Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
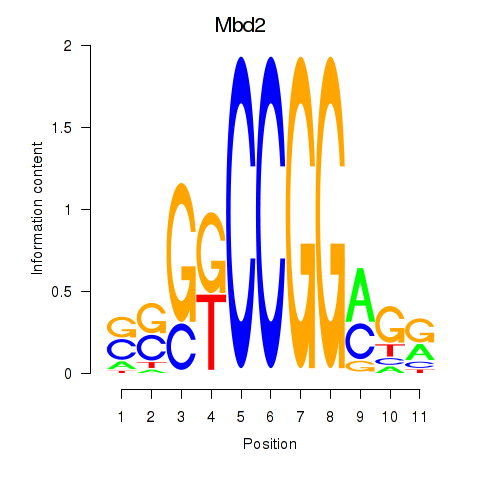
Results for Mbd2
Z-value: 0.89
Transcription factors associated with Mbd2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Mbd2
|
ENSMUSG00000024513.10 | methyl-CpG binding domain protein 2 |
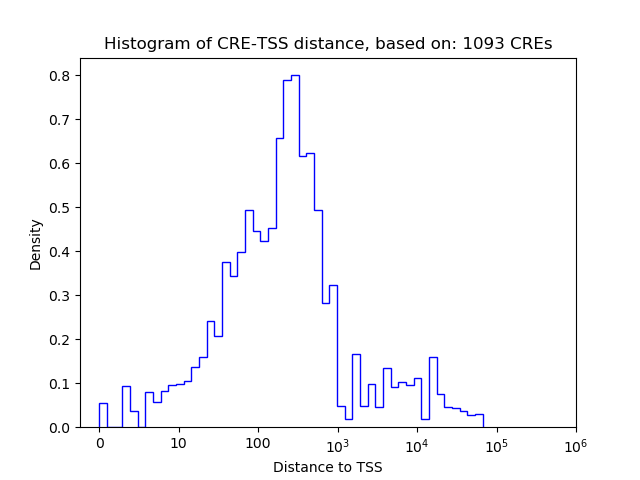
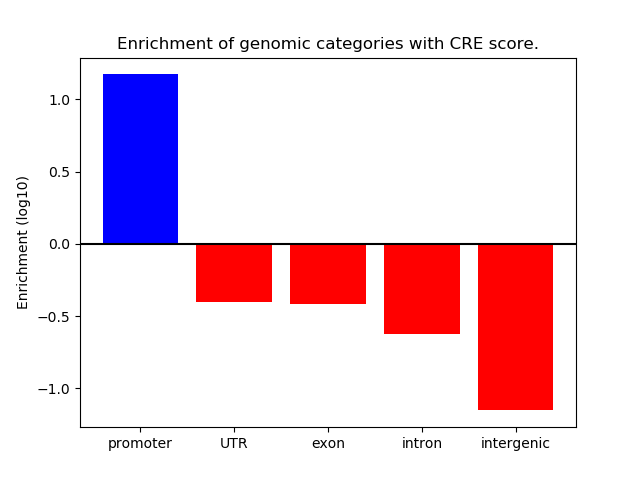
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr18_70567012_70567163 | Mbd2 | 1102 | 0.509881 | 0.90 | 1.5e-02 | Click! |
| chr18_70573928_70574153 | Mbd2 | 5706 | 0.208347 | 0.84 | 3.7e-02 | Click! |
| chr18_70620204_70620393 | Mbd2 | 2525 | 0.295051 | 0.80 | 5.4e-02 | Click! |
| chr18_70567312_70567486 | Mbd2 | 790 | 0.641316 | 0.80 | 5.6e-02 | Click! |
| chr18_70574437_70574595 | Mbd2 | 6182 | 0.205413 | 0.78 | 6.9e-02 | Click! |
Activity of the Mbd2 motif across conditions
Conditions sorted by the z-value of the Mbd2 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
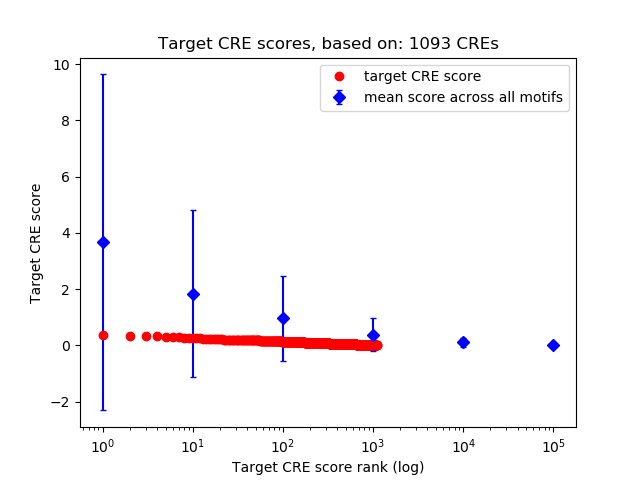
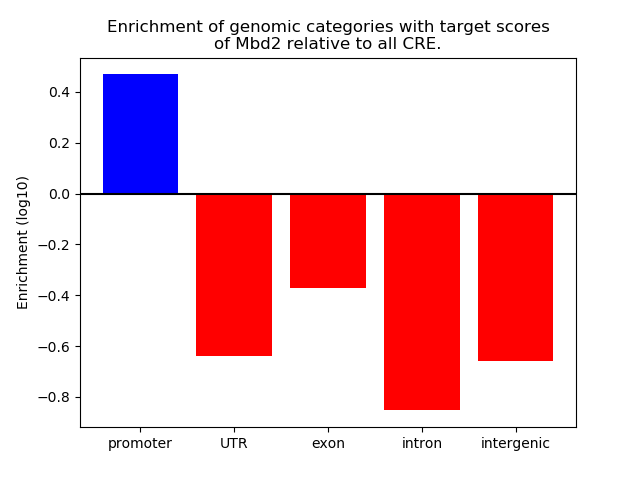
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr11_98588166_98588531 | 0.37 |
Ormdl3 |
ORM1-like 3 (S. cerevisiae) |
980 |
0.38 |
| chr2_130450428_130450784 | 0.32 |
Ptpra |
protein tyrosine phosphatase, receptor type, A |
62 |
0.95 |
| chr16_17131597_17132007 | 0.32 |
Sdf2l1 |
stromal cell-derived factor 2-like 1 |
581 |
0.49 |
| chr8_122550942_122551215 | 0.31 |
Piezo1 |
piezo-type mechanosensitive ion channel component 1 |
251 |
0.84 |
| chr16_17928117_17928461 | 0.29 |
Slc25a1 |
solute carrier family 25 (mitochondrial carrier, citrate transporter), member 1 |
70 |
0.94 |
| chr16_45843772_45843939 | 0.28 |
Phldb2 |
pleckstrin homology like domain, family B, member 2 |
379 |
0.88 |
| chr9_44083899_44084207 | 0.28 |
Usp2 |
ubiquitin specific peptidase 2 |
886 |
0.32 |
| chr10_61715458_61715732 | 0.27 |
Aifm2 |
apoptosis-inducing factor, mitochondrion-associated 2 |
263 |
0.88 |
| chr17_28518297_28518448 | 0.27 |
Fkbp5 |
FK506 binding protein 5 |
845 |
0.32 |
| chr8_95633615_95633766 | 0.26 |
Gins3 |
GINS complex subunit 3 (Psf3 homolog) |
136 |
0.93 |
| chr18_3507262_3507648 | 0.26 |
Bambi |
BMP and activin membrane-bound inhibitor |
468 |
0.77 |
| chr17_28485541_28485711 | 0.24 |
Fkbp5 |
FK506 binding protein 5 |
467 |
0.65 |
| chr11_107548058_107548221 | 0.24 |
Helz |
helicase with zinc finger domain |
171 |
0.57 |
| chr12_102704764_102704984 | 0.24 |
Itpk1 |
inositol 1,3,4-triphosphate 5/6 kinase |
40 |
0.95 |
| chr6_113604837_113605061 | 0.23 |
Brk1 |
BRICK1, SCAR/WAVE actin-nucleating complex subunit |
177 |
0.85 |
| chr9_95559372_95559553 | 0.22 |
Paqr9 |
progestin and adipoQ receptor family member IX |
195 |
0.91 |
| chr8_70905885_70906054 | 0.22 |
Map1s |
microtubule-associated protein 1S |
13 |
0.94 |
| chr7_139188415_139188566 | 0.21 |
Stk32c |
serine/threonine kinase 32C |
27 |
0.97 |
| chr7_39487472_39487798 | 0.21 |
Gm28455 |
predicted gene 28455 |
1803 |
0.25 |
| chr15_102090068_102090474 | 0.21 |
Eif4b |
eukaryotic translation initiation factor 4B |
4247 |
0.14 |
| chr16_32867691_32868221 | 0.21 |
Rubcn |
RUN domain and cysteine-rich domain containing, Beclin 1-interacting protein |
382 |
0.81 |
| chr14_18238484_18238667 | 0.21 |
Nr1d2 |
nuclear receptor subfamily 1, group D, member 2 |
436 |
0.77 |
| chr1_89940488_89940663 | 0.21 |
Gbx2 |
gastrulation brain homeobox 2 |
9396 |
0.19 |
| chr16_20301821_20302010 | 0.20 |
Parl |
presenilin associated, rhomboid-like |
102 |
0.96 |
| chr1_88702033_88702468 | 0.20 |
Arl4c |
ADP-ribosylation factor-like 4C |
29 |
0.97 |
| chr11_78343717_78343868 | 0.20 |
Unc119 |
unc-119 lipid binding chaperone |
247 |
0.83 |
| chrX_52988531_52988855 | 0.20 |
Hprt |
hypoxanthine guanine phosphoribosyl transferase |
556 |
0.67 |
| chr13_54504145_54504307 | 0.20 |
Simc1 |
SUMO-interacting motifs containing 1 |
417 |
0.77 |
| chr2_153444307_153444698 | 0.19 |
Nol4l |
nucleolar protein 4-like |
23 |
0.98 |
| chr8_81014588_81014762 | 0.19 |
Usp38 |
ubiquitin specific peptidase 38 |
238 |
0.71 |
| chr5_122284523_122284674 | 0.19 |
Pptc7 |
PTC7 protein phosphatase homolog |
233 |
0.88 |
| chr17_87107670_87107838 | 0.19 |
Socs5 |
suppressor of cytokine signaling 5 |
63 |
0.97 |
| chr4_119539797_119539948 | 0.19 |
Foxj3 |
forkhead box J3 |
93 |
0.77 |
| chr1_128103472_128103650 | 0.19 |
R3hdm1 |
R3H domain containing 1 |
230 |
0.7 |
| chr15_59633802_59633963 | 0.19 |
Trib1 |
tribbles pseudokinase 1 |
14468 |
0.19 |
| chr8_35725273_35725452 | 0.19 |
Gm35021 |
predicted gene, 35021 |
82 |
0.96 |
| chr16_33381353_33381732 | 0.19 |
Zfp148 |
zinc finger protein 148 |
76 |
0.94 |
| chr10_115385093_115385251 | 0.19 |
Zfc3h1 |
zinc finger, C3H1-type containing |
213 |
0.82 |
| chr13_56134831_56134982 | 0.19 |
Macroh2a1 |
macroH2A.1 histone |
81 |
0.97 |
| chr4_117886820_117887048 | 0.18 |
Atp6v0b |
ATPase, H+ transporting, lysosomal V0 subunit B |
298 |
0.78 |
| chr8_83699110_83699282 | 0.18 |
Pkn1 |
protein kinase N1 |
17 |
0.96 |
| chr2_70509001_70509542 | 0.18 |
Erich2os |
glutamate rich 2, opposite strand |
72 |
0.66 |
| chr1_152386380_152386577 | 0.18 |
Tsen15 |
tRNA splicing endonuclease subunit 15 |
114 |
0.97 |
| chr13_54574971_54575187 | 0.18 |
Arl10 |
ADP-ribosylation factor-like 10 |
32 |
0.95 |
| chr2_155473861_155474012 | 0.18 |
Ncoa6 |
nuclear receptor coactivator 6 |
42 |
0.96 |
| chr1_135375014_135375173 | 0.18 |
Shisa4 |
shisa family member 4 |
16 |
0.96 |
| chr1_156673428_156673871 | 0.18 |
Tor3a |
torsin family 3, member A |
665 |
0.65 |
| chr2_26588564_26589214 | 0.17 |
Egfl7 |
EGF-like domain 7 |
346 |
0.71 |
| chr1_180569064_180569225 | 0.17 |
Parp1 |
poly (ADP-ribose) polymerase family, member 1 |
82 |
0.97 |
| chr1_93754083_93754286 | 0.17 |
Thap4 |
THAP domain containing 4 |
466 |
0.56 |
| chr14_65098086_65098260 | 0.17 |
Extl3 |
exostosin-like glycosyltransferase 3 |
67 |
0.98 |
| chr2_26503007_26503238 | 0.17 |
Notch1 |
notch 1 |
700 |
0.54 |
| chr2_32846890_32847041 | 0.17 |
Stxbp1 |
syntaxin binding protein 1 |
237 |
0.78 |
| chr5_90589028_90589209 | 0.17 |
Gm42109 |
predicted gene, 42109 |
41 |
0.97 |
| chr10_57486792_57486966 | 0.17 |
Hsf2 |
heat shock factor 2 |
454 |
0.51 |
| chr11_119056746_119056911 | 0.17 |
Cbx8 |
chromobox 8 |
15859 |
0.14 |
| chr9_119143653_119143908 | 0.17 |
Acaa1b |
acetyl-Coenzyme A acyltransferase 1B |
6283 |
0.12 |
| chr12_86421804_86421960 | 0.17 |
Esrrb |
estrogen related receptor, beta |
1 |
0.98 |
| chr8_35588423_35588931 | 0.17 |
Mfhas1 |
malignant fibrous histiocytoma amplified sequence 1 |
279 |
0.5 |
| chr1_190149690_190149867 | 0.17 |
Gm28172 |
predicted gene 28172 |
18892 |
0.18 |
| chr7_27452561_27452934 | 0.16 |
Blvrb |
biliverdin reductase B (flavin reductase (NADPH)) |
303 |
0.79 |
| chr11_60913758_60913958 | 0.16 |
Natd1 |
N-acetyltransferase domain containing 1 |
46 |
0.96 |
| chr7_34653059_34653454 | 0.16 |
Kctd15 |
potassium channel tetramerisation domain containing 15 |
56 |
0.51 |
| chr4_134767914_134768168 | 0.16 |
Ldlrap1 |
low density lipoprotein receptor adaptor protein 1 |
17 |
0.98 |
| chr4_132345674_132345841 | 0.16 |
Rcc1 |
regulator of chromosome condensation 1 |
10 |
0.93 |
| chr4_147442937_147443122 | 0.16 |
Gm13161 |
predicted gene 13161 |
442 |
0.71 |
| chr16_26643088_26643295 | 0.16 |
Il1rap |
interleukin 1 receptor accessory protein |
19035 |
0.26 |
| chr2_26389257_26389428 | 0.16 |
Pmpca |
peptidase (mitochondrial processing) alpha |
3 |
0.49 |
| chr5_90931414_90931780 | 0.16 |
Mthfd2l |
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 2-like |
317 |
0.85 |
| chr7_16273015_16273208 | 0.16 |
Inafm1 |
InaF motif containing 1 |
506 |
0.63 |
| chr7_49778522_49778862 | 0.16 |
Prmt3 |
protein arginine N-methyltransferase 3 |
148 |
0.96 |
| chr3_87617470_87617634 | 0.15 |
Arhgef11 |
Rho guanine nucleotide exchange factor (GEF) 11 |
7 |
0.66 |
| chr5_88600656_88600807 | 0.15 |
Rufy3 |
RUN and FYVE domain containing 3 |
16937 |
0.16 |
| chr19_47580113_47580699 | 0.15 |
Slk |
STE20-like kinase |
387 |
0.82 |
| chr16_45158641_45158816 | 0.15 |
Slc35a5 |
solute carrier family 35, member A5 |
22 |
0.52 |
| chr11_58168082_58168288 | 0.15 |
Gemin5 |
gem nuclear organelle associated protein 5 |
354 |
0.77 |
| chrX_20426243_20426394 | 0.15 |
Jade3 |
jade family PHD finger 3 |
530 |
0.73 |
| chr1_98095204_98095355 | 0.15 |
Pam |
peptidylglycine alpha-amidating monooxygenase |
342 |
0.88 |
| chr11_88161368_88161548 | 0.15 |
Cuedc1 |
CUE domain containing 1 |
803 |
0.62 |
| chr9_56160728_56160881 | 0.15 |
Tspan3 |
tetraspanin 3 |
15 |
0.98 |
| chr8_109982243_109982440 | 0.15 |
Gm45795 |
predicted gene 45795 |
7989 |
0.12 |
| chr14_55721616_55722015 | 0.15 |
Rabggta |
Rab geranylgeranyl transferase, a subunit |
361 |
0.66 |
| chr18_67800248_67800418 | 0.15 |
Cep192 |
centrosomal protein 192 |
199 |
0.68 |
| chr3_142765428_142765623 | 0.15 |
Gtf2b |
general transcription factor IIB |
302 |
0.84 |
| chr11_75173089_75173918 | 0.14 |
Mir212 |
microRNA 212 |
115 |
0.56 |
| chr9_18292420_18292623 | 0.14 |
Chordc1 |
cysteine and histidine-rich domain (CHORD)-containing, zinc-binding protein 1 |
133 |
0.95 |
| chr17_26932920_26933100 | 0.14 |
Phf1 |
PHD finger protein 1 |
42 |
0.92 |
| chr15_93398257_93398420 | 0.14 |
Zcrb1 |
zinc finger CCHC-type and RNA binding motif 1 |
4 |
0.5 |
| chr4_148038831_148039191 | 0.14 |
Mthfr |
methylenetetrahydrofolate reductase |
66 |
0.66 |
| chr4_117289529_117289862 | 0.14 |
Rnf220 |
ring finger protein 220 |
135 |
0.93 |
| chr8_88362172_88362513 | 0.14 |
Brd7 |
bromodomain containing 7 |
148 |
0.96 |
| chrX_12761746_12762400 | 0.14 |
Med14 |
mediator complex subunit 14 |
0 |
0.73 |
| chr6_116193076_116193281 | 0.14 |
Tmcc1 |
transmembrane and coiled coil domains 1 |
308 |
0.82 |
| chr1_138856758_138856925 | 0.14 |
2310009B15Rik |
RIKEN cDNA 2310009B15 gene |
6 |
0.97 |
| chr9_64253292_64253498 | 0.14 |
Map2k1 |
mitogen-activated protein kinase kinase 1 |
236 |
0.86 |
| chr6_128843572_128843953 | 0.14 |
Gm44066 |
predicted gene, 44066 |
591 |
0.53 |
| chr10_110745240_110745533 | 0.14 |
E2f7 |
E2F transcription factor 7 |
53 |
0.98 |
| chr11_84828922_84829202 | 0.14 |
Dhrs11 |
dehydrogenase/reductase (SDR family) member 11 |
68 |
0.56 |
| chr1_136625470_136625621 | 0.13 |
Zfp281 |
zinc finger protein 281 |
644 |
0.47 |
| chr2_30062412_30062574 | 0.13 |
Set |
SET nuclear oncogene |
108 |
0.94 |
| chrX_10718554_10718705 | 0.13 |
Gm14493 |
predicted gene 14493 |
81 |
0.95 |
| chr8_109705810_109706060 | 0.13 |
Zfp821 |
zinc finger protein 821 |
153 |
0.94 |
| chr1_187609518_187609683 | 0.13 |
Esrrg |
estrogen-related receptor gamma |
96 |
0.98 |
| chr2_93334519_93334694 | 0.13 |
Tspan18 |
tetraspanin 18 |
101 |
0.97 |
| chr15_96288000_96288151 | 0.13 |
Arid2 |
AT rich interactive domain 2 (ARID, RFX-like) |
502 |
0.79 |
| chr6_88198537_88198710 | 0.13 |
Gata2 |
GATA binding protein 2 |
289 |
0.86 |
| chr6_83135074_83135242 | 0.13 |
Rtkn |
rhotekin |
305 |
0.69 |
| chr6_90716112_90716407 | 0.13 |
Iqsec1 |
IQ motif and Sec7 domain 1 |
270 |
0.9 |
| chr7_49778222_49778382 | 0.13 |
Prmt3 |
protein arginine N-methyltransferase 3 |
44 |
0.98 |
| chr7_127345321_127345481 | 0.13 |
Zfp768 |
zinc finger protein 768 |
22 |
0.94 |
| chr14_46832312_46832505 | 0.13 |
Cgrrf1 |
cell growth regulator with ring finger domain 1 |
164 |
0.71 |
| chr12_112950230_112950445 | 0.13 |
Gm26583 |
predicted gene, 26583 |
175 |
0.89 |
| chr8_83998442_83998628 | 0.13 |
Samd1 |
sterile alpha motif domain containing 1 |
111 |
0.9 |
| chr14_34317951_34318429 | 0.12 |
Glud1 |
glutamate dehydrogenase 1 |
7463 |
0.1 |
| chr19_38055072_38055331 | 0.12 |
Cep55 |
centrosomal protein 55 |
112 |
0.89 |
| chr18_60750592_60750964 | 0.12 |
Ndst1 |
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1 |
2128 |
0.23 |
| chr7_63919537_63919733 | 0.12 |
E030018B13Rik |
RIKEN cDNA E030018B13 gene |
2778 |
0.19 |
| chr18_25168691_25169056 | 0.12 |
AW554918 |
expressed sequence AW554918 |
126 |
0.51 |
| chr4_129461813_129461998 | 0.12 |
Bsdc1 |
BSD domain containing 1 |
69 |
0.94 |
| chrX_60547467_60547618 | 0.12 |
Gm715 |
predicted gene 715 |
477 |
0.75 |
| chr15_39944302_39944559 | 0.12 |
Lrp12 |
low density lipoprotein-related protein 12 |
436 |
0.82 |
| chr7_46919659_46919883 | 0.12 |
Tsg101 |
tumor susceptibility gene 101 |
98 |
0.93 |
| chr17_65772581_65772765 | 0.12 |
Rab31 |
RAB31, member RAS oncogene family |
33 |
0.98 |
| chr7_30957117_30957268 | 0.12 |
Gm4673 |
predicted gene 4673 |
217 |
0.61 |
| chr3_89907720_89908002 | 0.12 |
Gm42809 |
predicted gene 42809 |
5055 |
0.12 |
| chr10_79706208_79706359 | 0.12 |
Bsg |
basigin |
1713 |
0.15 |
| chr17_12389802_12390011 | 0.12 |
Plg |
plasminogen |
11247 |
0.16 |
| chr10_121739546_121739745 | 0.12 |
BC048403 |
cDNA sequence BC048403 |
292 |
0.91 |
| chr6_51325666_51325826 | 0.12 |
Gm32479 |
predicted gene, 32479 |
37891 |
0.14 |
| chr4_134287574_134287752 | 0.12 |
Pdik1l |
PDLIM1 interacting kinase 1 like |
232 |
0.86 |
| chr14_45657978_45658304 | 0.12 |
Ddhd1 |
DDHD domain containing 1 |
1 |
0.56 |
| chr5_135888324_135888475 | 0.12 |
Hspb1 |
heat shock protein 1 |
381 |
0.75 |
| chr4_89311006_89311177 | 0.12 |
Cdkn2b |
cyclin dependent kinase inhibitor 2B |
52 |
0.97 |
| chr7_89337730_89338159 | 0.12 |
Gm44995 |
predicted gene 44995 |
426 |
0.63 |
| chr15_76069223_76069568 | 0.12 |
Scrib |
scribbled planar cell polarity |
359 |
0.68 |
| chrX_9272438_9272819 | 0.11 |
Xk |
X-linked Kx blood group |
128 |
0.94 |
| chr1_133024897_133025230 | 0.11 |
Mdm4 |
transformed mouse 3T3 cell double minute 4 |
267 |
0.78 |
| chr11_103360146_103360491 | 0.11 |
Arhgap27 |
Rho GTPase activating protein 27 |
582 |
0.61 |
| chr7_89403924_89404083 | 0.11 |
Tmem135 |
transmembrane protein 135 |
219 |
0.64 |
| chr4_123411698_123411892 | 0.11 |
Macf1 |
microtubule-actin crosslinking factor 1 |
433 |
0.82 |
| chr8_84832176_84832355 | 0.11 |
Dand5 |
DAN domain family member 5, BMP antagonist |
144 |
0.83 |
| chr9_47582784_47583204 | 0.11 |
Cadm1 |
cell adhesion molecule 1 |
52621 |
0.13 |
| chr11_101070221_101070380 | 0.11 |
Naglu |
alpha-N-acetylglucosaminidase (Sanfilippo disease IIIB) |
288 |
0.79 |
| chr7_44645890_44646064 | 0.11 |
Gm18256 |
predicted gene, 18256 |
549 |
0.58 |
| chr5_102069359_102069519 | 0.11 |
Wdfy3 |
WD repeat and FYVE domain containing 3 |
482 |
0.61 |
| chr9_69989339_69989508 | 0.11 |
Bnip2 |
BCL2/adenovirus E1B interacting protein 2 |
43 |
0.9 |
| chr4_63216102_63216460 | 0.11 |
Col27a1 |
collagen, type XXVII, alpha 1 |
846 |
0.59 |
| chr5_93206054_93206360 | 0.11 |
Ccni |
cyclin I |
221 |
0.59 |
| chr19_7196055_7196232 | 0.11 |
Otub1 |
OTU domain, ubiquitin aldehyde binding 1 |
4321 |
0.15 |
| chr5_74068640_74068846 | 0.11 |
Usp46 |
ubiquitin specific peptidase 46 |
312 |
0.82 |
| chr10_9901181_9901520 | 0.11 |
Gm46210 |
predicted gene, 46210 |
37 |
0.79 |
| chr8_120668006_120668174 | 0.11 |
Emc8 |
ER membrane protein complex subunit 8 |
3 |
0.38 |
| chr6_83109169_83109423 | 0.11 |
Mrpl53 |
mitochondrial ribosomal protein L53 |
176 |
0.53 |
| chr6_125131489_125131640 | 0.11 |
Nop2 |
NOP2 nucleolar protein |
345 |
0.7 |
| chr7_97081394_97081770 | 0.11 |
Gab2 |
growth factor receptor bound protein 2-associated protein 2 |
4 |
0.98 |
| chr5_104218998_104219158 | 0.11 |
Dmp1 |
dentin matrix protein 1 |
16465 |
0.15 |
| chr18_74064019_74064179 | 0.11 |
Mapk4 |
mitogen-activated protein kinase 4 |
293 |
0.83 |
| chr17_88441455_88441615 | 0.11 |
Foxn2 |
forkhead box N2 |
760 |
0.66 |
| chr8_85555040_85555462 | 0.11 |
Dnaja2 |
DnaJ heat shock protein family (Hsp40) member A2 |
93 |
0.97 |
| chr4_135656378_135656550 | 0.11 |
1700029M20Rik |
RIKEN cDNA 1700029M20 gene |
29211 |
0.11 |
| chr3_152396303_152396479 | 0.11 |
Zzz3 |
zinc finger, ZZ domain containing 3 |
259 |
0.84 |
| chr6_140626846_140626997 | 0.11 |
Aebp2 |
AE binding protein 2 |
39 |
0.98 |
| chr5_136961771_136961940 | 0.11 |
Fis1 |
fission, mitochondrial 1 |
222 |
0.86 |
| chr5_25499515_25499834 | 0.10 |
Kmt2c |
lysine (K)-specific methyltransferase 2C |
891 |
0.47 |
| chr15_76710766_76710918 | 0.10 |
Lrrc14 |
leucine rich repeat containing 14 |
111 |
0.7 |
| chr12_31949810_31949961 | 0.10 |
Hbp1 |
high mobility group box transcription factor 1 |
277 |
0.92 |
| chr4_99120849_99121001 | 0.10 |
Dock7 |
dedicator of cytokinesis 7 |
10 |
0.97 |
| chr5_143527474_143527638 | 0.10 |
Rac1 |
Rac family small GTPase 1 |
437 |
0.73 |
| chr17_71552178_71552344 | 0.10 |
Spdya |
speedy/RINGO cell cycle regulator family, member A |
189 |
0.9 |
| chr4_155492144_155492592 | 0.10 |
Gnb1 |
guanine nucleotide binding protein (G protein), beta 1 |
541 |
0.64 |
| chr6_101377934_101378107 | 0.10 |
Pdzrn3 |
PDZ domain containing RING finger 3 |
123 |
0.96 |
| chr19_37695856_37696027 | 0.10 |
Cyp26a1 |
cytochrome P450, family 26, subfamily a, polypeptide 1 |
1857 |
0.32 |
| chr8_34965417_34965568 | 0.10 |
Tnks |
tankyrase, TRF1-interacting ankyrin-related ADP-ribose polymerase |
198 |
0.72 |
| chr11_94321873_94322587 | 0.10 |
Luc7l3 |
LUC7-like 3 (S. cerevisiae) |
242 |
0.9 |
| chr3_69126767_69126936 | 0.10 |
Kpna4 |
karyopherin (importin) alpha 4 |
221 |
0.9 |
| chr17_24470568_24470947 | 0.10 |
Pgp |
phosphoglycolate phosphatase |
322 |
0.51 |
| chr12_108269801_108270015 | 0.10 |
Ccdc85c |
coiled-coil domain containing 85C |
5225 |
0.2 |
| chr8_108745611_108745766 | 0.10 |
Gm38042 |
predicted gene, 38042 |
8095 |
0.24 |
| chr12_16894847_16894998 | 0.10 |
Rock2 |
Rho-associated coiled-coil containing protein kinase 2 |
27 |
0.97 |
| chr1_82815899_82816254 | 0.10 |
Gm28942 |
predicted gene 28942 |
3759 |
0.11 |
| chr13_31559937_31560122 | 0.10 |
A530084C06Rik |
RIKEN cDNA A530084C06 gene |
696 |
0.58 |
| chr11_105181263_105181626 | 0.10 |
1700052K11Rik |
RIKEN cDNA 1700052K11 gene |
11 |
0.56 |
| chr2_167349082_167349264 | 0.10 |
B4galt5 |
UDP-Gal:betaGlcNAc beta 1,4-galactosyltransferase, polypeptide 5 |
10 |
0.98 |
| chr1_132008229_132008400 | 0.10 |
Elk4 |
ELK4, member of ETS oncogene family |
17 |
0.97 |
| chr8_24554600_24554765 | 0.10 |
Ido2 |
indoleamine 2,3-dioxygenase 2 |
85 |
0.97 |
| chr4_58912380_58912600 | 0.10 |
Ecpas |
Ecm29 proteasome adaptor and scaffold |
214 |
0.93 |
| chr15_41789183_41789334 | 0.10 |
Oxr1 |
oxidation resistance 1 |
123 |
0.77 |
| chr1_166309098_166309279 | 0.10 |
5330438I03Rik |
RIKEN cDNA 5330438I03 gene |
397 |
0.85 |
| chr16_44724144_44724299 | 0.10 |
Nepro |
nucleolus and neural progenitor protein |
80 |
0.97 |
| chr2_105016740_105016924 | 0.10 |
Eif3m |
eukaryotic translation initiation factor 3, subunit M |
211 |
0.59 |
| chr8_84200664_84200922 | 0.10 |
Gm37352 |
predicted gene, 37352 |
76 |
0.9 |
| chr10_71344349_71344511 | 0.09 |
Cisd1 |
CDGSH iron sulfur domain 1 |
524 |
0.7 |
| chr17_44736318_44736469 | 0.09 |
Runx2 |
runt related transcription factor 2 |
217 |
0.78 |
| chr15_84923081_84923412 | 0.09 |
Nup50 |
nucleoporin 50 |
165 |
0.92 |
| chr15_98871705_98871879 | 0.09 |
Kmt2d |
lysine (K)-specific methyltransferase 2D |
588 |
0.34 |
| chr4_133633066_133633255 | 0.09 |
Zdhhc18 |
zinc finger, DHHC domain containing 18 |
309 |
0.83 |
| chr8_47674837_47675003 | 0.09 |
Ing2 |
inhibitor of growth family, member 2 |
238 |
0.69 |
| chr19_7240751_7240928 | 0.09 |
Naa40 |
N(alpha)-acetyltransferase 40, NatD catalytic subunit |
195 |
0.89 |
| chr5_130029440_130029773 | 0.09 |
Crcp |
calcitonin gene-related peptide-receptor component protein |
263 |
0.57 |
| chr15_12117744_12117919 | 0.09 |
Zfr |
zinc finger RNA binding protein |
0 |
0.97 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.1 | 0.3 | GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 0.1 | 0.3 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.1 | 0.2 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.1 | 0.2 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.1 | 0.1 | GO:0010725 | regulation of primitive erythrocyte differentiation(GO:0010725) |
| 0.1 | 0.2 | GO:1900186 | negative regulation of clathrin-mediated endocytosis(GO:1900186) |
| 0.1 | 0.2 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 0.1 | 0.2 | GO:1900060 | negative regulation of ceramide biosynthetic process(GO:1900060) |
| 0.0 | 0.1 | GO:0046098 | guanine metabolic process(GO:0046098) |
| 0.0 | 0.1 | GO:2001027 | negative regulation of endothelial cell chemotaxis(GO:2001027) |
| 0.0 | 0.1 | GO:0071930 | negative regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071930) |
| 0.0 | 0.1 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.0 | 0.1 | GO:0042167 | heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.0 | 0.2 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.0 | 0.1 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.0 | 0.2 | GO:0000032 | cell wall mannoprotein biosynthetic process(GO:0000032) mannoprotein metabolic process(GO:0006056) mannoprotein biosynthetic process(GO:0006057) cell wall glycoprotein biosynthetic process(GO:0031506) cell wall biogenesis(GO:0042546) cell wall macromolecule biosynthetic process(GO:0044038) chain elongation of O-linked mannose residue(GO:0044845) cellular component macromolecule biosynthetic process(GO:0070589) |
| 0.0 | 0.2 | GO:0010825 | positive regulation of centrosome duplication(GO:0010825) |
| 0.0 | 0.2 | GO:0071638 | negative regulation of monocyte chemotactic protein-1 production(GO:0071638) |
| 0.0 | 0.1 | GO:1903774 | positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 0.0 | 0.3 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 0.1 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.0 | 0.1 | GO:0045200 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.0 | 0.1 | GO:1900108 | negative regulation of nodal signaling pathway(GO:1900108) |
| 0.0 | 0.1 | GO:0070827 | chromatin maintenance(GO:0070827) |
| 0.0 | 0.1 | GO:0045726 | positive regulation of integrin biosynthetic process(GO:0045726) |
| 0.0 | 0.1 | GO:0010961 | cellular magnesium ion homeostasis(GO:0010961) |
| 0.0 | 0.1 | GO:0006227 | dUDP biosynthetic process(GO:0006227) pyrimidine nucleoside diphosphate biosynthetic process(GO:0009139) pyrimidine deoxyribonucleoside diphosphate metabolic process(GO:0009196) pyrimidine deoxyribonucleoside diphosphate biosynthetic process(GO:0009197) dUDP metabolic process(GO:0046077) |
| 0.0 | 0.2 | GO:0071447 | cellular response to hydroperoxide(GO:0071447) |
| 0.0 | 0.1 | GO:1900041 | negative regulation of interleukin-2 secretion(GO:1900041) |
| 0.0 | 0.1 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.0 | 0.1 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.0 | 0.1 | GO:0051599 | response to hydrostatic pressure(GO:0051599) |
| 0.0 | 0.0 | GO:0019676 | ammonia assimilation cycle(GO:0019676) |
| 0.0 | 0.1 | GO:0030382 | sperm mitochondrion organization(GO:0030382) |
| 0.0 | 0.2 | GO:0080182 | histone H3-K4 trimethylation(GO:0080182) |
| 0.0 | 0.1 | GO:0021747 | cochlear nucleus development(GO:0021747) |
| 0.0 | 0.0 | GO:0048320 | axial mesoderm formation(GO:0048320) |
| 0.0 | 0.1 | GO:0031086 | nuclear-transcribed mRNA catabolic process, deadenylation-independent decay(GO:0031086) deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.0 | 0.2 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 0.1 | GO:0006547 | histidine metabolic process(GO:0006547) |
| 0.0 | 0.1 | GO:0046208 | spermine catabolic process(GO:0046208) |
| 0.0 | 0.2 | GO:0032957 | inositol trisphosphate metabolic process(GO:0032957) |
| 0.0 | 0.1 | GO:0032439 | endosome localization(GO:0032439) |
| 0.0 | 0.1 | GO:0032367 | intracellular cholesterol transport(GO:0032367) |
| 0.0 | 0.1 | GO:0070858 | negative regulation of bile acid biosynthetic process(GO:0070858) negative regulation of bile acid metabolic process(GO:1904252) |
| 0.0 | 0.1 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.0 | 0.1 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.2 | GO:0035413 | positive regulation of catenin import into nucleus(GO:0035413) |
| 0.0 | 0.2 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.0 | 0.1 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.0 | 0.1 | GO:0070272 | proton-transporting ATP synthase complex assembly(GO:0043461) proton-transporting ATP synthase complex biogenesis(GO:0070272) |
| 0.0 | 0.0 | GO:1901187 | regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.0 | 0.1 | GO:1904469 | positive regulation of tumor necrosis factor secretion(GO:1904469) |
| 0.0 | 0.1 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.0 | 0.0 | GO:2001055 | positive regulation of mesenchymal cell apoptotic process(GO:2001055) |
| 0.0 | 0.1 | GO:1902459 | positive regulation of stem cell population maintenance(GO:1902459) |
| 0.0 | 0.2 | GO:0014041 | regulation of neuron maturation(GO:0014041) |
| 0.0 | 0.0 | GO:0060672 | epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 0.0 | 0.0 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.0 | 0.1 | GO:0033129 | positive regulation of histone phosphorylation(GO:0033129) |
| 0.0 | 0.0 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.0 | 0.1 | GO:0090188 | negative regulation of pancreatic juice secretion(GO:0090188) |
| 0.0 | 0.1 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.0 | 0.3 | GO:0048714 | positive regulation of oligodendrocyte differentiation(GO:0048714) |
| 0.0 | 0.2 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.0 | GO:0009189 | deoxyribonucleoside diphosphate biosynthetic process(GO:0009189) |
| 0.0 | 0.1 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.0 | 0.1 | GO:0045656 | negative regulation of monocyte differentiation(GO:0045656) |
| 0.0 | 0.0 | GO:0046881 | positive regulation of follicle-stimulating hormone secretion(GO:0046881) |
| 0.0 | 0.1 | GO:1904869 | protein localization to nuclear body(GO:1903405) protein localization to Cajal body(GO:1904867) regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) protein localization to nucleoplasm(GO:1990173) |
| 0.0 | 0.1 | GO:0048133 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.0 | 0.0 | GO:1900107 | regulation of nodal signaling pathway(GO:1900107) |
| 0.0 | 0.0 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.0 | 0.0 | GO:0000432 | regulation of transcription from RNA polymerase II promoter by glucose(GO:0000430) positive regulation of transcription from RNA polymerase II promoter by glucose(GO:0000432) |
| 0.0 | 0.0 | GO:0035793 | positive regulation of metanephric mesenchymal cell migration by platelet-derived growth factor receptor-beta signaling pathway(GO:0035793) regulation of metanephric mesenchymal cell migration by platelet-derived growth factor receptor-beta signaling pathway(GO:1900238) positive regulation of metanephric mesenchymal cell migration(GO:2000591) |
| 0.0 | 0.2 | GO:0090201 | negative regulation of release of cytochrome c from mitochondria(GO:0090201) |
| 0.0 | 0.1 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.0 | 0.1 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.0 | 0.1 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.0 | 0.0 | GO:0045658 | regulation of neutrophil differentiation(GO:0045658) |
| 0.0 | 0.1 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 | 0.0 | GO:0016103 | diterpenoid catabolic process(GO:0016103) retinoic acid catabolic process(GO:0034653) |
| 0.0 | 0.2 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.1 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.0 | 0.0 | GO:2000828 | regulation of parathyroid hormone secretion(GO:2000828) |
| 0.0 | 0.0 | GO:0002025 | vasodilation by norepinephrine-epinephrine involved in regulation of systemic arterial blood pressure(GO:0002025) |
| 0.0 | 0.2 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.0 | 0.0 | GO:1901492 | positive regulation of lymphangiogenesis(GO:1901492) |
| 0.0 | 0.1 | GO:0046599 | regulation of centriole replication(GO:0046599) |
| 0.0 | 0.0 | GO:0006787 | porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) |
| 0.0 | 0.0 | GO:0016259 | selenocysteine metabolic process(GO:0016259) |
| 0.0 | 0.1 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.0 | 0.0 | GO:2000645 | negative regulation of receptor catabolic process(GO:2000645) |
| 0.0 | 0.1 | GO:0038033 | positive regulation of endothelial cell chemotaxis by VEGF-activated vascular endothelial growth factor receptor signaling pathway(GO:0038033) |
| 0.0 | 0.2 | GO:0043306 | positive regulation of mast cell activation involved in immune response(GO:0033008) positive regulation of mast cell degranulation(GO:0043306) |
| 0.0 | 0.0 | GO:0033566 | gamma-tubulin complex localization(GO:0033566) |
| 0.0 | 0.0 | GO:0001555 | oocyte growth(GO:0001555) |
| 0.0 | 0.1 | GO:0007035 | vacuolar acidification(GO:0007035) |
| 0.0 | 0.0 | GO:0072602 | interleukin-4 secretion(GO:0072602) |
| 0.0 | 0.0 | GO:0043974 | histone H3-K27 acetylation(GO:0043974) regulation of histone H3-K27 acetylation(GO:1901674) |
| 0.0 | 0.0 | GO:1900127 | positive regulation of hyaluronan biosynthetic process(GO:1900127) |
| 0.0 | 0.1 | GO:0030205 | dermatan sulfate metabolic process(GO:0030205) |
| 0.0 | 0.0 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0031502 | dolichyl-phosphate-mannose-protein mannosyltransferase complex(GO:0031502) |
| 0.0 | 0.2 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.0 | 0.2 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.0 | 0.1 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.0 | 0.1 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 0.1 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.0 | 0.1 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.0 | 0.2 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 0.1 | GO:0034750 | Scrib-APC-beta-catenin complex(GO:0034750) |
| 0.0 | 0.1 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 0.1 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 0.1 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.0 | 0.1 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.0 | 0.1 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.0 | 0.2 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.1 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.1 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.1 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.1 | GO:0098536 | deuterosome(GO:0098536) |
| 0.0 | 0.0 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.0 | 0.2 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.0 | 0.1 | GO:0032797 | SMN complex(GO:0032797) |
| 0.0 | 0.1 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 0.2 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.0 | 0.0 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.0 | 0.1 | GO:0031616 | spindle pole centrosome(GO:0031616) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.1 | 0.2 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.0 | 0.1 | GO:0004487 | methenyltetrahydrofolate cyclohydrolase activity(GO:0004477) methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
| 0.0 | 0.1 | GO:0016649 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.0 | 0.2 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.2 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.0 | 0.1 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 0.1 | GO:0000171 | ribonuclease MRP activity(GO:0000171) |
| 0.0 | 0.2 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.0 | 0.1 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.0 | 0.2 | GO:0035242 | protein-arginine omega-N asymmetric methyltransferase activity(GO:0035242) |
| 0.0 | 0.1 | GO:0018558 | 2,3-dihydroxy DDT 1,2-dioxygenase activity(GO:0018542) phenanthrene dioxygenase activity(GO:0018555) 2,2',3-trihydroxybiphenyl dioxygenase activity(GO:0018556) 1,2-dihydroxyfluorene 1,1-alpha-dioxygenase activity(GO:0018557) 5,6-dihydroxy-3-methyl-2-oxo-1,2-dihydroquinoline dioxygenase activity(GO:0018558) 1,1-dichloro-2-(dihydroxy-4-chlorophenyl)-(4-chlorophenyl)ethene 1,2-dioxygenase activity(GO:0018559) protocatechuate 3,4-dioxygenase type II activity(GO:0018560) 2'-aminobiphenyl-2,3-diol 1,2-dioxygenase activity(GO:0018561) 3,4-dihydroxyfluorene 4,4-alpha-dioxygenase activity(GO:0018562) 2,3-dihydroxy-ethylbenzene 1,2-dioxygenase activity(GO:0018563) carbazole 1,9a-dioxygenase activity(GO:0018564) dihydroxydibenzothiophene dioxygenase activity(GO:0018565) 1,2-dihydroxynaphthalene-6-sulfonate 1,8a-dioxygenase activity(GO:0018566) styrene dioxygenase activity(GO:0018567) 3,4-dihydroxyphenanthrene dioxygenase activity(GO:0018568) hydroquinone 1,2-dioxygenase activity(GO:0018569) p-cumate 2,3-dioxygenase activity(GO:0018570) 2,3-dihydroxy-p-cumate dioxygenase activity(GO:0018571) 3,5-dichlorocatechol 1,2-dioxygenase activity(GO:0018572) 2-aminophenol 1,6-dioxygenase activity(GO:0018573) 2,6-dichloro-p-hydroquinone 1,2-dioxygenase activity(GO:0018574) chlorocatechol 1,2-dioxygenase activity(GO:0018575) catechol dioxygenase activity(GO:0019114) dihydroxyfluorene dioxygenase activity(GO:0019117) 5-aminosalicylate dioxygenase activity(GO:0034543) 3-hydroxy-2-naphthoate 2,3-dioxygenase activity(GO:0034803) benzo(a)pyrene 11,12-dioxygenase activity(GO:0034806) benzo(a)pyrene 4,5-dioxygenase activity(GO:0034808) 4,5-dihydroxybenzo(a)pyrene dioxygenase activity(GO:0034810) benzo(a)pyrene 9,10-dioxygenase activity(GO:0034811) 9,10-dihydroxybenzo(a)pyrene dioxygenase activity(GO:0034812) benzo(a)pyrene 7,8-dioxygenase activity(GO:0034813) 7,8-dihydroxy benzo(a)pyrene dioxygenase activity(GO:0034814) 1,2-dihydroxy-5,6,7,8-tetrahydronaphthalene extradiol dioxygenase activity(GO:0034827) 2-mercaptobenzothiazole dioxygenase activity(GO:0034834) pyridine-3,4-diol dioxygenase activity(GO:0034895) pyrene dioxygenase activity(GO:0034920) 4,5-dihydroxypyrene dioxygenase activity(GO:0034922) phenanthrene-4-carboxylate dioxygenase activity(GO:0034934) tetrachlorobenzene dioxygenase activity(GO:0034935) 4,6-dichloro-3-methylcatechol 1,2-dioxygenase activity(GO:0034936) 2,3-dihydroxydiphenyl ether dioxygenase activity(GO:0034955) diphenyl ether 1,2-dioxygenase activity(GO:0034956) arachidonate 8(S)-lipoxygenase activity(GO:0036403) 4-hydroxycatechol 1,2-dioxygenase activity(GO:0047074) |
| 0.0 | 0.1 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.0 | 0.2 | GO:0003909 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 0.1 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.0 | 0.1 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.0 | 0.1 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.0 | 0.1 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.0 | 0.1 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.0 | 0.1 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 0.0 | 0.1 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.0 | 0.1 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.1 | GO:0102007 | lactonohydrolase activity(GO:0046573) acyl-L-homoserine-lactone lactonohydrolase activity(GO:0102007) |
| 0.0 | 0.1 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.0 | 0.2 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.1 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.0 | 0.1 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.0 | 0.1 | GO:0048763 | calcium-induced calcium release activity(GO:0048763) |
| 0.0 | 0.1 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.0 | 0.1 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 0.1 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.0 | 0.0 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.0 | 0.1 | GO:0051022 | Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.0 | 0.0 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.0 | 0.0 | GO:0035175 | histone kinase activity (H3-S10 specific)(GO:0035175) |
| 0.0 | 0.2 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.1 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.0 | 0.1 | GO:0033592 | RNA strand annealing activity(GO:0033592) |
| 0.0 | 0.0 | GO:0015087 | cobalt ion transmembrane transporter activity(GO:0015087) |
| 0.0 | 0.1 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.0 | 0.1 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.0 | 0.1 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.0 | 0.2 | GO:0051765 | inositol tetrakisphosphate kinase activity(GO:0051765) |
| 0.0 | 0.2 | GO:0016646 | oxidoreductase activity, acting on the CH-NH group of donors, NAD or NADP as acceptor(GO:0016646) |
| 0.0 | 0.0 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) |
| 0.0 | 0.1 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 0.2 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.1 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.0 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.1 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.0 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.1 | GO:0097642 | calcitonin family receptor activity(GO:0097642) |
| 0.0 | 0.1 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 0.2 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.1 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.0 | 0.0 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.3 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.1 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.0 | 0.3 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.1 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.0 | 0.2 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.5 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.2 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |