Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
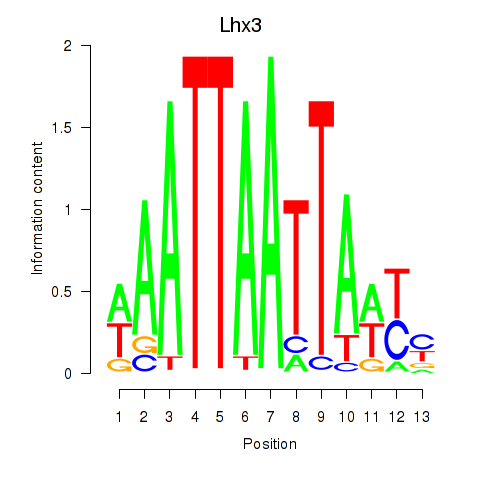
Results for Lhx3
Z-value: 1.65
Transcription factors associated with Lhx3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Lhx3
|
ENSMUSG00000026934.9 | LIM homeobox protein 3 |
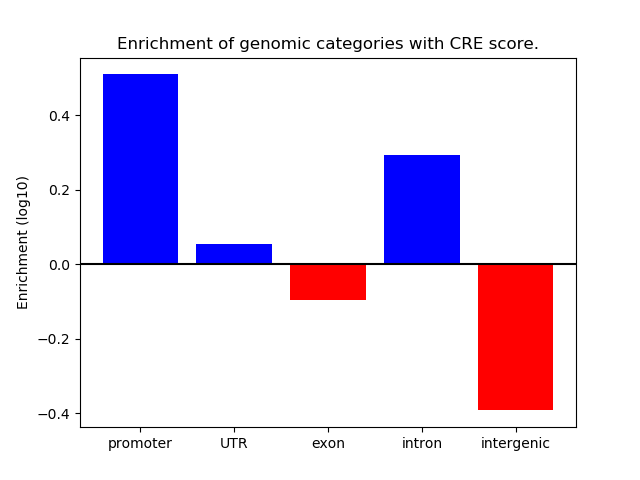
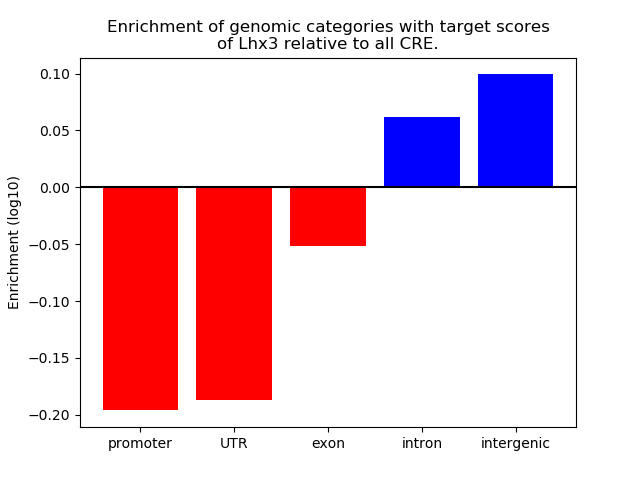
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr2_26214445_26214599 | Lhx3 | 6233 | 0.119378 | 0.82 | 4.5e-02 | Click! |
| chr2_26208213_26208385 | Lhx3 | 10 | 0.953015 | -0.38 | 4.6e-01 | Click! |
| chr2_26214666_26214824 | Lhx3 | 6456 | 0.118619 | -0.09 | 8.6e-01 | Click! |
| chr2_26208483_26208654 | Lhx3 | 279 | 0.829873 | 0.04 | 9.4e-01 | Click! |
Activity of the Lhx3 motif across conditions
Conditions sorted by the z-value of the Lhx3 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
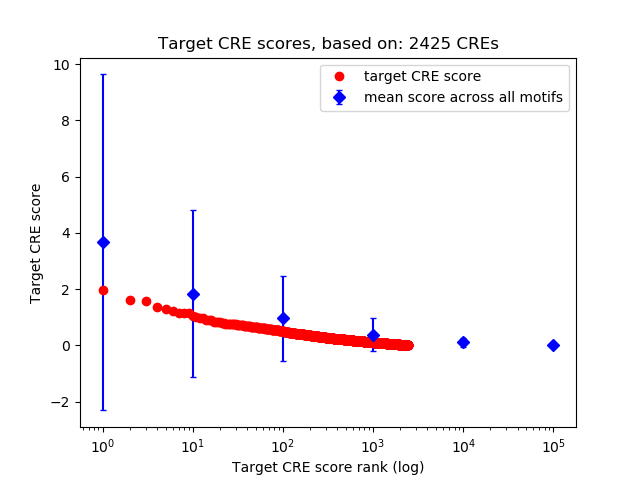
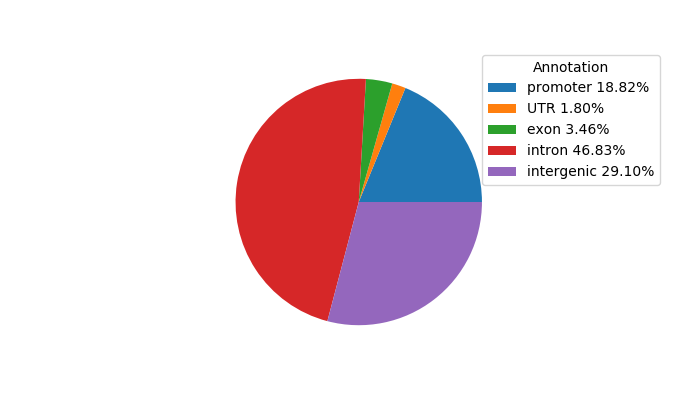
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr10_108612751_108612908 | 1.97 |
Syt1 |
synaptotagmin I |
24135 |
0.23 |
| chr6_35874026_35874908 | 1.63 |
Gm43442 |
predicted gene 43442 |
52244 |
0.17 |
| chr7_139559759_139559933 | 1.57 |
Nkx6-2 |
NK6 homeobox 2 |
22944 |
0.17 |
| chr8_4335762_4335913 | 1.35 |
Ccl25 |
chemokine (C-C motif) ligand 25 |
3578 |
0.14 |
| chr2_14099979_14100130 | 1.30 |
Stam |
signal transducing adaptor molecule (SH3 domain and ITAM motif) 1 |
25627 |
0.12 |
| chr10_115337191_115337351 | 1.21 |
Tmem19 |
transmembrane protein 19 |
11793 |
0.14 |
| chr11_111605518_111605669 | 1.16 |
Gm11676 |
predicted gene 11676 |
7713 |
0.32 |
| chr11_74821059_74821337 | 1.14 |
Mnt |
max binding protein |
9722 |
0.13 |
| chr12_8643213_8643385 | 1.14 |
Pum2 |
pumilio RNA-binding family member 2 |
30835 |
0.16 |
| chr11_70713419_70713657 | 1.06 |
Mir6925 |
microRNA 6925 |
7548 |
0.08 |
| chr2_155539663_155540189 | 1.02 |
Mipep-ps |
mitochondrial intermediate peptidase, pseudogene |
2721 |
0.13 |
| chr18_64325483_64325793 | 0.98 |
St8sia3os |
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 3, opposite strand |
8068 |
0.16 |
| chr15_79690269_79690868 | 0.98 |
Gtpbp1 |
GTP binding protein 1 |
277 |
0.78 |
| chr3_94338287_94338715 | 0.89 |
Gm43743 |
predicted gene 43743 |
3169 |
0.1 |
| chr15_31279226_31279377 | 0.89 |
Gm49296 |
predicted gene, 49296 |
2788 |
0.19 |
| chr5_117293923_117294074 | 0.89 |
Gm3786 |
predicted gene 3786 |
408 |
0.73 |
| chr19_44027305_44027461 | 0.84 |
Cyp2c23 |
cytochrome P450, family 2, subfamily c, polypeptide 23 |
1816 |
0.28 |
| chr16_27391982_27392165 | 0.84 |
Ccdc50 |
coiled-coil domain containing 50 |
2105 |
0.33 |
| chr2_119566293_119566470 | 0.83 |
Chp1 |
calcineurin-like EF hand protein 1 |
224 |
0.89 |
| chr1_60862632_60862783 | 0.83 |
Gm38137 |
predicted gene, 38137 |
17631 |
0.12 |
| chr4_142088876_142089143 | 0.80 |
Tmem51os1 |
Tmem51 opposite strand 1 |
4153 |
0.16 |
| chr15_54947760_54948020 | 0.79 |
Gm26684 |
predicted gene, 26684 |
4180 |
0.19 |
| chr15_25942169_25942359 | 0.77 |
Retreg1 |
reticulophagy regulator 1 |
415 |
0.85 |
| chr8_128687947_128688299 | 0.76 |
Itgb1 |
integrin beta 1 (fibronectin receptor beta) |
2253 |
0.29 |
| chr18_61398040_61398397 | 0.75 |
Mir378a |
microRNA 378a |
318 |
0.54 |
| chr4_116720481_116720689 | 0.75 |
Tesk2 |
testis-specific kinase 2 |
363 |
0.78 |
| chr19_43767883_43768034 | 0.75 |
Cutc |
cutC copper transporter |
3085 |
0.18 |
| chr16_24394028_24394581 | 0.75 |
Lpp |
LIM domain containing preferred translocation partner in lipoma |
566 |
0.73 |
| chr7_72345699_72346292 | 0.75 |
Mctp2 |
multiple C2 domains, transmembrane 2 |
39387 |
0.21 |
| chr2_166522580_166522731 | 0.75 |
Gm14267 |
predicted gene 14267 |
282 |
0.93 |
| chr3_23937769_23938040 | 0.73 |
Naaladl2 |
N-acetylated alpha-linked acidic dipeptidase-like 2 |
1573 |
0.58 |
| chr18_55095856_55096021 | 0.73 |
AC163347.1 |
novel transcript |
422 |
0.83 |
| chr3_80058876_80059038 | 0.72 |
A330069K06Rik |
RIKEN cDNA A330069K06 gene |
4065 |
0.25 |
| chr9_79213455_79213848 | 0.72 |
Gm47499 |
predicted gene, 47499 |
11644 |
0.17 |
| chr4_132129267_132129447 | 0.71 |
Oprd1 |
opioid receptor, delta 1 |
15129 |
0.1 |
| chr2_69228328_69228964 | 0.71 |
G6pc2 |
glucose-6-phosphatase, catalytic, 2 |
8618 |
0.15 |
| chr6_71236030_71236205 | 0.69 |
Smyd1 |
SET and MYND domain containing 1 |
19243 |
0.1 |
| chr4_48315210_48315516 | 0.69 |
Gm12435 |
predicted gene 12435 |
29867 |
0.16 |
| chr2_15096629_15097014 | 0.69 |
Arl5b |
ADP-ribosylation factor-like 5B |
21822 |
0.14 |
| chr1_119253363_119253514 | 0.69 |
Gm3508 |
predicted gene 3508 |
10685 |
0.22 |
| chr11_109475236_109475399 | 0.69 |
Slc16a6 |
solute carrier family 16 (monocarboxylic acid transporters), member 6 |
1719 |
0.22 |
| chr5_8928809_8928960 | 0.68 |
Abcb4 |
ATP-binding cassette, sub-family B (MDR/TAP), member 4 |
158 |
0.94 |
| chr3_93589735_93589907 | 0.68 |
S100a10 |
S100 calcium binding protein A10 (calpactin) |
29094 |
0.09 |
| chr11_78120991_78121457 | 0.67 |
Fam222b |
family with sequence similarity 222, member B |
5618 |
0.09 |
| chr3_28736802_28736989 | 0.66 |
1700112D23Rik |
RIKEN cDNA 1700112D23 gene |
5345 |
0.16 |
| chr17_73134412_73134584 | 0.65 |
Lclat1 |
lysocardiolipin acyltransferase 1 |
8071 |
0.23 |
| chr10_53348308_53348525 | 0.65 |
Cep85l |
centrosomal protein 85-like |
1407 |
0.3 |
| chr17_87445917_87446172 | 0.65 |
Calm2 |
calmodulin 2 |
804 |
0.49 |
| chr4_97099326_97100044 | 0.64 |
Gm27521 |
predicted gene, 27521 |
182665 |
0.03 |
| chr2_6611640_6611812 | 0.64 |
Celf2 |
CUGBP, Elav-like family member 2 |
4454 |
0.32 |
| chr13_25083444_25083634 | 0.64 |
Dcdc2a |
doublecortin domain containing 2a |
12808 |
0.18 |
| chr17_29381584_29381926 | 0.63 |
Fgd2 |
FYVE, RhoGEF and PH domain containing 2 |
5254 |
0.14 |
| chr12_33302809_33302960 | 0.63 |
Atxn7l1 |
ataxin 7-like 1 |
369 |
0.89 |
| chr9_66414274_66414486 | 0.63 |
Gm28379 |
predicted gene 28379 |
6261 |
0.21 |
| chr9_103363870_103364326 | 0.63 |
Cdv3 |
carnitine deficiency-associated gene expressed in ventricle 3 |
799 |
0.56 |
| chr5_87091073_87091224 | 0.63 |
Ugt2b36 |
UDP glucuronosyltransferase 2 family, polypeptide B36 |
9 |
0.96 |
| chr12_35294499_35294705 | 0.62 |
Gm44396 |
predicted gene, 44396 |
14289 |
0.25 |
| chr9_20730600_20730850 | 0.62 |
Olfm2 |
olfactomedin 2 |
2496 |
0.23 |
| chr6_128504140_128504367 | 0.61 |
Pzp |
PZP, alpha-2-macroglobulin like |
9030 |
0.09 |
| chr12_109991265_109991416 | 0.61 |
Gm34667 |
predicted gene, 34667 |
32533 |
0.1 |
| chr10_53753298_53753451 | 0.61 |
Fam184a |
family with sequence similarity 184, member A |
2251 |
0.35 |
| chr10_108074024_108074345 | 0.61 |
Gm47999 |
predicted gene, 47999 |
68248 |
0.12 |
| chr6_71173018_71173198 | 0.60 |
Gm44427 |
predicted gene, 44427 |
8309 |
0.13 |
| chr4_150685463_150685649 | 0.60 |
Gm16079 |
predicted gene 16079 |
6764 |
0.21 |
| chr7_14430188_14430339 | 0.59 |
Sult2a8 |
sulfotransferase family 2A, dehydroepiandrosterone (DHEA)-preferring, member 8 |
6184 |
0.17 |
| chr5_113041682_113041833 | 0.58 |
Gm22740 |
predicted gene, 22740 |
2387 |
0.2 |
| chr2_157536788_157536939 | 0.58 |
Gm14287 |
predicted gene 14287 |
7878 |
0.12 |
| chr4_155200341_155200507 | 0.57 |
Ski |
ski sarcoma viral oncogene homolog (avian) |
12128 |
0.17 |
| chr4_19696850_19697018 | 0.57 |
Wwp1 |
WW domain containing E3 ubiquitin protein ligase 1 |
12059 |
0.21 |
| chr3_10298177_10298449 | 0.57 |
Fabp12 |
fatty acid binding protein 12 |
2861 |
0.14 |
| chr10_71332846_71333050 | 0.57 |
Cisd1 |
CDGSH iron sulfur domain 1 |
12006 |
0.12 |
| chrX_38451090_38451246 | 0.57 |
Lamp2 |
lysosomal-associated membrane protein 2 |
5246 |
0.2 |
| chr8_71405347_71405498 | 0.56 |
Ankle1 |
ankyrin repeat and LEM domain containing 1 |
588 |
0.54 |
| chr5_30187138_30187289 | 0.56 |
Hadhb |
hydroxyacyl-CoA dehydrogenase trifunctional multienzyme complex subunit beta |
6981 |
0.12 |
| chr10_69165945_69166107 | 0.56 |
Rhobtb1 |
Rho-related BTB domain containing 1 |
14592 |
0.16 |
| chr1_163243306_163243486 | 0.56 |
Gm20471 |
predicted gene 20471 |
36960 |
0.15 |
| chr18_21372400_21372792 | 0.55 |
Gm22886 |
predicted gene, 22886 |
8335 |
0.2 |
| chr6_50180364_50180523 | 0.55 |
Mpp6 |
membrane protein, palmitoylated 6 (MAGUK p55 subfamily member 6) |
143 |
0.97 |
| chr19_36735506_36735662 | 0.55 |
Ppp1r3c |
protein phosphatase 1, regulatory subunit 3C |
1069 |
0.56 |
| chr5_104513210_104513369 | 0.54 |
Thoc2l |
THO complex subunit 2-like |
4937 |
0.19 |
| chr17_36032663_36032990 | 0.54 |
H2-T23 |
histocompatibility 2, T region locus 23 |
29 |
0.92 |
| chr12_77290130_77290290 | 0.54 |
Fut8 |
fucosyltransferase 8 |
51145 |
0.13 |
| chr17_5012388_5012845 | 0.53 |
Arid1b |
AT rich interactive domain 1B (SWI-like) |
16187 |
0.23 |
| chr7_16604272_16604437 | 0.53 |
Gm29443 |
predicted gene 29443 |
9470 |
0.09 |
| chr16_23319629_23319807 | 0.53 |
St6gal1 |
beta galactoside alpha 2,6 sialyltransferase 1 |
29248 |
0.15 |
| chr15_100654416_100654567 | 0.53 |
Bin2 |
bridging integrator 2 |
2831 |
0.13 |
| chr19_23024753_23025259 | 0.53 |
Gm50136 |
predicted gene, 50136 |
36448 |
0.17 |
| chr17_29502998_29503149 | 0.53 |
Pim1 |
proviral integration site 1 |
9666 |
0.1 |
| chr15_85668222_85668373 | 0.53 |
Lncppara |
long noncoding RNA near Ppara |
14681 |
0.14 |
| chr18_20868985_20869147 | 0.53 |
Trappc8 |
trafficking protein particle complex 8 |
6143 |
0.22 |
| chr12_118887325_118887612 | 0.51 |
Abcb5 |
ATP-binding cassette, sub-family B (MDR/TAP), member 5 |
3291 |
0.3 |
| chr18_69669013_69669180 | 0.51 |
Tcf4 |
transcription factor 4 |
10603 |
0.29 |
| chr6_141629422_141629807 | 0.51 |
Slco1b2 |
solute carrier organic anion transporter family, member 1b2 |
96 |
0.98 |
| chr5_115490406_115490588 | 0.51 |
Gm24407 |
predicted gene, 24407 |
243 |
0.76 |
| chr15_43615203_43615437 | 0.51 |
Emc2 |
ER membrane protein complex subunit 2 |
120410 |
0.06 |
| chr5_38480616_38480794 | 0.51 |
Slc2a9 |
solute carrier family 2 (facilitated glucose transporter), member 9 |
399 |
0.85 |
| chr7_75472656_75473119 | 0.51 |
Gm44834 |
predicted gene 44834 |
9112 |
0.16 |
| chr14_93654246_93654648 | 0.50 |
Gm48967 |
predicted gene, 48967 |
35601 |
0.23 |
| chr14_46545888_46546175 | 0.50 |
E130120K24Rik |
RIKEN cDNA E130120K24 gene |
10272 |
0.13 |
| chr10_68268511_68268720 | 0.50 |
Arid5b |
AT rich interactive domain 5B (MRF1-like) |
10106 |
0.23 |
| chr1_129250759_129250920 | 0.50 |
Thsd7b |
thrombospondin, type I, domain containing 7B |
22463 |
0.22 |
| chr6_17554780_17555139 | 0.49 |
Met |
met proto-oncogene |
7986 |
0.24 |
| chr3_116936111_116936286 | 0.49 |
Gm42892 |
predicted gene 42892 |
8132 |
0.15 |
| chr11_35015732_35015915 | 0.49 |
Slit3 |
slit guidance ligand 3 |
105401 |
0.07 |
| chr2_3412724_3412892 | 0.49 |
Meig1 |
meiosis expressed gene 1 |
5773 |
0.13 |
| chr15_100304113_100304275 | 0.48 |
Mettl7a1 |
methyltransferase like 7A1 |
12 |
0.96 |
| chr19_30195816_30196018 | 0.48 |
Gldc |
glycine decarboxylase |
20488 |
0.17 |
| chr1_183382689_183382840 | 0.48 |
Taf1a |
TATA-box binding protein associated factor, RNA polymerase I, A |
6217 |
0.13 |
| chr5_24696900_24697051 | 0.48 |
Gm26648 |
predicted gene, 26648 |
7715 |
0.13 |
| chr6_72259555_72259838 | 0.47 |
Atoh8 |
atonal bHLH transcription factor 8 |
24119 |
0.13 |
| chr10_111756586_111756764 | 0.47 |
Gm46193 |
predicted gene, 46193 |
11103 |
0.15 |
| chr10_111936910_111937296 | 0.47 |
Krr1 |
KRR1, small subunit (SSU) processome component, homolog (yeast) |
35561 |
0.12 |
| chr17_29504253_29504439 | 0.46 |
Pim1 |
proviral integration site 1 |
10939 |
0.1 |
| chr19_55166765_55167101 | 0.46 |
Tectb |
tectorin beta |
13800 |
0.17 |
| chr16_10972025_10972219 | 0.46 |
Litaf |
LPS-induced TN factor |
3457 |
0.14 |
| chr16_86808963_86809164 | 0.46 |
Gm32461 |
predicted gene, 32461 |
2550 |
0.28 |
| chr2_48442189_48442365 | 0.46 |
Gm13481 |
predicted gene 13481 |
14968 |
0.23 |
| chr3_133765600_133766533 | 0.45 |
Gm6135 |
prediticted gene 6135 |
25438 |
0.2 |
| chr8_72216672_72216850 | 0.45 |
Fam32a |
family with sequence similarity 32, member A |
2969 |
0.12 |
| chr6_121874198_121874349 | 0.45 |
Mug1 |
murinoglobulin 1 |
11304 |
0.19 |
| chr14_116448226_116448591 | 0.44 |
Gm38045 |
predicted gene, 38045 |
301807 |
0.01 |
| chr4_97757677_97758345 | 0.44 |
Gm12676 |
predicted gene 12676 |
622 |
0.75 |
| chr6_17255791_17255960 | 0.44 |
Cav2 |
caveolin 2 |
25310 |
0.17 |
| chr5_145464782_145464950 | 0.44 |
Cyp3a16 |
cytochrome P450, family 3, subfamily a, polypeptide 16 |
4857 |
0.19 |
| chr15_79553946_79554106 | 0.44 |
Ddx17 |
DEAD (Asp-Glu-Ala-Asp) box polypeptide 17 |
7285 |
0.11 |
| chr2_40852081_40852246 | 0.44 |
Gm13462 |
predicted gene 13462 |
106228 |
0.07 |
| chr18_39827979_39828276 | 0.43 |
Pabpc2 |
poly(A) binding protein, cytoplasmic 2 |
54630 |
0.14 |
| chr17_73839522_73839982 | 0.43 |
Gm4948 |
predicted gene 4948 |
31598 |
0.14 |
| chr8_86154549_86154716 | 0.43 |
Gm23904 |
predicted gene, 23904 |
79222 |
0.1 |
| chr2_165921904_165922055 | 0.43 |
Gm11461 |
predicted gene 11461 |
3862 |
0.16 |
| chr1_23163206_23163464 | 0.43 |
Ppp1r14bl |
protein phosphatase 1, regulatory inhibitor subunit 14B like |
61082 |
0.1 |
| chr1_174921018_174921617 | 0.43 |
Grem2 |
gremlin 2, DAN family BMP antagonist |
502 |
0.88 |
| chr6_108254587_108254738 | 0.43 |
Itpr1 |
inositol 1,4,5-trisphosphate receptor 1 |
1704 |
0.47 |
| chr1_136229022_136229181 | 0.43 |
Inava |
innate immunity activator |
918 |
0.4 |
| chr9_115403471_115403629 | 0.43 |
Gm9487 |
predicted gene 9487 |
1599 |
0.28 |
| chr13_64237836_64238013 | 0.43 |
Cdc14b |
CDC14 cell division cycle 14B |
10718 |
0.1 |
| chr17_10237978_10238400 | 0.42 |
Qk |
quaking |
24615 |
0.24 |
| chr5_138106488_138106739 | 0.42 |
Gm43493 |
predicted gene 43493 |
294 |
0.77 |
| chr6_13287887_13288060 | 0.42 |
AC121573.1 |
predicted 725 |
24228 |
0.22 |
| chr2_45114016_45114230 | 0.41 |
Zeb2 |
zinc finger E-box binding homeobox 2 |
615 |
0.66 |
| chr8_34506104_34506386 | 0.41 |
Gm8268 |
predicted gene 8268 |
962 |
0.54 |
| chr6_120023528_120024062 | 0.41 |
Mir706 |
microRNA 706 |
10516 |
0.16 |
| chr5_86725030_86725390 | 0.41 |
Gm16579 |
predicted gene 16579 |
3689 |
0.15 |
| chr12_99418906_99419292 | 0.41 |
Foxn3 |
forkhead box N3 |
6494 |
0.18 |
| chr14_27876257_27876427 | 0.41 |
Erc2 |
ELKS/RAB6-interacting/CAST family member 2 |
72 |
0.57 |
| chr5_30926172_30926329 | 0.41 |
Khk |
ketohexokinase |
4364 |
0.09 |
| chrX_60347501_60348092 | 0.41 |
Mir505 |
microRNA 505 |
46696 |
0.14 |
| chr4_82643627_82643778 | 0.40 |
Gm11268 |
predicted gene 11268 |
1302 |
0.49 |
| chr14_55682185_55682383 | 0.40 |
Gm49067 |
predicted gene, 49067 |
366 |
0.43 |
| chr1_133251724_133252043 | 0.40 |
Gm19461 |
predicted gene, 19461 |
2409 |
0.23 |
| chr12_31539489_31539733 | 0.40 |
Slc26a4 |
solute carrier family 26, member 4 |
3038 |
0.19 |
| chr5_134992454_134992629 | 0.40 |
Cldn3 |
claudin 3 |
6327 |
0.08 |
| chr13_22020926_22021324 | 0.40 |
Gm11290 |
predicted gene 11290 |
3619 |
0.06 |
| chr7_73538202_73538544 | 0.40 |
Gm44686 |
predicted gene 44686 |
360 |
0.65 |
| chr1_58112957_58113355 | 0.40 |
Aox3 |
aldehyde oxidase 3 |
18 |
0.98 |
| chr11_100819438_100819589 | 0.40 |
Stat5b |
signal transducer and activator of transcription 5B |
3015 |
0.18 |
| chr8_36820142_36820568 | 0.39 |
Dlc1 |
deleted in liver cancer 1 |
87301 |
0.09 |
| chr12_57514709_57514889 | 0.39 |
Gm2568 |
predicted gene 2568 |
5825 |
0.17 |
| chr2_44105283_44105434 | 0.39 |
Arhgap15os |
Rho GTPase activating protein 15, opposite strand |
40034 |
0.19 |
| chr11_84865098_84865523 | 0.39 |
Ggnbp2 |
gametogenetin binding protein 2 |
4651 |
0.13 |
| chr1_187214930_187215194 | 0.39 |
Spata17 |
spermatogenesis associated 17 |
359 |
0.55 |
| chr17_86896356_86896548 | 0.39 |
Tmem247 |
transmembrane protein 247 |
20896 |
0.13 |
| chr9_50501355_50501515 | 0.39 |
Plet1os |
placenta expressed transcript 1, opposite strand |
3370 |
0.16 |
| chr10_96854885_96855043 | 0.39 |
Gm24587 |
predicted gene, 24587 |
706 |
0.71 |
| chr10_108454541_108454850 | 0.39 |
Gm36283 |
predicted gene, 36283 |
8093 |
0.2 |
| chr19_34518935_34519125 | 0.39 |
Lipa |
lysosomal acid lipase A |
8050 |
0.13 |
| chr15_98609856_98610015 | 0.39 |
Adcy6 |
adenylate cyclase 6 |
97 |
0.93 |
| chr8_91381593_91381994 | 0.39 |
Fto |
fat mass and obesity associated |
8762 |
0.16 |
| chr1_30946725_30947101 | 0.39 |
Ptp4a1 |
protein tyrosine phosphatase 4a1 |
487 |
0.77 |
| chr13_4034887_4035107 | 0.39 |
Gm18857 |
predicted gene, 18857 |
248 |
0.89 |
| chr15_78804542_78804702 | 0.38 |
Card10 |
caspase recruitment domain family, member 10 |
1580 |
0.24 |
| chr8_82249699_82250126 | 0.38 |
Gm38590 |
predicted gene, 38590 |
42377 |
0.17 |
| chr10_111133507_111133671 | 0.38 |
Gm22101 |
predicted gene, 22101 |
4380 |
0.16 |
| chr12_21142640_21142894 | 0.38 |
Asap2 |
ArfGAP with SH3 domain, ankyrin repeat and PH domain 2 |
30813 |
0.16 |
| chr11_101675430_101675600 | 0.38 |
Arl4d |
ADP-ribosylation factor-like 4D |
9974 |
0.1 |
| chr8_127169398_127169549 | 0.38 |
Pard3 |
par-3 family cell polarity regulator |
1931 |
0.45 |
| chr3_50931112_50931300 | 0.37 |
Gm37843 |
predicted gene, 37843 |
9681 |
0.2 |
| chr4_148085280_148085458 | 0.37 |
Agtrap |
angiotensin II, type I receptor-associated protein |
593 |
0.56 |
| chr4_40698726_40698877 | 0.37 |
Aptx |
aprataxin |
3915 |
0.15 |
| chr11_119558053_119558232 | 0.37 |
Gm11762 |
predicted gene 11762 |
9512 |
0.15 |
| chr6_58989254_58989420 | 0.37 |
Gm45061 |
predicted gene 45061 |
16140 |
0.13 |
| chr9_62118138_62118584 | 0.37 |
Mir5133 |
microRNA 5133 |
4233 |
0.22 |
| chr19_53889976_53890197 | 0.37 |
Pdcd4 |
programmed cell death 4 |
2145 |
0.25 |
| chr15_34510659_34511041 | 0.37 |
Pop1 |
processing of precursor 1, ribonuclease P/MRP family, (S. cerevisiae) |
943 |
0.43 |
| chr4_35101216_35101367 | 0.37 |
Ifnk |
interferon kappa |
50765 |
0.12 |
| chr11_101395003_101395405 | 0.36 |
Gm25475 |
predicted gene, 25475 |
2564 |
0.09 |
| chr1_187175642_187176011 | 0.36 |
Spata17 |
spermatogenesis associated 17 |
32729 |
0.12 |
| chr4_21739391_21739542 | 0.36 |
Ccnc |
cyclin C |
11699 |
0.16 |
| chr3_82275838_82275989 | 0.36 |
Map9 |
microtubule-associated protein 9 |
82131 |
0.1 |
| chr18_11714442_11714593 | 0.36 |
Rbbp8 |
retinoblastoma binding protein 8, endonuclease |
7704 |
0.21 |
| chr16_32643661_32643943 | 0.36 |
Tnk2 |
tyrosine kinase, non-receptor, 2 |
72 |
0.96 |
| chr3_98055157_98055314 | 0.36 |
Gm42819 |
predicted gene 42819 |
24548 |
0.16 |
| chr19_4213519_4213699 | 0.36 |
Clcf1 |
cardiotrophin-like cytokine factor 1 |
629 |
0.3 |
| chr19_21107216_21107509 | 0.36 |
4930554I06Rik |
RIKEN cDNA 4930554I06 gene |
2652 |
0.34 |
| chr2_181487353_181487516 | 0.36 |
Abhd16b |
abhydrolase domain containing 16B |
5772 |
0.11 |
| chr1_88165845_88166009 | 0.35 |
Ugt1a5 |
UDP glucuronosyltransferase 1 family, polypeptide A5 |
85 |
0.91 |
| chr1_41555238_41555655 | 0.35 |
Gm28634 |
predicted gene 28634 |
25903 |
0.27 |
| chr10_68355501_68355861 | 0.35 |
4930545H06Rik |
RIKEN cDNA 4930545H06 gene |
34539 |
0.17 |
| chr14_69628830_69628981 | 0.35 |
Gm27177 |
predicted gene 27177 |
11369 |
0.11 |
| chr6_16701529_16701791 | 0.35 |
Gm36669 |
predicted gene, 36669 |
75864 |
0.11 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0061092 | regulation of phospholipid translocation(GO:0061091) positive regulation of phospholipid translocation(GO:0061092) |
| 0.2 | 0.7 | GO:0032417 | positive regulation of sodium:proton antiporter activity(GO:0032417) |
| 0.2 | 0.5 | GO:2000566 | positive regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000566) |
| 0.1 | 0.4 | GO:0010912 | regulation of isomerase activity(GO:0010911) positive regulation of isomerase activity(GO:0010912) |
| 0.1 | 0.4 | GO:0060300 | regulation of cytokine activity(GO:0060300) |
| 0.1 | 0.3 | GO:0021912 | regulation of transcription from RNA polymerase II promoter involved in spinal cord motor neuron fate specification(GO:0021912) |
| 0.1 | 0.4 | GO:0009115 | xanthine catabolic process(GO:0009115) |
| 0.1 | 0.3 | GO:0007161 | calcium-independent cell-matrix adhesion(GO:0007161) |
| 0.1 | 0.2 | GO:0002282 | microglial cell activation involved in immune response(GO:0002282) |
| 0.1 | 0.2 | GO:0061502 | early endosome to recycling endosome transport(GO:0061502) |
| 0.1 | 0.5 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.1 | 0.2 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.1 | 0.3 | GO:1900378 | positive regulation of melanin biosynthetic process(GO:0048023) positive regulation of secondary metabolite biosynthetic process(GO:1900378) |
| 0.1 | 0.2 | GO:0015772 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.1 | 0.4 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.1 | 0.2 | GO:0003289 | atrial septum primum morphogenesis(GO:0003289) |
| 0.1 | 0.2 | GO:1902953 | positive regulation of ER to Golgi vesicle-mediated transport(GO:1902953) |
| 0.1 | 0.3 | GO:0060689 | cell differentiation involved in salivary gland development(GO:0060689) |
| 0.1 | 0.2 | GO:0010482 | epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 0.1 | 0.2 | GO:0097503 | sialylation(GO:0097503) |
| 0.1 | 0.1 | GO:1990705 | cholangiocyte proliferation(GO:1990705) |
| 0.1 | 0.4 | GO:0035811 | negative regulation of urine volume(GO:0035811) |
| 0.1 | 0.3 | GO:0002591 | antigen processing and presentation of endogenous peptide antigen via MHC class Ib via ER pathway(GO:0002488) antigen processing and presentation of endogenous peptide antigen via MHC class Ib via ER pathway, TAP-dependent(GO:0002489) positive regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002591) |
| 0.1 | 0.6 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.1 | 0.4 | GO:0060613 | fat pad development(GO:0060613) |
| 0.0 | 0.1 | GO:0046618 | drug export(GO:0046618) |
| 0.0 | 0.1 | GO:1901492 | positive regulation of lymphangiogenesis(GO:1901492) |
| 0.0 | 0.3 | GO:0045040 | protein import into mitochondrial outer membrane(GO:0045040) |
| 0.0 | 0.2 | GO:0046292 | formaldehyde metabolic process(GO:0046292) |
| 0.0 | 0.2 | GO:0032364 | oxygen homeostasis(GO:0032364) |
| 0.0 | 0.2 | GO:0051790 | short-chain fatty acid biosynthetic process(GO:0051790) |
| 0.0 | 0.2 | GO:0035564 | regulation of kidney size(GO:0035564) |
| 0.0 | 0.1 | GO:0070672 | response to interleukin-15(GO:0070672) |
| 0.0 | 0.1 | GO:0042732 | D-xylose metabolic process(GO:0042732) |
| 0.0 | 0.2 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.0 | 0.1 | GO:0060051 | negative regulation of protein glycosylation(GO:0060051) |
| 0.0 | 0.1 | GO:0075509 | receptor-mediated endocytosis of virus by host cell(GO:0019065) endocytosis involved in viral entry into host cell(GO:0075509) |
| 0.0 | 0.2 | GO:0010825 | positive regulation of centrosome duplication(GO:0010825) |
| 0.0 | 0.1 | GO:0046341 | CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.0 | 0.2 | GO:2000467 | positive regulation of glycogen (starch) synthase activity(GO:2000467) |
| 0.0 | 0.9 | GO:0061014 | positive regulation of mRNA catabolic process(GO:0061014) |
| 0.0 | 0.1 | GO:0010909 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.0 | 0.1 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.1 | GO:0034395 | regulation of transcription from RNA polymerase II promoter in response to iron(GO:0034395) |
| 0.0 | 0.3 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.0 | 0.1 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.0 | 0.1 | GO:0045590 | negative regulation of regulatory T cell differentiation(GO:0045590) |
| 0.0 | 0.1 | GO:0090230 | regulation of centromere complex assembly(GO:0090230) |
| 0.0 | 0.1 | GO:1901859 | negative regulation of mitochondrial DNA metabolic process(GO:1901859) |
| 0.0 | 0.1 | GO:0042997 | negative regulation of Golgi to plasma membrane protein transport(GO:0042997) |
| 0.0 | 0.1 | GO:1902809 | regulation of skeletal muscle fiber differentiation(GO:1902809) |
| 0.0 | 0.1 | GO:0035860 | glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.0 | 0.3 | GO:2000392 | regulation of lamellipodium morphogenesis(GO:2000392) |
| 0.0 | 0.1 | GO:0044375 | regulation of peroxisome size(GO:0044375) |
| 0.0 | 0.1 | GO:0032907 | transforming growth factor beta3 production(GO:0032907) regulation of transforming growth factor beta3 production(GO:0032910) |
| 0.0 | 0.1 | GO:0046499 | S-adenosylmethioninamine metabolic process(GO:0046499) |
| 0.0 | 0.3 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.0 | 0.1 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.1 | GO:0051599 | response to hydrostatic pressure(GO:0051599) |
| 0.0 | 0.1 | GO:0051902 | negative regulation of mitochondrial depolarization(GO:0051902) |
| 0.0 | 0.1 | GO:0033578 | protein glycosylation in Golgi(GO:0033578) |
| 0.0 | 0.2 | GO:0016078 | tRNA catabolic process(GO:0016078) |
| 0.0 | 0.1 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.0 | 0.0 | GO:0001306 | age-dependent response to oxidative stress(GO:0001306) age-dependent general metabolic decline(GO:0007571) |
| 0.0 | 0.2 | GO:1903551 | regulation of extracellular exosome assembly(GO:1903551) |
| 0.0 | 0.1 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.1 | GO:0046552 | eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate commitment(GO:0046552) |
| 0.0 | 0.2 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.1 | GO:0006681 | galactosylceramide metabolic process(GO:0006681) |
| 0.0 | 0.1 | GO:0034499 | late endosome to Golgi transport(GO:0034499) |
| 0.0 | 0.1 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.0 | 0.1 | GO:0086023 | adrenergic receptor signaling pathway involved in heart process(GO:0086023) |
| 0.0 | 0.1 | GO:0071286 | cellular response to magnesium ion(GO:0071286) |
| 0.0 | 0.1 | GO:0030997 | regulation of centriole-centriole cohesion(GO:0030997) |
| 0.0 | 0.2 | GO:0038166 | angiotensin-activated signaling pathway(GO:0038166) |
| 0.0 | 0.1 | GO:0090259 | regulation of retinal ganglion cell axon guidance(GO:0090259) |
| 0.0 | 0.1 | GO:0071635 | negative regulation of transforming growth factor beta production(GO:0071635) |
| 0.0 | 0.1 | GO:0048295 | positive regulation of isotype switching to IgE isotypes(GO:0048295) |
| 0.0 | 0.1 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.0 | 0.2 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.0 | 0.1 | GO:0070350 | white fat cell proliferation(GO:0070343) regulation of white fat cell proliferation(GO:0070350) |
| 0.0 | 0.1 | GO:0016344 | meiotic chromosome movement towards spindle pole(GO:0016344) |
| 0.0 | 0.1 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.0 | 0.1 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.0 | 0.6 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.0 | 0.1 | GO:0036216 | response to stem cell factor(GO:0036215) cellular response to stem cell factor stimulus(GO:0036216) Kit signaling pathway(GO:0038109) |
| 0.0 | 0.2 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.1 | GO:0006546 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.0 | 0.1 | GO:0060971 | embryonic heart tube left/right pattern formation(GO:0060971) |
| 0.0 | 0.1 | GO:0061684 | chaperone-mediated autophagy(GO:0061684) |
| 0.0 | 0.0 | GO:0043134 | hindgut contraction(GO:0043133) regulation of hindgut contraction(GO:0043134) |
| 0.0 | 0.2 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.0 | 0.1 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.0 | 0.0 | GO:0014016 | neuroblast differentiation(GO:0014016) |
| 0.0 | 0.1 | GO:0046881 | positive regulation of follicle-stimulating hormone secretion(GO:0046881) |
| 0.0 | 0.1 | GO:0060083 | smooth muscle contraction involved in micturition(GO:0060083) |
| 0.0 | 0.1 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.0 | 0.1 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.0 | 0.1 | GO:0042536 | negative regulation of tumor necrosis factor biosynthetic process(GO:0042536) |
| 0.0 | 0.1 | GO:0090292 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.0 | GO:0045608 | negative regulation of auditory receptor cell differentiation(GO:0045608) |
| 0.0 | 0.1 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.0 | 0.0 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 0.0 | 0.0 | GO:0090210 | regulation of establishment of blood-brain barrier(GO:0090210) |
| 0.0 | 0.1 | GO:0070537 | histone H2A K63-linked deubiquitination(GO:0070537) |
| 0.0 | 0.0 | GO:0010847 | regulation of chromatin assembly(GO:0010847) |
| 0.0 | 0.1 | GO:1903223 | positive regulation of oxidative stress-induced neuron death(GO:1903223) |
| 0.0 | 0.0 | GO:0072300 | positive regulation of metanephric glomerulus development(GO:0072300) |
| 0.0 | 0.2 | GO:0046838 | phosphorylated carbohydrate dephosphorylation(GO:0046838) inositol phosphate dephosphorylation(GO:0046855) |
| 0.0 | 0.0 | GO:0046391 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.0 | 0.1 | GO:0045901 | positive regulation of translational elongation(GO:0045901) |
| 0.0 | 0.2 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.0 | 0.1 | GO:1901674 | histone H3-K27 acetylation(GO:0043974) regulation of histone H3-K27 acetylation(GO:1901674) |
| 0.0 | 0.1 | GO:0051660 | establishment of centrosome localization(GO:0051660) |
| 0.0 | 0.1 | GO:1902459 | positive regulation of stem cell population maintenance(GO:1902459) |
| 0.0 | 0.1 | GO:2000643 | positive regulation of early endosome to late endosome transport(GO:2000643) |
| 0.0 | 0.0 | GO:0046504 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) cellular lipid biosynthetic process(GO:0097384) ether biosynthetic process(GO:1901503) |
| 0.0 | 0.1 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.0 | 0.0 | GO:0034140 | negative regulation of toll-like receptor 3 signaling pathway(GO:0034140) |
| 0.0 | 0.2 | GO:0060294 | cilium movement involved in cell motility(GO:0060294) |
| 0.0 | 0.2 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.0 | 0.2 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 0.0 | 0.0 | GO:0060700 | regulation of ribonuclease activity(GO:0060700) |
| 0.0 | 0.1 | GO:0021555 | midbrain-hindbrain boundary morphogenesis(GO:0021555) |
| 0.0 | 0.1 | GO:0060160 | negative regulation of dopamine receptor signaling pathway(GO:0060160) |
| 0.0 | 0.1 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.0 | 0.1 | GO:0035633 | maintenance of blood-brain barrier(GO:0035633) |
| 0.0 | 0.2 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.1 | GO:0006991 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.0 | 0.1 | GO:0035336 | long-chain fatty-acyl-CoA metabolic process(GO:0035336) |
| 0.0 | 0.0 | GO:0035262 | gonad morphogenesis(GO:0035262) |
| 0.0 | 0.2 | GO:0031269 | pseudopodium assembly(GO:0031269) |
| 0.0 | 0.0 | GO:1902309 | negative regulation of peptidyl-serine dephosphorylation(GO:1902309) |
| 0.0 | 0.0 | GO:0002904 | positive regulation of B cell apoptotic process(GO:0002904) |
| 0.0 | 0.1 | GO:2000323 | negative regulation of glucocorticoid receptor signaling pathway(GO:2000323) |
| 0.0 | 0.0 | GO:0036112 | medium-chain fatty-acyl-CoA metabolic process(GO:0036112) |
| 0.0 | 0.2 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.0 | 0.2 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.0 | 0.0 | GO:0003166 | bundle of His development(GO:0003166) |
| 0.0 | 0.0 | GO:0033762 | response to glucagon(GO:0033762) |
| 0.0 | 0.1 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 0.0 | 0.1 | GO:0061304 | retinal blood vessel morphogenesis(GO:0061304) |
| 0.0 | 0.1 | GO:0048633 | positive regulation of skeletal muscle tissue growth(GO:0048633) |
| 0.0 | 0.1 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.0 | 0.0 | GO:0018171 | peptidyl-cysteine oxidation(GO:0018171) |
| 0.0 | 0.0 | GO:1990314 | cellular response to insulin-like growth factor stimulus(GO:1990314) |
| 0.0 | 0.1 | GO:0046146 | tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.0 | 0.0 | GO:1900041 | negative regulation of interleukin-2 secretion(GO:1900041) |
| 0.0 | 0.0 | GO:0021938 | smoothened signaling pathway involved in regulation of cerebellar granule cell precursor cell proliferation(GO:0021938) |
| 0.0 | 0.1 | GO:0010801 | negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 0.0 | 0.0 | GO:0030210 | heparin biosynthetic process(GO:0030210) |
| 0.0 | 0.1 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.0 | 0.1 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.0 | 0.0 | GO:0090071 | negative regulation of ribosome biogenesis(GO:0090071) |
| 0.0 | 0.2 | GO:0035879 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) plasma membrane lactate transport(GO:0035879) |
| 0.0 | 0.1 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 0.2 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 0.0 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.0 | 0.7 | GO:0048041 | cell-substrate adherens junction assembly(GO:0007045) focal adhesion assembly(GO:0048041) |
| 0.0 | 0.1 | GO:2000391 | regulation of neutrophil extravasation(GO:2000389) positive regulation of neutrophil extravasation(GO:2000391) |
| 0.0 | 0.0 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 0.1 | GO:0018406 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.0 | 0.1 | GO:0090188 | negative regulation of pancreatic juice secretion(GO:0090188) |
| 0.0 | 0.0 | GO:0021699 | cerebellar cortex maturation(GO:0021699) |
| 0.0 | 0.0 | GO:0006106 | fumarate metabolic process(GO:0006106) |
| 0.0 | 0.0 | GO:0006001 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) glycolytic process through fructose-1-phosphate(GO:0061625) |
| 0.0 | 0.0 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.0 | 0.0 | GO:0032906 | transforming growth factor beta2 production(GO:0032906) regulation of transforming growth factor beta2 production(GO:0032909) |
| 0.0 | 0.1 | GO:0035457 | cellular response to interferon-alpha(GO:0035457) |
| 0.0 | 0.2 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.0 | 0.1 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) |
| 0.0 | 0.1 | GO:0090435 | protein localization to nuclear envelope(GO:0090435) |
| 0.0 | 0.0 | GO:2000667 | positive regulation of interleukin-5 secretion(GO:2000664) positive regulation of interleukin-13 secretion(GO:2000667) |
| 0.0 | 0.1 | GO:0010815 | bradykinin catabolic process(GO:0010815) |
| 0.0 | 0.3 | GO:0019585 | uronic acid metabolic process(GO:0006063) glucuronate metabolic process(GO:0019585) |
| 0.0 | 0.1 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.0 | 0.0 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 0.0 | 0.0 | GO:0070458 | detoxification of nitrogen compound(GO:0051410) cellular detoxification of nitrogen compound(GO:0070458) |
| 0.0 | 0.1 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.0 | 0.0 | GO:1900126 | negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.0 | 0.1 | GO:2001241 | positive regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001241) |
| 0.0 | 0.0 | GO:2001025 | positive regulation of response to drug(GO:2001025) |
| 0.0 | 0.0 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.0 | 0.1 | GO:0042760 | very long-chain fatty acid catabolic process(GO:0042760) |
| 0.0 | 0.0 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 0.0 | 0.1 | GO:0090520 | sphingolipid mediated signaling pathway(GO:0090520) |
| 0.0 | 0.0 | GO:0032513 | negative regulation of protein phosphatase type 2B activity(GO:0032513) |
| 0.0 | 0.1 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) |
| 0.0 | 0.0 | GO:1901727 | positive regulation of histone deacetylase activity(GO:1901727) |
| 0.0 | 0.0 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 0.0 | 0.0 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.0 | 0.0 | GO:0061643 | chemorepulsion of axon(GO:0061643) |
| 0.0 | 0.1 | GO:1904668 | positive regulation of ubiquitin protein ligase activity(GO:1904668) |
| 0.0 | 0.0 | GO:0010871 | negative regulation of receptor biosynthetic process(GO:0010871) |
| 0.0 | 0.0 | GO:0061052 | negative regulation of cell growth involved in cardiac muscle cell development(GO:0061052) |
| 0.0 | 0.0 | GO:0021914 | negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021914) |
| 0.0 | 0.0 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 0.0 | 0.1 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.0 | 0.1 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.2 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
| 0.0 | 0.0 | GO:0002125 | maternal aggressive behavior(GO:0002125) |
| 0.0 | 0.0 | GO:2000418 | positive regulation of eosinophil migration(GO:2000418) |
| 0.0 | 0.0 | GO:0044034 | negative stranded viral RNA replication(GO:0039689) multi-organism biosynthetic process(GO:0044034) |
| 0.0 | 0.1 | GO:0071257 | cellular response to electrical stimulus(GO:0071257) |
| 0.0 | 0.1 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.0 | 0.0 | GO:0007529 | establishment of synaptic specificity at neuromuscular junction(GO:0007529) |
| 0.0 | 0.0 | GO:1900108 | negative regulation of nodal signaling pathway(GO:1900108) |
| 0.0 | 0.0 | GO:0070100 | negative regulation of chemokine-mediated signaling pathway(GO:0070100) |
| 0.0 | 0.0 | GO:0007182 | common-partner SMAD protein phosphorylation(GO:0007182) |
| 0.0 | 0.0 | GO:0097106 | postsynaptic density organization(GO:0097106) postsynaptic density assembly(GO:0097107) |
| 0.0 | 0.0 | GO:0043987 | histone H3-S10 phosphorylation(GO:0043987) |
| 0.0 | 0.0 | GO:0007042 | lysosomal lumen acidification(GO:0007042) |
| 0.0 | 0.0 | GO:0048294 | negative regulation of isotype switching to IgE isotypes(GO:0048294) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.2 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.1 | 0.2 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.1 | 0.2 | GO:0034679 | integrin alpha9-beta1 complex(GO:0034679) |
| 0.1 | 0.3 | GO:0071203 | WASH complex(GO:0071203) |
| 0.1 | 0.2 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.1 | 0.4 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.1 | 0.2 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.1 | 0.2 | GO:0010009 | cytoplasmic side of endosome membrane(GO:0010009) |
| 0.0 | 0.7 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 0.4 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 0.1 | GO:0097543 | ciliary inversin compartment(GO:0097543) |
| 0.0 | 0.5 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.0 | 0.3 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.0 | 0.3 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.1 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.0 | 0.1 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 0.5 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 0.2 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.0 | 0.3 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.1 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 0.1 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.0 | 0.1 | GO:0005767 | secondary lysosome(GO:0005767) |
| 0.0 | 0.1 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.0 | 0.2 | GO:0045179 | apical cortex(GO:0045179) |
| 0.0 | 0.1 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.1 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 0.0 | 0.1 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 0.2 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.1 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.0 | 0.1 | GO:0020018 | ciliary pocket(GO:0020016) ciliary pocket membrane(GO:0020018) |
| 0.0 | 0.0 | GO:0043511 | inhibin complex(GO:0043511) |
| 0.0 | 0.2 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.0 | 0.2 | GO:0036513 | Derlin-1 retrotranslocation complex(GO:0036513) |
| 0.0 | 0.1 | GO:0002142 | stereocilia ankle link complex(GO:0002142) |
| 0.0 | 0.1 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.0 | 0.2 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.1 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.1 | GO:0070187 | telosome(GO:0070187) |
| 0.0 | 0.0 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.0 | 0.1 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.0 | GO:0034750 | Scrib-APC-beta-catenin complex(GO:0034750) |
| 0.0 | 0.1 | GO:0044194 | cytolytic granule(GO:0044194) |
| 0.0 | 0.3 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.1 | GO:0071437 | invadopodium(GO:0071437) |
| 0.0 | 0.2 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.0 | 0.1 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.0 | 0.1 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.0 | 0.0 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.0 | 0.2 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.0 | 0.1 | GO:0060091 | kinocilium(GO:0060091) |
| 0.0 | 0.0 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.0 | 0.1 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 0.1 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.0 | 0.4 | GO:0098636 | protein complex involved in cell adhesion(GO:0098636) |
| 0.0 | 0.1 | GO:0098651 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.0 | 0.1 | GO:0016942 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) insulin-like growth factor ternary complex(GO:0042567) |
| 0.0 | 0.1 | GO:0020003 | symbiont-containing vacuole(GO:0020003) |
| 0.0 | 0.1 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 0.1 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.0 | 0.2 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) |
| 0.0 | 0.0 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.0 | 0.1 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.1 | GO:0000235 | astral microtubule(GO:0000235) |
| 0.0 | 0.1 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.1 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.1 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0008525 | phosphatidylcholine transporter activity(GO:0008525) |
| 0.1 | 0.4 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.1 | 0.3 | GO:0031800 | type 3 metabotropic glutamate receptor binding(GO:0031800) |
| 0.1 | 0.6 | GO:0015198 | oligopeptide transporter activity(GO:0015198) |
| 0.1 | 0.4 | GO:0030151 | molybdenum ion binding(GO:0030151) |
| 0.1 | 0.4 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.1 | 0.6 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.1 | 0.2 | GO:0016823 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.1 | 0.2 | GO:0015154 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.1 | 0.2 | GO:0000171 | ribonuclease MRP activity(GO:0000171) |
| 0.1 | 0.2 | GO:0031735 | CCR10 chemokine receptor binding(GO:0031735) |
| 0.1 | 0.2 | GO:0016508 | long-chain-enoyl-CoA hydratase activity(GO:0016508) |
| 0.1 | 0.4 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.0 | 0.6 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.0 | 0.2 | GO:0004945 | angiotensin type II receptor activity(GO:0004945) |
| 0.0 | 0.2 | GO:0019960 | C-X3-C chemokine binding(GO:0019960) |
| 0.0 | 0.2 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.0 | 0.1 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.0 | 0.2 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.0 | 0.1 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.3 | GO:0036122 | BMP binding(GO:0036122) |
| 0.0 | 0.8 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.1 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) DNA (cytosine-5-)-methyltransferase activity, acting on CpG substrates(GO:0051718) |
| 0.0 | 0.2 | GO:0047391 | alkylglycerophosphoethanolamine phosphodiesterase activity(GO:0047391) |
| 0.0 | 0.1 | GO:0050309 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.0 | 0.2 | GO:0034889 | alkyl sulfatase activity(GO:0018741) endosulfan hemisulfate sulfatase activity(GO:0034889) endosulfan sulfate hydrolase activity(GO:0034902) |
| 0.0 | 0.1 | GO:0070996 | type 1 melanocortin receptor binding(GO:0070996) |
| 0.0 | 0.1 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.0 | 0.3 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.0 | 0.1 | GO:0046848 | hydroxyapatite binding(GO:0046848) |
| 0.0 | 0.1 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 0.1 | GO:0017002 | activin-activated receptor activity(GO:0017002) |
| 0.0 | 0.1 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 0.1 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 0.0 | 0.1 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.0 | 0.1 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.1 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 0.3 | GO:0015266 | protein channel activity(GO:0015266) |
| 0.0 | 0.1 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.0 | 0.1 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 0.0 | 0.1 | GO:0033695 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 0.0 | 0.1 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.0 | 0.1 | GO:0015111 | iodide transmembrane transporter activity(GO:0015111) |
| 0.0 | 0.1 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.5 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.2 | GO:0001091 | RNA polymerase II basal transcription factor binding(GO:0001091) |
| 0.0 | 0.1 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.0 | 0.1 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.0 | 0.1 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.0 | 0.1 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.0 | 0.1 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.0 | 0.1 | GO:0043398 | HLH domain binding(GO:0043398) |
| 0.0 | 0.1 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.0 | 0.9 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 0.1 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.0 | 0.2 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.1 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 0.0 | 0.1 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.0 | 0.1 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.0 | 0.0 | GO:0004698 | calcium-dependent protein kinase C activity(GO:0004698) |
| 0.0 | 0.1 | GO:0005072 | transforming growth factor beta receptor, cytoplasmic mediator activity(GO:0005072) |
| 0.0 | 0.2 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.0 | 0.1 | GO:0052833 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.0 | 0.1 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.0 | 0.1 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.0 | 0.0 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.0 | 0.5 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 0.1 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.1 | GO:0098518 | polynucleotide phosphatase activity(GO:0098518) |
| 0.0 | 0.1 | GO:0050649 | testosterone 6-beta-hydroxylase activity(GO:0050649) |
| 0.0 | 0.2 | GO:0016920 | pyroglutamyl-peptidase activity(GO:0016920) |
| 0.0 | 0.1 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.0 | 0.2 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.1 | GO:0005290 | L-histidine transmembrane transporter activity(GO:0005290) |
| 0.0 | 0.1 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 0.1 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 0.1 | GO:0005534 | galactose binding(GO:0005534) |
| 0.0 | 0.1 | GO:0031748 | D1 dopamine receptor binding(GO:0031748) |
| 0.0 | 0.0 | GO:0070905 | serine binding(GO:0070905) |
| 0.0 | 0.2 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.1 | GO:0015238 | drug transmembrane transporter activity(GO:0015238) |
| 0.0 | 0.2 | GO:0051765 | inositol tetrakisphosphate kinase activity(GO:0051765) |
| 0.0 | 0.1 | GO:0031750 | D3 dopamine receptor binding(GO:0031750) |
| 0.0 | 0.1 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.0 | 0.0 | GO:0036435 | K48-linked polyubiquitin binding(GO:0036435) |
| 0.0 | 0.1 | GO:0003840 | gamma-glutamyltransferase activity(GO:0003840) glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.1 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 0.1 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) |
| 0.0 | 0.1 | GO:0016661 | oxidoreductase activity, acting on other nitrogenous compounds as donors(GO:0016661) |
| 0.0 | 0.0 | GO:0030519 | snoRNP binding(GO:0030519) |
| 0.0 | 0.2 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 0.2 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 0.1 | GO:0008312 | 7S RNA binding(GO:0008312) |
| 0.0 | 0.0 | GO:0035514 | DNA demethylase activity(GO:0035514) |
| 0.0 | 0.2 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.0 | 0.1 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
| 0.0 | 0.1 | GO:1990932 | 5.8S rRNA binding(GO:1990932) |
| 0.0 | 0.1 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.0 | 0.1 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 0.0 | 0.1 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.0 | 0.1 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.0 | 0.2 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.1 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.0 | 0.1 | GO:0002060 | purine nucleobase binding(GO:0002060) |
| 0.0 | 0.0 | GO:0004104 | cholinesterase activity(GO:0004104) |
| 0.0 | 0.1 | GO:0004030 | aldehyde dehydrogenase [NAD(P)+] activity(GO:0004030) |
| 0.0 | 0.0 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.0 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.0 | 0.0 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.0 | 0.1 | GO:0048531 | beta-1,3-galactosyltransferase activity(GO:0048531) |
| 0.0 | 0.0 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.0 | 0.1 | GO:0035252 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 0.0 | 0.6 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.0 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.0 | 0.1 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.0 | 0.0 | GO:0070139 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.0 | 0.5 | GO:0050694 | galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.0 | 0.0 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.0 | GO:0005415 | nucleoside:sodium symporter activity(GO:0005415) |
| 0.0 | 0.1 | GO:0038085 | vascular endothelial growth factor binding(GO:0038085) |
| 0.0 | 0.0 | GO:0045340 | mercury ion binding(GO:0045340) |
| 0.0 | 0.1 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 0.3 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.1 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.0 | 0.0 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 0.0 | GO:0047105 | aminobutyraldehyde dehydrogenase activity(GO:0019145) 4-trimethylammoniobutyraldehyde dehydrogenase activity(GO:0047105) |
| 0.0 | 0.1 | GO:0005536 | glucose binding(GO:0005536) |
| 0.0 | 0.0 | GO:0001031 | RNA polymerase III type 1 promoter DNA binding(GO:0001030) RNA polymerase III type 2 promoter DNA binding(GO:0001031) RNA polymerase III type 3 promoter DNA binding(GO:0001032) 5S rDNA binding(GO:0080084) |
| 0.0 | 0.0 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.0 | 0.2 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 0.1 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.0 | 0.0 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.0 | 0.1 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.0 | 0.1 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.0 | 0.0 | GO:0005345 | purine nucleobase transmembrane transporter activity(GO:0005345) pyrimidine nucleobase transmembrane transporter activity(GO:0005350) nucleobase transmembrane transporter activity(GO:0015205) |
| 0.0 | 0.1 | GO:0004630 | phospholipase D activity(GO:0004630) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.6 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.1 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.0 | 0.2 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.5 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.1 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.0 | 0.1 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 0.5 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.0 | 0.2 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.1 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.1 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 0.0 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 0.1 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 0.1 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 0.2 | ST GAQ PATHWAY | G alpha q Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.4 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.0 | 0.4 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.3 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.4 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.3 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.2 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.4 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.0 | 0.1 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.2 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 0.4 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.1 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.1 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.1 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.3 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.0 | 0.2 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.1 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 0.0 | 0.1 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 0.3 | REACTOME SEMA4D IN SEMAPHORIN SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.0 | 0.1 | REACTOME TRAF6 MEDIATED INDUCTION OF TAK1 COMPLEX | Genes involved in TRAF6 mediated induction of TAK1 complex |
| 0.0 | 0.1 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.1 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.1 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.2 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.1 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 0.0 | REACTOME CDT1 ASSOCIATION WITH THE CDC6 ORC ORIGIN COMPLEX | Genes involved in CDT1 association with the CDC6:ORC:origin complex |
| 0.0 | 0.1 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.1 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.1 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |