Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
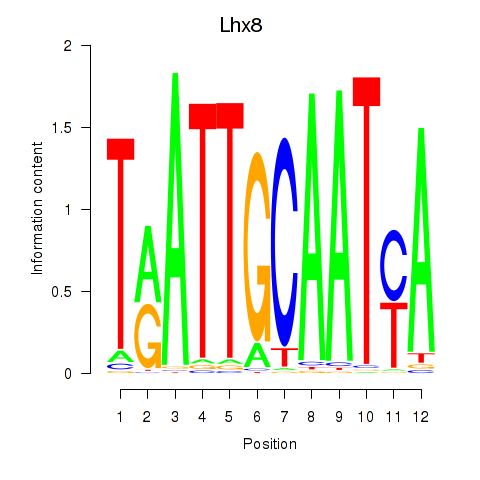
Results for Lhx8
Z-value: 1.95
Transcription factors associated with Lhx8
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Lhx8
|
ENSMUSG00000096225.2 | LIM homeobox protein 8 |
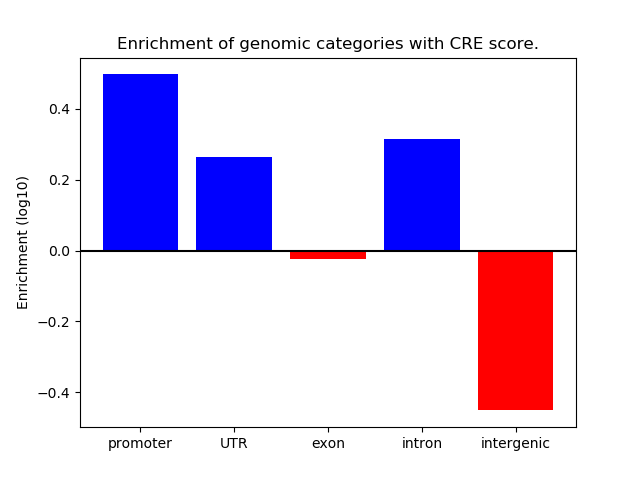
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr3_154297576_154297741 | Lhx8 | 27225 | 0.151037 | -0.25 | 6.3e-01 | Click! |
Activity of the Lhx8 motif across conditions
Conditions sorted by the z-value of the Lhx8 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
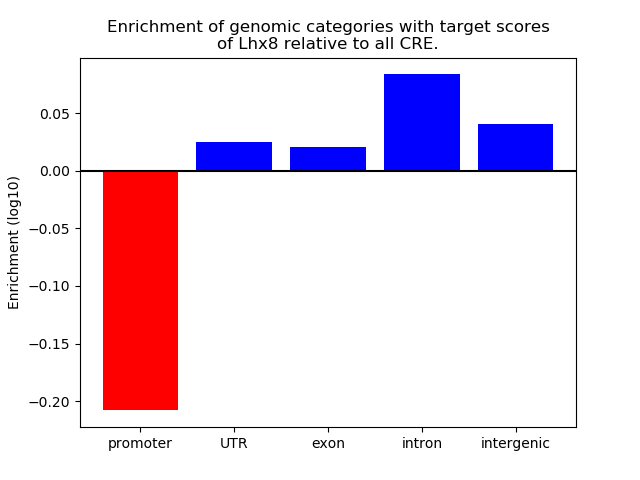
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr9_74893395_74893777 | 2.28 |
Onecut1 |
one cut domain, family member 1 |
27102 |
0.13 |
| chr7_63922331_63923024 | 1.48 |
Klf13 |
Kruppel-like factor 13 |
2193 |
0.22 |
| chr1_67169083_67169234 | 1.44 |
Cps1 |
carbamoyl-phosphate synthetase 1 |
46132 |
0.15 |
| chr7_72323573_72323742 | 1.35 |
Mctp2 |
multiple C2 domains, transmembrane 2 |
17049 |
0.28 |
| chr8_93176962_93177159 | 1.33 |
Ces1d |
carboxylesterase 1D |
1771 |
0.27 |
| chr1_107939897_107940125 | 1.32 |
D830032E09Rik |
RIKEN cDNA D830032E09 gene |
3302 |
0.23 |
| chr2_42022364_42022535 | 1.30 |
Gm13461 |
predicted gene 13461 |
33620 |
0.23 |
| chr11_16805061_16805227 | 1.27 |
Egfros |
epidermal growth factor receptor, opposite strand |
25558 |
0.18 |
| chr2_81270139_81270299 | 1.25 |
Gm23900 |
predicted gene, 23900 |
273123 |
0.01 |
| chr8_105095555_105095947 | 1.22 |
Ces3b |
carboxylesterase 3B |
7132 |
0.11 |
| chr9_109966945_109967106 | 1.21 |
Map4 |
microtubule-associated protein 4 |
1992 |
0.21 |
| chr11_16589661_16589834 | 1.18 |
Gm12663 |
predicted gene 12663 |
46319 |
0.12 |
| chr1_21261382_21261702 | 1.15 |
Gsta3 |
glutathione S-transferase, alpha 3 |
8021 |
0.11 |
| chr6_149344317_149344633 | 1.14 |
Gm15782 |
predicted gene 15782 |
9351 |
0.14 |
| chr16_43408501_43408652 | 1.11 |
Gm15713 |
predicted gene 15713 |
11620 |
0.2 |
| chr5_150362303_150362454 | 1.04 |
Fry |
FRY microtubule binding protein |
37275 |
0.13 |
| chr5_87254769_87254962 | 1.03 |
Ugt2b37 |
UDP glucuronosyltransferase 2 family, polypeptide B37 |
61 |
0.96 |
| chr19_20695080_20695252 | 1.02 |
Aldh1a7 |
aldehyde dehydrogenase family 1, subfamily A7 |
32396 |
0.2 |
| chr9_64383785_64383941 | 1.01 |
Megf11 |
multiple EGF-like-domains 11 |
1763 |
0.37 |
| chr18_38370754_38370932 | 1.00 |
Gm4949 |
predicted gene 4949 |
10305 |
0.12 |
| chr16_19655438_19655589 | 1.00 |
Gm49596 |
predicted gene, 49596 |
23611 |
0.13 |
| chr8_114151087_114151399 | 0.98 |
Nudt7 |
nudix (nucleoside diphosphate linked moiety X)-type motif 7 |
17601 |
0.25 |
| chr8_114155044_114155305 | 0.98 |
Nudt7 |
nudix (nucleoside diphosphate linked moiety X)-type motif 7 |
21532 |
0.24 |
| chr9_94704515_94704673 | 0.98 |
Gm16262 |
predicted gene 16262 |
2275 |
0.28 |
| chr5_100419251_100419681 | 0.95 |
Sec31a |
Sec31 homolog A (S. cerevisiae) |
3232 |
0.18 |
| chr3_60408325_60408501 | 0.93 |
Mbnl1 |
muscleblind like splicing factor 1 |
64417 |
0.12 |
| chr1_21264707_21265188 | 0.92 |
Gm28836 |
predicted gene 28836 |
6646 |
0.11 |
| chr8_114147681_114147946 | 0.91 |
Nudt7 |
nudix (nucleoside diphosphate linked moiety X)-type motif 7 |
14171 |
0.26 |
| chr2_177470070_177470297 | 0.90 |
Zfp970 |
zinc finger protein 970 |
5337 |
0.17 |
| chr3_100189024_100189191 | 0.90 |
Gdap2 |
ganglioside-induced differentiation-associated-protein 2 |
5473 |
0.23 |
| chr11_16811935_16812291 | 0.90 |
Egfros |
epidermal growth factor receptor, opposite strand |
18589 |
0.2 |
| chr17_81368572_81368728 | 0.88 |
Gm50044 |
predicted gene, 50044 |
2183 |
0.4 |
| chr14_8034997_8035230 | 0.88 |
Abhd6 |
abhydrolase domain containing 6 |
32147 |
0.13 |
| chr4_103933550_103933744 | 0.87 |
Gm12719 |
predicted gene 12719 |
5894 |
0.25 |
| chr12_104339796_104339996 | 0.86 |
Serpina3k |
serine (or cysteine) peptidase inhibitor, clade A, member 3K |
1410 |
0.24 |
| chr8_26351792_26351988 | 0.86 |
Gm31784 |
predicted gene, 31784 |
39556 |
0.12 |
| chr3_18210346_18210497 | 0.85 |
Gm23686 |
predicted gene, 23686 |
32796 |
0.17 |
| chr13_93719775_93719928 | 0.85 |
AW495222 |
expressed sequence AW495222 |
676 |
0.6 |
| chr4_80920883_80921053 | 0.83 |
Lurap1l |
leucine rich adaptor protein 1-like |
10322 |
0.26 |
| chr9_57711329_57711506 | 0.81 |
Edc3 |
enhancer of mRNA decapping 3 |
2850 |
0.18 |
| chr2_113790326_113790491 | 0.79 |
Grem1 |
gremlin 1, DAN family BMP antagonist |
31762 |
0.15 |
| chr3_115819747_115820078 | 0.79 |
Dph5 |
diphthamide biosynthesis 5 |
67925 |
0.08 |
| chr13_115653702_115653853 | 0.79 |
Gm47892 |
predicted gene, 47892 |
65722 |
0.13 |
| chr9_74695756_74695907 | 0.78 |
Gm27233 |
predicted gene 27233 |
13431 |
0.23 |
| chr19_44394517_44394772 | 0.75 |
Scd1 |
stearoyl-Coenzyme A desaturase 1 |
12046 |
0.14 |
| chr15_58961550_58961701 | 0.75 |
Mtss1 |
MTSS I-BAR domain containing 1 |
4785 |
0.16 |
| chr6_72155996_72156358 | 0.74 |
Gm38832 |
predicted gene, 38832 |
6672 |
0.15 |
| chr6_144709069_144709350 | 0.74 |
Sox5os4 |
SRY (sex determining region Y)-box 5, opposite strand 4 |
11702 |
0.16 |
| chr15_55117700_55118060 | 0.73 |
Gm9920 |
predicted gene 9920 |
4203 |
0.17 |
| chr9_122357372_122357607 | 0.73 |
Abhd5 |
abhydrolase domain containing 5 |
5700 |
0.15 |
| chr6_149089096_149089247 | 0.73 |
Dennd5b |
DENN/MADD domain containing 5B |
3808 |
0.13 |
| chr18_9832346_9832497 | 0.72 |
Gm23637 |
predicted gene, 23637 |
5302 |
0.16 |
| chr19_40180796_40180958 | 0.72 |
Cyp2c70 |
cytochrome P450, family 2, subfamily c, polypeptide 70 |
6409 |
0.16 |
| chr13_63665698_63665976 | 0.71 |
Gm47387 |
predicted gene, 47387 |
4040 |
0.18 |
| chr18_33299657_33299808 | 0.71 |
Gm5503 |
predicted gene 5503 |
85223 |
0.09 |
| chr2_34775525_34776380 | 0.71 |
Hspa5 |
heat shock protein 5 |
1105 |
0.4 |
| chr15_73408554_73408705 | 0.71 |
Gm7908 |
predicted gene 7908 |
390 |
0.81 |
| chr19_12685349_12685500 | 0.71 |
Gm49772 |
predicted gene, 49772 |
3653 |
0.11 |
| chr1_67192772_67192960 | 0.70 |
Gm15668 |
predicted gene 15668 |
56334 |
0.12 |
| chr14_17728323_17728474 | 0.70 |
Gm48320 |
predicted gene, 48320 |
42724 |
0.19 |
| chr8_57653465_57653731 | 0.70 |
Galnt7 |
polypeptide N-acetylgalactosaminyltransferase 7 |
566 |
0.41 |
| chr4_44920931_44921086 | 0.70 |
Zcchc7 |
zinc finger, CCHC domain containing 7 |
2110 |
0.25 |
| chr7_118146005_118146156 | 0.70 |
Smg1 |
SMG1 homolog, phosphatidylinositol 3-kinase-related kinase (C. elegans) |
4281 |
0.18 |
| chr16_42998647_42998825 | 0.69 |
Ndufs5-ps |
NADH:ubiquinone oxidoreductase core subunit S5, pseudogene |
43105 |
0.16 |
| chr13_115067681_115067870 | 0.68 |
Gm10734 |
predicted gene 10734 |
21726 |
0.14 |
| chr18_44453653_44453866 | 0.68 |
Mcc |
mutated in colorectal cancers |
65757 |
0.11 |
| chr10_28228240_28228413 | 0.68 |
Gm22370 |
predicted gene, 22370 |
14205 |
0.29 |
| chr6_119108994_119109155 | 0.68 |
Cacna1c |
calcium channel, voltage-dependent, L type, alpha 1C subunit |
501 |
0.86 |
| chr1_67215139_67215608 | 0.68 |
Gm15668 |
predicted gene 15668 |
33827 |
0.17 |
| chr2_134539298_134539479 | 0.67 |
Hao1 |
hydroxyacid oxidase 1, liver |
14919 |
0.28 |
| chr9_106261948_106262435 | 0.67 |
Alas1 |
aminolevulinic acid synthase 1 |
13537 |
0.1 |
| chr13_115096076_115096227 | 0.67 |
Gm49395 |
predicted gene, 49395 |
5473 |
0.14 |
| chr4_109458656_109458807 | 0.66 |
Rnf11 |
ring finger protein 11 |
17944 |
0.15 |
| chr9_103118235_103118386 | 0.66 |
Gm37166 |
predicted gene, 37166 |
2058 |
0.27 |
| chr4_148616552_148616759 | 0.66 |
Tardbp |
TAR DNA binding protein |
325 |
0.81 |
| chr11_86599706_86599919 | 0.66 |
Vmp1 |
vacuole membrane protein 1 |
2451 |
0.23 |
| chr11_60199358_60199583 | 0.66 |
Mir6921 |
microRNA 6921 |
1147 |
0.3 |
| chr9_122857713_122857864 | 0.65 |
Zfp445 |
zinc finger protein 445 |
288 |
0.82 |
| chr16_42908893_42909236 | 0.65 |
Zbtb20 |
zinc finger and BTB domain containing 20 |
1412 |
0.39 |
| chr1_172168265_172168469 | 0.65 |
Dcaf8 |
DDB1 and CUL4 associated factor 8 |
5587 |
0.11 |
| chr4_6056446_6056603 | 0.65 |
Gm11797 |
predicted gene 11797 |
13350 |
0.28 |
| chr5_40690611_40690784 | 0.65 |
Gm23022 |
predicted gene, 23022 |
284768 |
0.01 |
| chr6_37510078_37510463 | 0.64 |
Akr1d1 |
aldo-keto reductase family 1, member D1 |
19903 |
0.21 |
| chr11_50324646_50324797 | 0.64 |
Canx |
calnexin |
952 |
0.43 |
| chr8_22888536_22888711 | 0.64 |
Gm45555 |
predicted gene 45555 |
15034 |
0.15 |
| chr19_10181326_10181635 | 0.64 |
Fads1 |
fatty acid desaturase 1 |
1408 |
0.26 |
| chr4_3321136_3321319 | 0.64 |
Gm11786 |
predicted gene 11786 |
5948 |
0.22 |
| chr3_141967234_141967427 | 0.64 |
Bmpr1b |
bone morphogenetic protein receptor, type 1B |
14934 |
0.29 |
| chr5_33465562_33465766 | 0.63 |
Gm43851 |
predicted gene 43851 |
28200 |
0.15 |
| chr13_9060425_9060705 | 0.63 |
Gm36264 |
predicted gene, 36264 |
15886 |
0.13 |
| chr7_39841684_39841835 | 0.63 |
4930558N11Rik |
RIKEN cDNA 4930558N11 gene |
44017 |
0.11 |
| chr1_51914046_51914411 | 0.63 |
Myo1b |
myosin IB |
1684 |
0.29 |
| chr6_136906748_136907166 | 0.63 |
Erp27 |
endoplasmic reticulum protein 27 |
15183 |
0.11 |
| chr5_101742329_101742522 | 0.62 |
Cds1 |
CDP-diacylglycerol synthase 1 |
22705 |
0.14 |
| chr10_42232281_42232611 | 0.62 |
Foxo3 |
forkhead box O3 |
25920 |
0.22 |
| chr6_16551146_16551546 | 0.62 |
Gm36503 |
predicted gene, 36503 |
959 |
0.7 |
| chr3_121643612_121643813 | 0.62 |
A730020M07Rik |
RIKEN cDNA A730020M07 gene |
227 |
0.92 |
| chr3_32562720_32562907 | 0.61 |
Mfn1 |
mitofusin 1 |
95 |
0.96 |
| chr4_22498530_22498747 | 0.61 |
Gm30731 |
predicted gene, 30731 |
8090 |
0.16 |
| chr17_24236940_24237256 | 0.61 |
Mir5134 |
microRNA 5134 |
2494 |
0.12 |
| chr3_48846432_48846591 | 0.60 |
Gm37190 |
predicted gene, 37190 |
85562 |
0.1 |
| chr17_83843520_83843705 | 0.60 |
Haao |
3-hydroxyanthranilate 3,4-dioxygenase |
3137 |
0.22 |
| chrX_42223380_42223551 | 0.60 |
Stag2 |
stromal antigen 2 |
71285 |
0.1 |
| chr12_73861884_73862127 | 0.59 |
Gm15283 |
predicted gene 15283 |
7607 |
0.18 |
| chr1_164543147_164543444 | 0.59 |
D630023O14Rik |
RIKEN cDNA D630023O14 gene |
32160 |
0.13 |
| chr18_33436230_33436484 | 0.59 |
Nrep |
neuronal regeneration related protein |
27078 |
0.18 |
| chr11_16761103_16761540 | 0.59 |
Egfr |
epidermal growth factor receptor |
9091 |
0.2 |
| chr12_91560214_91560422 | 0.57 |
Gm25776 |
predicted gene, 25776 |
13254 |
0.16 |
| chr1_21240946_21241097 | 0.57 |
Gsta3 |
glutathione S-transferase, alpha 3 |
392 |
0.76 |
| chr18_39237141_39237343 | 0.57 |
Arhgap26 |
Rho GTPase activating protein 26 |
9874 |
0.26 |
| chr5_89352355_89352713 | 0.57 |
Gc |
vitamin D binding protein |
83094 |
0.1 |
| chr4_135787434_135787634 | 0.57 |
Myom3 |
myomesin family, member 3 |
10656 |
0.14 |
| chr6_146125298_146125516 | 0.56 |
Itpr2 |
inositol 1,4,5-triphosphate receptor 2 |
21801 |
0.23 |
| chr16_11135224_11135513 | 0.56 |
Txndc11 |
thioredoxin domain containing 11 |
718 |
0.49 |
| chr15_67925007_67925168 | 0.56 |
Gm49409 |
predicted gene, 49409 |
17717 |
0.25 |
| chr6_143857220_143857571 | 0.55 |
Sox5 |
SRY (sex determining region Y)-box 5 |
89693 |
0.09 |
| chr7_134372608_134372831 | 0.55 |
D7Ertd443e |
DNA segment, Chr 7, ERATO Doi 443, expressed |
4106 |
0.34 |
| chr14_88463900_88464058 | 0.54 |
Pcdh20 |
protocadherin 20 |
7367 |
0.23 |
| chr13_20403622_20403794 | 0.54 |
Elmo1 |
engulfment and cell motility 1 |
69018 |
0.1 |
| chr3_146260064_146260215 | 0.54 |
Lpar3 |
lysophosphatidic acid receptor 3 |
39176 |
0.13 |
| chr10_68327935_68328096 | 0.54 |
4930545H06Rik |
RIKEN cDNA 4930545H06 gene |
6873 |
0.22 |
| chr10_99222701_99222877 | 0.53 |
Gm34574 |
predicted gene, 34574 |
277 |
0.85 |
| chr18_38923142_38923385 | 0.53 |
Fgf1 |
fibroblast growth factor 1 |
4492 |
0.24 |
| chr13_4077186_4077340 | 0.53 |
Akr1c14 |
aldo-keto reductase family 1, member C14 |
991 |
0.44 |
| chr2_122206384_122206535 | 0.53 |
Gm14050 |
predicted gene 14050 |
1461 |
0.25 |
| chr2_177839201_177839672 | 0.53 |
Gm14325 |
predicted gene 14325 |
882 |
0.56 |
| chr6_31453535_31453686 | 0.52 |
Mkln1 |
muskelin 1, intracellular mediator containing kelch motifs |
14510 |
0.19 |
| chr4_76366772_76366969 | 0.52 |
Ptprd |
protein tyrosine phosphatase, receptor type, D |
22627 |
0.21 |
| chr6_119584715_119585189 | 0.52 |
Gm44317 |
predicted gene, 44317 |
14011 |
0.2 |
| chr15_99038783_99039270 | 0.52 |
Tuba1c |
tubulin, alpha 1C |
8705 |
0.09 |
| chr11_106667727_106667890 | 0.52 |
Pecam1 |
platelet/endothelial cell adhesion molecule 1 |
4482 |
0.2 |
| chr13_94803722_94803937 | 0.51 |
Tbca |
tubulin cofactor A |
13905 |
0.18 |
| chr8_84761164_84761339 | 0.51 |
Nfix |
nuclear factor I/X |
12145 |
0.11 |
| chr2_71212098_71212533 | 0.51 |
Dync1i2 |
dynein cytoplasmic 1 intermediate chain 2 |
311 |
0.92 |
| chr8_85504540_85504770 | 0.51 |
Gpt2 |
glutamic pyruvate transaminase (alanine aminotransferase) 2 |
11322 |
0.16 |
| chr11_28702228_28702389 | 0.51 |
2810471M01Rik |
RIKEN cDNA 2810471M01 gene |
20744 |
0.15 |
| chr2_42039612_42039773 | 0.51 |
Gm13461 |
predicted gene 13461 |
50863 |
0.18 |
| chr13_28877494_28877645 | 0.50 |
2610307P16Rik |
RIKEN cDNA 2610307P16 gene |
5887 |
0.2 |
| chr12_34997914_34998088 | 0.50 |
Prps1l1 |
phosphoribosyl pyrophosphate synthetase 1-like 1 |
13240 |
0.2 |
| chr3_79845512_79845692 | 0.50 |
Tmem144 |
transmembrane protein 144 |
2690 |
0.24 |
| chr11_16779721_16779963 | 0.50 |
Egfr |
epidermal growth factor receptor |
27612 |
0.17 |
| chr6_141633750_141633901 | 0.50 |
Slco1b2 |
solute carrier organic anion transporter family, member 1b2 |
633 |
0.81 |
| chr2_77522021_77522181 | 0.49 |
Zfp385b |
zinc finger protein 385B |
2483 |
0.38 |
| chr10_34326839_34326990 | 0.49 |
Nt5dc1 |
5'-nucleotidase domain containing 1 |
13231 |
0.13 |
| chr6_22341838_22341989 | 0.49 |
Fam3c |
family with sequence similarity 3, member C |
2000 |
0.42 |
| chr11_120238015_120238347 | 0.49 |
Bahcc1 |
BAH domain and coiled-coil containing 1 |
1482 |
0.22 |
| chr10_68131050_68131381 | 0.49 |
Arid5b |
AT rich interactive domain 5B (MRF1-like) |
5411 |
0.28 |
| chr8_104787830_104787982 | 0.48 |
Gm45782 |
predicted gene 45782 |
2078 |
0.17 |
| chr19_44386173_44386338 | 0.48 |
Scd1 |
stearoyl-Coenzyme A desaturase 1 |
20435 |
0.13 |
| chr4_121168042_121168205 | 0.48 |
Rlf |
rearranged L-myc fusion sequence |
20411 |
0.12 |
| chr11_67359982_67360339 | 0.48 |
Myh13 |
myosin, heavy polypeptide 13, skeletal muscle |
4625 |
0.26 |
| chr3_30609113_30609310 | 0.48 |
Mynn |
myoneurin |
4300 |
0.13 |
| chr15_77307896_77308074 | 0.48 |
Rbfox2 |
RNA binding protein, fox-1 homolog (C. elegans) 2 |
981 |
0.5 |
| chr6_124455872_124456023 | 0.48 |
Clstn3 |
calsyntenin 3 |
1938 |
0.2 |
| chr13_4230959_4231234 | 0.48 |
Akr1c19 |
aldo-keto reductase family 1, member C19 |
2396 |
0.22 |
| chr3_18877514_18877665 | 0.47 |
Gm30341 |
predicted gene, 30341 |
124512 |
0.06 |
| chr2_32254540_32254691 | 0.47 |
Uck1 |
uridine-cytidine kinase 1 |
2054 |
0.14 |
| chr13_43548363_43548529 | 0.47 |
Mcur1 |
mitochondrial calcium uniporter regulator 1 |
2326 |
0.24 |
| chr2_58766481_58766674 | 0.47 |
Upp2 |
uridine phosphorylase 2 |
1252 |
0.49 |
| chr7_84417589_84417755 | 0.47 |
Arnt2 |
aryl hydrocarbon receptor nuclear translocator 2 |
7496 |
0.17 |
| chr2_72178406_72178610 | 0.47 |
Rapgef4 |
Rap guanine nucleotide exchange factor (GEF) 4 |
1185 |
0.5 |
| chr16_43234125_43234650 | 0.47 |
Zbtb20 |
zinc finger and BTB domain containing 20 |
1493 |
0.45 |
| chr9_53359307_53359484 | 0.47 |
Exph5 |
exophilin 5 |
18206 |
0.15 |
| chr18_39459109_39459298 | 0.47 |
Nr3c1 |
nuclear receptor subfamily 3, group C, member 1 |
28029 |
0.2 |
| chr6_28118749_28118957 | 0.46 |
Grm8 |
glutamate receptor, metabotropic 8 |
7272 |
0.25 |
| chr5_87499331_87499512 | 0.46 |
Ugt2a1 |
UDP glucuronosyltransferase 2 family, polypeptide A1 |
8550 |
0.12 |
| chr8_34157897_34158198 | 0.46 |
Saraf |
store-operated calcium entry-associated regulatory factor |
434 |
0.71 |
| chr10_95413803_95413974 | 0.46 |
Socs2 |
suppressor of cytokine signaling 2 |
779 |
0.53 |
| chr11_86614634_86614813 | 0.46 |
Vmp1 |
vacuole membrane protein 1 |
12428 |
0.15 |
| chr13_112486027_112486262 | 0.46 |
Il6st |
interleukin 6 signal transducer |
11098 |
0.17 |
| chr4_53596359_53596666 | 0.46 |
Slc44a1 |
solute carrier family 44, member 1 |
407 |
0.85 |
| chr10_3109374_3109527 | 0.46 |
Gm30674 |
predicted gene, 30674 |
5331 |
0.15 |
| chr11_80974682_80974867 | 0.46 |
Gm11416 |
predicted gene 11416 |
72020 |
0.1 |
| chr13_46058970_46059151 | 0.45 |
Gm45949 |
predicted gene, 45949 |
1537 |
0.49 |
| chr15_42717783_42717947 | 0.45 |
Angpt1 |
angiopoietin 1 |
40888 |
0.18 |
| chr4_33310735_33311130 | 0.45 |
Rngtt |
RNA guanylyltransferase and 5'-phosphatase |
604 |
0.51 |
| chr18_37761572_37761738 | 0.45 |
Pcdhgb8 |
protocadherin gamma subfamily B, 8 |
146 |
0.83 |
| chr2_177397223_177397387 | 0.45 |
Gm14418 |
predicted gene 14418 |
1014 |
0.51 |
| chr9_70446864_70447026 | 0.45 |
Rnf111 |
ring finger 111 |
6901 |
0.16 |
| chr2_4895477_4895652 | 0.45 |
Sephs1 |
selenophosphate synthetase 1 |
11180 |
0.15 |
| chr18_12668617_12668967 | 0.45 |
Gm41668 |
predicted gene, 41668 |
20373 |
0.13 |
| chr17_5261920_5262119 | 0.45 |
Gm29050 |
predicted gene 29050 |
126744 |
0.05 |
| chr8_77199905_77200064 | 0.45 |
Gm23260 |
predicted gene, 23260 |
5289 |
0.24 |
| chr1_67204437_67204883 | 0.45 |
Gm15668 |
predicted gene 15668 |
44540 |
0.15 |
| chr11_106468981_106469217 | 0.45 |
Ern1 |
endoplasmic reticulum (ER) to nucleus signalling 1 |
18697 |
0.14 |
| chr1_67136982_67137399 | 0.45 |
Cps1 |
carbamoyl-phosphate synthetase 1 |
14164 |
0.23 |
| chr15_51796404_51796577 | 0.45 |
Eif3h |
eukaryotic translation initiation factor 3, subunit H |
5941 |
0.26 |
| chr11_51595976_51596127 | 0.45 |
Phykpl |
5-phosphohydroxy-L-lysine phospholyase |
3216 |
0.16 |
| chr3_118487666_118487837 | 0.44 |
Gm26871 |
predicted gene, 26871 |
30092 |
0.12 |
| chr9_42259997_42260195 | 0.44 |
Sc5d |
sterol-C5-desaturase |
4104 |
0.2 |
| chr2_176635310_176635480 | 0.44 |
Gm14308 |
predicted gene 14308 |
923 |
0.55 |
| chr13_112331091_112331250 | 0.44 |
Ankrd55 |
ankyrin repeat domain 55 |
12716 |
0.17 |
| chr3_143103418_143103577 | 0.44 |
Gm43614 |
predicted gene 43614 |
4451 |
0.28 |
| chr2_175738745_175738952 | 0.44 |
Gm2007 |
predicted gene 2007 |
2689 |
0.22 |
| chr3_133348031_133348200 | 0.44 |
Ppa2 |
pyrophosphatase (inorganic) 2 |
39 |
0.98 |
| chr9_79762117_79762289 | 0.44 |
Cox7a2 |
cytochrome c oxidase subunit 7A2 |
2325 |
0.25 |
| chr11_89613753_89613946 | 0.43 |
Ankfn1 |
ankyrin-repeat and fibronectin type III domain containing 1 |
75293 |
0.12 |
| chr11_111998446_111998808 | 0.43 |
Gm11679 |
predicted gene 11679 |
44951 |
0.19 |
| chr1_136629198_136629381 | 0.43 |
Zfp281 |
zinc finger protein 281 |
4388 |
0.14 |
| chr17_15050243_15050467 | 0.43 |
Ermard |
ER membrane associated RNA degradation |
10 |
0.97 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.2 | 0.6 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.2 | 1.0 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.1 | 0.6 | GO:0086045 | membrane depolarization during AV node cell action potential(GO:0086045) |
| 0.1 | 0.3 | GO:0006768 | biotin metabolic process(GO:0006768) |
| 0.1 | 0.6 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.1 | 0.3 | GO:0021538 | epithalamus development(GO:0021538) habenula development(GO:0021986) |
| 0.1 | 0.3 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) platelet activating factor metabolic process(GO:0046469) |
| 0.1 | 0.3 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.1 | 0.3 | GO:0019482 | beta-alanine metabolic process(GO:0019482) |
| 0.1 | 0.9 | GO:0009404 | toxin metabolic process(GO:0009404) |
| 0.1 | 0.5 | GO:0015871 | choline transport(GO:0015871) |
| 0.1 | 0.2 | GO:1990314 | cellular response to insulin-like growth factor stimulus(GO:1990314) |
| 0.1 | 0.5 | GO:0034310 | primary alcohol catabolic process(GO:0034310) |
| 0.1 | 0.3 | GO:0006987 | activation of signaling protein activity involved in unfolded protein response(GO:0006987) |
| 0.1 | 0.3 | GO:1903966 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.1 | 0.3 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.1 | 0.2 | GO:0070103 | regulation of interleukin-6-mediated signaling pathway(GO:0070103) |
| 0.1 | 0.3 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.1 | 0.9 | GO:0009109 | coenzyme catabolic process(GO:0009109) |
| 0.1 | 0.3 | GO:0032364 | oxygen homeostasis(GO:0032364) |
| 0.1 | 0.1 | GO:2000017 | positive regulation of determination of dorsal identity(GO:2000017) |
| 0.1 | 0.1 | GO:0090027 | negative regulation of monocyte chemotaxis(GO:0090027) |
| 0.1 | 0.2 | GO:0021827 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) postnatal olfactory bulb interneuron migration(GO:0021827) |
| 0.1 | 0.2 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.1 | 0.3 | GO:0032055 | negative regulation of translation in response to stress(GO:0032055) |
| 0.1 | 0.2 | GO:0071926 | cannabinoid signaling pathway(GO:0038171) endocannabinoid signaling pathway(GO:0071926) |
| 0.1 | 0.4 | GO:1902222 | L-phenylalanine catabolic process(GO:0006559) erythrose 4-phosphate/phosphoenolpyruvate family amino acid catabolic process(GO:1902222) |
| 0.1 | 0.2 | GO:0003383 | apical constriction(GO:0003383) |
| 0.1 | 0.2 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.1 | 0.2 | GO:0031087 | nuclear-transcribed mRNA catabolic process, deadenylation-independent decay(GO:0031086) deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.1 | 0.3 | GO:0060916 | mesenchymal cell proliferation involved in lung development(GO:0060916) |
| 0.1 | 0.3 | GO:1900094 | nodal signaling pathway involved in determination of left/right asymmetry(GO:0038107) regulation of transcription from RNA polymerase II promoter involved in determination of left/right symmetry(GO:1900094) nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900164) |
| 0.1 | 0.2 | GO:0003062 | regulation of heart rate by chemical signal(GO:0003062) |
| 0.1 | 0.4 | GO:0001547 | antral ovarian follicle growth(GO:0001547) |
| 0.0 | 0.1 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.0 | 0.1 | GO:0002036 | regulation of L-glutamate transport(GO:0002036) |
| 0.0 | 1.1 | GO:0009812 | flavonoid metabolic process(GO:0009812) |
| 0.0 | 0.3 | GO:0046874 | quinolinate metabolic process(GO:0046874) |
| 0.0 | 0.1 | GO:1902474 | positive regulation of protein localization to synapse(GO:1902474) |
| 0.0 | 0.1 | GO:0090283 | regulation of protein glycosylation in Golgi(GO:0090283) |
| 0.0 | 0.1 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.0 | 0.1 | GO:0015770 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.0 | 0.2 | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.0 | 0.2 | GO:0018214 | protein carboxylation(GO:0018214) |
| 0.0 | 0.1 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.0 | 0.2 | GO:0000480 | endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.0 | 0.2 | GO:0032488 | Cdc42 protein signal transduction(GO:0032488) |
| 0.0 | 0.1 | GO:0008050 | female courtship behavior(GO:0008050) |
| 0.0 | 0.3 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.0 | 0.1 | GO:1901844 | regulation of cell communication by electrical coupling involved in cardiac conduction(GO:1901844) |
| 0.0 | 0.5 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.0 | 0.3 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.0 | 0.2 | GO:0042985 | negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 0.0 | 0.1 | GO:0042126 | nitrate metabolic process(GO:0042126) |
| 0.0 | 0.1 | GO:0046618 | drug export(GO:0046618) |
| 0.0 | 0.1 | GO:0019065 | receptor-mediated endocytosis of virus by host cell(GO:0019065) endocytosis involved in viral entry into host cell(GO:0075509) |
| 0.0 | 0.1 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.0 | 0.1 | GO:1901475 | pyruvate transmembrane transport(GO:1901475) |
| 0.0 | 0.4 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.0 | 0.1 | GO:1990036 | calcium ion import into sarcoplasmic reticulum(GO:1990036) |
| 0.0 | 0.1 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.0 | 0.0 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.0 | 1.0 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 0.1 | GO:0035910 | ascending aorta development(GO:0035905) ascending aorta morphogenesis(GO:0035910) |
| 0.0 | 0.2 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.0 | 0.2 | GO:0055091 | phospholipid homeostasis(GO:0055091) |
| 0.0 | 0.1 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.0 | 0.1 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.0 | 0.1 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.0 | 0.3 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.0 | 0.1 | GO:0014722 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) |
| 0.0 | 0.1 | GO:0014835 | myoblast differentiation involved in skeletal muscle regeneration(GO:0014835) |
| 0.0 | 0.3 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 0.1 | GO:0006046 | N-acetylglucosamine catabolic process(GO:0006046) |
| 0.0 | 0.1 | GO:0010891 | negative regulation of sequestering of triglyceride(GO:0010891) |
| 0.0 | 0.1 | GO:0071816 | protein insertion into ER membrane(GO:0045048) tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 0.0 | 0.2 | GO:0098908 | regulation of neuronal action potential(GO:0098908) |
| 0.0 | 0.1 | GO:0032380 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.0 | 0.2 | GO:0006655 | phosphatidylglycerol biosynthetic process(GO:0006655) |
| 0.0 | 0.1 | GO:0061010 | gall bladder development(GO:0061010) |
| 0.0 | 0.3 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.0 | 0.1 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.0 | 0.6 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.0 | 0.2 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.0 | 0.1 | GO:1900135 | positive regulation of renin secretion into blood stream(GO:1900135) |
| 0.0 | 0.2 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.0 | 0.1 | GO:0061072 | iris morphogenesis(GO:0061072) |
| 0.0 | 0.0 | GO:0055005 | ventricular cardiac myofibril assembly(GO:0055005) |
| 0.0 | 0.2 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) |
| 0.0 | 0.1 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.0 | 0.1 | GO:0072393 | microtubule anchoring at microtubule organizing center(GO:0072393) |
| 0.0 | 0.0 | GO:1903999 | negative regulation of eating behavior(GO:1903999) |
| 0.0 | 0.1 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 0.2 | GO:1902855 | regulation of nonmotile primary cilium assembly(GO:1902855) |
| 0.0 | 0.1 | GO:2000211 | regulation of glutamate metabolic process(GO:2000211) |
| 0.0 | 0.1 | GO:0043651 | linoleic acid metabolic process(GO:0043651) |
| 0.0 | 0.1 | GO:0050904 | diapedesis(GO:0050904) |
| 0.0 | 0.0 | GO:1904251 | regulation of bile acid metabolic process(GO:1904251) |
| 0.0 | 0.1 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.0 | 0.1 | GO:0016561 | protein import into peroxisome matrix, translocation(GO:0016561) |
| 0.0 | 0.1 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.0 | 0.1 | GO:0048633 | positive regulation of skeletal muscle tissue growth(GO:0048633) |
| 0.0 | 0.0 | GO:2000828 | regulation of parathyroid hormone secretion(GO:2000828) |
| 0.0 | 0.1 | GO:0072602 | interleukin-4 secretion(GO:0072602) |
| 0.0 | 0.1 | GO:0015747 | urate transport(GO:0015747) |
| 0.0 | 0.1 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.0 | 0.1 | GO:0010040 | response to iron(II) ion(GO:0010040) |
| 0.0 | 0.1 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.0 | 0.1 | GO:1900378 | positive regulation of melanin biosynthetic process(GO:0048023) positive regulation of secondary metabolite biosynthetic process(GO:1900378) |
| 0.0 | 0.1 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.2 | GO:0097105 | presynaptic membrane assembly(GO:0097105) |
| 0.0 | 0.1 | GO:2000609 | regulation of thyroid hormone generation(GO:2000609) |
| 0.0 | 0.3 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.1 | GO:0006499 | N-terminal protein myristoylation(GO:0006499) |
| 0.0 | 0.1 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.0 | GO:1900133 | regulation of renin secretion into blood stream(GO:1900133) |
| 0.0 | 0.1 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.0 | 0.1 | GO:0034756 | regulation of iron ion transport(GO:0034756) |
| 0.0 | 0.1 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 0.0 | 0.2 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.0 | 0.0 | GO:0003419 | growth plate cartilage chondrocyte proliferation(GO:0003419) |
| 0.0 | 0.1 | GO:0002439 | chronic inflammatory response to antigenic stimulus(GO:0002439) |
| 0.0 | 0.1 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.0 | 0.1 | GO:0070447 | positive regulation of oligodendrocyte progenitor proliferation(GO:0070447) |
| 0.0 | 0.1 | GO:0030382 | sperm mitochondrion organization(GO:0030382) |
| 0.0 | 0.0 | GO:2000370 | positive regulation of clathrin-mediated endocytosis(GO:2000370) |
| 0.0 | 0.3 | GO:0006699 | bile acid biosynthetic process(GO:0006699) |
| 0.0 | 0.1 | GO:0010612 | regulation of cardiac muscle adaptation(GO:0010612) regulation of cardiac muscle hypertrophy in response to stress(GO:1903242) |
| 0.0 | 0.2 | GO:1902715 | positive regulation of interferon-gamma secretion(GO:1902715) |
| 0.0 | 0.1 | GO:0010572 | positive regulation of platelet activation(GO:0010572) |
| 0.0 | 0.1 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.0 | 0.1 | GO:0048752 | semicircular canal morphogenesis(GO:0048752) |
| 0.0 | 0.1 | GO:0060355 | positive regulation of cell adhesion molecule production(GO:0060355) |
| 0.0 | 0.0 | GO:0014732 | skeletal muscle atrophy(GO:0014732) |
| 0.0 | 0.0 | GO:1903347 | negative regulation of bicellular tight junction assembly(GO:1903347) |
| 0.0 | 0.1 | GO:1901164 | negative regulation of trophoblast cell migration(GO:1901164) |
| 0.0 | 0.1 | GO:0072643 | interferon-gamma secretion(GO:0072643) |
| 0.0 | 0.1 | GO:0008655 | pyrimidine-containing compound salvage(GO:0008655) pyrimidine nucleoside salvage(GO:0043097) |
| 0.0 | 0.0 | GO:0010693 | negative regulation of alkaline phosphatase activity(GO:0010693) |
| 0.0 | 0.1 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.0 | 0.1 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.0 | 0.2 | GO:0090005 | negative regulation of establishment of protein localization to plasma membrane(GO:0090005) |
| 0.0 | 0.1 | GO:1904479 | negative regulation of intestinal absorption(GO:1904479) |
| 0.0 | 0.1 | GO:0018211 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.0 | 0.1 | GO:1900103 | positive regulation of endoplasmic reticulum unfolded protein response(GO:1900103) |
| 0.0 | 0.0 | GO:1903279 | regulation of calcium:sodium antiporter activity(GO:1903279) |
| 0.0 | 0.0 | GO:0018364 | peptidyl-glutamine methylation(GO:0018364) |
| 0.0 | 0.0 | GO:0032898 | neurotrophin production(GO:0032898) |
| 0.0 | 0.0 | GO:0006172 | ADP biosynthetic process(GO:0006172) |
| 0.0 | 0.1 | GO:1903069 | regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903069) |
| 0.0 | 0.2 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.0 | GO:0003162 | atrioventricular node development(GO:0003162) |
| 0.0 | 0.1 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.0 | 0.0 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.0 | 0.1 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.0 | 0.1 | GO:0007567 | parturition(GO:0007567) |
| 0.0 | 0.2 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 0.0 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 0.0 | 0.2 | GO:0045056 | transcytosis(GO:0045056) |
| 0.0 | 0.1 | GO:0098904 | regulation of AV node cell action potential(GO:0098904) |
| 0.0 | 0.1 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 | 0.0 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.0 | 0.0 | GO:0072361 | regulation of glycolytic process by regulation of transcription from RNA polymerase II promoter(GO:0072361) |
| 0.0 | 0.0 | GO:0021912 | regulation of transcription from RNA polymerase II promoter involved in spinal cord motor neuron fate specification(GO:0021912) |
| 0.0 | 0.1 | GO:0048671 | negative regulation of collateral sprouting(GO:0048671) |
| 0.0 | 0.0 | GO:2000686 | regulation of rubidium ion transmembrane transporter activity(GO:2000686) |
| 0.0 | 0.1 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.0 | 0.0 | GO:0050883 | musculoskeletal movement, spinal reflex action(GO:0050883) |
| 0.0 | 0.0 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.0 | 0.0 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.0 | 0.1 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.0 | 0.0 | GO:0016557 | peroxisome membrane biogenesis(GO:0016557) |
| 0.0 | 0.1 | GO:0090400 | stress-induced premature senescence(GO:0090400) |
| 0.0 | 0.0 | GO:1900126 | negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.0 | 0.1 | GO:0071447 | cellular response to hydroperoxide(GO:0071447) |
| 0.0 | 0.0 | GO:0000087 | mitotic M phase(GO:0000087) |
| 0.0 | 0.1 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.0 | 0.0 | GO:0009188 | ribonucleoside diphosphate biosynthetic process(GO:0009188) |
| 0.0 | 0.1 | GO:0021684 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.0 | 0.1 | GO:1902897 | regulation of postsynaptic density protein 95 clustering(GO:1902897) |
| 0.0 | 0.1 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.0 | 0.1 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.0 | 0.1 | GO:0006104 | succinyl-CoA metabolic process(GO:0006104) |
| 0.0 | 0.0 | GO:0040031 | snRNA modification(GO:0040031) |
| 0.0 | 0.0 | GO:0044828 | negative regulation by host of viral genome replication(GO:0044828) |
| 0.0 | 0.0 | GO:0006069 | ethanol metabolic process(GO:0006067) ethanol oxidation(GO:0006069) |
| 0.0 | 0.0 | GO:0071494 | cellular response to UV-C(GO:0071494) |
| 0.0 | 0.2 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.0 | 0.0 | GO:0060336 | negative regulation of response to interferon-gamma(GO:0060331) negative regulation of interferon-gamma-mediated signaling pathway(GO:0060336) |
| 0.0 | 0.1 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 0.0 | 0.1 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 0.0 | 0.1 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.0 | GO:0048382 | mesendoderm development(GO:0048382) |
| 0.0 | 0.0 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.1 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.0 | 0.1 | GO:0098719 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.2 | GO:0032516 | positive regulation of phosphoprotein phosphatase activity(GO:0032516) |
| 0.0 | 0.0 | GO:0010571 | positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.0 | 0.0 | GO:1902202 | regulation of hepatocyte growth factor receptor signaling pathway(GO:1902202) |
| 0.0 | 0.1 | GO:0015867 | ATP transport(GO:0015867) |
| 0.0 | 0.0 | GO:0032058 | positive regulation of translational initiation in response to stress(GO:0032058) |
| 0.0 | 0.2 | GO:0007096 | regulation of exit from mitosis(GO:0007096) |
| 0.0 | 0.1 | GO:0046606 | negative regulation of centrosome duplication(GO:0010826) negative regulation of centrosome cycle(GO:0046606) |
| 0.0 | 0.1 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.0 | 0.1 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.0 | 0.1 | GO:0090503 | RNA phosphodiester bond hydrolysis, exonucleolytic(GO:0090503) |
| 0.0 | 0.0 | GO:0070197 | meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.0 | 0.1 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 0.0 | GO:0060353 | regulation of cell adhesion molecule production(GO:0060353) |
| 0.0 | 0.1 | GO:0007135 | meiosis II(GO:0007135) |
| 0.0 | 0.0 | GO:0090206 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.0 | 0.1 | GO:0006537 | glutamate biosynthetic process(GO:0006537) |
| 0.0 | 0.2 | GO:0019430 | removal of superoxide radicals(GO:0019430) |
| 0.0 | 0.1 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 | 0.0 | GO:0043974 | histone H3-K27 acetylation(GO:0043974) regulation of histone H3-K27 acetylation(GO:1901674) |
| 0.0 | 0.0 | GO:2000645 | negative regulation of receptor catabolic process(GO:2000645) |
| 0.0 | 0.0 | GO:1903423 | positive regulation of synaptic vesicle recycling(GO:1903423) |
| 0.0 | 0.1 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.0 | 0.1 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.2 | GO:0006308 | DNA catabolic process(GO:0006308) |
| 0.0 | 0.1 | GO:0019370 | leukotriene biosynthetic process(GO:0019370) |
| 0.0 | 0.1 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 0.0 | 0.3 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 0.1 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 0.0 | 0.1 | GO:0002024 | diet induced thermogenesis(GO:0002024) adaptive thermogenesis(GO:1990845) |
| 0.0 | 0.1 | GO:0051974 | negative regulation of telomerase activity(GO:0051974) |
| 0.0 | 0.0 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.0 | 0.0 | GO:0031547 | brain-derived neurotrophic factor receptor signaling pathway(GO:0031547) |
| 0.0 | 0.1 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 0.0 | 0.1 | GO:0021960 | anterior commissure morphogenesis(GO:0021960) |
| 0.0 | 0.0 | GO:0061624 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) glycolytic process through fructose-1-phosphate(GO:0061625) |
| 0.0 | 0.0 | GO:0086017 | Purkinje myocyte action potential(GO:0086017) |
| 0.0 | 0.0 | GO:0006971 | hypotonic response(GO:0006971) |
| 0.0 | 0.1 | GO:0031269 | pseudopodium assembly(GO:0031269) |
| 0.0 | 0.0 | GO:0036135 | Schwann cell migration(GO:0036135) regulation of Schwann cell migration(GO:1900147) |
| 0.0 | 0.0 | GO:0035519 | protein K29-linked ubiquitination(GO:0035519) |
| 0.0 | 0.0 | GO:0006063 | uronic acid metabolic process(GO:0006063) glucuronate metabolic process(GO:0019585) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.1 | 0.6 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.1 | 0.6 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.1 | 0.2 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.1 | 0.2 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.1 | 0.2 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.1 | 0.2 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.0 | 0.2 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.0 | 0.1 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.1 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 0.2 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.0 | 0.3 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 0.1 | GO:0071256 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.0 | 0.1 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 0.1 | GO:0071818 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.0 | 0.2 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.1 | GO:0016939 | kinesin II complex(GO:0016939) |
| 0.0 | 0.6 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.1 | GO:0034666 | integrin alpha2-beta1 complex(GO:0034666) |
| 0.0 | 0.2 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.1 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.0 | 0.4 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.3 | GO:0031332 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.0 | 0.1 | GO:1990597 | AIP1-IRE1 complex(GO:1990597) |
| 0.0 | 0.3 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 1.0 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.1 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.0 | 0.2 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.1 | GO:0033648 | host intracellular organelle(GO:0033647) host intracellular membrane-bounded organelle(GO:0033648) |
| 0.0 | 0.1 | GO:0044308 | axonal spine(GO:0044308) |
| 0.0 | 0.1 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 0.4 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 0.1 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.0 | 0.2 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.1 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.0 | 0.3 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.1 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.0 | GO:0097413 | Lewy body(GO:0097413) |
| 0.0 | 0.1 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 0.1 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 0.2 | GO:0098533 | ATPase dependent transmembrane transport complex(GO:0098533) |
| 0.0 | 0.1 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.0 | 0.1 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.1 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.2 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.3 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.1 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.0 | 0.0 | GO:0031502 | dolichyl-phosphate-mannose-protein mannosyltransferase complex(GO:0031502) |
| 0.0 | 0.0 | GO:0090661 | box H/ACA telomerase RNP complex(GO:0090661) |
| 0.0 | 0.2 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 0.1 | GO:0000782 | telomere cap complex(GO:0000782) nuclear telomere cap complex(GO:0000783) |
| 0.0 | 0.1 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.0 | 0.2 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.2 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.0 | 0.1 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.0 | 0.0 | GO:0005593 | FACIT collagen trimer(GO:0005593) |
| 0.0 | 0.1 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.0 | 0.1 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.0 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.0 | 0.1 | GO:0030062 | mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0016743 | carboxyl- or carbamoyltransferase activity(GO:0016743) |
| 0.3 | 0.9 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.2 | 0.7 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.2 | 0.6 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.1 | 0.4 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.1 | 0.4 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.1 | 0.4 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.1 | 0.4 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.1 | 0.2 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 0.1 | 0.5 | GO:0015198 | oligopeptide transporter activity(GO:0015198) |
| 0.1 | 0.2 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.1 | 0.2 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.1 | 0.3 | GO:0032896 | palmitoyl-CoA 9-desaturase activity(GO:0032896) |
| 0.1 | 0.3 | GO:0047023 | androsterone dehydrogenase activity(GO:0047023) |
| 0.1 | 0.3 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.1 | 0.1 | GO:0000009 | alpha-1,6-mannosyltransferase activity(GO:0000009) |
| 0.1 | 0.2 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.1 | 0.2 | GO:0016899 | oxidoreductase activity, acting on the CH-OH group of donors, oxygen as acceptor(GO:0016899) |
| 0.1 | 0.2 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.1 | 0.2 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.1 | 0.2 | GO:0098518 | polynucleotide phosphatase activity(GO:0098518) |
| 0.1 | 0.6 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.1 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.0 | 0.2 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.0 | 0.1 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.0 | 0.1 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.0 | 0.2 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.0 | 0.1 | GO:0008515 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.0 | 0.1 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.1 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.0 | 0.2 | GO:0005072 | transforming growth factor beta receptor, cytoplasmic mediator activity(GO:0005072) |
| 0.0 | 0.2 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.0 | 0.3 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.1 | GO:0030350 | iron-responsive element binding(GO:0030350) |
| 0.0 | 0.2 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.0 | 0.2 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.0 | 0.1 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.0 | 0.3 | GO:0034796 | 3-oxo-2-(2'-pentenyl)cyclopentane-1-octanoic acid CoA ligase activity(GO:0010435) 3-isopropenyl-6-oxoheptanoyl-CoA synthetase activity(GO:0018854) 2-oxo-delta3-4,5,5-trimethylcyclopentenylacetyl-CoA synthetase activity(GO:0018855) benzoyl acetate-CoA ligase activity(GO:0018856) 2,4-dichlorobenzoate-CoA ligase activity(GO:0018857) pivalate-CoA ligase activity(GO:0034783) cyclopropanecarboxylate-CoA ligase activity(GO:0034793) adipate-CoA ligase activity(GO:0034796) citronellyl-CoA ligase activity(GO:0034823) mentha-1,3-dione-CoA ligase activity(GO:0034841) thiophene-2-carboxylate-CoA ligase activity(GO:0034842) 2,4,4-trimethylpentanoate-CoA ligase activity(GO:0034865) cis-2-methyl-5-isopropylhexa-2,5-dienoate-CoA ligase activity(GO:0034942) trans-2-methyl-5-isopropylhexa-2,5-dienoate-CoA ligase activity(GO:0034943) branched-chain acyl-CoA synthetase (ADP-forming) activity(GO:0043759) aryl-CoA synthetase (ADP-forming) activity(GO:0043762) 3-hydroxypropionyl-CoA synthetase activity(GO:0043955) perillic acid:CoA ligase (ADP-forming) activity(GO:0052685) perillic acid:CoA ligase (AMP-forming) activity(GO:0052686) (3R)-3-isopropenyl-6-oxoheptanoate:CoA ligase (ADP-forming) activity(GO:0052687) (3R)-3-isopropenyl-6-oxoheptanoate:CoA ligase (AMP-forming) activity(GO:0052688) pristanate-CoA ligase activity(GO:0070251) malonyl-CoA synthetase activity(GO:0090409) |
| 0.0 | 0.2 | GO:0016717 | oxidoreductase activity, acting on paired donors, with oxidation of a pair of donors resulting in the reduction of molecular oxygen to two molecules of water(GO:0016717) |
| 0.0 | 0.2 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.0 | 0.1 | GO:0001642 | group III metabotropic glutamate receptor activity(GO:0001642) |
| 0.0 | 0.2 | GO:0050072 | m7G(5')pppN diphosphatase activity(GO:0050072) |
| 0.0 | 0.1 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.0 | 0.1 | GO:0035671 | enone reductase activity(GO:0035671) |
| 0.0 | 1.0 | GO:0080031 | prenylcysteine methylesterase activity(GO:0010296) 1-oxa-2-oxocycloheptane lactonase activity(GO:0018731) sulfolactone hydrolase activity(GO:0018732) butyrolactone hydrolase activity(GO:0018734) endosulfan lactone lactonase activity(GO:0034892) L-ascorbate 6-phosphate lactonase activity(GO:0035460) Ser-tRNA(Thr) hydrolase activity(GO:0043905) Ala-tRNA(Pro) hydrolase activity(GO:0043906) Cys-tRNA(Pro) hydrolase activity(GO:0043907) Ser(Gly)-tRNA(Ala) hydrolase activity(GO:0043908) all-trans-retinyl-palmitate hydrolase, all-trans-retinol forming activity(GO:0047376) mannosyl-oligosaccharide 1,6-alpha-mannosidase activity(GO:0052767) mannosyl-oligosaccharide 1,3-alpha-mannosidase activity(GO:0052768) methyl indole-3-acetate esterase activity(GO:0080030) methyl salicylate esterase activity(GO:0080031) methyl jasmonate esterase activity(GO:0080032) |
| 0.0 | 0.1 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 0.0 | 0.2 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.1 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.1 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.0 | 0.0 | GO:0051723 | protein methylesterase activity(GO:0051723) |
| 0.0 | 0.1 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.1 | GO:0030572 | cardiolipin synthase activity(GO:0008808) phosphatidyltransferase activity(GO:0030572) |
| 0.0 | 0.1 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.1 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.1 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 0.1 | GO:0000171 | ribonuclease MRP activity(GO:0000171) |
| 0.0 | 0.1 | GO:0097001 | ceramide binding(GO:0097001) |
| 0.0 | 0.4 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.2 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.0 | 0.1 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.0 | 0.1 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.2 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.0 | 0.2 | GO:1900750 | glutathione binding(GO:0043295) oligopeptide binding(GO:1900750) |
| 0.0 | 0.1 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 0.1 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.1 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 0.1 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 0.1 | GO:0070191 | methionine-R-sulfoxide reductase activity(GO:0070191) |
| 0.0 | 0.4 | GO:0003756 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.0 | 0.1 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.0 | 0.6 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 0.2 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.1 | GO:1990239 | steroid hormone binding(GO:1990239) |
| 0.0 | 0.1 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) |
| 0.0 | 0.1 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.0 | 0.1 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.3 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.0 | 0.2 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.0 | 0.6 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.3 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.2 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.0 | 0.1 | GO:0016882 | cyclo-ligase activity(GO:0016882) |
| 0.0 | 0.1 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.0 | 0.1 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.0 | 0.2 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 0.1 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.0 | 0.0 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.0 | 0.1 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.0 | 0.1 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.1 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.0 | 0.1 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 0.1 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 0.1 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.0 | 0.0 | GO:0055100 | adiponectin binding(GO:0055100) |
| 0.0 | 0.1 | GO:0018573 | 2,3-dihydroxy DDT 1,2-dioxygenase activity(GO:0018542) phenanthrene dioxygenase activity(GO:0018555) 2,2',3-trihydroxybiphenyl dioxygenase activity(GO:0018556) 1,2-dihydroxyfluorene 1,1-alpha-dioxygenase activity(GO:0018557) 5,6-dihydroxy-3-methyl-2-oxo-1,2-dihydroquinoline dioxygenase activity(GO:0018558) 1,1-dichloro-2-(dihydroxy-4-chlorophenyl)-(4-chlorophenyl)ethene 1,2-dioxygenase activity(GO:0018559) protocatechuate 3,4-dioxygenase type II activity(GO:0018560) 2'-aminobiphenyl-2,3-diol 1,2-dioxygenase activity(GO:0018561) 3,4-dihydroxyfluorene 4,4-alpha-dioxygenase activity(GO:0018562) 2,3-dihydroxy-ethylbenzene 1,2-dioxygenase activity(GO:0018563) carbazole 1,9a-dioxygenase activity(GO:0018564) dihydroxydibenzothiophene dioxygenase activity(GO:0018565) 1,2-dihydroxynaphthalene-6-sulfonate 1,8a-dioxygenase activity(GO:0018566) styrene dioxygenase activity(GO:0018567) 3,4-dihydroxyphenanthrene dioxygenase activity(GO:0018568) hydroquinone 1,2-dioxygenase activity(GO:0018569) p-cumate 2,3-dioxygenase activity(GO:0018570) 2,3-dihydroxy-p-cumate dioxygenase activity(GO:0018571) 3,5-dichlorocatechol 1,2-dioxygenase activity(GO:0018572) 2-aminophenol 1,6-dioxygenase activity(GO:0018573) 2,6-dichloro-p-hydroquinone 1,2-dioxygenase activity(GO:0018574) chlorocatechol 1,2-dioxygenase activity(GO:0018575) catechol dioxygenase activity(GO:0019114) dihydroxyfluorene dioxygenase activity(GO:0019117) 5-aminosalicylate dioxygenase activity(GO:0034543) 3-hydroxy-2-naphthoate 2,3-dioxygenase activity(GO:0034803) benzo(a)pyrene 11,12-dioxygenase activity(GO:0034806) benzo(a)pyrene 4,5-dioxygenase activity(GO:0034808) 4,5-dihydroxybenzo(a)pyrene dioxygenase activity(GO:0034810) benzo(a)pyrene 9,10-dioxygenase activity(GO:0034811) 9,10-dihydroxybenzo(a)pyrene dioxygenase activity(GO:0034812) benzo(a)pyrene 7,8-dioxygenase activity(GO:0034813) 7,8-dihydroxy benzo(a)pyrene dioxygenase activity(GO:0034814) 1,2-dihydroxy-5,6,7,8-tetrahydronaphthalene extradiol dioxygenase activity(GO:0034827) 2-mercaptobenzothiazole dioxygenase activity(GO:0034834) pyridine-3,4-diol dioxygenase activity(GO:0034895) pyrene dioxygenase activity(GO:0034920) 4,5-dihydroxypyrene dioxygenase activity(GO:0034922) phenanthrene-4-carboxylate dioxygenase activity(GO:0034934) tetrachlorobenzene dioxygenase activity(GO:0034935) 4,6-dichloro-3-methylcatechol 1,2-dioxygenase activity(GO:0034936) 2,3-dihydroxydiphenyl ether dioxygenase activity(GO:0034955) diphenyl ether 1,2-dioxygenase activity(GO:0034956) arachidonate 8(S)-lipoxygenase activity(GO:0036403) 4-hydroxycatechol 1,2-dioxygenase activity(GO:0047074) |
| 0.0 | 0.1 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.0 | 0.1 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 0.1 | GO:0004558 | alpha-1,4-glucosidase activity(GO:0004558) |
| 0.0 | 0.1 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.0 | 0.1 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.2 | GO:0016303 | 1-phosphatidylinositol-3-kinase activity(GO:0016303) |
| 0.0 | 0.1 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 0.2 | GO:0016888 | endodeoxyribonuclease activity, producing 5'-phosphomonoesters(GO:0016888) |
| 0.0 | 0.0 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 0.0 | 1.4 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.1 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.0 | 0.0 | GO:0070538 | oleic acid binding(GO:0070538) |
| 0.0 | 0.2 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.0 | 0.1 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.1 | GO:0016679 | oxidoreductase activity, acting on diphenols and related substances as donors(GO:0016679) |
| 0.0 | 0.1 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.1 | GO:0043140 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) |
| 0.0 | 0.2 | GO:0016893 | endonuclease activity, active with either ribo- or deoxyribonucleic acids and producing 5'-phosphomonoesters(GO:0016893) |
| 0.0 | 0.1 | GO:0047105 | aminobutyraldehyde dehydrogenase activity(GO:0019145) 4-trimethylammoniobutyraldehyde dehydrogenase activity(GO:0047105) |
| 0.0 | 0.0 | GO:0016880 | acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.0 | 0.1 | GO:0032557 | pyrimidine ribonucleotide binding(GO:0032557) |
| 0.0 | 0.1 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 0.0 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.0 | 0.1 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.0 | 0.1 | GO:0016401 | palmitoyl-CoA oxidase activity(GO:0016401) |
| 0.0 | 0.2 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.3 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.1 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.0 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 0.0 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.0 | 0.1 | GO:0071614 | linoleic acid epoxygenase activity(GO:0071614) |
| 0.0 | 0.1 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.0 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.0 | 0.0 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.0 | 0.1 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.0 | 0.1 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.0 | 0.1 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.0 | 0.1 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.0 | 0.2 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.0 | 0.0 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 0.0 | GO:0008260 | 3-oxoacid CoA-transferase activity(GO:0008260) |
| 0.0 | 0.0 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 0.0 | 0.0 | GO:0016822 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.0 | 0.5 | GO:0016763 | transferase activity, transferring pentosyl groups(GO:0016763) |
| 0.0 | 0.1 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.0 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 0.0 | 0.1 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 0.0 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.1 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.1 | GO:0048019 | receptor antagonist activity(GO:0048019) |
| 0.0 | 0.1 | GO:0015385 | sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.1 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 0.0 | GO:0004920 | interleukin-10 receptor activity(GO:0004920) |
| 0.0 | 0.1 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.0 | 0.0 | GO:0042171 | lysophosphatidic acid acyltransferase activity(GO:0042171) |
| 0.0 | 0.1 | GO:0008601 | protein phosphatase type 2A regulator activity(GO:0008601) |
| 0.0 | 0.1 | GO:0008641 | small protein activating enzyme activity(GO:0008641) |
| 0.0 | 0.0 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 0.1 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.0 | 0.0 | GO:0019797 | procollagen-proline 3-dioxygenase activity(GO:0019797) |
| 0.0 | 0.0 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 0.0 | 0.1 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 0.1 | GO:0016661 | oxidoreductase activity, acting on other nitrogenous compounds as donors(GO:0016661) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.1 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.4 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.4 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.0 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 0.4 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.4 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.4 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.2 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 0.4 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.0 | 0.3 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
| 0.0 | 0.1 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.2 | ST B CELL ANTIGEN RECEPTOR | B Cell Antigen Receptor |
| 0.0 | 0.1 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.0 | 0.7 | REACTOME SHC1 EVENTS IN EGFR SIGNALING | Genes involved in SHC1 events in EGFR signaling |
| 0.0 | 0.6 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.7 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.3 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.3 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.0 | REACTOME SIGNALING BY WNT | Genes involved in Signaling by Wnt |
| 0.0 | 0.1 | REACTOME ER PHAGOSOME PATHWAY | Genes involved in ER-Phagosome pathway |
| 0.0 | 0.4 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.5 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.0 | REACTOME BILE ACID AND BILE SALT METABOLISM | Genes involved in Bile acid and bile salt metabolism |
| 0.0 | 0.5 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.4 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.0 | 0.4 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.3 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 0.2 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.1 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.1 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.1 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.3 | REACTOME PERK REGULATED GENE EXPRESSION | Genes involved in PERK regulated gene expression |
| 0.0 | 0.2 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.0 | 0.1 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 0.3 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.0 | 0.2 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.2 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.1 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.0 | 0.0 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.0 | 0.1 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 0.2 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.1 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 0.1 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.1 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.1 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.5 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.3 | REACTOME SEMA4D IN SEMAPHORIN SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.0 | 0.1 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.1 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |