Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
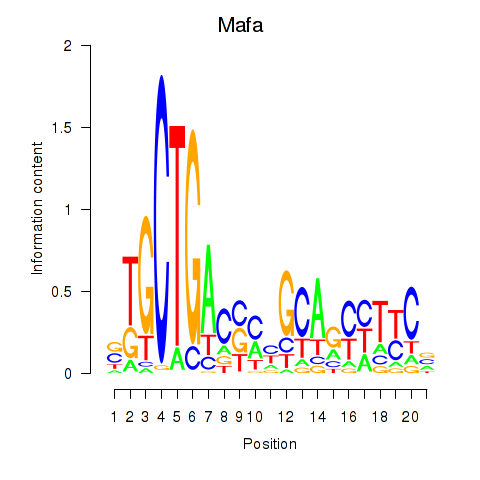
Results for Mafa
Z-value: 0.93
Transcription factors associated with Mafa
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Mafa
|
ENSMUSG00000047591.4 | v-maf musculoaponeurotic fibrosarcoma oncogene family, protein A (avian) |
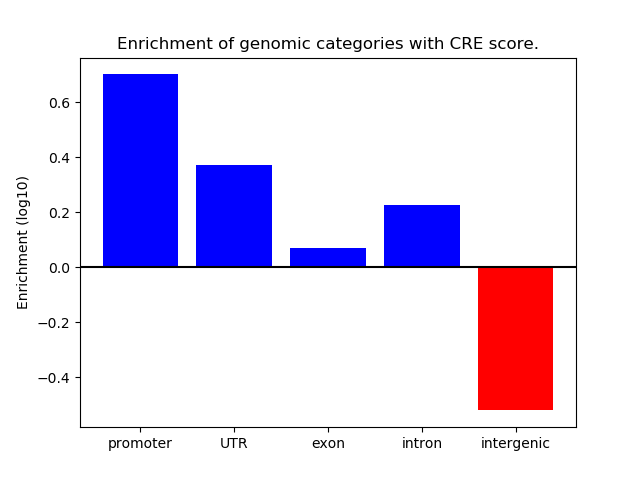
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr15_75748699_75748884 | Mafa | 563 | 0.657377 | -0.76 | 8.1e-02 | Click! |
| chr15_75748359_75748528 | Mafa | 215 | 0.899459 | -0.57 | 2.4e-01 | Click! |
| chr15_75748161_75748312 | Mafa | 8 | 0.964253 | -0.52 | 2.9e-01 | Click! |
Activity of the Mafa motif across conditions
Conditions sorted by the z-value of the Mafa motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
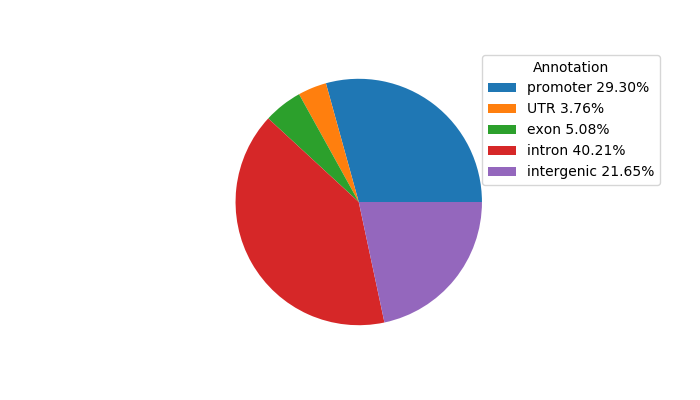
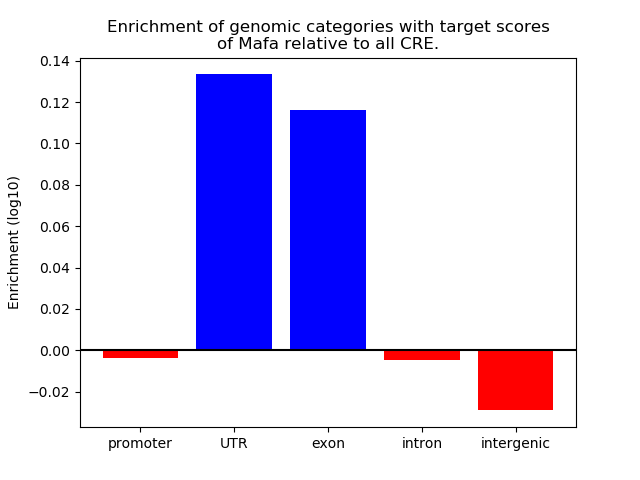
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr15_27466172_27467167 | 0.52 |
Ank |
progressive ankylosis |
8 |
0.97 |
| chr11_4378478_4378660 | 0.51 |
Hormad2 |
HORMA domain containing 2 |
48693 |
0.1 |
| chr17_46052256_46052407 | 0.48 |
Vegfa |
vascular endothelial growth factor A |
19962 |
0.13 |
| chr13_45717276_45717427 | 0.42 |
Gm47460 |
predicted gene, 47460 |
32221 |
0.22 |
| chr19_40155227_40155577 | 0.38 |
Cyp2c70 |
cytochrome P450, family 2, subfamily c, polypeptide 70 |
31884 |
0.13 |
| chr3_52649145_52649332 | 0.37 |
Gm10293 |
predicted pseudogene 10293 |
36403 |
0.17 |
| chr8_69090324_69090475 | 0.37 |
Slc18a1 |
solute carrier family 18 (vesicular monoamine), member 1 |
1164 |
0.38 |
| chr2_44113422_44113573 | 0.36 |
Arhgap15os |
Rho GTPase activating protein 15, opposite strand |
48173 |
0.17 |
| chr17_28761439_28761590 | 0.35 |
Mapk13 |
mitogen-activated protein kinase 13 |
7783 |
0.12 |
| chr11_82527093_82527250 | 0.34 |
Gm24612 |
predicted gene, 24612 |
69008 |
0.09 |
| chr11_83721035_83721187 | 0.34 |
Wfdc18 |
WAP four-disulfide core domain 18 |
11986 |
0.08 |
| chr5_53278040_53278387 | 0.34 |
Smim20 |
small integral membrane protein 20 |
1095 |
0.48 |
| chr2_153444307_153444698 | 0.33 |
Nol4l |
nucleolar protein 4-like |
23 |
0.98 |
| chr19_4493105_4493260 | 0.33 |
2010003K11Rik |
RIKEN cDNA 2010003K11 gene |
5401 |
0.13 |
| chr7_90099485_90099651 | 0.32 |
Gm5341 |
predicted pseudogene 5341 |
8400 |
0.13 |
| chrX_36823954_36824173 | 0.32 |
Gm14549 |
predicted gene 14549 |
9292 |
0.12 |
| chr5_122320451_122320627 | 0.32 |
Gm15842 |
predicted gene 15842 |
18117 |
0.09 |
| chr17_8769269_8769442 | 0.31 |
Gm15426 |
predicted gene 15426 |
3428 |
0.29 |
| chr17_25127135_25127453 | 0.31 |
Clcn7 |
chloride channel, voltage-sensitive 7 |
6097 |
0.1 |
| chr19_16437834_16438025 | 0.30 |
Gna14 |
guanine nucleotide binding protein, alpha 14 |
1371 |
0.4 |
| chr3_52531720_52531871 | 0.30 |
Gm30173 |
predicted gene, 30173 |
14831 |
0.23 |
| chr13_63293654_63293805 | 0.28 |
Aopep |
aminopeptidase O |
5064 |
0.09 |
| chr6_51325666_51325826 | 0.28 |
Gm32479 |
predicted gene, 32479 |
37891 |
0.14 |
| chr6_90568297_90568448 | 0.27 |
Aldh1l1 |
aldehyde dehydrogenase 1 family, member L1 |
666 |
0.63 |
| chr4_108362975_108363166 | 0.27 |
Shisal2a |
shisa like 2A |
20279 |
0.11 |
| chr13_112331307_112331468 | 0.27 |
Ankrd55 |
ankyrin repeat domain 55 |
12933 |
0.17 |
| chr15_61098917_61099257 | 0.27 |
Gm38563 |
predicted gene, 38563 |
58782 |
0.13 |
| chr6_144709069_144709350 | 0.26 |
Sox5os4 |
SRY (sex determining region Y)-box 5, opposite strand 4 |
11702 |
0.16 |
| chr2_165442563_165442717 | 0.26 |
Gm14437 |
predicted gene 14437 |
376 |
0.82 |
| chr1_131278227_131278383 | 0.26 |
Ikbke |
inhibitor of kappaB kinase epsilon |
1301 |
0.32 |
| chr5_129846397_129846548 | 0.24 |
Sumf2 |
sulfatase modifying factor 2 |
514 |
0.59 |
| chr9_94705415_94705619 | 0.24 |
Gm16262 |
predicted gene 16262 |
3198 |
0.23 |
| chr17_81389463_81389664 | 0.23 |
Gm50044 |
predicted gene, 50044 |
18730 |
0.23 |
| chr3_51251932_51252083 | 0.23 |
Elf2 |
E74-like factor 2 |
8234 |
0.13 |
| chr7_27452561_27452934 | 0.23 |
Blvrb |
biliverdin reductase B (flavin reductase (NADPH)) |
303 |
0.79 |
| chr1_88702033_88702468 | 0.22 |
Arl4c |
ADP-ribosylation factor-like 4C |
29 |
0.97 |
| chr1_21247881_21248070 | 0.22 |
Gsta3 |
glutathione S-transferase, alpha 3 |
5546 |
0.12 |
| chr2_52580732_52580891 | 0.22 |
Cacnb4 |
calcium channel, voltage-dependent, beta 4 subunit |
22244 |
0.17 |
| chr3_51231004_51231169 | 0.22 |
Gm38357 |
predicted gene, 38357 |
831 |
0.53 |
| chr14_86346221_86346400 | 0.22 |
Gm4350 |
predicted gene 4350 |
26025 |
0.13 |
| chr3_27562067_27562227 | 0.22 |
Gm43344 |
predicted gene 43344 |
15506 |
0.22 |
| chr8_126928052_126928350 | 0.21 |
Gm26397 |
predicted gene, 26397 |
16779 |
0.14 |
| chr18_32271671_32271889 | 0.21 |
Ercc3 |
excision repair cross-complementing rodent repair deficiency, complementation group 3 |
14511 |
0.2 |
| chr11_120817696_120817936 | 0.21 |
Fasn |
fatty acid synthase |
2361 |
0.15 |
| chrY_90792035_90792186 | 0.21 |
Gm47283 |
predicted gene, 47283 |
1659 |
0.36 |
| chr6_28755592_28755775 | 0.21 |
Snd1 |
staphylococcal nuclease and tudor domain containing 1 |
9007 |
0.22 |
| chrX_170017443_170017594 | 0.21 |
Erdr1 |
erythroid differentiation regulator 1 |
6774 |
0.18 |
| chr11_115898937_115899098 | 0.21 |
Smim5 |
small integral membrane protein 5 |
949 |
0.35 |
| chr3_52985515_52985857 | 0.21 |
Cog6 |
component of oligomeric golgi complex 6 |
8999 |
0.16 |
| chr9_106238560_106238771 | 0.20 |
Alas1 |
aminolevulinic acid synthase 1 |
65 |
0.95 |
| chr11_64842929_64843090 | 0.20 |
Gm12292 |
predicted gene 12292 |
744 |
0.76 |
| chr3_51230122_51230437 | 0.20 |
Gm38357 |
predicted gene, 38357 |
1638 |
0.29 |
| chrX_12792421_12792582 | 0.19 |
Gm14634 |
predicted gene 14634 |
29957 |
0.15 |
| chr16_30968839_30969241 | 0.19 |
Gm15742 |
predicted gene 15742 |
4478 |
0.21 |
| chr2_44106392_44106564 | 0.19 |
Arhgap15os |
Rho GTPase activating protein 15, opposite strand |
41154 |
0.18 |
| chr9_44087034_44087185 | 0.19 |
Usp2 |
ubiquitin specific peptidase 2 |
81 |
0.91 |
| chr10_5424273_5424447 | 0.19 |
Syne1 |
spectrin repeat containing, nuclear envelope 1 |
55874 |
0.14 |
| chr9_122027805_122027998 | 0.19 |
Gm47117 |
predicted gene, 47117 |
15761 |
0.1 |
| chr1_151243904_151244075 | 0.19 |
Gm24402 |
predicted gene, 24402 |
16925 |
0.12 |
| chr1_162891657_162891835 | 0.19 |
Fmo2 |
flavin containing monooxygenase 2 |
5231 |
0.19 |
| chr1_133370822_133370973 | 0.19 |
Etnk2 |
ethanolamine kinase 2 |
3610 |
0.17 |
| chr7_52391946_52392105 | 0.18 |
Gm20074 |
predicted gene, 20074 |
19428 |
0.28 |
| chr10_4599676_4599928 | 0.18 |
Esr1 |
estrogen receptor 1 (alpha) |
11791 |
0.21 |
| chr9_61911741_61911892 | 0.18 |
Rplp1 |
ribosomal protein, large, P1 |
2726 |
0.27 |
| chr5_115515109_115515270 | 0.18 |
Gm13840 |
predicted gene 13840 |
74 |
0.92 |
| chr3_117078252_117078413 | 0.18 |
1700061I17Rik |
RIKEN cDNA 1700061I17 gene |
567 |
0.75 |
| chr14_25312206_25312357 | 0.18 |
Gm26660 |
predicted gene, 26660 |
66354 |
0.1 |
| chr6_83797123_83797297 | 0.18 |
Nagk |
N-acetylglucosamine kinase |
178 |
0.88 |
| chr13_107469451_107469885 | 0.18 |
AI197445 |
expressed sequence AI197445 |
154 |
0.97 |
| chr5_120481198_120481714 | 0.18 |
Gm15690 |
predicted gene 15690 |
553 |
0.54 |
| chr17_14988060_14988226 | 0.17 |
9030025P20Rik |
RIKEN cDNA 9030025P20 gene |
8644 |
0.11 |
| chr6_90746922_90747105 | 0.17 |
Iqsec1 |
IQ motif and Sec7 domain 1 |
10326 |
0.17 |
| chr1_21166525_21166783 | 0.17 |
Gm2693 |
predicted gene 2693 |
12334 |
0.16 |
| chr16_90283788_90283943 | 0.17 |
Scaf4 |
SR-related CTD-associated factor 4 |
443 |
0.8 |
| chr11_16900676_16901084 | 0.17 |
Egfr |
epidermal growth factor receptor |
4305 |
0.23 |
| chr8_25636930_25637092 | 0.17 |
Nsd3 |
nuclear receptor binding SET domain protein 3 |
3273 |
0.13 |
| chr17_87302277_87302563 | 0.17 |
Ttc7 |
tetratricopeptide repeat domain 7 |
6912 |
0.15 |
| chr12_31499269_31499433 | 0.17 |
Cbll1 |
Casitas B-lineage lymphoma-like 1 |
204 |
0.88 |
| chr5_121564754_121565431 | 0.17 |
Aldh2 |
aldehyde dehydrogenase 2, mitochondrial |
6095 |
0.1 |
| chr11_120568518_120568669 | 0.16 |
P4hb |
prolyl 4-hydroxylase, beta polypeptide |
4248 |
0.07 |
| chr13_44735230_44735426 | 0.16 |
Jarid2 |
jumonji, AT rich interactive domain 2 |
1288 |
0.56 |
| chr17_5849675_5849852 | 0.16 |
Snx9 |
sorting nexin 9 |
8411 |
0.15 |
| chr9_61348025_61348190 | 0.16 |
Gm10655 |
predicted gene 10655 |
23520 |
0.16 |
| chr8_84842789_84843130 | 0.16 |
Calr |
calreticulin |
1957 |
0.13 |
| chr17_47976252_47976712 | 0.16 |
Gm14871 |
predicted gene 14871 |
27090 |
0.11 |
| chr13_8865143_8865294 | 0.16 |
Wdr37 |
WD repeat domain 37 |
3993 |
0.12 |
| chr11_6026043_6026414 | 0.16 |
Camk2b |
calcium/calmodulin-dependent protein kinase II, beta |
25780 |
0.15 |
| chr8_121973410_121973561 | 0.16 |
Gm17709 |
predicted gene, 17709 |
337 |
0.72 |
| chr11_67142292_67142463 | 0.16 |
2310065F04Rik |
RIKEN cDNA 2310065F04 gene |
22297 |
0.12 |
| chr8_109969231_109969458 | 0.16 |
Gm45795 |
predicted gene 45795 |
5008 |
0.14 |
| chr15_58927649_58927810 | 0.16 |
Tatdn1 |
TatD DNase domain containing 1 |
5639 |
0.14 |
| chr1_52631028_52631204 | 0.16 |
Nemp2 |
nuclear envelope integral membrane protein 2 |
319 |
0.86 |
| chr17_48598725_48598879 | 0.16 |
Gm24071 |
predicted gene, 24071 |
19021 |
0.19 |
| chr4_143021381_143021546 | 0.16 |
6330411D24Rik |
RIKEN cDNA 6330411D24 gene |
52303 |
0.14 |
| chr6_15828886_15829037 | 0.16 |
Gm43990 |
predicted gene, 43990 |
7128 |
0.25 |
| chr3_18251077_18251277 | 0.16 |
Cyp7b1 |
cytochrome P450, family 7, subfamily b, polypeptide 1 |
7839 |
0.24 |
| chr5_36390576_36390854 | 0.16 |
Sorcs2 |
sortilin-related VPS10 domain containing receptor 2 |
7392 |
0.21 |
| chr4_36014169_36014515 | 0.16 |
Gm12369 |
predicted gene 12369 |
10163 |
0.27 |
| chr11_5524215_5524690 | 0.16 |
Xbp1 |
X-box binding protein 1 |
2529 |
0.2 |
| chr9_121555731_121555882 | 0.15 |
Gm47095 |
predicted gene, 47095 |
19655 |
0.13 |
| chr10_87880307_87880601 | 0.15 |
Igf1os |
insulin-like growth factor 1, opposite strand |
17073 |
0.18 |
| chr5_123907365_123907778 | 0.15 |
Denr |
density-regulated protein |
82 |
0.95 |
| chr1_186250377_186250604 | 0.15 |
Gm37272 |
predicted gene, 37272 |
68233 |
0.1 |
| chr17_28010581_28010732 | 0.15 |
Anks1 |
ankyrin repeat and SAM domain containing 1 |
3311 |
0.15 |
| chr7_126932811_126932965 | 0.15 |
Kctd13 |
potassium channel tetramerisation domain containing 13 |
2967 |
0.08 |
| chr5_77309423_77309744 | 0.15 |
Noa1 |
nitric oxide associated 1 |
476 |
0.48 |
| chr15_59016273_59016565 | 0.15 |
Mtss1 |
MTSS I-BAR domain containing 1 |
24177 |
0.16 |
| chr11_120820263_120820477 | 0.15 |
Fasn |
fatty acid synthase |
3410 |
0.12 |
| chr3_41614429_41614618 | 0.15 |
Jade1 |
jade family PHD finger 1 |
12093 |
0.16 |
| chr15_27585587_27585755 | 0.15 |
Mir7117 |
microRNA 7117 |
14177 |
0.15 |
| chr18_12671580_12671783 | 0.15 |
Ttc39c |
tetratricopeptide repeat domain 39C |
18047 |
0.13 |
| chr19_38015663_38015814 | 0.15 |
Myof |
myoferlin |
27613 |
0.14 |
| chr11_97363051_97363241 | 0.15 |
Socs7 |
suppressor of cytokine signaling 7 |
595 |
0.66 |
| chr15_100228265_100228661 | 0.15 |
Atf1 |
activating transcription factor 1 |
200 |
0.92 |
| chr17_46139458_46139771 | 0.14 |
Rsph9 |
radial spoke head 9 homolog (Chlamydomonas) |
2784 |
0.15 |
| chr9_44478965_44479362 | 0.14 |
C030014I23Rik |
RIKEN cDNA C030014I23 gene |
385 |
0.49 |
| chr10_81408031_81408226 | 0.14 |
Nfic |
nuclear factor I/C |
487 |
0.49 |
| chr6_142788740_142788928 | 0.14 |
Sult6b2 |
sulfotransferase family 6B, member 2 |
15632 |
0.16 |
| chr8_93108415_93108795 | 0.14 |
Ces1c |
carboxylesterase 1C |
15924 |
0.14 |
| chr2_134534653_134534804 | 0.14 |
Hao1 |
hydroxyacid oxidase 1, liver |
19579 |
0.27 |
| chr7_122066462_122066699 | 0.14 |
Ears2 |
glutamyl-tRNA synthetase 2, mitochondrial |
431 |
0.53 |
| chr2_34777307_34777554 | 0.14 |
Hspa5 |
heat shock protein 5 |
2583 |
0.19 |
| chr18_76305906_76306080 | 0.14 |
Gm50360 |
predicted gene, 50360 |
15831 |
0.21 |
| chr3_97635501_97635936 | 0.14 |
Fmo5 |
flavin containing monooxygenase 5 |
6835 |
0.14 |
| chr4_117126763_117126969 | 0.14 |
Tctex1d4 |
Tctex1 domain containing 4 |
53 |
0.9 |
| chr14_65759139_65759314 | 0.14 |
Pbk |
PDZ binding kinase |
46611 |
0.13 |
| chr2_58786750_58786901 | 0.14 |
Upp2 |
uridine phosphorylase 2 |
21500 |
0.19 |
| chr5_4918481_4918656 | 0.14 |
Cdk14 |
cyclin-dependent kinase 14 |
29656 |
0.14 |
| chr6_29815607_29815784 | 0.14 |
Ahcyl2 |
S-adenosylhomocysteine hydrolase-like 2 |
13836 |
0.18 |
| chr5_123022889_123023101 | 0.14 |
Orai1 |
ORAI calcium release-activated calcium modulator 1 |
7657 |
0.09 |
| chr6_134517015_134517190 | 0.14 |
Lrp6 |
low density lipoprotein receptor-related protein 6 |
24612 |
0.17 |
| chr1_162892140_162892544 | 0.14 |
Fmo2 |
flavin containing monooxygenase 2 |
5827 |
0.19 |
| chr3_94697991_94698327 | 0.14 |
Selenbp2 |
selenium binding protein 2 |
4500 |
0.13 |
| chr2_20666086_20666239 | 0.14 |
Gm13362 |
predicted gene 13362 |
47072 |
0.16 |
| chr3_21371390_21371542 | 0.14 |
Gm29137 |
predicted gene 29137 |
102376 |
0.08 |
| chr13_109536856_109537007 | 0.14 |
Pde4d |
phosphodiesterase 4D, cAMP specific |
44474 |
0.21 |
| chr11_28698214_28698534 | 0.14 |
2810471M01Rik |
RIKEN cDNA 2810471M01 gene |
16810 |
0.16 |
| chr11_60256374_60256525 | 0.14 |
Tom1l2 |
target of myb1-like 2 (chicken) |
10780 |
0.12 |
| chr11_87456055_87456225 | 0.14 |
Rnu3b4 |
U3B small nuclear RNA 4 |
6146 |
0.09 |
| chr15_76208375_76208737 | 0.14 |
Plec |
plectin |
97 |
0.92 |
| chr3_94937900_94938079 | 0.14 |
Selenbp1 |
selenium binding protein 1 |
4830 |
0.1 |
| chr13_37831757_37832025 | 0.14 |
Rreb1 |
ras responsive element binding protein 1 |
4498 |
0.22 |
| chr13_84221796_84222007 | 0.13 |
A230107N01Rik |
RIKEN cDNA A230107N01 gene |
199 |
0.71 |
| chr13_63520249_63520412 | 0.13 |
Ptch1 |
patched 1 |
8301 |
0.15 |
| chr12_104087488_104087669 | 0.13 |
Serpina4-ps1 |
serine (or cysteine) peptidase inhibitor, clade A, member 4, pseudogene 1 |
6929 |
0.1 |
| chr3_89140809_89141081 | 0.13 |
Pklr |
pyruvate kinase liver and red blood cell |
4322 |
0.08 |
| chr12_28629267_28629434 | 0.13 |
Rps7 |
ribosomal protein S7 |
2567 |
0.2 |
| chr7_80030553_80030704 | 0.13 |
Zfp710 |
zinc finger protein 710 |
2814 |
0.17 |
| chr7_34226952_34227127 | 0.13 |
Gpi1 |
glucose-6-phosphate isomerase 1 |
1958 |
0.17 |
| chr4_63251381_63251799 | 0.13 |
Mir455 |
microRNA 455 |
5261 |
0.19 |
| chr9_92350909_92351360 | 0.13 |
1700057G04Rik |
RIKEN cDNA 1700057G04 gene |
49 |
0.97 |
| chr11_104441511_104441826 | 0.13 |
Kansl1 |
KAT8 regulatory NSL complex subunit 1 |
34 |
0.97 |
| chr2_181044162_181044343 | 0.13 |
Chrna4 |
cholinergic receptor, nicotinic, alpha polypeptide 4 |
706 |
0.59 |
| chr17_71215570_71215721 | 0.13 |
Lpin2 |
lipin 2 |
10969 |
0.16 |
| chr1_58382672_58382837 | 0.13 |
Gm37607 |
predicted gene, 37607 |
3380 |
0.17 |
| chr9_45095657_45095825 | 0.13 |
Jaml |
junction adhesion molecule like |
7053 |
0.09 |
| chr10_21678793_21678948 | 0.13 |
Gm5420 |
predicted gene 5420 |
7541 |
0.23 |
| chr6_72360494_72360859 | 0.13 |
Rnf181 |
ring finger protein 181 |
391 |
0.72 |
| chr13_48972078_48972229 | 0.13 |
Fam120a |
family with sequence similarity 120, member A |
4136 |
0.28 |
| chr19_10046514_10046840 | 0.13 |
Fads3 |
fatty acid desaturase 3 |
4945 |
0.14 |
| chr2_160249478_160249680 | 0.12 |
Gm826 |
predicted gene 826 |
64037 |
0.13 |
| chr8_84723357_84723534 | 0.12 |
G430095P16Rik |
RIKEN cDNA G430095P16 gene |
438 |
0.7 |
| chr9_45205284_45205435 | 0.12 |
Tmprss4 |
transmembrane protease, serine 4 |
1267 |
0.29 |
| chr13_55211103_55211935 | 0.12 |
Nsd1 |
nuclear receptor-binding SET-domain protein 1 |
22 |
0.97 |
| chr13_44291297_44291488 | 0.12 |
Gm29676 |
predicted gene, 29676 |
10943 |
0.21 |
| chr3_116162254_116162405 | 0.12 |
Gm26544 |
predicted gene, 26544 |
11850 |
0.13 |
| chr4_3937763_3937914 | 0.12 |
Plag1 |
pleiomorphic adenoma gene 1 |
544 |
0.59 |
| chr2_70798855_70799007 | 0.12 |
Tlk1 |
tousled-like kinase 1 |
26297 |
0.18 |
| chr16_95605411_95605586 | 0.12 |
Erg |
ETS transcription factor |
18905 |
0.24 |
| chr6_13677230_13677408 | 0.12 |
Bmt2 |
base methyltransferase of 25S rRNA 2 |
619 |
0.8 |
| chr18_60217937_60218088 | 0.12 |
Gm5970 |
predicted gene 5970 |
2831 |
0.21 |
| chr5_118649962_118650113 | 0.12 |
Gm43274 |
predicted gene 43274 |
5664 |
0.18 |
| chr13_93710471_93710659 | 0.12 |
Dmgdh |
dimethylglycine dehydrogenase precursor |
974 |
0.45 |
| chr10_39209250_39209435 | 0.12 |
Gm6477 |
predicted gene 6477 |
10804 |
0.16 |
| chr17_15019435_15019586 | 0.12 |
Gm3435 |
predicted gene 3435 |
9175 |
0.12 |
| chr15_85720937_85721131 | 0.12 |
Mirlet7b |
microRNA let7b |
13715 |
0.13 |
| chr6_71458715_71458866 | 0.12 |
Gm44172 |
predicted gene, 44172 |
6798 |
0.11 |
| chr19_30092253_30092507 | 0.12 |
Uhrf2 |
ubiquitin-like, containing PHD and RING finger domains 2 |
419 |
0.86 |
| chr7_139703566_139703726 | 0.12 |
Rpl21-ps13 |
ribosomal protein L21-ps13 |
15753 |
0.15 |
| chr8_126876394_126876573 | 0.12 |
Gm31718 |
predicted gene, 31718 |
45 |
0.98 |
| chr11_60251360_60251526 | 0.12 |
Tom1l2 |
target of myb1-like 2 (chicken) |
5774 |
0.13 |
| chr11_102404567_102404909 | 0.12 |
Slc25a39 |
solute carrier family 25, member 39 |
245 |
0.47 |
| chrX_60013443_60013609 | 0.12 |
F9 |
coagulation factor IX |
14062 |
0.2 |
| chr19_21599131_21599282 | 0.12 |
1110059E24Rik |
RIKEN cDNA 1110059E24 gene |
5113 |
0.24 |
| chr17_33932704_33933052 | 0.12 |
Rgl2 |
ral guanine nucleotide dissociation stimulator-like 2 |
747 |
0.27 |
| chr17_29431315_29431488 | 0.12 |
Gm36199 |
predicted gene, 36199 |
1434 |
0.29 |
| chr13_55212302_55212785 | 0.12 |
Nsd1 |
nuclear receptor-binding SET-domain protein 1 |
157 |
0.94 |
| chr3_94399233_94399394 | 0.12 |
Lingo4 |
leucine rich repeat and Ig domain containing 4 |
796 |
0.33 |
| chr13_97138128_97138479 | 0.12 |
Gfm2 |
G elongation factor, mitochondrial 2 |
263 |
0.59 |
| chr8_105084813_105085337 | 0.12 |
Ces3b |
carboxylesterase 3B |
1312 |
0.28 |
| chr9_121799708_121799875 | 0.12 |
Hhatl |
hedgehog acyltransferase-like |
7284 |
0.09 |
| chr1_134764237_134764440 | 0.11 |
Ppp1r12b |
protein phosphatase 1, regulatory subunit 12B |
3985 |
0.18 |
| chr19_30229320_30229513 | 0.11 |
Mbl2 |
mannose-binding lectin (protein C) 2 |
3526 |
0.25 |
| chr5_24502879_24503079 | 0.11 |
Agap3 |
ArfGAP with GTPase domain, ankyrin repeat and PH domain 3 |
22807 |
0.08 |
| chr6_29523600_29523797 | 0.11 |
Irf5 |
interferon regulatory factor 5 |
2927 |
0.15 |
| chr17_28436263_28436419 | 0.11 |
Fkbp5 |
FK506 binding protein 5 |
4669 |
0.12 |
| chr1_82158565_82158766 | 0.11 |
Mir6344 |
microRNA 6344 |
26749 |
0.19 |
| chr9_48604518_48604682 | 0.11 |
Nnmt |
nicotinamide N-methyltransferase |
477 |
0.86 |
| chr12_80231463_80231884 | 0.11 |
Gm47767 |
predicted gene, 47767 |
22430 |
0.13 |
| chr8_36265736_36265923 | 0.11 |
Lonrf1 |
LON peptidase N-terminal domain and ring finger 1 |
16313 |
0.2 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.1 | 0.2 | GO:0042167 | heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.1 | 0.3 | GO:0009256 | 10-formyltetrahydrofolate metabolic process(GO:0009256) |
| 0.0 | 0.2 | GO:1903964 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.0 | 0.2 | GO:0033159 | negative regulation of protein import into nucleus, translocation(GO:0033159) |
| 0.0 | 0.1 | GO:0072718 | response to cisplatin(GO:0072718) |
| 0.0 | 0.2 | GO:0000414 | regulation of histone H3-K36 methylation(GO:0000414) |
| 0.0 | 0.1 | GO:0097195 | pilomotor reflex(GO:0097195) |
| 0.0 | 0.1 | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.0 | 0.1 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.1 | GO:1901525 | negative regulation of macromitophagy(GO:1901525) |
| 0.0 | 0.2 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.0 | 0.1 | GO:0072361 | regulation of glycolytic process by regulation of transcription from RNA polymerase II promoter(GO:0072361) |
| 0.0 | 0.1 | GO:1903799 | negative regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903799) |
| 0.0 | 0.1 | GO:0051581 | negative regulation of neurotransmitter uptake(GO:0051581) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.0 | 0.1 | GO:0003062 | regulation of heart rate by chemical signal(GO:0003062) |
| 0.0 | 0.1 | GO:1904016 | response to Thyroglobulin triiodothyronine(GO:1904016) cellular response to Thyroglobulin triiodothyronine(GO:1904017) |
| 0.0 | 0.1 | GO:0000101 | sulfur amino acid transport(GO:0000101) |
| 0.0 | 0.1 | GO:0010046 | response to mycotoxin(GO:0010046) |
| 0.0 | 0.1 | GO:0036166 | phenotypic switching(GO:0036166) |
| 0.0 | 0.1 | GO:0070447 | positive regulation of oligodendrocyte progenitor proliferation(GO:0070447) |
| 0.0 | 0.1 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.0 | 0.1 | GO:0006987 | activation of signaling protein activity involved in unfolded protein response(GO:0006987) |
| 0.0 | 0.1 | GO:0061084 | negative regulation of protein refolding(GO:0061084) |
| 0.0 | 0.1 | GO:0018197 | peptidyl-aspartic acid modification(GO:0018197) |
| 0.0 | 0.1 | GO:1904430 | negative regulation of t-circle formation(GO:1904430) |
| 0.0 | 0.1 | GO:0036112 | medium-chain fatty-acyl-CoA metabolic process(GO:0036112) |
| 0.0 | 0.0 | GO:0032058 | positive regulation of translational initiation in response to stress(GO:0032058) |
| 0.0 | 0.1 | GO:0070094 | positive regulation of glucagon secretion(GO:0070094) |
| 0.0 | 0.1 | GO:0050427 | 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) |
| 0.0 | 0.1 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.0 | 0.0 | GO:0048290 | isotype switching to IgA isotypes(GO:0048290) regulation of isotype switching to IgA isotypes(GO:0048296) |
| 0.0 | 0.1 | GO:0003431 | growth plate cartilage chondrocyte development(GO:0003431) |
| 0.0 | 0.1 | GO:1903798 | regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903798) |
| 0.0 | 0.1 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.0 | 0.1 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.0 | 0.2 | GO:0061032 | visceral serous pericardium development(GO:0061032) |
| 0.0 | 0.1 | GO:0048619 | embryonic hindgut morphogenesis(GO:0048619) |
| 0.0 | 0.1 | GO:0051031 | tRNA transport(GO:0051031) |
| 0.0 | 0.1 | GO:0060059 | embryonic retina morphogenesis in camera-type eye(GO:0060059) |
| 0.0 | 0.1 | GO:2001260 | regulation of semaphorin-plexin signaling pathway(GO:2001260) |
| 0.0 | 0.0 | GO:0006296 | nucleotide-excision repair, DNA incision, 5'-to lesion(GO:0006296) |
| 0.0 | 0.0 | GO:0046271 | phenylpropanoid catabolic process(GO:0046271) |
| 0.0 | 0.0 | GO:0033122 | negative regulation of purine nucleotide catabolic process(GO:0033122) |
| 0.0 | 0.1 | GO:0006407 | rRNA export from nucleus(GO:0006407) |
| 0.0 | 0.0 | GO:0006172 | ADP biosynthetic process(GO:0006172) |
| 0.0 | 0.0 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) |
| 0.0 | 0.1 | GO:0070417 | cellular response to cold(GO:0070417) |
| 0.0 | 0.1 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.0 | 0.0 | GO:0006701 | progesterone biosynthetic process(GO:0006701) |
| 0.0 | 0.1 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 0.1 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.0 | 0.1 | GO:0006573 | valine metabolic process(GO:0006573) |
| 0.0 | 0.0 | GO:0007089 | traversing start control point of mitotic cell cycle(GO:0007089) |
| 0.0 | 0.0 | GO:1904380 | endoplasmic reticulum mannose trimming(GO:1904380) |
| 0.0 | 0.0 | GO:0002572 | pro-T cell differentiation(GO:0002572) |
| 0.0 | 0.0 | GO:0045658 | regulation of neutrophil differentiation(GO:0045658) |
| 0.0 | 0.1 | GO:0021942 | radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.0 | 0.3 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.0 | 0.1 | GO:0045719 | negative regulation of glycogen biosynthetic process(GO:0045719) |
| 0.0 | 0.0 | GO:0001996 | positive regulation of heart rate by epinephrine-norepinephrine(GO:0001996) |
| 0.0 | 0.0 | GO:0002071 | glandular epithelial cell maturation(GO:0002071) |
| 0.0 | 0.0 | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 0.0 | 0.1 | GO:0070127 | tRNA aminoacylation for mitochondrial protein translation(GO:0070127) |
| 0.0 | 0.0 | GO:0032485 | regulation of Ral protein signal transduction(GO:0032485) |
| 0.0 | 0.0 | GO:0014722 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) |
| 0.0 | 0.1 | GO:0006561 | proline biosynthetic process(GO:0006561) |
| 0.0 | 0.0 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.0 | 0.0 | GO:0035522 | monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.0 | 0.0 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.0 | GO:0021943 | formation of radial glial scaffolds(GO:0021943) |
| 0.0 | 0.0 | GO:0060988 | lipid tube assembly(GO:0060988) |
| 0.0 | 0.0 | GO:0030167 | proteoglycan catabolic process(GO:0030167) |
| 0.0 | 0.0 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.0 | 0.0 | GO:1900041 | negative regulation of interleukin-2 secretion(GO:1900041) |
| 0.0 | 0.0 | GO:1901187 | regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.0 | 0.0 | GO:2001184 | positive regulation of interleukin-12 secretion(GO:2001184) |
| 0.0 | 0.0 | GO:0000087 | mitotic M phase(GO:0000087) |
| 0.0 | 0.1 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.0 | 0.1 | GO:0035356 | cellular triglyceride homeostasis(GO:0035356) |
| 0.0 | 0.0 | GO:0061043 | regulation of vascular wound healing(GO:0061043) |
| 0.0 | 0.1 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.0 | 0.2 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.0 | 0.0 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.0 | 0.0 | GO:0046469 | platelet activating factor biosynthetic process(GO:0006663) platelet activating factor metabolic process(GO:0046469) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.0 | 0.2 | GO:0031094 | platelet dense tubular network(GO:0031094) |
| 0.0 | 0.1 | GO:0097433 | dense body(GO:0097433) |
| 0.0 | 0.1 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.0 | 0.1 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.1 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 0.1 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.0 | 0.1 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.0 | 0.1 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.0 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.0 | 0.0 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.0 | 0.0 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.0 | 0.0 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 0.1 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.0 | 0.0 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.0 | 0.1 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 0.1 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.0 | 0.1 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.0 | 0.1 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.0 | 0.0 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.1 | 0.3 | GO:0019145 | aminobutyraldehyde dehydrogenase activity(GO:0019145) 4-trimethylammoniobutyraldehyde dehydrogenase activity(GO:0047105) |
| 0.0 | 0.1 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.2 | GO:0032896 | palmitoyl-CoA 9-desaturase activity(GO:0032896) |
| 0.0 | 0.1 | GO:0015222 | serotonin transmembrane transporter activity(GO:0015222) |
| 0.0 | 0.2 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 0.1 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 0.1 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.0 | 0.1 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.0 | 0.1 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 0.3 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.1 | GO:0032564 | dATP binding(GO:0032564) |
| 0.0 | 0.1 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.0 | 0.1 | GO:0008240 | tripeptidyl-peptidase activity(GO:0008240) |
| 0.0 | 0.1 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.0 | 0.1 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.0 | 0.1 | GO:0047760 | butyrate-CoA ligase activity(GO:0047760) |
| 0.0 | 0.1 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.0 | 0.1 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 0.1 | GO:0030943 | mitochondrion targeting sequence binding(GO:0030943) |
| 0.0 | 0.0 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) |
| 0.0 | 0.1 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.0 | 0.1 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.0 | 0.1 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.0 | 0.2 | GO:0016646 | oxidoreductase activity, acting on the CH-NH group of donors, NAD or NADP as acceptor(GO:0016646) |
| 0.0 | 0.1 | GO:0019960 | C-X3-C chemokine binding(GO:0019960) |
| 0.0 | 0.0 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.1 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.0 | 0.0 | GO:0003941 | L-serine ammonia-lyase activity(GO:0003941) |
| 0.0 | 0.1 | GO:0000099 | sulfur amino acid transmembrane transporter activity(GO:0000099) |
| 0.0 | 0.1 | GO:1990932 | 5.8S rRNA binding(GO:1990932) |
| 0.0 | 0.1 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 0.0 | GO:0051033 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.0 | 0.1 | GO:0009374 | biotin binding(GO:0009374) |
| 0.0 | 0.0 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.0 | 0.1 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.0 | 0.1 | GO:0043996 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.0 | 0.1 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 0.1 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.0 | 0.1 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.0 | GO:0071614 | linoleic acid epoxygenase activity(GO:0071614) |
| 0.0 | 0.2 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.1 | GO:0005024 | transforming growth factor beta-activated receptor activity(GO:0005024) |
| 0.0 | 0.0 | GO:0005275 | amine transmembrane transporter activity(GO:0005275) |
| 0.0 | 0.0 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 0.1 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.0 | GO:0008898 | S-adenosylmethionine-homocysteine S-methyltransferase activity(GO:0008898) |
| 0.0 | 0.3 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.1 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.2 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.1 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.1 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.1 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.2 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |