Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
Results for Lhx5_Lmx1b_Lhx1
Z-value: 1.35
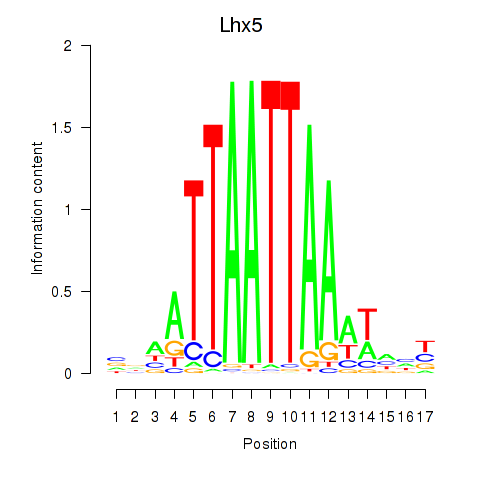
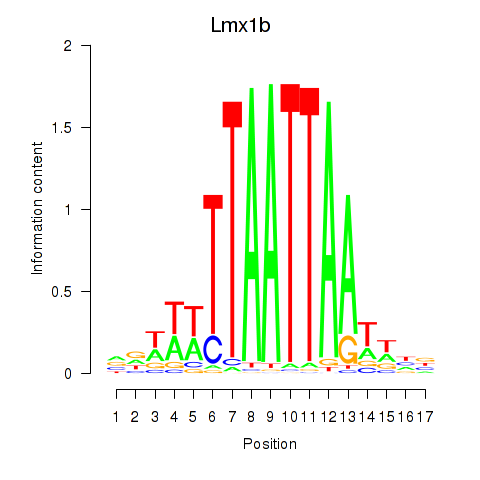
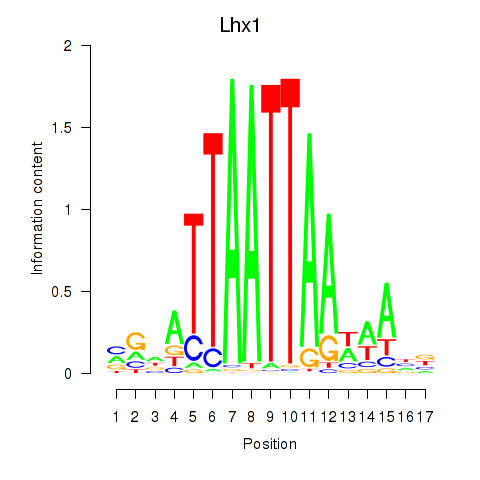
Transcription factors associated with Lhx5_Lmx1b_Lhx1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Lhx5
|
ENSMUSG00000029595.7 | LIM homeobox protein 5 |
|
Lmx1b
|
ENSMUSG00000038765.7 | LIM homeobox transcription factor 1 beta |
|
Lhx1
|
ENSMUSG00000018698.9 | LIM homeobox protein 1 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr11_84522955_84523135 | Lhx1 | 334 | 0.868084 | 0.70 | 1.2e-01 | Click! |
| chr11_84523271_84523458 | Lhx1 | 653 | 0.669544 | 0.63 | 1.8e-01 | Click! |
| chr11_84523501_84523678 | Lhx1 | 878 | 0.542340 | 0.49 | 3.3e-01 | Click! |
| chr11_84522677_84522841 | Lhx1 | 48 | 0.972621 | 0.45 | 3.7e-01 | Click! |
| chr11_84523882_84524217 | Lhx1 | 1338 | 0.356147 | 0.25 | 6.3e-01 | Click! |
Activity of the Lhx5_Lmx1b_Lhx1 motif across conditions
Conditions sorted by the z-value of the Lhx5_Lmx1b_Lhx1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
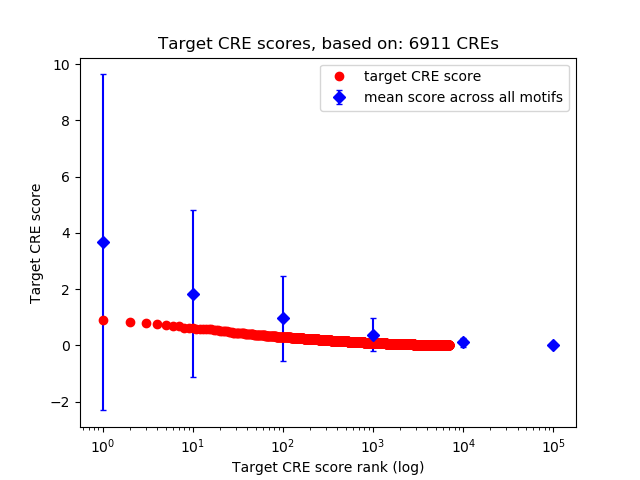
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr3_60495047_60495368 | 0.91 |
Mbnl1 |
muscleblind like splicing factor 1 |
5728 |
0.26 |
| chr16_84807428_84807749 | 0.82 |
Jam2 |
junction adhesion molecule 2 |
1819 |
0.26 |
| chr2_34049474_34049625 | 0.81 |
C230014O12Rik |
RIKEN cDNA C230014O12 gene |
58180 |
0.11 |
| chr19_31886956_31887132 | 0.77 |
A1cf |
APOBEC1 complementation factor |
18263 |
0.2 |
| chr10_125785978_125786129 | 0.73 |
Lrig3 |
leucine-rich repeats and immunoglobulin-like domains 3 |
180115 |
0.03 |
| chr14_30530847_30531018 | 0.67 |
Dcp1a |
decapping mRNA 1A |
7225 |
0.15 |
| chr1_73954752_73954963 | 0.67 |
Tns1 |
tensin 1 |
8186 |
0.24 |
| chr2_127608341_127608498 | 0.63 |
Mrps5 |
mitochondrial ribosomal protein S5 |
8386 |
0.13 |
| chr9_15301981_15302132 | 0.61 |
4931406C07Rik |
RIKEN cDNA 4931406C07 gene |
500 |
0.46 |
| chr10_28178649_28178847 | 0.60 |
Gm22370 |
predicted gene, 22370 |
35373 |
0.21 |
| chr11_90184166_90184514 | 0.59 |
n-R5s72 |
nuclear encoded rRNA 5S 72 |
787 |
0.57 |
| chr3_157024330_157024498 | 0.58 |
Gm43527 |
predicted gene 43527 |
23997 |
0.2 |
| chr9_106264996_106265377 | 0.58 |
Poc1a |
POC1 centriolar protein A |
15875 |
0.1 |
| chr10_37324159_37324310 | 0.58 |
Gm26535 |
predicted gene, 26535 |
13252 |
0.25 |
| chr3_69115488_69115663 | 0.57 |
Gm37558 |
predicted gene, 37558 |
7149 |
0.14 |
| chr11_21121403_21121623 | 0.56 |
Peli1 |
pellino 1 |
13956 |
0.18 |
| chr12_55420887_55421038 | 0.55 |
Psma6 |
proteasome subunit alpha 6 |
13579 |
0.16 |
| chr19_48883437_48883771 | 0.55 |
Gm50436 |
predicted gene, 50436 |
72468 |
0.11 |
| chr13_5229559_5229735 | 0.54 |
Gm47451 |
predicted gene, 47451 |
13469 |
0.25 |
| chr4_63738674_63738850 | 0.52 |
Tnfsf15 |
tumor necrosis factor (ligand) superfamily, member 15 |
6351 |
0.22 |
| chr6_100267052_100267222 | 0.51 |
Gm44044 |
predicted gene, 44044 |
6561 |
0.17 |
| chr5_92607049_92607221 | 0.51 |
Stbd1 |
starch binding domain 1 |
4067 |
0.19 |
| chr13_117745874_117746186 | 0.50 |
4933413L06Rik |
RIKEN cDNA 4933413L06 gene |
26019 |
0.26 |
| chr12_73341772_73342027 | 0.49 |
Slc38a6 |
solute carrier family 38, member 6 |
2468 |
0.25 |
| chr11_98339208_98339359 | 0.49 |
Ppp1r1b |
protein phosphatase 1, regulatory inhibitor subunit 1B |
9121 |
0.09 |
| chr19_7500165_7500363 | 0.48 |
Atl3 |
atlastin GTPase 3 |
5729 |
0.15 |
| chr3_10211563_10211714 | 0.46 |
Fabp4 |
fatty acid binding protein 4, adipocyte |
3062 |
0.13 |
| chr11_111605518_111605669 | 0.45 |
Gm11676 |
predicted gene 11676 |
7713 |
0.32 |
| chr14_64444658_64444809 | 0.45 |
Msra |
methionine sulfoxide reductase A |
3816 |
0.31 |
| chr7_45480279_45480444 | 0.45 |
Dhdh |
dihydrodiol dehydrogenase (dimeric) |
430 |
0.52 |
| chr19_32544319_32544470 | 0.43 |
Gm36419 |
predicted gene, 36419 |
1046 |
0.53 |
| chr19_27328502_27328670 | 0.43 |
Kcnv2 |
potassium channel, subfamily V, member 2 |
5998 |
0.19 |
| chr9_54139319_54139478 | 0.43 |
Cyp19a1 |
cytochrome P450, family 19, subfamily a, polypeptide 1 |
42676 |
0.14 |
| chr2_76970830_76970996 | 0.43 |
Ttn |
titin |
9269 |
0.25 |
| chr2_75605802_75605972 | 0.42 |
Gm13655 |
predicted gene 13655 |
27495 |
0.15 |
| chr17_32257244_32257401 | 0.42 |
Brd4 |
bromodomain containing 4 |
15043 |
0.12 |
| chr5_99976807_99976958 | 0.42 |
Hnrnpd |
heterogeneous nuclear ribonucleoprotein D |
331 |
0.73 |
| chr12_4844936_4845087 | 0.41 |
Wdcp |
WD repeat and coiled coil containing |
1617 |
0.23 |
| chr9_20504907_20505058 | 0.41 |
Zfp426 |
zinc finger protein 426 |
12236 |
0.11 |
| chr1_39133288_39133558 | 0.41 |
Gm37091 |
predicted gene, 37091 |
50085 |
0.11 |
| chr14_100201613_100201764 | 0.41 |
4930517O19Rik |
RIKEN cDNA 4930517O19 gene |
11454 |
0.2 |
| chr17_13821264_13821648 | 0.40 |
Afdn |
afadin, adherens junction formation factor |
29506 |
0.14 |
| chr1_9968620_9968771 | 0.40 |
Tcf24 |
transcription factor 24 |
763 |
0.41 |
| chr2_155378239_155378390 | 0.40 |
Trp53inp2 |
transformation related protein 53 inducible nuclear protein 2 |
2745 |
0.19 |
| chr14_62736570_62736721 | 0.40 |
Ints6 |
integrator complex subunit 6 |
24467 |
0.14 |
| chr18_10461512_10461756 | 0.40 |
Greb1l |
growth regulation by estrogen in breast cancer-like |
3409 |
0.24 |
| chr13_104035617_104035778 | 0.39 |
Nln |
neurolysin (metallopeptidase M3 family) |
870 |
0.63 |
| chr7_115674458_115674622 | 0.38 |
Sox6 |
SRY (sex determining region Y)-box 6 |
12175 |
0.3 |
| chr6_146730929_146731080 | 0.38 |
Stk38l |
serine/threonine kinase 38 like |
5692 |
0.17 |
| chr4_123419684_123419835 | 0.37 |
Macf1 |
microtubule-actin crosslinking factor 1 |
7531 |
0.17 |
| chr18_5013827_5013978 | 0.37 |
Svil |
supervillin |
18966 |
0.27 |
| chr19_60570173_60570324 | 0.37 |
Gm25238 |
predicted gene, 25238 |
10281 |
0.18 |
| chr2_73276147_73276506 | 0.37 |
Sp9 |
trans-acting transcription factor 9 |
4360 |
0.18 |
| chr1_90915370_90915560 | 0.36 |
Mlph |
melanophilin |
380 |
0.84 |
| chr4_35101216_35101367 | 0.36 |
Ifnk |
interferon kappa |
50765 |
0.12 |
| chr17_42859720_42859889 | 0.36 |
Cd2ap |
CD2-associated protein |
16585 |
0.26 |
| chr12_51348476_51348659 | 0.35 |
G2e3 |
G2/M-phase specific E3 ubiquitin ligase |
195 |
0.95 |
| chr1_172700535_172700739 | 0.35 |
Crp |
C-reactive protein, pentraxin-related |
2581 |
0.25 |
| chr16_43014052_43014241 | 0.35 |
Ndufs5-ps |
NADH:ubiquinone oxidoreductase core subunit S5, pseudogene |
58515 |
0.13 |
| chr13_34555664_34555830 | 0.35 |
Gm36758 |
predicted gene, 36758 |
6239 |
0.19 |
| chr6_28587807_28587958 | 0.35 |
Gm37978 |
predicted gene, 37978 |
19810 |
0.16 |
| chr13_104044189_104044340 | 0.35 |
Nln |
neurolysin (metallopeptidase M3 family) |
6622 |
0.22 |
| chr1_112947623_112947888 | 0.35 |
Gm28190 |
predicted gene 28190 |
19176 |
0.27 |
| chr16_27403508_27403659 | 0.35 |
Ccdc50 |
coiled-coil domain containing 50 |
13615 |
0.21 |
| chr19_60812093_60812292 | 0.35 |
Fam45a |
family with sequence similarity 45, member A |
152 |
0.92 |
| chr11_77770194_77770550 | 0.34 |
Myo18a |
myosin XVIIIA |
1779 |
0.27 |
| chr15_77251530_77251759 | 0.34 |
Rbfox2 |
RNA binding protein, fox-1 homolog (C. elegans) 2 |
6058 |
0.17 |
| chr14_21095037_21095192 | 0.34 |
Adk |
adenosine kinase |
18962 |
0.21 |
| chr3_79379701_79379957 | 0.34 |
Gm17359 |
predicted gene, 17359 |
34450 |
0.17 |
| chr8_33905969_33906196 | 0.33 |
Rbpms |
RNA binding protein gene with multiple splicing |
14318 |
0.16 |
| chr16_37606843_37607024 | 0.33 |
Gm46559 |
predicted gene, 46559 |
10743 |
0.15 |
| chr10_59521015_59521234 | 0.33 |
Mcu |
mitochondrial calcium uniporter |
62964 |
0.1 |
| chr3_131485378_131485553 | 0.33 |
Sgms2 |
sphingomyelin synthase 2 |
5014 |
0.28 |
| chr11_121550191_121550342 | 0.33 |
Tbcd |
tubulin-specific chaperone d |
155 |
0.96 |
| chr2_62386509_62386700 | 0.32 |
Dpp4 |
dipeptidylpeptidase 4 |
8356 |
0.19 |
| chr7_24600475_24600626 | 0.32 |
Phldb3 |
pleckstrin homology like domain, family B, member 3 |
10213 |
0.08 |
| chr6_141634890_141635041 | 0.32 |
Slco1b2 |
solute carrier organic anion transporter family, member 1b2 |
507 |
0.86 |
| chr10_87868820_87868971 | 0.32 |
Igf1os |
insulin-like growth factor 1, opposite strand |
5514 |
0.22 |
| chr19_55111923_55112097 | 0.32 |
Gpam |
glycerol-3-phosphate acyltransferase, mitochondrial |
8919 |
0.2 |
| chr5_138080510_138080792 | 0.32 |
Zkscan1 |
zinc finger with KRAB and SCAN domains 1 |
4433 |
0.1 |
| chr4_63773592_63773743 | 0.32 |
Tnfsf15 |
tumor necrosis factor (ligand) superfamily, member 15 |
28554 |
0.18 |
| chr2_72309170_72309345 | 0.32 |
Map3k20 |
mitogen-activated protein kinase kinase kinase 20 |
11356 |
0.19 |
| chr1_119646750_119646988 | 0.32 |
Epb41l5 |
erythrocyte membrane protein band 4.1 like 5 |
1748 |
0.32 |
| chr16_37530514_37530882 | 0.31 |
Gtf2e1 |
general transcription factor II E, polypeptide 1 (alpha subunit) |
9078 |
0.16 |
| chr1_21315523_21315674 | 0.31 |
Gm21909 |
predicted gene, 21909 |
722 |
0.5 |
| chr11_106583145_106583311 | 0.31 |
Tex2 |
testis expressed gene 2 |
2628 |
0.28 |
| chr12_12940858_12941058 | 0.31 |
Mycn |
v-myc avian myelocytomatosis viral related oncogene, neuroblastoma derived |
342 |
0.83 |
| chr3_133042582_133042733 | 0.31 |
Gm42874 |
predicted gene 42874 |
9738 |
0.14 |
| chr17_84990758_84990958 | 0.31 |
Ppm1b |
protein phosphatase 1B, magnesium dependent, beta isoform |
2815 |
0.25 |
| chr18_12202319_12202472 | 0.31 |
Npc1 |
NPC intracellular cholesterol transporter 1 |
6988 |
0.14 |
| chr13_28718314_28718531 | 0.31 |
Mir6368 |
microRNA 6368 |
7549 |
0.28 |
| chr6_101095542_101095693 | 0.30 |
Gm43948 |
predicted gene, 43948 |
36446 |
0.13 |
| chr12_79404609_79405013 | 0.30 |
Rad51b |
RAD51 paralog B |
77458 |
0.1 |
| chr10_12645976_12646209 | 0.30 |
Utrn |
utrophin |
31261 |
0.22 |
| chr5_129669529_129669717 | 0.30 |
Zfp11 |
zinc finger protein 11 |
476 |
0.73 |
| chr6_8862762_8862913 | 0.30 |
Ica1 |
islet cell autoantigen 1 |
84349 |
0.1 |
| chr11_111067244_111067395 | 0.30 |
Kcnj2 |
potassium inwardly-rectifying channel, subfamily J, member 2 |
1155 |
0.62 |
| chr12_3811591_3811763 | 0.30 |
Dnmt3a |
DNA methyltransferase 3A |
4517 |
0.2 |
| chr5_123975512_123975863 | 0.30 |
Hip1r |
huntingtin interacting protein 1 related |
2059 |
0.19 |
| chr6_108836766_108836917 | 0.30 |
Edem1 |
ER degradation enhancer, mannosidase alpha-like 1 |
431 |
0.86 |
| chr4_57160943_57161117 | 0.29 |
Gm12530 |
predicted gene 12530 |
11041 |
0.2 |
| chr15_25942169_25942359 | 0.29 |
Retreg1 |
reticulophagy regulator 1 |
415 |
0.85 |
| chr4_87899011_87899212 | 0.29 |
Mllt3 |
myeloid/lymphoid or mixed-lineage leukemia; translocated to, 3 |
24569 |
0.24 |
| chr7_118151770_118151921 | 0.29 |
Gm25683 |
predicted gene, 25683 |
1635 |
0.32 |
| chr9_46479855_46480133 | 0.29 |
Gm47144 |
predicted gene, 47144 |
25079 |
0.15 |
| chr11_80247133_80247304 | 0.29 |
Rhot1 |
ras homolog family member T1 |
2456 |
0.28 |
| chr19_37507552_37507703 | 0.29 |
Exoc6 |
exocyst complex component 6 |
13252 |
0.16 |
| chr12_103388315_103388482 | 0.29 |
Otub2 |
OTU domain, ubiquitin aldehyde binding 2 |
284 |
0.82 |
| chr2_115449849_115450246 | 0.29 |
3110099E03Rik |
RIKEN cDNA 3110099E03 gene |
62154 |
0.12 |
| chr12_78266965_78267134 | 0.29 |
Gm48225 |
predicted gene, 48225 |
5756 |
0.17 |
| chr14_114517521_114517864 | 0.29 |
Gm19829 |
predicted gene, 19829 |
206889 |
0.02 |
| chr1_84132307_84132510 | 0.29 |
Gm37058 |
predicted gene, 37058 |
2102 |
0.37 |
| chr17_50370553_50370865 | 0.29 |
Gm49906 |
predicted gene, 49906 |
31457 |
0.19 |
| chr5_135776480_135776870 | 0.29 |
Styxl1 |
serine/threonine/tyrosine interacting-like 1 |
1563 |
0.22 |
| chr2_124852123_124852274 | 0.29 |
Gm13994 |
predicted gene 13994 |
143543 |
0.04 |
| chr9_121353230_121353406 | 0.28 |
Gm47092 |
predicted gene, 47092 |
6762 |
0.17 |
| chr14_21599276_21599451 | 0.28 |
Kat6b |
K(lysine) acetyltransferase 6B |
23179 |
0.21 |
| chr10_117123042_117123236 | 0.28 |
Frs2 |
fibroblast growth factor receptor substrate 2 |
7707 |
0.15 |
| chr18_38524013_38524201 | 0.28 |
Gm50349 |
predicted gene, 50349 |
42804 |
0.11 |
| chr10_89502299_89502578 | 0.28 |
Nr1h4 |
nuclear receptor subfamily 1, group H, member 4 |
4211 |
0.26 |
| chr2_67956563_67957019 | 0.28 |
Gm37964 |
predicted gene, 37964 |
58195 |
0.14 |
| chr4_40698726_40698877 | 0.28 |
Aptx |
aprataxin |
3915 |
0.15 |
| chr16_90266479_90266654 | 0.28 |
Scaf4 |
SR-related CTD-associated factor 4 |
7766 |
0.17 |
| chr11_75734405_75734749 | 0.28 |
Ywhae |
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, epsilon polypeptide |
1429 |
0.37 |
| chr7_123124873_123125354 | 0.28 |
Tnrc6a |
trinucleotide repeat containing 6a |
857 |
0.66 |
| chr5_22506386_22506555 | 0.27 |
Orc5 |
origin recognition complex, subunit 5 |
16196 |
0.12 |
| chr3_63413482_63413662 | 0.27 |
Gm34240 |
predicted gene, 34240 |
25679 |
0.19 |
| chr14_8035648_8035848 | 0.27 |
Abhd6 |
abhydrolase domain containing 6 |
32782 |
0.13 |
| chr18_64619314_64619465 | 0.27 |
Gm6978 |
predicted gene 6978 |
6290 |
0.17 |
| chr11_120186847_120187216 | 0.27 |
2810410L24Rik |
RIKEN cDNA 2810410L24 gene |
1720 |
0.22 |
| chr17_88614606_88614762 | 0.27 |
Gm9406 |
predicted gene 9406 |
6140 |
0.17 |
| chr14_21165960_21166111 | 0.27 |
Adk |
adenosine kinase |
89883 |
0.08 |
| chr13_115653702_115653853 | 0.26 |
Gm47892 |
predicted gene, 47892 |
65722 |
0.13 |
| chr14_27402254_27402522 | 0.26 |
Gm23633 |
predicted gene, 23633 |
21538 |
0.16 |
| chr8_34506104_34506386 | 0.26 |
Gm8268 |
predicted gene 8268 |
962 |
0.54 |
| chr15_88205866_88206049 | 0.26 |
B230214G05Rik |
RIKEN cDNA B230214G05 gene |
108917 |
0.07 |
| chr16_94606252_94606403 | 0.26 |
Gm15971 |
predicted gene 15971 |
14050 |
0.19 |
| chr1_88764125_88764295 | 0.26 |
Platr5 |
pluripotency associated transcript 5 |
9256 |
0.2 |
| chrX_85856039_85856199 | 0.26 |
5430427O19Rik |
RIKEN cDNA 5430427O19 gene |
14235 |
0.16 |
| chr8_89407033_89407242 | 0.26 |
Gm26331 |
predicted gene, 26331 |
182404 |
0.03 |
| chr8_93223476_93223647 | 0.26 |
Ces1e |
carboxylesterase 1E |
4129 |
0.16 |
| chr15_39870011_39870207 | 0.26 |
Dpys |
dihydropyrimidinase |
12639 |
0.21 |
| chr19_36629275_36629447 | 0.26 |
Hectd2os |
Hectd2, opposite strand |
3337 |
0.28 |
| chr15_16844404_16844562 | 0.26 |
Gm49127 |
predicted gene, 49127 |
6481 |
0.3 |
| chr11_88618341_88618520 | 0.26 |
Msi2 |
musashi RNA-binding protein 2 |
28283 |
0.2 |
| chr5_87124253_87124404 | 0.26 |
Gm42795 |
predicted gene 42795 |
9719 |
0.12 |
| chr10_100031523_100031679 | 0.25 |
Kitl |
kit ligand |
15685 |
0.19 |
| chr9_118689127_118689278 | 0.25 |
Itga9 |
integrin alpha 9 |
3367 |
0.3 |
| chr17_10187170_10187336 | 0.25 |
Qk |
quaking |
26321 |
0.25 |
| chr18_11614667_11614838 | 0.25 |
Gm50067 |
predicted gene, 50067 |
10163 |
0.2 |
| chr15_55823868_55824092 | 0.25 |
Sntb1 |
syntrophin, basic 1 |
82330 |
0.08 |
| chr13_16760766_16760929 | 0.25 |
Gm7537 |
predicted gene 7537 |
121357 |
0.06 |
| chr18_68292618_68292974 | 0.25 |
Fam210a |
family with sequence similarity 210, member A |
7406 |
0.16 |
| chr14_67900348_67900512 | 0.25 |
Dock5 |
dedicator of cytokinesis 5 |
33002 |
0.17 |
| chr1_131465972_131466299 | 0.25 |
Gm29487 |
predicted gene 29487 |
18440 |
0.14 |
| chr19_34852544_34852695 | 0.25 |
Pank1 |
pantothenate kinase 1 |
25230 |
0.13 |
| chr17_24818767_24819039 | 0.25 |
Meiob |
meiosis specific with OB domains |
14544 |
0.08 |
| chr6_51456047_51456211 | 0.25 |
Hnrnpa2b1 |
heterogeneous nuclear ribonucleoprotein A2/B1 |
7055 |
0.17 |
| chr7_71419354_71419520 | 0.25 |
Gm29328 |
predicted gene 29328 |
49104 |
0.14 |
| chr7_49764294_49764503 | 0.25 |
Htatip2 |
HIV-1 Tat interactive protein 2 |
5245 |
0.23 |
| chr9_112123334_112123526 | 0.25 |
Mir128-2 |
microRNA 128-2 |
4719 |
0.29 |
| chr11_88277047_88277211 | 0.25 |
Ccdc182 |
coiled-coil domain containing 182 |
16929 |
0.15 |
| chr14_27897472_27897637 | 0.25 |
Erc2 |
ELKS/RAB6-interacting/CAST family member 2 |
21140 |
0.22 |
| chr8_93397874_93398136 | 0.25 |
Gm45764 |
predicted gene 45764 |
900 |
0.48 |
| chr13_93889949_93890100 | 0.25 |
Gm25534 |
predicted gene, 25534 |
33287 |
0.15 |
| chr14_52196558_52196748 | 0.25 |
Supt16 |
SPT16, facilitates chromatin remodeling subunit |
763 |
0.35 |
| chr7_68285442_68285612 | 0.24 |
Gm16157 |
predicted gene 16157 |
8933 |
0.14 |
| chr12_34238298_34238489 | 0.24 |
Gm18025 |
predicted gene, 18025 |
52742 |
0.18 |
| chr6_122148793_122148971 | 0.24 |
Gm10319 |
predicted pseudogene 10319 |
12150 |
0.17 |
| chr13_101016552_101016703 | 0.24 |
Gm6114 |
predicted gene 6114 |
2641 |
0.28 |
| chr4_134130965_134131164 | 0.24 |
Cep85 |
centrosomal protein 85 |
1578 |
0.21 |
| chr5_66054307_66054470 | 0.24 |
Rbm47 |
RNA binding motif protein 47 |
164 |
0.93 |
| chr15_102191817_102192017 | 0.24 |
Csad |
cysteine sulfinic acid decarboxylase |
142 |
0.92 |
| chr10_22004298_22004461 | 0.24 |
Sgk1 |
serum/glucocorticoid regulated kinase 1 |
7788 |
0.15 |
| chr16_95141689_95141853 | 0.24 |
Gm49642 |
predicted gene, 49642 |
29739 |
0.18 |
| chr12_8641402_8641553 | 0.24 |
Pum2 |
pumilio RNA-binding family member 2 |
32657 |
0.16 |
| chr3_14787462_14787685 | 0.24 |
Car1 |
carbonic anhydrase 1 |
9114 |
0.18 |
| chr1_131504400_131504768 | 0.24 |
Srgap2 |
SLIT-ROBO Rho GTPase activating protein 2 |
218 |
0.91 |
| chr8_109803384_109803547 | 0.23 |
Ap1g1 |
adaptor protein complex AP-1, gamma 1 subunit |
24559 |
0.12 |
| chr18_60385124_60385313 | 0.23 |
Iigp1 |
interferon inducible GTPase 1 |
2535 |
0.22 |
| chr19_56336755_56336948 | 0.23 |
Habp2 |
hyaluronic acid binding protein 2 |
23903 |
0.15 |
| chr12_40561876_40562268 | 0.23 |
Dock4 |
dedicator of cytokinesis 4 |
115736 |
0.06 |
| chr10_92231237_92231404 | 0.23 |
Gm8512 |
predicted gene 8512 |
7846 |
0.21 |
| chr13_60267432_60267612 | 0.23 |
Gm24999 |
predicted gene, 24999 |
26150 |
0.16 |
| chr9_68809615_68809769 | 0.23 |
Rora |
RAR-related orphan receptor alpha |
154389 |
0.04 |
| chr3_85265616_85265908 | 0.23 |
1700036G14Rik |
RIKEN cDNA 1700036G14 gene |
51757 |
0.14 |
| chr5_86114424_86114626 | 0.23 |
Stap1 |
signal transducing adaptor family member 1 |
28067 |
0.13 |
| chr4_126764112_126764263 | 0.23 |
AU040320 |
expressed sequence AU040320 |
10361 |
0.13 |
| chr3_108496483_108496708 | 0.23 |
Gm23336 |
predicted gene, 23336 |
1523 |
0.23 |
| chr7_71467703_71467871 | 0.23 |
Gm29328 |
predicted gene 29328 |
97454 |
0.07 |
| chr4_124809297_124809476 | 0.23 |
Mtf1 |
metal response element binding transcription factor 1 |
6708 |
0.1 |
| chr6_72120521_72121047 | 0.23 |
4933431G14Rik |
RIKEN cDNA 4933431G14 gene |
2156 |
0.2 |
| chr17_64607771_64607970 | 0.23 |
Man2a1 |
mannosidase 2, alpha 1 |
7134 |
0.26 |
| chr14_45487800_45488006 | 0.23 |
Fermt2 |
fermitin family member 2 |
3934 |
0.14 |
| chr1_160996279_160996443 | 0.23 |
Gm37072 |
predicted gene, 37072 |
5484 |
0.07 |
| chr7_75467233_75467387 | 0.23 |
Akap13 |
A kinase (PRKA) anchor protein 13 |
11442 |
0.16 |
| chr13_31779124_31779297 | 0.23 |
Gm11379 |
predicted gene 11379 |
18329 |
0.16 |
| chr18_3465417_3465568 | 0.23 |
Gm50088 |
predicted gene, 50088 |
1010 |
0.43 |
| chr11_60782796_60782947 | 0.23 |
Smcr8 |
Smith-Magenis syndrome chromosome region, candidate 8 homolog (human) |
5346 |
0.09 |
| chr11_53418931_53419109 | 0.22 |
Leap2 |
liver-expressed antimicrobial peptide 2 |
4150 |
0.1 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.1 | 0.3 | GO:0045472 | response to ether(GO:0045472) |
| 0.1 | 0.3 | GO:1904380 | endoplasmic reticulum mannose trimming(GO:1904380) |
| 0.1 | 0.3 | GO:1902809 | regulation of skeletal muscle fiber differentiation(GO:1902809) |
| 0.1 | 0.3 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.1 | 0.2 | GO:0042732 | D-xylose metabolic process(GO:0042732) |
| 0.1 | 0.2 | GO:0046439 | cysteine catabolic process(GO:0009093) L-cysteine catabolic process(GO:0019448) L-cysteine metabolic process(GO:0046439) |
| 0.1 | 0.2 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.1 | 0.2 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.1 | 0.2 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.1 | 0.2 | GO:0046271 | phenylpropanoid catabolic process(GO:0046271) |
| 0.1 | 0.2 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.0 | 0.2 | GO:0003383 | apical constriction(GO:0003383) |
| 0.0 | 0.2 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.1 | GO:0030241 | skeletal muscle myosin thick filament assembly(GO:0030241) striated muscle myosin thick filament assembly(GO:0071688) |
| 0.0 | 0.2 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.0 | 0.2 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.0 | 0.1 | GO:1901078 | negative regulation of relaxation of muscle(GO:1901078) |
| 0.0 | 0.1 | GO:0061724 | lipophagy(GO:0061724) |
| 0.0 | 0.2 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.2 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.0 | 0.2 | GO:1902035 | positive regulation of hematopoietic stem cell proliferation(GO:1902035) |
| 0.0 | 0.1 | GO:0014834 | skeletal muscle satellite cell maintenance involved in skeletal muscle regeneration(GO:0014834) |
| 0.0 | 0.1 | GO:0032911 | negative regulation of transforming growth factor beta1 production(GO:0032911) |
| 0.0 | 0.1 | GO:0010909 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.0 | 0.1 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.0 | 0.1 | GO:0006208 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) thymine metabolic process(GO:0019859) |
| 0.0 | 0.1 | GO:0044004 | killing by symbiont of host cells(GO:0001907) disruption by symbiont of host cell(GO:0044004) |
| 0.0 | 0.1 | GO:1904339 | negative regulation of dopaminergic neuron differentiation(GO:1904339) |
| 0.0 | 0.1 | GO:0006808 | regulation of nitrogen utilization(GO:0006808) |
| 0.0 | 0.3 | GO:0045607 | regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.0 | 0.1 | GO:0086046 | membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.0 | 0.1 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.0 | 0.2 | GO:0086013 | membrane repolarization during cardiac muscle cell action potential(GO:0086013) |
| 0.0 | 0.1 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.0 | 0.1 | GO:0014045 | establishment of endothelial blood-brain barrier(GO:0014045) |
| 0.0 | 0.1 | GO:0031087 | nuclear-transcribed mRNA catabolic process, deadenylation-independent decay(GO:0031086) deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.0 | 0.1 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.0 | 0.2 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.0 | 0.1 | GO:0006701 | progesterone biosynthetic process(GO:0006701) |
| 0.0 | 0.1 | GO:0033031 | positive regulation of neutrophil apoptotic process(GO:0033031) |
| 0.0 | 0.1 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.1 | GO:0006537 | glutamate biosynthetic process(GO:0006537) |
| 0.0 | 0.1 | GO:0010835 | regulation of protein ADP-ribosylation(GO:0010835) |
| 0.0 | 0.1 | GO:0003213 | cardiac right atrium morphogenesis(GO:0003213) |
| 0.0 | 0.1 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 0.0 | 0.1 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.0 | 0.2 | GO:0051409 | response to nitrosative stress(GO:0051409) |
| 0.0 | 0.1 | GO:0006681 | galactosylceramide metabolic process(GO:0006681) |
| 0.0 | 0.1 | GO:0003331 | regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) |
| 0.0 | 0.1 | GO:0002414 | immunoglobulin transcytosis in epithelial cells(GO:0002414) |
| 0.0 | 0.1 | GO:0006499 | N-terminal protein myristoylation(GO:0006499) |
| 0.0 | 0.0 | GO:0032489 | regulation of Cdc42 protein signal transduction(GO:0032489) |
| 0.0 | 0.1 | GO:0034499 | late endosome to Golgi transport(GO:0034499) |
| 0.0 | 0.2 | GO:2001223 | negative regulation of neuron migration(GO:2001223) |
| 0.0 | 0.1 | GO:1990168 | protein K29-linked deubiquitination(GO:0035523) protein K33-linked deubiquitination(GO:1990168) |
| 0.0 | 0.1 | GO:0046813 | receptor-mediated virion attachment to host cell(GO:0046813) |
| 0.0 | 0.1 | GO:1902564 | negative regulation of neutrophil activation(GO:1902564) |
| 0.0 | 0.1 | GO:0051790 | short-chain fatty acid biosynthetic process(GO:0051790) |
| 0.0 | 0.1 | GO:0042524 | negative regulation of tyrosine phosphorylation of Stat5 protein(GO:0042524) |
| 0.0 | 0.1 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.0 | 0.1 | GO:0070782 | phosphatidylserine exposure on apoptotic cell surface(GO:0070782) |
| 0.0 | 0.1 | GO:0034395 | regulation of transcription from RNA polymerase II promoter in response to iron(GO:0034395) |
| 0.0 | 0.0 | GO:0035962 | response to interleukin-13(GO:0035962) cellular response to interleukin-13(GO:0035963) |
| 0.0 | 0.2 | GO:0055091 | phospholipid homeostasis(GO:0055091) |
| 0.0 | 0.1 | GO:1900246 | positive regulation of RIG-I signaling pathway(GO:1900246) |
| 0.0 | 0.1 | GO:0072095 | regulation of branch elongation involved in ureteric bud branching(GO:0072095) |
| 0.0 | 0.1 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.0 | 0.1 | GO:0090296 | regulation of mitochondrial DNA replication(GO:0090296) |
| 0.0 | 0.0 | GO:0044860 | protein localization to plasma membrane raft(GO:0044860) |
| 0.0 | 0.1 | GO:2000418 | positive regulation of eosinophil migration(GO:2000418) |
| 0.0 | 0.1 | GO:0071727 | response to triacyl bacterial lipopeptide(GO:0071725) cellular response to triacyl bacterial lipopeptide(GO:0071727) |
| 0.0 | 0.0 | GO:2000566 | positive regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000566) |
| 0.0 | 0.0 | GO:0070103 | regulation of interleukin-6-mediated signaling pathway(GO:0070103) |
| 0.0 | 0.1 | GO:2000860 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.0 | 0.1 | GO:0098902 | regulation of membrane depolarization during action potential(GO:0098902) |
| 0.0 | 0.1 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.1 | GO:0006987 | activation of signaling protein activity involved in unfolded protein response(GO:0006987) |
| 0.0 | 0.0 | GO:0060842 | arterial endothelial cell differentiation(GO:0060842) |
| 0.0 | 0.1 | GO:0018243 | protein O-linked glycosylation via threonine(GO:0018243) |
| 0.0 | 0.0 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.0 | 0.1 | GO:0048294 | negative regulation of isotype switching to IgE isotypes(GO:0048294) |
| 0.0 | 0.1 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.0 | 0.1 | GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 0.0 | 0.0 | GO:0009804 | coumarin metabolic process(GO:0009804) |
| 0.0 | 0.1 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 0.0 | 0.2 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.0 | 0.1 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.0 | 0.1 | GO:1902309 | negative regulation of peptidyl-serine dephosphorylation(GO:1902309) |
| 0.0 | 0.1 | GO:0021684 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.0 | 0.1 | GO:0061010 | gall bladder development(GO:0061010) |
| 0.0 | 0.0 | GO:0032741 | positive regulation of interleukin-18 production(GO:0032741) |
| 0.0 | 0.1 | GO:0045896 | regulation of transcription during mitosis(GO:0045896) positive regulation of transcription during mitosis(GO:0045897) |
| 0.0 | 0.1 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.0 | 0.1 | GO:2000562 | negative regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000562) |
| 0.0 | 0.0 | GO:0048022 | negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) |
| 0.0 | 0.1 | GO:0034139 | regulation of toll-like receptor 3 signaling pathway(GO:0034139) |
| 0.0 | 0.1 | GO:0035054 | embryonic heart tube anterior/posterior pattern specification(GO:0035054) |
| 0.0 | 0.1 | GO:0061470 | T follicular helper cell differentiation(GO:0061470) |
| 0.0 | 0.0 | GO:0000720 | pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) |
| 0.0 | 0.1 | GO:0043622 | cortical microtubule organization(GO:0043622) |
| 0.0 | 0.1 | GO:0006714 | sesquiterpenoid metabolic process(GO:0006714) |
| 0.0 | 0.1 | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.0 | 0.1 | GO:0009052 | pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 0.0 | 0.0 | GO:0070350 | white fat cell proliferation(GO:0070343) regulation of white fat cell proliferation(GO:0070350) |
| 0.0 | 0.1 | GO:0006578 | amino-acid betaine biosynthetic process(GO:0006578) |
| 0.0 | 0.1 | GO:0034773 | histone H4-K20 trimethylation(GO:0034773) |
| 0.0 | 0.1 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.0 | 0.1 | GO:0000966 | RNA 5'-end processing(GO:0000966) |
| 0.0 | 0.0 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 0.0 | 0.1 | GO:0045653 | negative regulation of megakaryocyte differentiation(GO:0045653) |
| 0.0 | 0.0 | GO:0003223 | ventricular compact myocardium morphogenesis(GO:0003223) |
| 0.0 | 0.1 | GO:0060501 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 0.0 | 0.1 | GO:0046499 | S-adenosylmethioninamine metabolic process(GO:0046499) |
| 0.0 | 0.1 | GO:0051775 | response to redox state(GO:0051775) |
| 0.0 | 0.1 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.0 | 0.0 | GO:0032788 | saturated monocarboxylic acid metabolic process(GO:0032788) unsaturated monocarboxylic acid metabolic process(GO:0032789) |
| 0.0 | 0.0 | GO:0039689 | negative stranded viral RNA replication(GO:0039689) multi-organism biosynthetic process(GO:0044034) |
| 0.0 | 0.1 | GO:0097066 | response to thyroid hormone(GO:0097066) |
| 0.0 | 0.1 | GO:1901409 | regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.0 | 0.3 | GO:0032400 | melanosome localization(GO:0032400) |
| 0.0 | 0.1 | GO:0045901 | positive regulation of translational elongation(GO:0045901) |
| 0.0 | 0.1 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.0 | 0.1 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 0.1 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.0 | 0.0 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.0 | 0.0 | GO:0097195 | pilomotor reflex(GO:0097195) |
| 0.0 | 0.0 | GO:2000828 | regulation of parathyroid hormone secretion(GO:2000828) |
| 0.0 | 0.2 | GO:0010971 | positive regulation of G2/M transition of mitotic cell cycle(GO:0010971) |
| 0.0 | 0.0 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 0.0 | 0.0 | GO:0045085 | negative regulation of interleukin-2 biosynthetic process(GO:0045085) |
| 0.0 | 0.1 | GO:1902459 | positive regulation of stem cell population maintenance(GO:1902459) |
| 0.0 | 0.1 | GO:0001302 | replicative cell aging(GO:0001302) |
| 0.0 | 0.0 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 0.0 | 0.0 | GO:0032439 | endosome localization(GO:0032439) |
| 0.0 | 0.0 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 0.0 | 0.1 | GO:0042904 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.0 | 0.0 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.0 | 0.0 | GO:1902075 | cellular response to salt(GO:1902075) |
| 0.0 | 0.1 | GO:1904354 | negative regulation of telomere capping(GO:1904354) |
| 0.0 | 0.1 | GO:0097105 | presynaptic membrane assembly(GO:0097105) |
| 0.0 | 0.1 | GO:0015816 | glycine transport(GO:0015816) |
| 0.0 | 0.1 | GO:0051547 | regulation of keratinocyte migration(GO:0051547) positive regulation of keratinocyte migration(GO:0051549) |
| 0.0 | 0.1 | GO:0060696 | regulation of phospholipid catabolic process(GO:0060696) |
| 0.0 | 0.1 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.0 | 0.0 | GO:2000302 | positive regulation of synaptic vesicle exocytosis(GO:2000302) |
| 0.0 | 0.0 | GO:0046618 | drug export(GO:0046618) |
| 0.0 | 0.1 | GO:0090520 | sphingolipid mediated signaling pathway(GO:0090520) |
| 0.0 | 0.1 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 0.0 | 0.1 | GO:0033578 | protein glycosylation in Golgi(GO:0033578) |
| 0.0 | 0.0 | GO:0061209 | cell proliferation involved in mesonephros development(GO:0061209) |
| 0.0 | 0.0 | GO:0050427 | 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) |
| 0.0 | 0.0 | GO:1901491 | negative regulation of lymphangiogenesis(GO:1901491) |
| 0.0 | 0.1 | GO:0032324 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) molybdopterin cofactor biosynthetic process(GO:0032324) molybdopterin cofactor metabolic process(GO:0043545) prosthetic group metabolic process(GO:0051189) |
| 0.0 | 0.0 | GO:0060596 | mammary placode formation(GO:0060596) |
| 0.0 | 0.0 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.0 | 0.1 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.0 | 0.1 | GO:0046602 | regulation of mitotic centrosome separation(GO:0046602) |
| 0.0 | 0.0 | GO:0048021 | regulation of melanin biosynthetic process(GO:0048021) regulation of secondary metabolite biosynthetic process(GO:1900376) |
| 0.0 | 0.0 | GO:0006553 | lysine metabolic process(GO:0006553) |
| 0.0 | 0.0 | GO:0051791 | medium-chain fatty acid metabolic process(GO:0051791) |
| 0.0 | 0.0 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.0 | 0.1 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.0 | 0.0 | GO:0046449 | creatinine metabolic process(GO:0046449) cellular lactam metabolic process(GO:0072338) |
| 0.0 | 0.0 | GO:0070427 | nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070427) |
| 0.0 | 0.1 | GO:0001842 | neural fold formation(GO:0001842) |
| 0.0 | 0.0 | GO:0036265 | RNA (guanine-N7)-methylation(GO:0036265) |
| 0.0 | 0.1 | GO:0033631 | cell-cell adhesion mediated by integrin(GO:0033631) |
| 0.0 | 0.0 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.0 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.0 | 0.0 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 0.0 | GO:0046874 | quinolinate metabolic process(GO:0046874) |
| 0.0 | 0.0 | GO:0002322 | B cell proliferation involved in immune response(GO:0002322) |
| 0.0 | 0.3 | GO:0046834 | lipid phosphorylation(GO:0046834) |
| 0.0 | 0.0 | GO:1903223 | positive regulation of oxidative stress-induced neuron death(GO:1903223) |
| 0.0 | 0.2 | GO:0001967 | suckling behavior(GO:0001967) |
| 0.0 | 0.0 | GO:0071798 | response to prostaglandin D(GO:0071798) cellular response to prostaglandin D stimulus(GO:0071799) |
| 0.0 | 0.1 | GO:0010992 | ubiquitin homeostasis(GO:0010992) |
| 0.0 | 0.2 | GO:0010569 | regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.0 | 0.1 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.0 | 0.0 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.0 | 0.1 | GO:0042737 | drug catabolic process(GO:0042737) |
| 0.0 | 0.0 | GO:0071830 | chylomicron remnant clearance(GO:0034382) triglyceride-rich lipoprotein particle clearance(GO:0071830) |
| 0.0 | 0.0 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.0 | 0.0 | GO:0051081 | membrane disassembly(GO:0030397) nuclear envelope disassembly(GO:0051081) |
| 0.0 | 0.1 | GO:0006528 | asparagine metabolic process(GO:0006528) |
| 0.0 | 0.1 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 0.1 | GO:1900264 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.0 | 0.0 | GO:1902109 | negative regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902109) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0045293 | mRNA editing complex(GO:0045293) |
| 0.0 | 0.3 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 0.1 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 0.1 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.0 | 0.2 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.0 | 0.1 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.1 | GO:0035355 | Toll-like receptor 2-Toll-like receptor 6 protein complex(GO:0035355) |
| 0.0 | 0.2 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.1 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.0 | 0.2 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.1 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.1 | GO:0005784 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.0 | 0.3 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.1 | GO:0000938 | GARP complex(GO:0000938) |
| 0.0 | 0.1 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.0 | 0.1 | GO:0043219 | lateral loop(GO:0043219) |
| 0.0 | 0.1 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.0 | 0.1 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.0 | 0.1 | GO:0016942 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) insulin-like growth factor ternary complex(GO:0042567) |
| 0.0 | 0.0 | GO:0097441 | basilar dendrite(GO:0097441) |
| 0.0 | 0.0 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.0 | 0.0 | GO:0035838 | growing cell tip(GO:0035838) |
| 0.0 | 0.1 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.0 | 0.0 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.0 | 0.1 | GO:0070419 | nonhomologous end joining complex(GO:0070419) |
| 0.0 | 0.1 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 0.0 | GO:1990597 | AIP1-IRE1 complex(GO:1990597) |
| 0.0 | 0.1 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.1 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.1 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.0 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.0 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.0 | 0.1 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.0 | 0.0 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 0.2 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.0 | GO:0030891 | VCB complex(GO:0030891) |
| 0.0 | 0.1 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.1 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 0.1 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.0 | 0.1 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.1 | 0.3 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.1 | 0.2 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.0 | 0.1 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.0 | 0.3 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.2 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.1 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.2 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.0 | 0.1 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.0 | 0.1 | GO:0070905 | serine binding(GO:0070905) |
| 0.0 | 0.2 | GO:0015198 | oligopeptide transporter activity(GO:0015198) |
| 0.0 | 0.1 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.0 | 0.1 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.0 | 0.0 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.0 | 0.1 | GO:0008525 | phosphatidylcholine transporter activity(GO:0008525) |
| 0.0 | 0.2 | GO:0086008 | voltage-gated potassium channel activity involved in cardiac muscle cell action potential repolarization(GO:0086008) |
| 0.0 | 0.2 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.0 | 0.1 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 0.1 | GO:0015173 | aromatic amino acid transmembrane transporter activity(GO:0015173) |
| 0.0 | 0.1 | GO:0004802 | transketolase activity(GO:0004802) |
| 0.0 | 0.1 | GO:0038181 | bile acid receptor activity(GO:0038181) |
| 0.0 | 0.1 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 0.1 | GO:0003846 | 2-acylglycerol O-acyltransferase activity(GO:0003846) |
| 0.0 | 0.3 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.3 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.2 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.0 | 0.2 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.1 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.0 | 0.2 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.0 | 0.1 | GO:0098518 | polynucleotide phosphatase activity(GO:0098518) |
| 0.0 | 0.1 | GO:0016635 | oxidoreductase activity, acting on the CH-CH group of donors, quinone or related compound as acceptor(GO:0016635) |
| 0.0 | 0.1 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 0.1 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.0 | 0.1 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.0 | 0.1 | GO:0001094 | TFIID-class transcription factor binding(GO:0001094) |
| 0.0 | 0.1 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.1 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) DNA (cytosine-5-)-methyltransferase activity, acting on CpG substrates(GO:0051718) |
| 0.0 | 0.1 | GO:0002054 | nucleobase binding(GO:0002054) |
| 0.0 | 0.1 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.0 | 0.1 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 0.0 | 0.1 | GO:0000171 | ribonuclease MRP activity(GO:0000171) |
| 0.0 | 0.1 | GO:0035663 | Toll-like receptor 2 binding(GO:0035663) |
| 0.0 | 0.1 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.0 | 0.2 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.0 | 0.1 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.1 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.0 | 0.1 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.1 | GO:0001226 | RNA polymerase II transcription corepressor binding(GO:0001226) |
| 0.0 | 0.1 | GO:0015252 | hydrogen ion channel activity(GO:0015252) |
| 0.0 | 0.0 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.0 | 0.1 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.0 | 0.0 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.0 | 0.1 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.1 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
| 0.0 | 0.0 | GO:0052658 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) |
| 0.0 | 0.0 | GO:0003941 | L-serine ammonia-lyase activity(GO:0003941) |
| 0.0 | 0.0 | GO:0016508 | long-chain-enoyl-CoA hydratase activity(GO:0016508) |
| 0.0 | 0.0 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.0 | 0.1 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 0.0 | 0.0 | GO:0035650 | AP-1 adaptor complex binding(GO:0035650) |
| 0.0 | 0.1 | GO:0070878 | primary miRNA binding(GO:0070878) |
| 0.0 | 0.1 | GO:0016899 | oxidoreductase activity, acting on the CH-OH group of donors, oxygen as acceptor(GO:0016899) |
| 0.0 | 0.0 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.0 | 0.1 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.1 | GO:0070326 | very-low-density lipoprotein particle receptor binding(GO:0070326) |
| 0.0 | 0.3 | GO:0016780 | phosphotransferase activity, for other substituted phosphate groups(GO:0016780) |
| 0.0 | 0.0 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.0 | 0.0 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.0 | 0.1 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.0 | 0.2 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.1 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.0 | GO:0019961 | interferon binding(GO:0019961) |
| 0.0 | 0.1 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.1 | GO:0034875 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 0.0 | 0.0 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.0 | 0.1 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.0 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.0 | 0.0 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.2 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.0 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.0 | 0.0 | GO:0102345 | 3-hydroxyacyl-CoA dehydratase activity(GO:0018812) 3-hydroxy-behenoyl-CoA dehydratase activity(GO:0102344) 3-hydroxy-lignoceroyl-CoA dehydratase activity(GO:0102345) |
| 0.0 | 0.1 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.2 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.0 | 0.1 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.0 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.0 | 0.0 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.0 | 0.1 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 0.1 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 0.0 | GO:0051033 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.0 | 0.0 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.4 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 0.0 | GO:0086007 | voltage-gated calcium channel activity involved in cardiac muscle cell action potential(GO:0086007) |
| 0.0 | 0.1 | GO:0008601 | protein phosphatase type 2A regulator activity(GO:0008601) |
| 0.0 | 0.1 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.0 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.0 | 0.0 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.0 | 0.1 | GO:0035242 | protein-arginine omega-N asymmetric methyltransferase activity(GO:0035242) |
| 0.0 | 0.1 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.4 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 0.0 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.0 | 0.0 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.0 | 0.2 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 0.1 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 0.1 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 0.2 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.0 | 0.2 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.3 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.3 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.2 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.0 | 0.2 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.1 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.0 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.0 | 0.1 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.2 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.1 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.1 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.0 | 0.1 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.1 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.0 | 0.5 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |