GO:1903238) |
| 0.0 | 0.2 | GO:0032464 | positive regulation of protein homooligomerization(GO:0032464) |
| 0.0 | 0.1 | GO:0001828 | inner cell mass cellular morphogenesis(GO:0001828) |
| 0.0 | 0.1 | GO:0039689 | negative stranded viral RNA replication(GO:0039689) multi-organism biosynthetic process(GO:0044034) |
| 0.0 | 0.4 | GO:0071285 | cellular response to lithium ion(GO:0071285) |
| 0.0 | 0.1 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.0 | 0.1 | GO:0043309 | regulation of eosinophil degranulation(GO:0043309) |
| 0.0 | 0.1 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.0 | 0.2 | GO:0070102 | interleukin-6-mediated signaling pathway(GO:0070102) |
| 0.0 | 0.1 | GO:1901078 | negative regulation of relaxation of muscle(GO:1901078) |
| 0.0 | 0.1 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.3 | GO:0006107 | oxaloacetate metabolic process(GO:0006107) |
| 0.0 | 0.1 | GO:0044375 | regulation of peroxisome size(GO:0044375) |
| 0.0 | 0.1 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.0 | 0.1 | GO:1903774 | positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 0.0 | 0.1 | GO:1901492 | positive regulation of lymphangiogenesis(GO:1901492) |
| 0.0 | 0.1 | GO:0002125 | maternal aggressive behavior(GO:0002125) |
| 0.0 | 0.1 | GO:0090160 | Golgi to lysosome transport(GO:0090160) |
| 0.0 | 0.2 | GO:0010310 | regulation of hydrogen peroxide metabolic process(GO:0010310) |
| 0.0 | 0.0 | GO:0033686 | positive regulation of luteinizing hormone secretion(GO:0033686) |
| 0.0 | 0.1 | GO:0045650 | negative regulation of macrophage differentiation(GO:0045650) |
| 0.0 | 0.1 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.0 | 0.2 | GO:0097084 | vascular smooth muscle cell development(GO:0097084) |
| 0.0 | 0.0 | GO:0044800 | fusion of virus membrane with host plasma membrane(GO:0019064) membrane fusion involved in viral entry into host cell(GO:0039663) multi-organism membrane fusion(GO:0044800) |
| 0.0 | 0.1 | GO:1900126 | negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.0 | 0.1 | GO:0002913 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.0 | 0.1 | GO:0010990 | regulation of SMAD protein complex assembly(GO:0010990) |
| 0.0 | 0.0 | GO:0003340 | negative regulation of mesenchymal to epithelial transition involved in metanephros morphogenesis(GO:0003340) |
| 0.0 | 0.1 | GO:0034214 | protein hexamerization(GO:0034214) |
| 0.0 | 0.1 | GO:0045656 | negative regulation of monocyte differentiation(GO:0045656) |
| 0.0 | 0.2 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.0 | 0.1 | GO:0006933 | negative regulation of cell adhesion involved in substrate-bound cell migration(GO:0006933) |
| 0.0 | 0.1 | GO:0015817 | histidine transport(GO:0015817) |
| 0.0 | 0.0 | GO:2000566 | positive regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000566) |
| 0.0 | 0.1 | GO:0042985 | negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 0.0 | 0.1 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.0 | 0.1 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.0 | 0.1 | GO:0001923 | B-1 B cell differentiation(GO:0001923) |
| 0.0 | 0.1 | GO:0035754 | B cell chemotaxis(GO:0035754) |
| 0.0 | 0.1 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.1 | GO:0060613 | fat pad development(GO:0060613) |
| 0.0 | 0.1 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.0 | 0.1 | GO:0009794 | regulation of mitotic cell cycle, embryonic(GO:0009794) mitotic cell cycle, embryonic(GO:0045448) |
| 0.0 | 0.1 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.0 | GO:0060137 | maternal process involved in parturition(GO:0060137) |
| 0.0 | 0.0 | GO:0003273 | cell migration involved in endocardial cushion formation(GO:0003273) |
| 0.0 | 0.1 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.0 | 0.2 | GO:0043518 | negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) |
| 0.0 | 0.1 | GO:1900864 | mitochondrial tRNA modification(GO:0070900) mitochondrial RNA modification(GO:1900864) |
| 0.0 | 0.1 | GO:0034382 | chylomicron remnant clearance(GO:0034382) triglyceride-rich lipoprotein particle clearance(GO:0071830) |
| 0.0 | 0.0 | GO:0002378 | immunoglobulin biosynthetic process(GO:0002378) |
| 0.0 | 0.0 | GO:0060084 | synaptic transmission involved in micturition(GO:0060084) |
| 0.0 | 0.0 | GO:1904706 | negative regulation of vascular smooth muscle cell proliferation(GO:1904706) |
| 0.0 | 0.0 | GO:0008050 | female courtship behavior(GO:0008050) |
| 0.0 | 0.1 | GO:1900246 | positive regulation of RIG-I signaling pathway(GO:1900246) |
| 0.0 | 0.1 | GO:0046110 | xanthine catabolic process(GO:0009115) xanthine metabolic process(GO:0046110) |
| 0.0 | 0.1 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.0 | 0.1 | GO:0035562 | negative regulation of chromatin binding(GO:0035562) |
| 0.0 | 0.0 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 0.0 | GO:0015744 | succinate transport(GO:0015744) |
| 0.0 | 0.0 | GO:0071926 | cannabinoid signaling pathway(GO:0038171) endocannabinoid signaling pathway(GO:0071926) |
| 0.0 | 0.1 | GO:0015808 | L-alanine transport(GO:0015808) |
| 0.0 | 0.0 | GO:1901164 | negative regulation of trophoblast cell migration(GO:1901164) |
| 0.0 | 0.2 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 0.0 | GO:0014873 | response to muscle activity involved in regulation of muscle adaptation(GO:0014873) |
| 0.0 | 0.1 | GO:1903867 | chorion development(GO:0060717) extraembryonic membrane development(GO:1903867) |
| 0.0 | 0.6 | GO:0001662 | behavioral fear response(GO:0001662) |
| 0.0 | 0.1 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.0 | 0.0 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.0 | 0.0 | GO:0070340 | detection of bacterial lipopeptide(GO:0070340) |
| 0.0 | 0.2 | GO:0042059 | negative regulation of epidermal growth factor receptor signaling pathway(GO:0042059) |
| 0.0 | 0.1 | GO:0006987 | activation of signaling protein activity involved in unfolded protein response(GO:0006987) |
| 0.0 | 0.0 | GO:0030997 | regulation of centriole-centriole cohesion(GO:0030997) |
| 0.0 | 0.1 | GO:0002441 | histamine production involved in inflammatory response(GO:0002349) histamine secretion involved in inflammatory response(GO:0002441) histamine secretion by mast cell(GO:0002553) |
| 0.0 | 0.1 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.0 | 0.0 | GO:0060336 | negative regulation of response to interferon-gamma(GO:0060331) negative regulation of interferon-gamma-mediated signaling pathway(GO:0060336) |
| 0.0 | 0.0 | GO:0019249 | lactate biosynthetic process from pyruvate(GO:0019244) lactate biosynthetic process(GO:0019249) |
| 0.0 | 0.0 | GO:0032079 | positive regulation of endodeoxyribonuclease activity(GO:0032079) |
| 0.0 | 0.0 | GO:0031999 | negative regulation of fatty acid beta-oxidation(GO:0031999) |
| 0.0 | 0.0 | GO:0032345 | negative regulation of aldosterone metabolic process(GO:0032345) negative regulation of aldosterone biosynthetic process(GO:0032348) negative regulation of cortisol biosynthetic process(GO:2000065) |
| 0.0 | 0.2 | GO:2001273 | glucose import in response to insulin stimulus(GO:0044381) regulation of glucose import in response to insulin stimulus(GO:2001273) |
| 0.0 | 0.0 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) |
| 0.0 | 0.1 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 0.0 | GO:0051088 | PMA-inducible membrane protein ectodomain proteolysis(GO:0051088) |
| 0.0 | 0.1 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 | 0.0 | GO:0045472 | response to ether(GO:0045472) |
| 0.0 | 0.0 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.0 | 0.1 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.1 | GO:0043922 | negative regulation by host of viral transcription(GO:0043922) |
| 0.0 | 0.0 | GO:0006393 | termination of mitochondrial transcription(GO:0006393) |
| 0.0 | 0.1 | GO:0032049 | cardiolipin biosynthetic process(GO:0032049) |
| 0.0 | 0.0 | GO:0030576 | Cajal body organization(GO:0030576) |
| 0.0 | 0.1 | GO:0001302 | replicative cell aging(GO:0001302) |
| 0.0 | 0.2 | GO:0006656 | phosphatidylcholine biosynthetic process(GO:0006656) |
| 0.0 | 0.0 | GO:0090324 | negative regulation of oxidative phosphorylation(GO:0090324) |
| 0.0 | 0.0 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.0 | 0.1 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.1 | 0.4 | GO:0036488 | CHOP-C/EBP complex(GO:0036488) |
| 0.1 | 0.4 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.1 | 0.2 | GO:0044352 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.0 | 0.2 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.0 | 0.1 | GO:0097651 | phosphatidylinositol 3-kinase complex, class I(GO:0097651) |
| 0.0 | 0.5 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.1 | GO:0097629 | extrinsic component of omegasome membrane(GO:0097629) |
| 0.0 | 0.1 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 0.2 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.1 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.0 | 0.1 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.0 | 0.1 | GO:0031523 | Myb complex(GO:0031523) |
| 0.0 | 0.1 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.0 | 0.1 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.0 | 0.1 | GO:0020016 | ciliary pocket(GO:0020016) ciliary pocket membrane(GO:0020018) |
| 0.0 | 0.1 | GO:0097433 | dense body(GO:0097433) |
| 0.0 | 0.1 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.0 | 0.1 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.0 | 0.0 | GO:0035355 | Toll-like receptor 2-Toll-like receptor 6 protein complex(GO:0035355) |
| 0.0 | 0.0 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.0 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.0 | 1.3 | GO:0043296 | apical junction complex(GO:0043296) |
| 0.0 | 0.1 | GO:0030132 | clathrin coat of coated pit(GO:0030132) |
| 0.0 | 0.0 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.0 | 0.1 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 0.2 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.0 | 0.1 | GO:0005883 | neurofilament(GO:0005883) |
| 0.0 | 0.0 | GO:0043293 | apoptosome(GO:0043293) |
| 0.0 | 0.0 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0015526 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.1 | 0.3 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.1 | 0.4 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.1 | 0.7 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.1 | 0.2 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) |
| 0.1 | 0.3 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.1 | 0.8 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.1 | 0.2 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.1 | 0.3 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.1 | 0.3 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.1 | 0.2 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 0.1 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.0 | 0.0 | GO:0015141 | succinate transmembrane transporter activity(GO:0015141) |
| 0.0 | 0.2 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.0 | 0.3 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 0.2 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.0 | 0.4 | GO:0001013 | RNA polymerase I regulatory region DNA binding(GO:0001013) |
| 0.0 | 0.2 | GO:0015252 | hydrogen ion channel activity(GO:0015252) |
| 0.0 | 0.1 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.1 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.0 | 0.2 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.0 | 0.1 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.0 | 0.1 | GO:0016823 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.0 | 0.1 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.0 | 0.1 | GO:0031893 | vasopressin receptor binding(GO:0031893) |
| 0.0 | 0.2 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.1 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.0 | 0.1 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 0.1 | GO:0017153 | sodium:dicarboxylate symporter activity(GO:0017153) |
| 0.0 | 0.1 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) |
| 0.0 | 0.1 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.0 | 0.2 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.0 | 0.0 | GO:0005290 | L-histidine transmembrane transporter activity(GO:0005290) |
| 0.0 | 0.1 | GO:0051022 | Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.0 | 0.1 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.0 | 0.1 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.1 | GO:0004854 | xanthine dehydrogenase activity(GO:0004854) oxidoreductase activity, acting on CH or CH2 groups, NAD or NADP as acceptor(GO:0016726) |
| 0.0 | 0.1 | GO:0032557 | pyrimidine ribonucleotide binding(GO:0032557) |
| 0.0 | 0.1 | GO:0015180 | L-alanine transmembrane transporter activity(GO:0015180) alanine transmembrane transporter activity(GO:0022858) |
| 0.0 | 0.1 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.0 | 0.1 | GO:0018646 | 3-(3-hydroxyphenyl)propionate hydroxylase activity(GO:0008688) 4-chlorobenzaldehyde oxidase activity(GO:0018471) 3,5-xylenol methylhydroxylase activity(GO:0018630) phenylacetate hydroxylase activity(GO:0018631) 4-nitrophenol 4-monooxygenase activity(GO:0018632) dimethyl sulfide monooxygenase activity(GO:0018633) alpha-pinene monooxygenase [NADH] activity(GO:0018634) 1-hydroxy-2-naphthoate hydroxylase activity(GO:0018637) toluene 4-monooxygenase activity(GO:0018638) xylene monooxygenase activity(GO:0018639) dibenzothiophene monooxygenase activity(GO:0018640) 6-hydroxy-3-methyl-2-oxo-1,2-dihydroquinoline 6-monooxygenase activity(GO:0018641) chlorophenol 4-monooxygenase activity(GO:0018642) carbon disulfide oxygenase activity(GO:0018643) toluene 2-monooxygenase activity(GO:0018644) 1-hydroxy-2-oxolimonene 1,2-monooxygenase activity(GO:0018646) phenanthrene 1,2-monooxygenase activity(GO:0018647) tetrahydrofuran hydroxylase activity(GO:0018649) styrene monooxygenase activity(GO:0018650) toluene-4-sulfonate monooxygenase activity(GO:0018651) toluene-sulfonate methyl-monooxygenase activity(GO:0018652) 3-methyl-2-oxo-1,2-dihydroquinoline 6-monooxygenase activity(GO:0018653) 2-hydroxy-phenylacetate hydroxylase activity(GO:0018654) 2-oxo-delta3-4,5,5-trimethylcyclopentenylacetyl-CoA 1,2-monooxygenase activity(GO:0018655) phenanthrene 3,4-monooxygenase activity(GO:0018656) toluene 3-monooxygenase activity(GO:0018657) 4-hydroxyphenylacetate,NADH:oxygen oxidoreductase (3-hydroxylating) activity(GO:0018660) limonene monooxygenase activity(GO:0019113) 2-methylnaphthalene hydroxylase activity(GO:0034526) 1-methylnaphthalene hydroxylase activity(GO:0034534) bisphenol A hydroxylase A activity(GO:0034560) salicylate 5-hydroxylase activity(GO:0034785) isobutylamine N-hydroxylase activity(GO:0034791) branched-chain dodecylbenzene sulfonate monooxygenase activity(GO:0034802) 3-HSA hydroxylase activity(GO:0034819) 4-hydroxypyridine-3-hydroxylase activity(GO:0034894) 2-octaprenyl-3-methyl-6-methoxy-1,4-benzoquinol hydroxylase activity(GO:0043719) 6-hydroxynicotinate 3-monooxygenase activity(GO:0043731) thalianol hydroxylase activity(GO:0080014) |
| 0.0 | 0.1 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.0 | 0.0 | GO:0008240 | tripeptidyl-peptidase activity(GO:0008240) |
| 0.0 | 0.1 | GO:0004897 | ciliary neurotrophic factor receptor activity(GO:0004897) |
| 0.0 | 0.1 | GO:0030572 | cardiolipin synthase activity(GO:0008808) phosphatidyltransferase activity(GO:0030572) |
| 0.0 | 0.1 | GO:0009374 | biotin binding(GO:0009374) |
| 0.0 | 0.1 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 0.0 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.1 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.0 | 0.0 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 0.0 | 0.1 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.0 | 0.0 | GO:0051733 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) |
| 0.0 | 0.0 | GO:0035175 | histone kinase activity (H3-S10 specific)(GO:0035175) |
| 0.0 | 0.1 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 0.0 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) |
| 0.0 | 0.1 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.0 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 0.1 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.1 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.1 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.0 | 0.1 | GO:0050649 | testosterone 6-beta-hydroxylase activity(GO:0050649) |
| 0.0 | 0.1 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.0 | 0.1 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.0 | 0.0 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.9 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.0 | 0.2 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.0 | 0.3 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.4 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.2 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.5 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.1 | 1.1 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.9 | REACTOME RORA ACTIVATES CIRCADIAN EXPRESSION | Genes involved in RORA Activates Circadian Expression |
| 0.0 | 0.2 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.0 | 0.2 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.3 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.0 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 0.2 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.1 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.0 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 0.5 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.0 | 0.2 | REACTOME FATTY ACYL COA BIOSYNTHESIS | Genes involved in Fatty Acyl-CoA Biosynthesis |
| 0.0 | 0.0 | REACTOME TCR SIGNALING | Genes involved in TCR signaling |
| 0.0 | 0.0 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.0 | 0.2 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 0.1 | REACTOME TRANSPORT OF MATURE MRNA DERIVED FROM AN INTRONLESS TRANSCRIPT | Genes involved in Transport of Mature mRNA Derived from an Intronless Transcript |
| 0.0 | 0.1 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.2 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.0 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.0 | 0.1 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.1 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.1 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 0.2 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |