Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
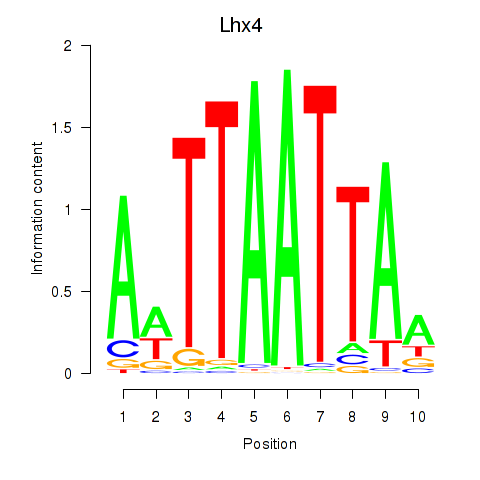
Results for Lhx4
Z-value: 0.70
Transcription factors associated with Lhx4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Lhx4
|
ENSMUSG00000026468.8 | LIM homeobox protein 4 |
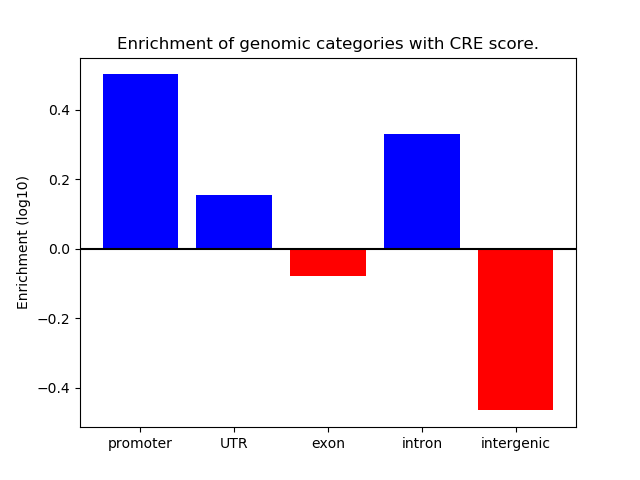
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr1_155721550_155721719 | Lhx4 | 20393 | 0.145526 | 0.45 | 3.7e-01 | Click! |
| chr1_155720147_155720298 | Lhx4 | 21805 | 0.143623 | -0.01 | 9.9e-01 | Click! |
Activity of the Lhx4 motif across conditions
Conditions sorted by the z-value of the Lhx4 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
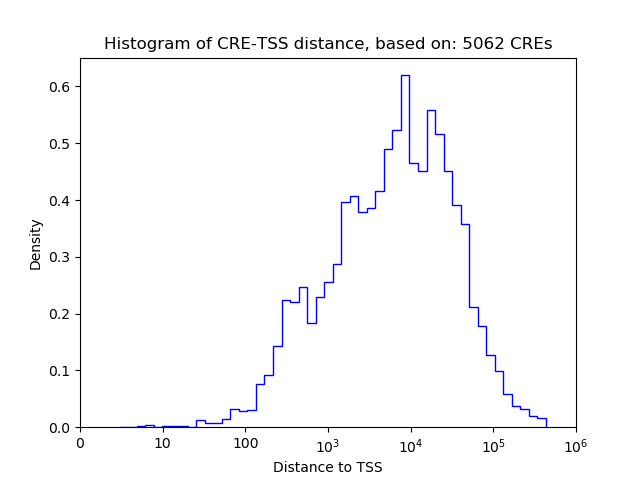
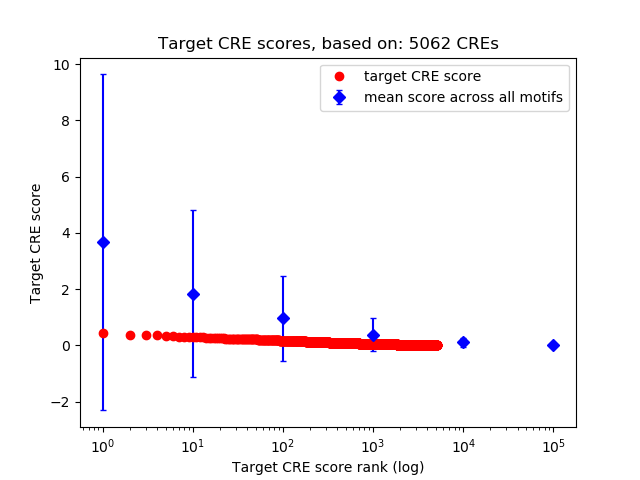
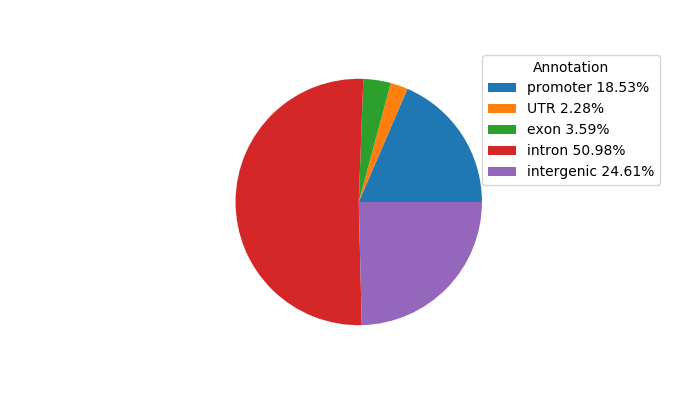
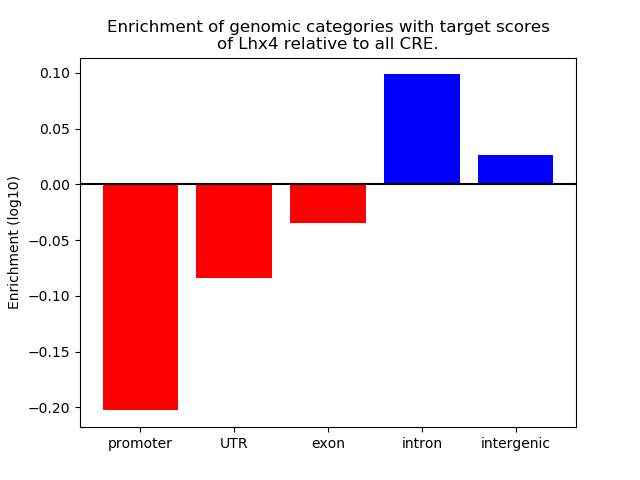
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr1_39133288_39133558 | 0.43 |
Gm37091 |
predicted gene, 37091 |
50085 |
0.11 |
| chr5_99976807_99976958 | 0.36 |
Hnrnpd |
heterogeneous nuclear ribonucleoprotein D |
331 |
0.73 |
| chr5_22506386_22506555 | 0.36 |
Orc5 |
origin recognition complex, subunit 5 |
16196 |
0.12 |
| chr2_34049474_34049625 | 0.35 |
C230014O12Rik |
RIKEN cDNA C230014O12 gene |
58180 |
0.11 |
| chr4_88889115_88889274 | 0.32 |
Ifne |
interferon epsilon |
8993 |
0.08 |
| chr9_106264996_106265377 | 0.32 |
Poc1a |
POC1 centriolar protein A |
15875 |
0.1 |
| chr10_125785978_125786129 | 0.31 |
Lrig3 |
leucine-rich repeats and immunoglobulin-like domains 3 |
180115 |
0.03 |
| chr2_31519719_31520357 | 0.31 |
Ass1 |
argininosuccinate synthetase 1 |
1548 |
0.36 |
| chr10_69208679_69208830 | 0.30 |
Rhobtb1 |
Rho-related BTB domain containing 1 |
202 |
0.94 |
| chr4_76367194_76367568 | 0.29 |
Ptprd |
protein tyrosine phosphatase, receptor type, D |
23138 |
0.21 |
| chr7_118151770_118151921 | 0.29 |
Gm25683 |
predicted gene, 25683 |
1635 |
0.32 |
| chr15_61562347_61562502 | 0.28 |
Gm49498 |
predicted gene, 49498 |
18241 |
0.28 |
| chr19_20628055_20628251 | 0.28 |
Aldh1a1 |
aldehyde dehydrogenase family 1, subfamily A1 |
26192 |
0.19 |
| chr1_84128867_84129018 | 0.28 |
Gm37058 |
predicted gene, 37058 |
5568 |
0.25 |
| chr5_20590046_20590233 | 0.28 |
Gm3544 |
predicted gene 3544 |
1543 |
0.45 |
| chr14_30530847_30531018 | 0.27 |
Dcp1a |
decapping mRNA 1A |
7225 |
0.15 |
| chr6_108836766_108836917 | 0.27 |
Edem1 |
ER degradation enhancer, mannosidase alpha-like 1 |
431 |
0.86 |
| chr11_35732740_35732891 | 0.26 |
Pank3 |
pantothenate kinase 3 |
36669 |
0.14 |
| chr6_100267052_100267222 | 0.26 |
Gm44044 |
predicted gene, 44044 |
6561 |
0.17 |
| chr19_7500165_7500363 | 0.25 |
Atl3 |
atlastin GTPase 3 |
5729 |
0.15 |
| chr2_154875946_154876097 | 0.25 |
Eif2s2 |
eukaryotic translation initiation factor 2, subunit 2 (beta) |
16761 |
0.19 |
| chr6_72120521_72121047 | 0.25 |
4933431G14Rik |
RIKEN cDNA 4933431G14 gene |
2156 |
0.2 |
| chr17_66754427_66754593 | 0.24 |
Gm47471 |
predicted gene, 47471 |
15756 |
0.14 |
| chr1_67222842_67222998 | 0.24 |
Gm15668 |
predicted gene 15668 |
26280 |
0.19 |
| chr10_37324159_37324310 | 0.23 |
Gm26535 |
predicted gene, 26535 |
13252 |
0.25 |
| chr10_89573725_89573906 | 0.23 |
Gm48087 |
predicted gene, 48087 |
18096 |
0.17 |
| chr9_55280024_55280330 | 0.23 |
Nrg4 |
neuregulin 4 |
3395 |
0.23 |
| chr11_21121403_21121623 | 0.23 |
Peli1 |
pellino 1 |
13956 |
0.18 |
| chr17_32257244_32257401 | 0.23 |
Brd4 |
bromodomain containing 4 |
15043 |
0.12 |
| chrX_49489499_49489695 | 0.23 |
Arhgap36 |
Rho GTPase activating protein 36 |
3922 |
0.27 |
| chr6_29571610_29572022 | 0.23 |
Tnpo3 |
transportin 3 |
297 |
0.87 |
| chr13_107645708_107645859 | 0.22 |
Gm32090 |
predicted gene, 32090 |
32934 |
0.17 |
| chr2_72309170_72309345 | 0.22 |
Map3k20 |
mitogen-activated protein kinase kinase kinase 20 |
11356 |
0.19 |
| chr10_87040962_87041138 | 0.22 |
1700113H08Rik |
RIKEN cDNA 1700113H08 gene |
16995 |
0.16 |
| chr15_6218493_6218848 | 0.22 |
Gm23139 |
predicted gene, 23139 |
67322 |
0.11 |
| chr4_63773592_63773743 | 0.22 |
Tnfsf15 |
tumor necrosis factor (ligand) superfamily, member 15 |
28554 |
0.18 |
| chr11_31875210_31875374 | 0.22 |
Cpeb4 |
cytoplasmic polyadenylation element binding protein 4 |
2017 |
0.35 |
| chr8_105069434_105069585 | 0.22 |
Ces3b |
carboxylesterase 3B |
14244 |
0.09 |
| chr3_60097287_60097456 | 0.22 |
Sucnr1 |
succinate receptor 1 |
15469 |
0.18 |
| chr2_70666883_70667034 | 0.22 |
Gorasp2 |
golgi reassembly stacking protein 2 |
4758 |
0.18 |
| chr1_100298746_100298897 | 0.22 |
Gm29667 |
predicted gene 29667 |
16030 |
0.18 |
| chr12_51274477_51274668 | 0.22 |
Rps11-ps4 |
ribosomal protein S11, pseudogene 4 |
22915 |
0.22 |
| chr5_90364825_90365036 | 0.22 |
Ankrd17 |
ankyrin repeat domain 17 |
1255 |
0.37 |
| chr4_123990393_123990580 | 0.22 |
Gm12902 |
predicted gene 12902 |
64252 |
0.08 |
| chr6_72897626_72897819 | 0.22 |
Kcmf1 |
potassium channel modulatory factor 1 |
1361 |
0.34 |
| chr19_60570173_60570324 | 0.21 |
Gm25238 |
predicted gene, 25238 |
10281 |
0.18 |
| chr6_49217272_49217620 | 0.21 |
Igf2bp3 |
insulin-like growth factor 2 mRNA binding protein 3 |
2489 |
0.23 |
| chr9_118689127_118689278 | 0.21 |
Itga9 |
integrin alpha 9 |
3367 |
0.3 |
| chr10_22004298_22004461 | 0.21 |
Sgk1 |
serum/glucocorticoid regulated kinase 1 |
7788 |
0.15 |
| chr2_127608341_127608498 | 0.21 |
Mrps5 |
mitochondrial ribosomal protein S5 |
8386 |
0.13 |
| chr1_67213641_67214357 | 0.21 |
Gm15668 |
predicted gene 15668 |
35201 |
0.17 |
| chr8_93189262_93189430 | 0.21 |
Gm45909 |
predicted gene 45909 |
2012 |
0.24 |
| chr19_27328502_27328670 | 0.21 |
Kcnv2 |
potassium channel, subfamily V, member 2 |
5998 |
0.19 |
| chr12_73341772_73342027 | 0.20 |
Slc38a6 |
solute carrier family 38, member 6 |
2468 |
0.25 |
| chr6_141634890_141635041 | 0.20 |
Slco1b2 |
solute carrier organic anion transporter family, member 1b2 |
507 |
0.86 |
| chr11_90184166_90184514 | 0.20 |
n-R5s72 |
nuclear encoded rRNA 5S 72 |
787 |
0.57 |
| chr1_77168235_77168428 | 0.20 |
Gm38265 |
predicted gene, 38265 |
21339 |
0.22 |
| chr11_28696253_28696446 | 0.19 |
2810471M01Rik |
RIKEN cDNA 2810471M01 gene |
14785 |
0.17 |
| chr6_101095542_101095693 | 0.19 |
Gm43948 |
predicted gene, 43948 |
36446 |
0.13 |
| chr9_9091904_9092129 | 0.19 |
Gm16485 |
predicted gene 16485 |
120225 |
0.06 |
| chr1_73954752_73954963 | 0.19 |
Tns1 |
tensin 1 |
8186 |
0.24 |
| chr11_98339208_98339359 | 0.19 |
Ppp1r1b |
protein phosphatase 1, regulatory inhibitor subunit 1B |
9121 |
0.09 |
| chr3_131269432_131269662 | 0.19 |
Hadh |
hydroxyacyl-Coenzyme A dehydrogenase |
2477 |
0.24 |
| chr1_127787130_127787281 | 0.19 |
Ccnt2 |
cyclin T2 |
9366 |
0.16 |
| chr4_131876493_131876847 | 0.19 |
A930004J17Rik |
RIKEN cDNA A930004J17 gene |
1958 |
0.19 |
| chr10_42211682_42211846 | 0.18 |
Foxo3 |
forkhead box O3 |
46602 |
0.16 |
| chr2_38823684_38823849 | 0.18 |
Nr6a1os |
nuclear receptor subfamily 6, group A, member 1, opposite strand |
425 |
0.74 |
| chr4_32300145_32300312 | 0.18 |
Bach2it1 |
BTB and CNC homology 2, intronic transcript 1 |
48467 |
0.14 |
| chr3_118588032_118588183 | 0.18 |
Dpyd |
dihydropyrimidine dehydrogenase |
25921 |
0.17 |
| chr4_134814659_134814853 | 0.18 |
Maco1 |
macoilin 1 |
3543 |
0.24 |
| chr19_31886956_31887132 | 0.18 |
A1cf |
APOBEC1 complementation factor |
18263 |
0.2 |
| chr9_54139319_54139478 | 0.18 |
Cyp19a1 |
cytochrome P450, family 19, subfamily a, polypeptide 1 |
42676 |
0.14 |
| chr1_183438610_183438761 | 0.18 |
Gm37214 |
predicted gene, 37214 |
18030 |
0.13 |
| chr3_158035265_158035416 | 0.18 |
Gm43362 |
predicted gene 43362 |
1069 |
0.28 |
| chr12_79436402_79436553 | 0.18 |
Rad51b |
RAD51 paralog B |
109124 |
0.06 |
| chr17_10187170_10187336 | 0.18 |
Qk |
quaking |
26321 |
0.25 |
| chr7_112825614_112825993 | 0.18 |
Tead1 |
TEA domain family member 1 |
13831 |
0.24 |
| chr4_123419684_123419835 | 0.18 |
Macf1 |
microtubule-actin crosslinking factor 1 |
7531 |
0.17 |
| chr3_70400102_70400273 | 0.18 |
Gm6631 |
predicted gene 6631 |
13132 |
0.29 |
| chr4_76435923_76436234 | 0.18 |
Ptprd |
protein tyrosine phosphatase, receptor type, D |
13902 |
0.21 |
| chr2_155378239_155378390 | 0.18 |
Trp53inp2 |
transformation related protein 53 inducible nuclear protein 2 |
2745 |
0.19 |
| chr19_55891664_55891815 | 0.18 |
Tcf7l2 |
transcription factor 7 like 2, T cell specific, HMG box |
1613 |
0.48 |
| chr3_108701624_108701775 | 0.18 |
Gpsm2 |
G-protein signalling modulator 2 (AGS3-like, C. elegans) |
814 |
0.52 |
| chr16_37606843_37607024 | 0.17 |
Gm46559 |
predicted gene, 46559 |
10743 |
0.15 |
| chr10_87936931_87937082 | 0.17 |
Tyms-ps |
thymidylate synthase, pseudogene |
29841 |
0.15 |
| chr5_87130645_87130796 | 0.17 |
Ugt2b5 |
UDP glucuronosyltransferase 2 family, polypeptide B5 |
9598 |
0.12 |
| chr9_115270461_115270612 | 0.17 |
Stmn1-rs1 |
stathmin 1, related sequence 1 |
11445 |
0.17 |
| chr13_31779124_31779297 | 0.17 |
Gm11379 |
predicted gene 11379 |
18329 |
0.16 |
| chr3_131485378_131485553 | 0.17 |
Sgms2 |
sphingomyelin synthase 2 |
5014 |
0.28 |
| chr19_39287276_39287531 | 0.17 |
Cyp2c29 |
cytochrome P450, family 2, subfamily c, polypeptide 29 |
329 |
0.91 |
| chr17_80022007_80022158 | 0.17 |
Gm22215 |
predicted gene, 22215 |
11432 |
0.14 |
| chr16_43375815_43375981 | 0.17 |
Zbtb20 |
zinc finger and BTB domain containing 20 |
11694 |
0.17 |
| chr15_12577123_12577293 | 0.17 |
Gm2606 |
predicted pseudogene 2606 |
2319 |
0.26 |
| chr8_89407033_89407242 | 0.17 |
Gm26331 |
predicted gene, 26331 |
182404 |
0.03 |
| chr6_13654409_13654594 | 0.17 |
Bmt2 |
base methyltransferase of 25S rRNA 2 |
23437 |
0.21 |
| chr4_63738674_63738850 | 0.17 |
Tnfsf15 |
tumor necrosis factor (ligand) superfamily, member 15 |
6351 |
0.22 |
| chr17_51511001_51511172 | 0.17 |
Gm31143 |
predicted gene, 31143 |
23772 |
0.19 |
| chr6_71999696_71999905 | 0.17 |
Gm26628 |
predicted gene, 26628 |
36025 |
0.1 |
| chr19_39460982_39461329 | 0.17 |
Cyp2c38 |
cytochrome P450, family 2, subfamily c, polypeptide 38 |
1920 |
0.42 |
| chr15_3721208_3721477 | 0.17 |
Gm4823 |
predicted gene 4823 |
25533 |
0.23 |
| chr17_79018863_79019084 | 0.17 |
Prkd3 |
protein kinase D3 |
1804 |
0.3 |
| chr13_102625798_102626135 | 0.17 |
Gm47014 |
predicted gene, 47014 |
21433 |
0.2 |
| chr2_48981052_48981464 | 0.16 |
Orc4 |
origin recognition complex, subunit 4 |
30981 |
0.18 |
| chr10_56437367_56437519 | 0.16 |
4930452L12Rik |
RIKEN cDNA 4930452L12 gene |
160 |
0.95 |
| chr3_108496483_108496708 | 0.16 |
Gm23336 |
predicted gene, 23336 |
1523 |
0.23 |
| chr14_64444658_64444809 | 0.16 |
Msra |
methionine sulfoxide reductase A |
3816 |
0.31 |
| chr11_77770194_77770550 | 0.16 |
Myo18a |
myosin XVIIIA |
1779 |
0.27 |
| chr1_124069161_124069328 | 0.16 |
Dpp10 |
dipeptidylpeptidase 10 |
23685 |
0.27 |
| chr19_48883437_48883771 | 0.16 |
Gm50436 |
predicted gene, 50436 |
72468 |
0.11 |
| chr4_57160943_57161117 | 0.16 |
Gm12530 |
predicted gene 12530 |
11041 |
0.2 |
| chr12_55420887_55421038 | 0.16 |
Psma6 |
proteasome subunit alpha 6 |
13579 |
0.16 |
| chr3_52271842_52271993 | 0.16 |
Gm20402 |
predicted gene 20402 |
2482 |
0.19 |
| chr5_86919547_86919811 | 0.16 |
Gm25211 |
predicted gene, 25211 |
3357 |
0.13 |
| chr1_15616194_15616345 | 0.16 |
Gm37138 |
predicted gene, 37138 |
60020 |
0.12 |
| chr9_15301981_15302132 | 0.16 |
4931406C07Rik |
RIKEN cDNA 4931406C07 gene |
500 |
0.46 |
| chr17_28283665_28283835 | 0.16 |
Ppard |
peroxisome proliferator activator receptor delta |
9496 |
0.11 |
| chr16_24892958_24893109 | 0.16 |
Gm22672 |
predicted gene, 22672 |
8860 |
0.25 |
| chr19_8373854_8374041 | 0.16 |
Slc22a30 |
solute carrier family 22, member 30 |
31113 |
0.16 |
| chr2_124852123_124852274 | 0.16 |
Gm13994 |
predicted gene 13994 |
143543 |
0.04 |
| chr6_41044689_41044854 | 0.16 |
2210010C04Rik |
RIKEN cDNA 2210010C04 gene |
9262 |
0.07 |
| chr2_136449372_136449559 | 0.16 |
Gm14054 |
predicted gene 14054 |
46043 |
0.13 |
| chr6_15283034_15283262 | 0.16 |
Foxp2 |
forkhead box P2 |
28865 |
0.25 |
| chr17_89653592_89653743 | 0.16 |
Gm50061 |
predicted gene, 50061 |
98859 |
0.09 |
| chr18_10707142_10707343 | 0.16 |
4930563E18Rik |
RIKEN cDNA 4930563E18 gene |
382 |
0.52 |
| chr1_13715965_13716146 | 0.16 |
Xkr9 |
X-linked Kx blood group related 9 |
47284 |
0.12 |
| chr4_15029409_15029632 | 0.16 |
Gm11844 |
predicted gene 11844 |
6938 |
0.27 |
| chr3_138223407_138223558 | 0.16 |
Adh7 |
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
2011 |
0.23 |
| chr10_87541080_87541552 | 0.16 |
Pah |
phenylalanine hydroxylase |
5357 |
0.23 |
| chr5_90322868_90323041 | 0.16 |
Ankrd17 |
ankyrin repeat domain 17 |
16789 |
0.19 |
| chr5_123975512_123975863 | 0.16 |
Hip1r |
huntingtin interacting protein 1 related |
2059 |
0.19 |
| chr10_12645976_12646209 | 0.16 |
Utrn |
utrophin |
31261 |
0.22 |
| chr13_62872928_62873142 | 0.16 |
Fbp1 |
fructose bisphosphatase 1 |
277 |
0.87 |
| chr8_35559883_35560059 | 0.16 |
Mfhas1 |
malignant fibrous histiocytoma amplified sequence 1 |
27827 |
0.15 |
| chr8_90907363_90907534 | 0.16 |
Gm6658 |
predicted gene 6658 |
967 |
0.37 |
| chr7_71467703_71467871 | 0.16 |
Gm29328 |
predicted gene 29328 |
97454 |
0.07 |
| chr11_88277047_88277211 | 0.16 |
Ccdc182 |
coiled-coil domain containing 182 |
16929 |
0.15 |
| chr10_128978991_128979196 | 0.15 |
9030616G12Rik |
RIKEN cDNA 9030616G12 gene |
7896 |
0.09 |
| chr1_80334794_80334975 | 0.15 |
Cul3 |
cullin 3 |
5395 |
0.16 |
| chr14_7835138_7835309 | 0.15 |
Flnb |
filamin, beta |
17266 |
0.15 |
| chr3_133750379_133750596 | 0.15 |
Gm6135 |
prediticted gene 6135 |
41017 |
0.15 |
| chr5_45491887_45492071 | 0.15 |
Lap3 |
leucine aminopeptidase 3 |
1395 |
0.31 |
| chr5_25247792_25247943 | 0.15 |
Galnt11 |
polypeptide N-acetylgalactosaminyltransferase 11 |
504 |
0.76 |
| chr16_70352273_70352442 | 0.15 |
Gbe1 |
glucan (1,4-alpha-), branching enzyme 1 |
38223 |
0.19 |
| chr7_115674458_115674622 | 0.15 |
Sox6 |
SRY (sex determining region Y)-box 6 |
12175 |
0.3 |
| chr13_58209928_58210079 | 0.15 |
Ubqln1 |
ubiquilin 1 |
5616 |
0.11 |
| chr5_40908767_40908928 | 0.15 |
4930519E07Rik |
RIKEN cDNA 4930519E07 gene |
372389 |
0.01 |
| chr9_7216362_7216525 | 0.15 |
Dcun1d5 |
DCN1, defective in cullin neddylation 1, domain containing 5 (S. cerevisiae) |
12958 |
0.17 |
| chr17_84990758_84990958 | 0.15 |
Ppm1b |
protein phosphatase 1B, magnesium dependent, beta isoform |
2815 |
0.25 |
| chr13_115653330_115653690 | 0.15 |
Gm47892 |
predicted gene, 47892 |
65989 |
0.13 |
| chr5_150650487_150650670 | 0.14 |
N4bp2l2 |
NEDD4 binding protein 2-like 2 |
2317 |
0.19 |
| chr10_117123042_117123236 | 0.14 |
Frs2 |
fibroblast growth factor receptor substrate 2 |
7707 |
0.15 |
| chr1_91258114_91258381 | 0.14 |
Ube2f |
ubiquitin-conjugating enzyme E2F (putative) |
6340 |
0.15 |
| chr14_21599276_21599451 | 0.14 |
Kat6b |
K(lysine) acetyltransferase 6B |
23179 |
0.21 |
| chr12_12940858_12941058 | 0.14 |
Mycn |
v-myc avian myelocytomatosis viral related oncogene, neuroblastoma derived |
342 |
0.83 |
| chr3_69115488_69115663 | 0.14 |
Gm37558 |
predicted gene, 37558 |
7149 |
0.14 |
| chr8_110644559_110644860 | 0.14 |
Vac14 |
Vac14 homolog (S. cerevisiae) |
871 |
0.63 |
| chr15_59231191_59231350 | 0.14 |
Gm7691 |
predicted gene 7691 |
7219 |
0.19 |
| chr5_53656682_53656871 | 0.14 |
Rbpj |
recombination signal binding protein for immunoglobulin kappa J region |
11038 |
0.24 |
| chr11_80247133_80247304 | 0.14 |
Rhot1 |
ras homolog family member T1 |
2456 |
0.28 |
| chr17_28428613_28428954 | 0.14 |
Fkbp5 |
FK506 binding protein 5 |
154 |
0.91 |
| chr10_28178649_28178847 | 0.14 |
Gm22370 |
predicted gene, 22370 |
35373 |
0.21 |
| chr18_65154932_65155123 | 0.14 |
Nedd4l |
neural precursor cell expressed, developmentally down-regulated gene 4-like |
707 |
0.74 |
| chr4_40698726_40698877 | 0.14 |
Aptx |
aprataxin |
3915 |
0.15 |
| chr4_72144165_72144326 | 0.14 |
Tle1 |
transducin-like enhancer of split 1 |
2932 |
0.31 |
| chr11_6025571_6025816 | 0.14 |
Camk2b |
calcium/calmodulin-dependent protein kinase II, beta |
25245 |
0.15 |
| chr16_72808058_72808236 | 0.14 |
Robo1 |
roundabout guidance receptor 1 |
144943 |
0.05 |
| chr16_24449611_24449762 | 0.14 |
Lpp |
LIM domain containing preferred translocation partner in lipoma |
1595 |
0.42 |
| chr12_83926450_83926603 | 0.14 |
Numb |
NUMB endocytic adaptor protein |
4592 |
0.13 |
| chr1_172700535_172700739 | 0.14 |
Crp |
C-reactive protein, pentraxin-related |
2581 |
0.25 |
| chr5_73970292_73970468 | 0.14 |
C78283 |
expressed sequence C78283 |
23999 |
0.14 |
| chr12_86641903_86642073 | 0.14 |
Gm8504 |
predicted gene 8504 |
8585 |
0.17 |
| chr3_65950089_65950240 | 0.14 |
Ccnl1 |
cyclin L1 |
814 |
0.48 |
| chr7_64186221_64186468 | 0.14 |
Trpm1 |
transient receptor potential cation channel, subfamily M, member 1 |
743 |
0.57 |
| chr1_119646750_119646988 | 0.14 |
Epb41l5 |
erythrocyte membrane protein band 4.1 like 5 |
1748 |
0.32 |
| chr6_8862762_8862913 | 0.14 |
Ica1 |
islet cell autoantigen 1 |
84349 |
0.1 |
| chr11_100975850_100976001 | 0.14 |
Cavin1 |
caveolae associated 1 |
5038 |
0.14 |
| chr14_62736570_62736721 | 0.14 |
Ints6 |
integrator complex subunit 6 |
24467 |
0.14 |
| chr2_144369139_144369688 | 0.14 |
Kat14 |
lysine acetyltransferase 14 |
414 |
0.43 |
| chr4_124809297_124809476 | 0.14 |
Mtf1 |
metal response element binding transcription factor 1 |
6708 |
0.1 |
| chr2_69724299_69724450 | 0.13 |
Ppig |
peptidyl-prolyl isomerase G (cyclophilin G) |
1048 |
0.4 |
| chr2_73276147_73276506 | 0.13 |
Sp9 |
trans-acting transcription factor 9 |
4360 |
0.18 |
| chr5_122060223_122060392 | 0.13 |
Cux2 |
cut-like homeobox 2 |
10205 |
0.14 |
| chr8_90945191_90945444 | 0.13 |
Chd9 |
chromodomain helicase DNA binding protein 9 |
9641 |
0.17 |
| chr1_9985468_9985649 | 0.13 |
Ppp1r42 |
protein phosphatase 1, regulatory subunit 42 |
134 |
0.92 |
| chr18_51150358_51150691 | 0.13 |
Prr16 |
proline rich 16 |
32786 |
0.23 |
| chr1_23258293_23258464 | 0.13 |
Gm6420 |
predicted gene 6420 |
1971 |
0.22 |
| chr5_86114424_86114626 | 0.13 |
Stap1 |
signal transducing adaptor family member 1 |
28067 |
0.13 |
| chr14_62843293_62843448 | 0.13 |
Mir8098 |
microRNA 8098 |
5649 |
0.13 |
| chr5_135776480_135776870 | 0.13 |
Styxl1 |
serine/threonine/tyrosine interacting-like 1 |
1563 |
0.22 |
| chr1_60098880_60099160 | 0.13 |
Wdr12 |
WD repeat domain 12 |
375 |
0.68 |
| chr3_24507903_24508125 | 0.13 |
Gm24704 |
predicted gene, 24704 |
54060 |
0.18 |
| chr14_76901248_76901599 | 0.13 |
Gm48969 |
predicted gene, 48969 |
39852 |
0.15 |
| chr17_64607771_64607970 | 0.13 |
Man2a1 |
mannosidase 2, alpha 1 |
7134 |
0.26 |
| chr5_114589239_114589390 | 0.13 |
Fam222a |
family with sequence similarity 222, member A |
21297 |
0.14 |
| chr19_34517768_34517926 | 0.13 |
Lipa |
lysosomal acid lipase A |
6867 |
0.14 |
| chr4_57140481_57140705 | 0.13 |
Epb41l4b |
erythrocyte membrane protein band 4.1 like 4b |
2401 |
0.32 |
| chr1_112947623_112947888 | 0.13 |
Gm28190 |
predicted gene 28190 |
19176 |
0.27 |
| chr16_16990792_16991110 | 0.13 |
Mapk1 |
mitogen-activated protein kinase 1 |
7384 |
0.11 |
| chr19_40153747_40153898 | 0.13 |
Cyp2c70 |
cytochrome P450, family 2, subfamily c, polypeptide 70 |
33464 |
0.13 |
| chr1_105738205_105738360 | 0.13 |
Relch |
RAB11 binding and LisH domain, coiled-coil and HEAT repeat containing |
1930 |
0.29 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0097167 | circadian regulation of translation(GO:0097167) |
| 0.1 | 0.2 | GO:1904154 | positive regulation of retrograde protein transport, ER to cytosol(GO:1904154) |
| 0.1 | 0.2 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.1 | GO:0010046 | response to mycotoxin(GO:0010046) |
| 0.0 | 0.1 | GO:0046271 | phenylpropanoid catabolic process(GO:0046271) |
| 0.0 | 0.1 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.0 | 0.2 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.0 | 0.1 | GO:0002386 | immune response in mucosal-associated lymphoid tissue(GO:0002386) |
| 0.0 | 0.1 | GO:2000211 | regulation of glutamate metabolic process(GO:2000211) |
| 0.0 | 0.1 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.0 | 0.1 | GO:0018243 | protein O-linked glycosylation via threonine(GO:0018243) |
| 0.0 | 0.1 | GO:0006987 | activation of signaling protein activity involved in unfolded protein response(GO:0006987) |
| 0.0 | 0.1 | GO:0010908 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.0 | 0.1 | GO:1902809 | regulation of skeletal muscle fiber differentiation(GO:1902809) |
| 0.0 | 0.1 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.0 | 0.0 | GO:0061724 | lipophagy(GO:0061724) |
| 0.0 | 0.1 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.0 | 0.1 | GO:0019086 | late viral transcription(GO:0019086) |
| 0.0 | 0.1 | GO:0003383 | apical constriction(GO:0003383) |
| 0.0 | 0.1 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.0 | 0.1 | GO:0046449 | creatinine metabolic process(GO:0046449) cellular lactam metabolic process(GO:0072338) |
| 0.0 | 0.1 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.0 | 0.1 | GO:0046104 | thymidine metabolic process(GO:0046104) |
| 0.0 | 0.1 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.0 | 0.1 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.0 | 0.1 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.0 | 0.1 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.0 | 0.1 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.0 | 0.1 | GO:0042737 | drug catabolic process(GO:0042737) |
| 0.0 | 0.1 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.0 | 0.1 | GO:0000720 | pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) |
| 0.0 | 0.1 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.0 | 0.0 | GO:0061317 | canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:0061317) |
| 0.0 | 0.1 | GO:1903333 | negative regulation of protein folding(GO:1903333) |
| 0.0 | 0.1 | GO:0097105 | presynaptic membrane assembly(GO:0097105) |
| 0.0 | 0.1 | GO:0071286 | cellular response to magnesium ion(GO:0071286) |
| 0.0 | 0.1 | GO:0060283 | negative regulation of oocyte development(GO:0060283) |
| 0.0 | 0.0 | GO:0033031 | positive regulation of neutrophil apoptotic process(GO:0033031) |
| 0.0 | 0.1 | GO:0070295 | renal water absorption(GO:0070295) |
| 0.0 | 0.0 | GO:0034310 | primary alcohol catabolic process(GO:0034310) |
| 0.0 | 0.1 | GO:0051890 | regulation of cardioblast differentiation(GO:0051890) |
| 0.0 | 0.1 | GO:0030638 | polyketide metabolic process(GO:0030638) aminoglycoside antibiotic metabolic process(GO:0030647) daunorubicin metabolic process(GO:0044597) doxorubicin metabolic process(GO:0044598) |
| 0.0 | 0.1 | GO:0006559 | L-phenylalanine catabolic process(GO:0006559) erythrose 4-phosphate/phosphoenolpyruvate family amino acid catabolic process(GO:1902222) |
| 0.0 | 0.1 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.0 | 0.1 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.0 | 0.1 | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.0 | 0.0 | GO:2000809 | positive regulation of synaptic vesicle clustering(GO:2000809) |
| 0.0 | 0.0 | GO:0071725 | response to triacyl bacterial lipopeptide(GO:0071725) cellular response to triacyl bacterial lipopeptide(GO:0071727) |
| 0.0 | 0.0 | GO:2001027 | negative regulation of endothelial cell chemotaxis(GO:2001027) |
| 0.0 | 0.1 | GO:0009404 | toxin metabolic process(GO:0009404) |
| 0.0 | 0.0 | GO:0098902 | regulation of membrane depolarization during action potential(GO:0098902) |
| 0.0 | 0.1 | GO:1902459 | positive regulation of stem cell population maintenance(GO:1902459) |
| 0.0 | 0.0 | GO:0042732 | D-xylose metabolic process(GO:0042732) |
| 0.0 | 0.1 | GO:1901407 | regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.0 | 0.1 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.0 | 0.1 | GO:0000960 | mitochondrial RNA catabolic process(GO:0000957) regulation of mitochondrial RNA catabolic process(GO:0000960) |
| 0.0 | 0.1 | GO:0042904 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.0 | 0.0 | GO:0030050 | vesicle transport along actin filament(GO:0030050) |
| 0.0 | 0.1 | GO:2000370 | positive regulation of clathrin-mediated endocytosis(GO:2000370) |
| 0.0 | 0.0 | GO:1901475 | pyruvate transmembrane transport(GO:1901475) |
| 0.0 | 0.1 | GO:0043415 | positive regulation of skeletal muscle tissue regeneration(GO:0043415) |
| 0.0 | 0.0 | GO:0051081 | membrane disassembly(GO:0030397) nuclear envelope disassembly(GO:0051081) |
| 0.0 | 0.1 | GO:2000980 | regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.0 | 0.0 | GO:1901740 | negative regulation of myoblast fusion(GO:1901740) |
| 0.0 | 0.1 | GO:1902035 | positive regulation of hematopoietic stem cell proliferation(GO:1902035) |
| 0.0 | 0.1 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.1 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.0 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.0 | 0.1 | GO:1901029 | negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 0.0 | 0.0 | GO:0046149 | heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.0 | 0.0 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 0.0 | 0.0 | GO:0060399 | positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.0 | 0.1 | GO:0051657 | maintenance of organelle location(GO:0051657) |
| 0.0 | 0.0 | GO:0018879 | biphenyl metabolic process(GO:0018879) |
| 0.0 | 0.0 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.0 | 0.0 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.0 | 0.0 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 0.0 | GO:0036015 | response to interleukin-3(GO:0036015) cellular response to interleukin-3(GO:0036016) |
| 0.0 | 0.1 | GO:0006307 | DNA dealkylation involved in DNA repair(GO:0006307) |
| 0.0 | 0.0 | GO:0060839 | endothelial cell fate commitment(GO:0060839) |
| 0.0 | 0.1 | GO:2001286 | regulation of caveolin-mediated endocytosis(GO:2001286) |
| 0.0 | 0.0 | GO:0006714 | sesquiterpenoid metabolic process(GO:0006714) |
| 0.0 | 0.0 | GO:0008228 | opsonization(GO:0008228) |
| 0.0 | 0.1 | GO:0052697 | xenobiotic glucuronidation(GO:0052697) |
| 0.0 | 0.0 | GO:0090365 | regulation of mRNA modification(GO:0090365) |
| 0.0 | 0.1 | GO:0032000 | positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.0 | 0.1 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) |
| 0.0 | 0.1 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 0.1 | GO:0060020 | Bergmann glial cell differentiation(GO:0060020) |
| 0.0 | 0.1 | GO:1901525 | negative regulation of macromitophagy(GO:1901525) |
| 0.0 | 0.0 | GO:0031086 | nuclear-transcribed mRNA catabolic process, deadenylation-independent decay(GO:0031086) deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.0 | 0.0 | GO:0006578 | amino-acid betaine biosynthetic process(GO:0006578) |
| 0.0 | 0.1 | GO:1902774 | late endosome to lysosome transport(GO:1902774) |
| 0.0 | 0.0 | GO:1901491 | negative regulation of lymphangiogenesis(GO:1901491) |
| 0.0 | 0.0 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.0 | GO:0045901 | positive regulation of translational elongation(GO:0045901) |
| 0.0 | 0.0 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 0.1 | GO:0035838 | growing cell tip(GO:0035838) |
| 0.0 | 0.1 | GO:0002177 | manchette(GO:0002177) |
| 0.0 | 0.1 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 0.1 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.0 | 0.1 | GO:0097441 | basilar dendrite(GO:0097441) |
| 0.0 | 0.2 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.0 | GO:0035355 | Toll-like receptor 2-Toll-like receptor 6 protein complex(GO:0035355) |
| 0.0 | 0.1 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.0 | 0.1 | GO:0034715 | pICln-Sm protein complex(GO:0034715) |
| 0.0 | 0.1 | GO:0045293 | mRNA editing complex(GO:0045293) |
| 0.0 | 0.0 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.0 | 0.1 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.0 | 0.0 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.0 | 0.1 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 0.1 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0034875 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 0.0 | 0.1 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.0 | 0.3 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.1 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.0 | 0.1 | GO:0071553 | uridine nucleotide receptor activity(GO:0015065) G-protein coupled pyrimidinergic nucleotide receptor activity(GO:0071553) |
| 0.0 | 0.2 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.1 | GO:0016743 | carboxyl- or carbamoyltransferase activity(GO:0016743) |
| 0.0 | 0.1 | GO:0038181 | bile acid receptor activity(GO:0038181) |
| 0.0 | 0.1 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.0 | 0.1 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.0 | 0.1 | GO:0031697 | beta-1 adrenergic receptor binding(GO:0031697) |
| 0.0 | 0.1 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.0 | 0.1 | GO:0003846 | 2-acylglycerol O-acyltransferase activity(GO:0003846) |
| 0.0 | 0.1 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.0 | 0.1 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.1 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.0 | 0.0 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.0 | 0.0 | GO:0035276 | ethanol binding(GO:0035276) |
| 0.0 | 0.1 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.1 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.0 | 0.1 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 0.0 | 0.1 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.0 | 0.0 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 0.0 | 0.0 | GO:0035663 | Toll-like receptor 2 binding(GO:0035663) |
| 0.0 | 0.1 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.1 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) |
| 0.0 | 0.1 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.1 | GO:0015198 | oligopeptide transporter activity(GO:0015198) |
| 0.0 | 0.0 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.0 | 0.1 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 0.1 | GO:0019763 | immunoglobulin receptor activity(GO:0019763) |
| 0.0 | 0.1 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.0 | 0.3 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.1 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.1 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 0.0 | GO:0016899 | oxidoreductase activity, acting on the CH-OH group of donors, oxygen as acceptor(GO:0016899) |
| 0.0 | 0.1 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.0 | 0.0 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.0 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.0 | 0.0 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.0 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.1 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.1 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.1 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.2 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.0 | 0.1 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |