Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
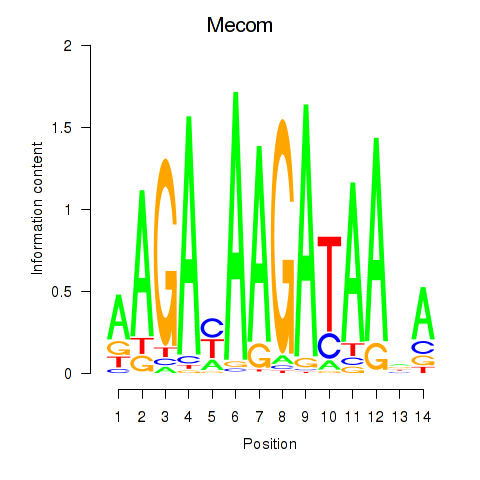
Results for Mecom
Z-value: 1.81
Transcription factors associated with Mecom
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Mecom
|
ENSMUSG00000027684.10 | MDS1 and EVI1 complex locus |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr3_30506438_30507337 | Mecom | 2600 | 0.198314 | 0.96 | 2.5e-03 | Click! |
| chr3_30507376_30507724 | Mecom | 1937 | 0.249845 | 0.95 | 3.1e-03 | Click! |
| chr3_30549571_30549744 | Mecom | 1649 | 0.260930 | -0.78 | 7.0e-02 | Click! |
| chr3_30101238_30101422 | Mecom | 39093 | 0.156518 | -0.77 | 7.2e-02 | Click! |
| chr3_30015349_30015519 | Mecom | 1929 | 0.306530 | 0.64 | 1.7e-01 | Click! |
Activity of the Mecom motif across conditions
Conditions sorted by the z-value of the Mecom motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
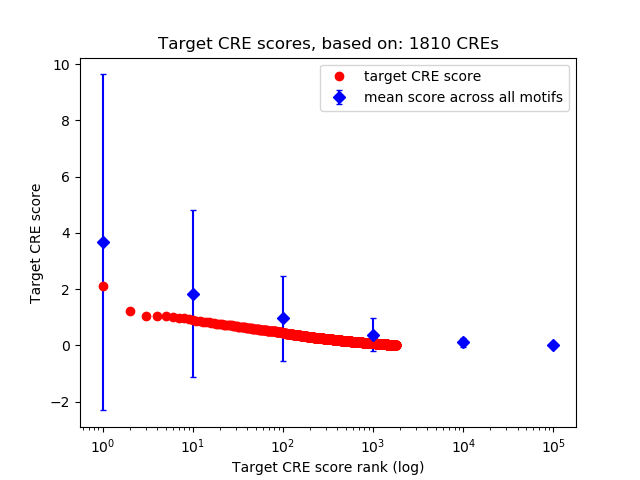
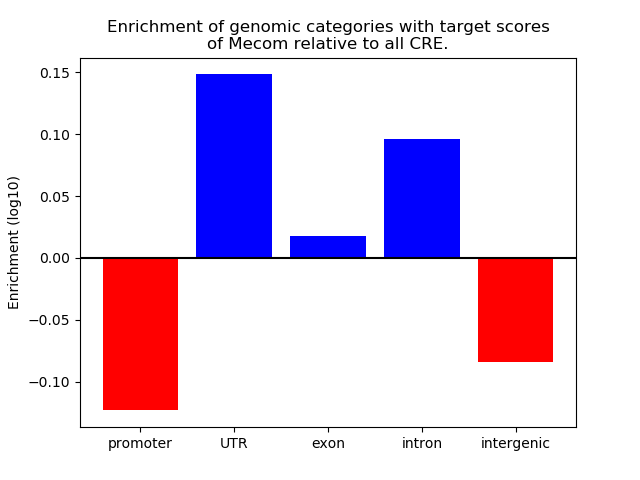
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr4_108075287_108075438 | 2.12 |
Scp2 |
sterol carrier protein 2, liver |
3943 |
0.17 |
| chr3_152357627_152357783 | 1.22 |
Usp33 |
ubiquitin specific peptidase 33 |
4193 |
0.14 |
| chr8_3217015_3217198 | 1.05 |
Insr |
insulin receptor |
24389 |
0.17 |
| chr5_87499331_87499512 | 1.03 |
Ugt2a1 |
UDP glucuronosyltransferase 2 family, polypeptide A1 |
8550 |
0.12 |
| chr16_42947764_42948084 | 1.03 |
Ndufs5-ps |
NADH:ubiquinone oxidoreductase core subunit S5, pseudogene |
7707 |
0.21 |
| chr11_106728824_106729001 | 0.99 |
Pecam1 |
platelet/endothelial cell adhesion molecule 1 |
13518 |
0.12 |
| chr5_66095093_66095575 | 0.99 |
Rbm47 |
RNA binding motif protein 47 |
2857 |
0.18 |
| chr2_121439577_121439771 | 0.97 |
Ell3 |
elongation factor RNA polymerase II-like 3 |
1477 |
0.18 |
| chr9_77753656_77753905 | 0.92 |
Gclc |
glutamate-cysteine ligase, catalytic subunit |
755 |
0.58 |
| chr11_120807385_120807536 | 0.89 |
Fasn |
fatty acid synthase |
1145 |
0.28 |
| chr7_46031049_46031458 | 0.87 |
Abcc6 |
ATP-binding cassette, sub-family C (CFTR/MRP), member 6 |
951 |
0.41 |
| chr8_46486311_46486711 | 0.86 |
Acsl1 |
acyl-CoA synthetase long-chain family member 1 |
4455 |
0.18 |
| chr6_32962622_32962773 | 0.84 |
Chchd3 |
coiled-coil-helix-coiled-coil-helix domain containing 3 |
65626 |
0.11 |
| chr5_134436362_134436513 | 0.84 |
Gm26340 |
predicted gene, 26340 |
12137 |
0.11 |
| chr17_49429803_49429983 | 0.82 |
Gm20540 |
predicted gene 20540 |
653 |
0.65 |
| chr14_76827348_76827510 | 0.79 |
Gm48968 |
predicted gene, 48968 |
5392 |
0.22 |
| chr15_3435423_3435589 | 0.78 |
Ghr |
growth hormone receptor |
36138 |
0.21 |
| chr3_58454505_58454707 | 0.77 |
Tsc22d2 |
TSC22 domain family, member 2 |
37122 |
0.13 |
| chr3_89149636_89149787 | 0.77 |
Hcn3 |
hyperpolarization-activated, cyclic nucleotide-gated K+ 3 |
1503 |
0.17 |
| chr4_137794057_137794257 | 0.76 |
Alpl |
alkaline phosphatase, liver/bone/kidney |
2073 |
0.34 |
| chr2_91782501_91782660 | 0.75 |
Ambra1 |
autophagy/beclin 1 regulator 1 |
9348 |
0.18 |
| chr4_106354740_106354963 | 0.74 |
Usp24 |
ubiquitin specific peptidase 24 |
38617 |
0.13 |
| chr11_94583519_94583672 | 0.73 |
Acsf2 |
acyl-CoA synthetase family member 2 |
12817 |
0.11 |
| chr7_144040112_144040314 | 0.73 |
Shank2 |
SH3 and multiple ankyrin repeat domains 2 |
8949 |
0.21 |
| chr16_24892958_24893109 | 0.71 |
Gm22672 |
predicted gene, 22672 |
8860 |
0.25 |
| chr6_6204143_6204472 | 0.71 |
Slc25a13 |
solute carrier family 25 (mitochondrial carrier, adenine nucleotide translocator), member 13 |
12811 |
0.23 |
| chr8_122943666_122943817 | 0.71 |
Ankrd11 |
ankyrin repeat domain 11 |
27754 |
0.11 |
| chr6_94653423_94653689 | 0.70 |
Lrig1 |
leucine-rich repeats and immunoglobulin-like domains 1 |
1969 |
0.38 |
| chr19_55252347_55252717 | 0.69 |
Acsl5 |
acyl-CoA synthetase long-chain family member 5 |
572 |
0.74 |
| chr2_117119777_117119942 | 0.68 |
Spred1 |
sprouty protein with EVH-1 domain 1, related sequence |
1515 |
0.45 |
| chr6_124688048_124688642 | 0.67 |
Lpcat3 |
lysophosphatidylcholine acyltransferase 3 |
13074 |
0.06 |
| chr14_67053186_67053337 | 0.67 |
Ppp2r2a |
protein phosphatase 2, regulatory subunit B, alpha |
9732 |
0.17 |
| chr18_39483316_39483539 | 0.66 |
Nr3c1 |
nuclear receptor subfamily 3, group C, member 1 |
3805 |
0.31 |
| chr15_6891673_6891824 | 0.66 |
Osmr |
oncostatin M receptor |
16779 |
0.26 |
| chr9_55214755_55215047 | 0.64 |
Fbxo22 |
F-box protein 22 |
1238 |
0.43 |
| chr14_65385019_65385170 | 0.64 |
Zfp395 |
zinc finger protein 395 |
9701 |
0.18 |
| chr10_61648098_61648328 | 0.64 |
Ppa1 |
pyrophosphatase (inorganic) 1 |
339 |
0.83 |
| chr17_91875617_91875801 | 0.63 |
Gm41654 |
predicted gene, 41654 |
3570 |
0.25 |
| chr3_75952417_75952705 | 0.62 |
Gm37685 |
predicted gene, 37685 |
437 |
0.81 |
| chr19_25343677_25343828 | 0.62 |
Gm34432 |
predicted gene, 34432 |
3451 |
0.3 |
| chr4_76368947_76369220 | 0.62 |
Gm11252 |
predicted gene 11252 |
24071 |
0.2 |
| chr8_116404224_116404375 | 0.61 |
1700018P08Rik |
RIKEN cDNA 1700018P08 gene |
47238 |
0.17 |
| chr4_15267631_15268095 | 0.61 |
Tmem64 |
transmembrane protein 64 |
2032 |
0.43 |
| chr18_65223520_65223956 | 0.61 |
Nedd4l |
neural precursor cell expressed, developmentally down-regulated gene 4-like |
15854 |
0.15 |
| chr1_119647745_119647910 | 0.60 |
Epb41l5 |
erythrocyte membrane protein band 4.1 like 5 |
790 |
0.6 |
| chr16_46848879_46849042 | 0.60 |
Gm6912 |
predicted gene 6912 |
225238 |
0.02 |
| chr5_87573106_87573296 | 0.60 |
Sult1d1 |
sulfotransferase family 1D, member 1 |
4174 |
0.13 |
| chr4_41331357_41331705 | 0.59 |
Gm26084 |
predicted gene, 26084 |
14420 |
0.1 |
| chr16_26698467_26698766 | 0.59 |
Il1rap |
interleukin 1 receptor accessory protein |
23818 |
0.24 |
| chr9_80672315_80672507 | 0.58 |
Gm39380 |
predicted gene, 39380 |
37584 |
0.19 |
| chr8_25603179_25603391 | 0.58 |
Nsd3 |
nuclear receptor binding SET domain protein 3 |
966 |
0.35 |
| chr6_14897330_14897481 | 0.58 |
Foxp2 |
forkhead box P2 |
3944 |
0.37 |
| chr11_75737051_75737202 | 0.57 |
Ywhae |
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, epsilon polypeptide |
3978 |
0.19 |
| chr14_14067010_14067161 | 0.57 |
Atxn7 |
ataxin 7 |
20384 |
0.17 |
| chr14_11567053_11567226 | 0.56 |
Ptprg |
protein tyrosine phosphatase, receptor type, G |
13558 |
0.22 |
| chr9_90270049_90270249 | 0.56 |
Tbc1d2b |
TBC1 domain family, member 2B |
620 |
0.71 |
| chr8_91494941_91495092 | 0.56 |
Gm45289 |
predicted gene 45289 |
1997 |
0.25 |
| chr18_58272927_58273088 | 0.56 |
Gm25660 |
predicted gene, 25660 |
41432 |
0.19 |
| chr2_32073660_32073977 | 0.56 |
Fam78a |
family with sequence similarity 78, member A |
5534 |
0.12 |
| chr12_73341772_73342027 | 0.55 |
Slc38a6 |
solute carrier family 38, member 6 |
2468 |
0.25 |
| chr12_104355745_104355928 | 0.54 |
Serpina3l-ps |
serine (or cysteine) peptidase inhibitor, clade A, member 3L, pseudogene |
2167 |
0.21 |
| chr5_122937277_122937753 | 0.54 |
Kdm2b |
lysine (K)-specific demethylase 2B |
10479 |
0.13 |
| chr4_76350562_76351008 | 0.54 |
Ptprd |
protein tyrosine phosphatase, receptor type, D |
6542 |
0.27 |
| chr10_71382555_71382706 | 0.54 |
Ipmk |
inositol polyphosphate multikinase |
881 |
0.56 |
| chr3_32364907_32365337 | 0.53 |
Zmat3 |
zinc finger matrin type 3 |
338 |
0.87 |
| chr6_85933884_85934041 | 0.53 |
Nat8b-ps |
N-acetyltransferase 8B, pseudogene |
583 |
0.52 |
| chr3_138286066_138286462 | 0.52 |
Adh1 |
alcohol dehydrogenase 1 (class I) |
8613 |
0.12 |
| chr19_61226929_61227456 | 0.52 |
Csf2ra |
colony stimulating factor 2 receptor, alpha, low-affinity (granulocyte-macrophage) |
44 |
0.96 |
| chr2_75773042_75773329 | 0.52 |
Gm13657 |
predicted gene 13657 |
4003 |
0.18 |
| chr6_117009658_117009825 | 0.51 |
Gm43929 |
predicted gene, 43929 |
27722 |
0.19 |
| chr2_115758350_115758524 | 0.51 |
Meis2 |
Meis homeobox 2 |
110430 |
0.06 |
| chr8_93164567_93164980 | 0.51 |
Ces1d |
carboxylesterase 1D |
5202 |
0.15 |
| chr5_77460215_77460391 | 0.50 |
1700017L05Rik |
RIKEN cDNA 1700017L05 gene |
5185 |
0.17 |
| chr2_155470676_155470827 | 0.50 |
Ncoa6 |
nuclear receptor coactivator 6 |
3055 |
0.15 |
| chr17_13018717_13018879 | 0.50 |
Sod2 |
superoxide dismutase 2, mitochondrial |
8458 |
0.11 |
| chr2_60122174_60122591 | 0.50 |
Gm13620 |
predicted gene 13620 |
2714 |
0.2 |
| chr11_55497781_55497965 | 0.50 |
Mir7652 |
microRNA 7652 |
3353 |
0.14 |
| chr19_4600545_4600696 | 0.50 |
Pcx |
pyruvate carboxylase |
6212 |
0.12 |
| chr13_112308580_112308731 | 0.50 |
Ankrd55 |
ankyrin repeat domain 55 |
9799 |
0.17 |
| chr6_90555718_90555877 | 0.49 |
Aldh1l1 |
aldehyde dehydrogenase 1 family, member L1 |
4949 |
0.16 |
| chr12_45069728_45070323 | 0.49 |
Stxbp6 |
syntaxin binding protein 6 (amisyn) |
4087 |
0.24 |
| chr7_66388868_66389043 | 0.49 |
Lrrk1 |
leucine-rich repeat kinase 1 |
605 |
0.64 |
| chr4_148644834_148645010 | 0.49 |
Gm572 |
predicted gene 572 |
1605 |
0.29 |
| chr5_134312402_134312749 | 0.49 |
Gtf2i |
general transcription factor II I |
553 |
0.67 |
| chr17_13774125_13774292 | 0.48 |
Tcte2 |
t-complex-associated testis expressed 2 |
12383 |
0.15 |
| chr4_134926867_134927018 | 0.48 |
Rsrp1 |
arginine/serine rich protein 1 |
1496 |
0.34 |
| chr2_70838613_70839186 | 0.48 |
Tlk1 |
tousled-like kinase 1 |
13171 |
0.18 |
| chr3_121795871_121796026 | 0.48 |
4633401B06Rik |
RIKEN cDNA 4633401B06 gene |
7376 |
0.13 |
| chr19_44380608_44380766 | 0.47 |
Scd1 |
stearoyl-Coenzyme A desaturase 1 |
26003 |
0.12 |
| chr8_25775115_25775327 | 0.47 |
Bag4 |
BCL2-associated athanogene 4 |
2361 |
0.16 |
| chr19_44403240_44403567 | 0.47 |
Scd1 |
stearoyl-Coenzyme A desaturase 1 |
3287 |
0.19 |
| chr15_82166714_82166896 | 0.47 |
Gm49502 |
predicted gene, 49502 |
18711 |
0.08 |
| chr9_100535718_100535869 | 0.46 |
Gm28586 |
predicted gene 28586 |
4722 |
0.16 |
| chr9_40801417_40801568 | 0.46 |
Hspa8 |
heat shock protein 8 |
31 |
0.94 |
| chr16_65617781_65617932 | 0.46 |
Gm49633 |
predicted gene, 49633 |
4437 |
0.27 |
| chr4_97779759_97779983 | 0.46 |
E130114P18Rik |
RIKEN cDNA E130114P18 gene |
1793 |
0.32 |
| chr4_132414912_132415080 | 0.46 |
Gm25707 |
predicted gene, 25707 |
6687 |
0.1 |
| chr19_36835196_36835389 | 0.45 |
Tnks2 |
tankyrase, TRF1-interacting ankyrin-related ADP-ribose polymerase 2 |
888 |
0.61 |
| chr3_60046790_60046945 | 0.45 |
Aadac |
arylacetamide deacetylase |
14991 |
0.15 |
| chr9_51313328_51313479 | 0.45 |
1810046K07Rik |
RIKEN cDNA 1810046K07 gene |
688 |
0.69 |
| chr6_108456531_108456780 | 0.45 |
Itpr1 |
inositol 1,4,5-trisphosphate receptor 1 |
2048 |
0.31 |
| chr3_149090724_149090881 | 0.44 |
Gm25127 |
predicted gene, 25127 |
61870 |
0.11 |
| chr5_53470110_53470463 | 0.44 |
Rbpj |
recombination signal binding protein for immunoglobulin kappa J region |
4134 |
0.24 |
| chr13_40851030_40851206 | 0.43 |
Gm35160 |
predicted gene, 35160 |
4379 |
0.12 |
| chr16_13320219_13320378 | 0.43 |
Mrtfb |
myocardin related transcription factor B |
38123 |
0.17 |
| chr12_40574749_40574900 | 0.43 |
Dock4 |
dedicator of cytokinesis 4 |
128488 |
0.05 |
| chr11_110242652_110242881 | 0.42 |
Abca6 |
ATP-binding cassette, sub-family A (ABC1), member 6 |
9010 |
0.26 |
| chr6_128843572_128843953 | 0.42 |
Gm44066 |
predicted gene, 44066 |
591 |
0.53 |
| chr19_4786678_4786855 | 0.42 |
Rbm4 |
RNA binding motif protein 4 |
7085 |
0.08 |
| chr3_58418942_58419336 | 0.42 |
Tsc22d2 |
TSC22 domain family, member 2 |
1655 |
0.37 |
| chr6_116068553_116068726 | 0.42 |
Tmcc1 |
transmembrane and coiled coil domains 1 |
4517 |
0.2 |
| chr11_28694849_28695038 | 0.41 |
2810471M01Rik |
RIKEN cDNA 2810471M01 gene |
13379 |
0.17 |
| chr5_66046377_66046556 | 0.41 |
Rbm47 |
RNA binding motif protein 47 |
8086 |
0.13 |
| chr5_125483749_125483900 | 0.40 |
Gm27551 |
predicted gene, 27551 |
4447 |
0.15 |
| chr2_103594624_103594957 | 0.40 |
Abtb2 |
ankyrin repeat and BTB (POZ) domain containing 2 |
28480 |
0.17 |
| chr12_84185436_84185743 | 0.40 |
Gm19327 |
predicted gene, 19327 |
2217 |
0.18 |
| chr3_52298742_52298941 | 0.40 |
Gm38034 |
predicted gene, 38034 |
16600 |
0.14 |
| chr12_71909107_71909265 | 0.40 |
Daam1 |
dishevelled associated activator of morphogenesis 1 |
19456 |
0.21 |
| chr12_87443355_87443540 | 0.40 |
Alkbh1 |
alkB homolog 1, histone H2A dioxygenase |
379 |
0.44 |
| chr17_29717187_29717383 | 0.39 |
1810014P07Rik |
RIKEN cDNA 1810014P07 gene |
213 |
0.52 |
| chr19_58398099_58398278 | 0.39 |
Gfra1 |
glial cell line derived neurotrophic factor family receptor alpha 1 |
56278 |
0.14 |
| chr4_88715181_88715384 | 0.39 |
Gm26566 |
predicted gene, 26566 |
6523 |
0.05 |
| chr18_61909540_61909715 | 0.39 |
Ablim3 |
actin binding LIM protein family, member 3 |
2196 |
0.31 |
| chr4_55242738_55242936 | 0.39 |
Gm12508 |
predicted gene 12508 |
11903 |
0.17 |
| chr4_97779546_97779733 | 0.39 |
E130114P18Rik |
RIKEN cDNA E130114P18 gene |
1561 |
0.34 |
| chr8_93227828_93228167 | 0.39 |
Ces1e |
carboxylesterase 1E |
1621 |
0.29 |
| chr17_86494572_86495073 | 0.39 |
Prkce |
protein kinase C, epsilon |
1495 |
0.46 |
| chr3_97632897_97633210 | 0.39 |
Fmo5 |
flavin containing monooxygenase 5 |
4170 |
0.16 |
| chr16_4932467_4932619 | 0.38 |
Mgrn1 |
mahogunin, ring finger 1 |
4865 |
0.11 |
| chr4_123990670_123991070 | 0.38 |
Gm12902 |
predicted gene 12902 |
64636 |
0.08 |
| chr4_76448425_76449174 | 0.38 |
Ptprd |
protein tyrosine phosphatase, receptor type, D |
1181 |
0.49 |
| chr16_25879136_25879302 | 0.38 |
Gm4524 |
predicted gene 4524 |
77256 |
0.1 |
| chr4_99037726_99038187 | 0.37 |
Angptl3 |
angiopoietin-like 3 |
4753 |
0.21 |
| chr2_166027566_166027720 | 0.37 |
Ncoa3 |
nuclear receptor coactivator 3 |
20275 |
0.14 |
| chr17_46110701_46110892 | 0.37 |
Mrps18a |
mitochondrial ribosomal protein S18A |
190 |
0.91 |
| chr6_38827332_38827516 | 0.37 |
Hipk2 |
homeodomain interacting protein kinase 2 |
9078 |
0.23 |
| chr19_37439255_37439863 | 0.37 |
Hhex |
hematopoietically expressed homeobox |
2820 |
0.17 |
| chr19_10065348_10065653 | 0.36 |
Fads2 |
fatty acid desaturase 2 |
5687 |
0.14 |
| chr16_26608378_26608529 | 0.36 |
Il1rap |
interleukin 1 receptor accessory protein |
15703 |
0.26 |
| chr3_151838300_151838467 | 0.36 |
Ptgfr |
prostaglandin F receptor |
753 |
0.58 |
| chr18_38176749_38176900 | 0.36 |
Pcdh1 |
protocadherin 1 |
26339 |
0.12 |
| chr13_9035178_9035364 | 0.36 |
Gtpbp4 |
GTP binding protein 4 |
39188 |
0.08 |
| chr16_43277669_43278048 | 0.36 |
Gm37946 |
predicted gene, 37946 |
30423 |
0.15 |
| chr1_131501686_131501867 | 0.36 |
Srgap2 |
SLIT-ROBO Rho GTPase activating protein 2 |
2590 |
0.21 |
| chr2_167325906_167326068 | 0.36 |
B4galt5 |
UDP-Gal:betaGlcNAc beta 1,4-galactosyltransferase, polypeptide 5 |
12087 |
0.18 |
| chr17_46505508_46505659 | 0.36 |
Cul9 |
cullin 9 |
4512 |
0.12 |
| chr7_45574336_45574519 | 0.36 |
Bcat2 |
branched chain aminotransferase 2, mitochondrial |
673 |
0.29 |
| chr17_46454562_46454932 | 0.36 |
Gm5093 |
predicted gene 5093 |
14650 |
0.09 |
| chr14_105253422_105253603 | 0.36 |
Gm22290 |
predicted gene, 22290 |
3502 |
0.18 |
| chr2_30413885_30414091 | 0.35 |
Gm16323 |
predicted gene 16323 |
272 |
0.54 |
| chr15_6448654_6448821 | 0.35 |
C9 |
complement component 9 |
3365 |
0.27 |
| chr2_152240729_152240889 | 0.35 |
Csnk2a1 |
casein kinase 2, alpha 1 polypeptide |
13896 |
0.14 |
| chr13_23574893_23575044 | 0.35 |
Gm44359 |
predicted gene, 44359 |
115 |
0.72 |
| chr18_11317026_11317247 | 0.35 |
Gata6 |
GATA binding protein 6 |
258089 |
0.02 |
| chr3_51416126_51416308 | 0.35 |
Naa15 |
N(alpha)-acetyltransferase 15, NatA auxiliary subunit |
79 |
0.94 |
| chr12_24614688_24615076 | 0.35 |
Gm6969 |
predicted pseudogene 6969 |
24128 |
0.13 |
| chr1_130830297_130830686 | 0.35 |
Pigr |
polymeric immunoglobulin receptor |
1481 |
0.28 |
| chr9_70234061_70234212 | 0.34 |
Myo1e |
myosin IE |
26768 |
0.19 |
| chr17_5017691_5017888 | 0.34 |
Arid1b |
AT rich interactive domain 1B (SWI-like) |
21360 |
0.22 |
| chr6_97304469_97304675 | 0.34 |
Frmd4b |
FERM domain containing 4B |
7808 |
0.22 |
| chr13_96746192_96746359 | 0.34 |
Ankrd31 |
ankyrin repeat domain 31 |
1997 |
0.31 |
| chr10_110708063_110708339 | 0.34 |
E2f7 |
E2F transcription factor 7 |
37238 |
0.16 |
| chr6_28482820_28482980 | 0.34 |
Snd1 |
staphylococcal nuclease and tudor domain containing 1 |
2471 |
0.22 |
| chr12_30348373_30348544 | 0.33 |
Sntg2 |
syntrophin, gamma 2 |
9225 |
0.26 |
| chr11_48838500_48838661 | 0.33 |
Trim7 |
tripartite motif-containing 7 |
66 |
0.94 |
| chr4_148143306_148143457 | 0.33 |
Mad2l2 |
MAD2 mitotic arrest deficient-like 2 |
1409 |
0.24 |
| chr10_108170850_108171005 | 0.33 |
Ppp1r12a |
protein phosphatase 1, regulatory subunit 12A |
8527 |
0.25 |
| chr17_15372798_15372972 | 0.33 |
Dll1 |
delta like canonical Notch ligand 1 |
1054 |
0.47 |
| chr15_93398531_93398907 | 0.33 |
Pphln1 |
periphilin 1 |
255 |
0.63 |
| chr15_82635338_82635489 | 0.33 |
Gm27463 |
predicted gene, 27463 |
6588 |
0.08 |
| chr7_73543304_73543467 | 0.33 |
Chd2 |
chromodomain helicase DNA binding protein 2 |
1555 |
0.24 |
| chr14_73255473_73255646 | 0.33 |
Rb1 |
RB transcriptional corepressor 1 |
8762 |
0.17 |
| chr10_33866082_33866287 | 0.32 |
Sult3a1 |
sulfotransferase family 3A, member 1 |
2249 |
0.21 |
| chr11_98206684_98206835 | 0.32 |
Cdk12 |
cyclin-dependent kinase 12 |
3424 |
0.14 |
| chr4_45799507_45799658 | 0.32 |
Aldh1b1 |
aldehyde dehydrogenase 1 family, member B1 |
461 |
0.76 |
| chr5_54043709_54043896 | 0.32 |
Stim2 |
stromal interaction molecule 2 |
30935 |
0.2 |
| chr10_29302371_29302533 | 0.32 |
Echdc1 |
enoyl Coenzyme A hydratase domain containing 1 |
10714 |
0.13 |
| chr12_111375253_111375404 | 0.32 |
Cdc42bpb |
CDC42 binding protein kinase beta |
2291 |
0.23 |
| chr6_50587768_50588064 | 0.32 |
4921507P07Rik |
RIKEN cDNA 4921507P07 gene |
8716 |
0.09 |
| chr4_144900352_144900520 | 0.32 |
Dhrs3 |
dehydrogenase/reductase (SDR family) member 3 |
7217 |
0.22 |
| chr16_94571169_94571354 | 0.32 |
Dyrk1a |
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1a |
354 |
0.88 |
| chr7_26916759_26917069 | 0.32 |
Cyp2a21-ps |
cytochrome P450, family 2, subfamily a, polypeptide 21, pseudogene |
467 |
0.74 |
| chr11_51739157_51739308 | 0.32 |
Gm25291 |
predicted gene, 25291 |
1310 |
0.37 |
| chr3_52257597_52258053 | 0.31 |
Foxo1 |
forkhead box O1 |
10511 |
0.12 |
| chr4_53095991_53096266 | 0.31 |
Abca1 |
ATP-binding cassette, sub-family A (ABC1), member 1 |
55202 |
0.11 |
| chr2_69205518_69205715 | 0.31 |
Spc25 |
SPC25, NDC80 kinetochore complex component, homolog (S. cerevisiae) |
401 |
0.81 |
| chr9_9180190_9180363 | 0.31 |
Gm16833 |
predicted gene, 16833 |
56012 |
0.14 |
| chr10_61444973_61445124 | 0.31 |
Gm48086 |
predicted gene, 48086 |
5669 |
0.11 |
| chr19_44458524_44458675 | 0.31 |
Gm35406 |
predicted gene, 35406 |
5582 |
0.16 |
| chr13_101779442_101779645 | 0.31 |
Pik3r1 |
phosphoinositide-3-kinase regulatory subunit 1 |
11326 |
0.2 |
| chr7_118698593_118698744 | 0.31 |
Gde1 |
glycerophosphodiester phosphodiesterase 1 |
6712 |
0.13 |
| chr17_32946227_32946412 | 0.31 |
Cyp4f13 |
cytochrome P450, family 4, subfamily f, polypeptide 13 |
1016 |
0.36 |
| chr2_15064726_15064962 | 0.31 |
Arl5b |
ADP-ribosylation factor-like 5B |
3295 |
0.18 |
| chr11_120695384_120695739 | 0.31 |
Aspscr1 |
alveolar soft part sarcoma chromosome region, candidate 1 (human) |
6251 |
0.07 |
| chr13_115090984_115091155 | 0.30 |
Pelo |
pelota mRNA surveillance and ribosome rescue factor |
883 |
0.44 |
| chr1_63115073_63115446 | 0.30 |
Ino80dos |
INO80 complex subunit D, opposite strand |
519 |
0.42 |
| chr19_34524861_34525231 | 0.30 |
Lipa |
lysosomal acid lipase A |
2365 |
0.2 |
| chr2_167422512_167422688 | 0.30 |
Slc9a8 |
solute carrier family 9 (sodium/hydrogen exchanger), member 8 |
858 |
0.6 |
| chr17_84268442_84268633 | 0.30 |
Gm24492 |
predicted gene, 24492 |
19210 |
0.18 |
| chr17_65577307_65577458 | 0.30 |
Gm49866 |
predicted gene, 49866 |
16606 |
0.14 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0006701 | progesterone biosynthetic process(GO:0006701) |
| 0.1 | 0.4 | GO:1903334 | positive regulation of protein folding(GO:1903334) |
| 0.1 | 0.4 | GO:0060847 | endothelial cell fate specification(GO:0060847) |
| 0.1 | 0.4 | GO:0009826 | unidimensional cell growth(GO:0009826) |
| 0.1 | 0.1 | GO:0006549 | isoleucine metabolic process(GO:0006549) |
| 0.1 | 0.5 | GO:0097011 | cellular response to granulocyte macrophage colony-stimulating factor stimulus(GO:0097011) |
| 0.1 | 0.3 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.1 | 0.3 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.1 | 0.3 | GO:0072338 | creatinine metabolic process(GO:0046449) cellular lactam metabolic process(GO:0072338) |
| 0.1 | 0.5 | GO:0010746 | regulation of plasma membrane long-chain fatty acid transport(GO:0010746) |
| 0.1 | 0.2 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.1 | 0.5 | GO:0097105 | presynaptic membrane assembly(GO:0097105) |
| 0.1 | 0.2 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.1 | 0.5 | GO:0051409 | response to nitrosative stress(GO:0051409) |
| 0.1 | 0.3 | GO:0072602 | interleukin-4 secretion(GO:0072602) |
| 0.1 | 0.3 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 0.1 | 0.1 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.1 | 0.3 | GO:1903964 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.1 | 0.2 | GO:0071799 | response to prostaglandin D(GO:0071798) cellular response to prostaglandin D stimulus(GO:0071799) |
| 0.1 | 0.1 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.1 | 0.3 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.1 | 0.2 | GO:0006166 | purine ribonucleoside salvage(GO:0006166) |
| 0.1 | 0.3 | GO:0010917 | negative regulation of mitochondrial membrane potential(GO:0010917) |
| 0.1 | 0.2 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.1 | 0.3 | GO:0044789 | modulation by host of viral release from host cell(GO:0044789) positive regulation by host of viral release from host cell(GO:0044791) |
| 0.1 | 0.3 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.1 | 0.2 | GO:0000957 | mitochondrial RNA catabolic process(GO:0000957) regulation of mitochondrial RNA catabolic process(GO:0000960) |
| 0.0 | 0.1 | GO:1902309 | negative regulation of peptidyl-serine dephosphorylation(GO:1902309) |
| 0.0 | 0.1 | GO:0006714 | sesquiterpenoid metabolic process(GO:0006714) |
| 0.0 | 0.1 | GO:0090365 | regulation of mRNA modification(GO:0090365) |
| 0.0 | 0.3 | GO:0006307 | DNA dealkylation involved in DNA repair(GO:0006307) |
| 0.0 | 0.2 | GO:0061073 | ciliary body morphogenesis(GO:0061073) |
| 0.0 | 0.1 | GO:0052490 | negative regulation by symbiont of host apoptotic process(GO:0033668) negative regulation by symbiont of host programmed cell death(GO:0052041) negative regulation by organism of programmed cell death in other organism involved in symbiotic interaction(GO:0052490) |
| 0.0 | 0.2 | GO:1903599 | positive regulation of mitophagy(GO:1903599) |
| 0.0 | 0.1 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.0 | 0.1 | GO:1901492 | positive regulation of lymphangiogenesis(GO:1901492) |
| 0.0 | 0.5 | GO:0010801 | negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 0.0 | 0.2 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) |
| 0.0 | 0.2 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.0 | 0.1 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.0 | 0.1 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) |
| 0.0 | 0.1 | GO:0052203 | modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) |
| 0.0 | 0.1 | GO:0070318 | positive regulation of G0 to G1 transition(GO:0070318) |
| 0.0 | 0.1 | GO:0061010 | gall bladder development(GO:0061010) |
| 0.0 | 0.2 | GO:0072675 | osteoclast fusion(GO:0072675) |
| 0.0 | 0.1 | GO:0006210 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) thymine metabolic process(GO:0019859) |
| 0.0 | 0.2 | GO:0019087 | transformation of host cell by virus(GO:0019087) |
| 0.0 | 0.1 | GO:0002337 | B-1a B cell differentiation(GO:0002337) |
| 0.0 | 0.1 | GO:0002386 | immune response in mucosal-associated lymphoid tissue(GO:0002386) |
| 0.0 | 0.1 | GO:2000370 | positive regulation of clathrin-mediated endocytosis(GO:2000370) |
| 0.0 | 0.1 | GO:0044027 | hypermethylation of CpG island(GO:0044027) |
| 0.0 | 0.1 | GO:0042360 | vitamin E metabolic process(GO:0042360) |
| 0.0 | 0.4 | GO:0014733 | regulation of skeletal muscle adaptation(GO:0014733) |
| 0.0 | 0.1 | GO:0051791 | medium-chain fatty acid metabolic process(GO:0051791) |
| 0.0 | 0.2 | GO:0070544 | histone H3-K36 demethylation(GO:0070544) |
| 0.0 | 0.1 | GO:0051987 | positive regulation of attachment of spindle microtubules to kinetochore(GO:0051987) |
| 0.0 | 0.1 | GO:0046602 | regulation of mitotic centrosome separation(GO:0046602) |
| 0.0 | 0.1 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.0 | 0.2 | GO:0060501 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 0.0 | 0.1 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.0 | 0.1 | GO:1903677 | regulation of cap-independent translational initiation(GO:1903677) positive regulation of cap-independent translational initiation(GO:1903679) regulation of cytoplasmic translational initiation(GO:1904688) positive regulation of cytoplasmic translational initiation(GO:1904690) |
| 0.0 | 0.1 | GO:1902488 | cholangiocyte apoptotic process(GO:1902488) regulation of cholangiocyte apoptotic process(GO:1904192) negative regulation of cholangiocyte apoptotic process(GO:1904193) |
| 0.0 | 0.1 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.0 | 0.1 | GO:2000467 | positive regulation of glycogen (starch) synthase activity(GO:2000467) |
| 0.0 | 0.1 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.0 | 0.3 | GO:0048742 | regulation of skeletal muscle fiber development(GO:0048742) |
| 0.0 | 0.1 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 0.2 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.0 | 0.1 | GO:0032513 | negative regulation of protein phosphatase type 2B activity(GO:0032513) |
| 0.0 | 0.1 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.0 | 0.1 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.0 | 0.4 | GO:0051923 | sulfation(GO:0051923) |
| 0.0 | 0.1 | GO:0036091 | positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 0.0 | 0.0 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.0 | 0.3 | GO:0036035 | osteoclast development(GO:0036035) |
| 0.0 | 0.1 | GO:0055093 | response to increased oxygen levels(GO:0036296) response to hyperoxia(GO:0055093) |
| 0.0 | 0.1 | GO:0006777 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) molybdopterin cofactor biosynthetic process(GO:0032324) molybdopterin cofactor metabolic process(GO:0043545) prosthetic group metabolic process(GO:0051189) |
| 0.0 | 0.1 | GO:2001286 | regulation of caveolin-mediated endocytosis(GO:2001286) |
| 0.0 | 0.0 | GO:0003162 | atrioventricular node development(GO:0003162) |
| 0.0 | 0.1 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.0 | 0.1 | GO:0070885 | negative regulation of calcineurin-NFAT signaling cascade(GO:0070885) |
| 0.0 | 0.1 | GO:0072310 | glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.0 | 0.0 | GO:2000809 | positive regulation of synaptic vesicle clustering(GO:2000809) |
| 0.0 | 0.1 | GO:0045200 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.0 | 0.1 | GO:0009642 | response to light intensity(GO:0009642) |
| 0.0 | 0.2 | GO:0042637 | catagen(GO:0042637) |
| 0.0 | 0.1 | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.0 | 0.1 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.0 | 0.0 | GO:1902809 | regulation of skeletal muscle fiber differentiation(GO:1902809) |
| 0.0 | 0.1 | GO:2000483 | negative regulation of interleukin-8 secretion(GO:2000483) |
| 0.0 | 0.1 | GO:0090219 | negative regulation of lipid kinase activity(GO:0090219) |
| 0.0 | 0.1 | GO:1990928 | response to amino acid starvation(GO:1990928) |
| 0.0 | 0.3 | GO:0015858 | nucleoside transport(GO:0015858) |
| 0.0 | 0.0 | GO:0060137 | maternal process involved in parturition(GO:0060137) |
| 0.0 | 0.1 | GO:0072592 | oxygen metabolic process(GO:0072592) |
| 0.0 | 0.1 | GO:0006678 | glucosylceramide metabolic process(GO:0006678) |
| 0.0 | 0.0 | GO:0038027 | apolipoprotein A-I-mediated signaling pathway(GO:0038027) |
| 0.0 | 0.1 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.0 | 0.1 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.0 | 0.1 | GO:0070947 | neutrophil mediated killing of fungus(GO:0070947) |
| 0.0 | 0.0 | GO:0044004 | killing by symbiont of host cells(GO:0001907) disruption by symbiont of host cell(GO:0044004) |
| 0.0 | 0.0 | GO:2000836 | androgen secretion(GO:0035935) regulation of androgen secretion(GO:2000834) positive regulation of androgen secretion(GO:2000836) |
| 0.0 | 0.1 | GO:0009972 | cytidine catabolic process(GO:0006216) cytidine deamination(GO:0009972) cytidine metabolic process(GO:0046087) |
| 0.0 | 0.0 | GO:0010694 | positive regulation of alkaline phosphatase activity(GO:0010694) |
| 0.0 | 0.1 | GO:0042795 | snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.0 | 0.1 | GO:1903999 | negative regulation of eating behavior(GO:1903999) |
| 0.0 | 0.1 | GO:0002439 | chronic inflammatory response to antigenic stimulus(GO:0002439) |
| 0.0 | 0.0 | GO:0033159 | negative regulation of protein import into nucleus, translocation(GO:0033159) |
| 0.0 | 0.1 | GO:0010387 | COP9 signalosome assembly(GO:0010387) |
| 0.0 | 0.1 | GO:0090241 | negative regulation of histone H4 acetylation(GO:0090241) |
| 0.0 | 0.3 | GO:0090312 | positive regulation of protein deacetylation(GO:0090312) |
| 0.0 | 0.1 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.1 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 | 0.2 | GO:0007183 | SMAD protein complex assembly(GO:0007183) |
| 0.0 | 0.1 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.0 | 0.2 | GO:0060346 | bone trabecula formation(GO:0060346) |
| 0.0 | 0.1 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.0 | 0.1 | GO:0046881 | positive regulation of follicle-stimulating hormone secretion(GO:0046881) |
| 0.0 | 0.1 | GO:0086013 | membrane repolarization during cardiac muscle cell action potential(GO:0086013) |
| 0.0 | 0.1 | GO:0032055 | negative regulation of translation in response to stress(GO:0032055) |
| 0.0 | 0.1 | GO:1901525 | regulation of macromitophagy(GO:1901524) negative regulation of macromitophagy(GO:1901525) |
| 0.0 | 0.5 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.0 | 0.1 | GO:0039536 | negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.0 | 0.1 | GO:0002155 | thyroid hormone mediated signaling pathway(GO:0002154) regulation of thyroid hormone mediated signaling pathway(GO:0002155) |
| 0.0 | 0.0 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.0 | 0.1 | GO:2000659 | regulation of interleukin-1-mediated signaling pathway(GO:2000659) |
| 0.0 | 0.3 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.0 | 0.1 | GO:0006207 | 'de novo' pyrimidine nucleobase biosynthetic process(GO:0006207) pyrimidine nucleobase biosynthetic process(GO:0019856) |
| 0.0 | 0.0 | GO:0002904 | positive regulation of B cell apoptotic process(GO:0002904) |
| 0.0 | 0.0 | GO:0086023 | adrenergic receptor signaling pathway involved in heart process(GO:0086023) |
| 0.0 | 0.1 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) |
| 0.0 | 0.0 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.0 | 0.0 | GO:0090259 | regulation of retinal ganglion cell axon guidance(GO:0090259) |
| 0.0 | 0.1 | GO:0042448 | progesterone metabolic process(GO:0042448) |
| 0.0 | 0.0 | GO:0035860 | glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.0 | 0.1 | GO:0035457 | cellular response to interferon-alpha(GO:0035457) |
| 0.0 | 0.2 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.0 | 0.1 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 0.0 | GO:0035609 | C-terminal protein deglutamylation(GO:0035609) |
| 0.0 | 0.1 | GO:0007097 | nuclear migration(GO:0007097) |
| 0.0 | 0.1 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 0.1 | GO:0008354 | germ cell migration(GO:0008354) |
| 0.0 | 0.1 | GO:0071044 | histone mRNA catabolic process(GO:0071044) |
| 0.0 | 0.1 | GO:0010571 | positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.0 | 0.0 | GO:0042726 | flavin-containing compound metabolic process(GO:0042726) |
| 0.0 | 0.2 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 | 0.3 | GO:0001958 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.0 | 0.1 | GO:0050904 | diapedesis(GO:0050904) |
| 0.0 | 0.0 | GO:0048696 | regulation of collateral sprouting in absence of injury(GO:0048696) |
| 0.0 | 0.1 | GO:0002538 | arachidonic acid metabolite production involved in inflammatory response(GO:0002538) |
| 0.0 | 0.1 | GO:0045714 | regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045714) |
| 0.0 | 0.1 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.0 | 0.1 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.0 | 0.0 | GO:1904861 | excitatory synapse assembly(GO:1904861) |
| 0.0 | 0.1 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.0 | 0.2 | GO:0071625 | vocalization behavior(GO:0071625) |
| 0.0 | 0.2 | GO:0006359 | regulation of transcription from RNA polymerase III promoter(GO:0006359) |
| 0.0 | 0.1 | GO:0060396 | growth hormone receptor signaling pathway(GO:0060396) cellular response to growth hormone stimulus(GO:0071378) |
| 0.0 | 0.0 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.0 | 0.0 | GO:1902897 | regulation of postsynaptic density protein 95 clustering(GO:1902897) |
| 0.0 | 0.1 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.0 | 0.0 | GO:0090310 | negative regulation of methylation-dependent chromatin silencing(GO:0090310) |
| 0.0 | 0.1 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.0 | 0.1 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.0 | 0.1 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.0 | 0.1 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.0 | 0.0 | GO:1904479 | negative regulation of intestinal absorption(GO:1904479) |
| 0.0 | 0.1 | GO:1902744 | negative regulation of lamellipodium organization(GO:1902744) |
| 0.0 | 0.1 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.0 | 0.1 | GO:0045908 | negative regulation of vasodilation(GO:0045908) |
| 0.0 | 0.2 | GO:0010758 | regulation of macrophage chemotaxis(GO:0010758) |
| 0.0 | 0.2 | GO:0060074 | synapse maturation(GO:0060074) |
| 0.0 | 0.2 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.0 | 0.2 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.0 | 0.0 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.1 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.1 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.0 | 0.0 | GO:0060744 | thelarche(GO:0042695) mammary gland branching involved in thelarche(GO:0060744) |
| 0.0 | 0.1 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.0 | 0.1 | GO:0042760 | very long-chain fatty acid catabolic process(GO:0042760) |
| 0.0 | 0.0 | GO:0008050 | female courtship behavior(GO:0008050) |
| 0.0 | 0.1 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 0.0 | 0.0 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.0 | 0.2 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.0 | 0.0 | GO:0070375 | ERK5 cascade(GO:0070375) |
| 0.0 | 0.1 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.0 | 0.0 | GO:0006982 | response to lipid hydroperoxide(GO:0006982) |
| 0.0 | 0.0 | GO:0031947 | negative regulation of glucocorticoid metabolic process(GO:0031944) negative regulation of glucocorticoid biosynthetic process(GO:0031947) |
| 0.0 | 0.0 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.0 | 0.1 | GO:0006878 | cellular copper ion homeostasis(GO:0006878) |
| 0.0 | 0.0 | GO:0003223 | ventricular compact myocardium morphogenesis(GO:0003223) |
| 0.0 | 0.1 | GO:0002072 | optic cup morphogenesis involved in camera-type eye development(GO:0002072) |
| 0.0 | 0.0 | GO:2000304 | positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 0.0 | 0.0 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.0 | 0.0 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.0 | 0.0 | GO:0036135 | Schwann cell migration(GO:0036135) regulation of Schwann cell migration(GO:1900147) |
| 0.0 | 0.0 | GO:0097527 | necroptotic signaling pathway(GO:0097527) |
| 0.0 | 0.0 | GO:0060066 | oviduct development(GO:0060066) |
| 0.0 | 0.1 | GO:0035507 | regulation of myosin-light-chain-phosphatase activity(GO:0035507) |
| 0.0 | 0.1 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.0 | 0.1 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.0 | 0.0 | GO:0002913 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.0 | 0.0 | GO:0055099 | response to high density lipoprotein particle(GO:0055099) |
| 0.0 | 0.0 | GO:0006971 | hypotonic response(GO:0006971) |
| 0.0 | 0.0 | GO:0016259 | selenocysteine metabolic process(GO:0016259) |
| 0.0 | 0.1 | GO:0009162 | deoxyribonucleoside monophosphate metabolic process(GO:0009162) |
| 0.0 | 0.0 | GO:0042732 | D-xylose metabolic process(GO:0042732) |
| 0.0 | 0.0 | GO:1990791 | dorsal root ganglion development(GO:1990791) |
| 0.0 | 0.1 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.0 | 0.1 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.0 | 0.2 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.1 | GO:0033004 | negative regulation of mast cell activation(GO:0033004) |
| 0.0 | 0.1 | GO:0045943 | positive regulation of transcription from RNA polymerase I promoter(GO:0045943) |
| 0.0 | 0.1 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
| 0.0 | 0.0 | GO:2000259 | positive regulation of complement activation(GO:0045917) positive regulation of protein activation cascade(GO:2000259) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.1 | 0.2 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.1 | 0.4 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.1 | 0.3 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.1 | 0.2 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.1 | 0.2 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.1 | 0.3 | GO:0031415 | NatA complex(GO:0031415) |
| 0.0 | 0.2 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.0 | 0.3 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 0.1 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 0.2 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.0 | 0.1 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.0 | 0.1 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.0 | 0.2 | GO:0005883 | neurofilament(GO:0005883) |
| 0.0 | 0.1 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 0.1 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.0 | 0.1 | GO:0036396 | MIS complex(GO:0036396) |
| 0.0 | 0.1 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.0 | 0.1 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 0.2 | GO:0065010 | extracellular membrane-bounded organelle(GO:0065010) |
| 0.0 | 0.1 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.0 | 0.1 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.1 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.1 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.0 | 0.2 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.0 | 0.1 | GO:0097433 | dense body(GO:0097433) |
| 0.0 | 0.0 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 0.1 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.1 | GO:0000835 | ER ubiquitin ligase complex(GO:0000835) Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.0 | 0.1 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.0 | 0.1 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 0.1 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.0 | 0.1 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 0.3 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.0 | 0.1 | GO:0090576 | RNA polymerase III transcription factor complex(GO:0090576) |
| 0.0 | 0.1 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 0.1 | GO:0070688 | MLL5-L complex(GO:0070688) |
| 0.0 | 0.1 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.1 | GO:0042825 | TAP complex(GO:0042825) |
| 0.0 | 0.0 | GO:0034666 | integrin alpha2-beta1 complex(GO:0034666) |
| 0.0 | 0.1 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 0.0 | GO:0033647 | host intracellular organelle(GO:0033647) host intracellular membrane-bounded organelle(GO:0033648) |
| 0.0 | 0.1 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.0 | 0.1 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 0.1 | GO:0002142 | stereocilia ankle link complex(GO:0002142) |
| 0.0 | 0.1 | GO:0001527 | microfibril(GO:0001527) |
| 0.0 | 0.1 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 0.1 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.2 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.3 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.0 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.0 | 0.1 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.0 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.0 | 0.1 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.0 | 0.2 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.1 | GO:0005869 | dynactin complex(GO:0005869) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.2 | 0.5 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.1 | 0.4 | GO:0031686 | A1 adenosine receptor binding(GO:0031686) |
| 0.1 | 0.7 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.1 | 0.2 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.1 | 0.3 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.1 | 0.3 | GO:0035276 | ethanol binding(GO:0035276) |
| 0.1 | 0.3 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.1 | 0.3 | GO:0032896 | palmitoyl-CoA 9-desaturase activity(GO:0032896) |
| 0.1 | 0.3 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.1 | 0.2 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.1 | 0.4 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.1 | 0.2 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.1 | 0.2 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.1 | 0.3 | GO:1990239 | steroid hormone binding(GO:1990239) |
| 0.1 | 0.2 | GO:0030984 | kininogen binding(GO:0030984) |
| 0.0 | 0.2 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.6 | GO:0016595 | glutamate binding(GO:0016595) |
| 0.0 | 0.2 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.0 | 0.3 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.0 | 0.2 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 0.2 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.0 | 0.2 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.0 | 0.1 | GO:0035175 | histone kinase activity (H3-S10 specific)(GO:0035175) |
| 0.0 | 0.1 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.0 | 0.2 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.3 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.2 | GO:0015198 | oligopeptide transporter activity(GO:0015198) |
| 0.0 | 0.1 | GO:0035515 | oxidative RNA demethylase activity(GO:0035515) |
| 0.0 | 0.2 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 0.1 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.1 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.0 | 0.4 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 0.2 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.1 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.0 | 0.3 | GO:0004312 | fatty acid synthase activity(GO:0004312) |
| 0.0 | 0.1 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.0 | 0.1 | GO:0016899 | oxidoreductase activity, acting on the CH-OH group of donors, oxygen as acceptor(GO:0016899) |
| 0.0 | 0.1 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.0 | 0.0 | GO:0032767 | copper-dependent protein binding(GO:0032767) |
| 0.0 | 0.1 | GO:0043125 | ErbB-3 class receptor binding(GO:0043125) |
| 0.0 | 0.1 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.0 | 0.2 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.8 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 0.0 | 0.0 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) |
| 0.0 | 0.0 | GO:0005350 | purine nucleobase transmembrane transporter activity(GO:0005345) pyrimidine nucleobase transmembrane transporter activity(GO:0005350) nucleobase transmembrane transporter activity(GO:0015205) |
| 0.0 | 0.1 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.0 | 0.1 | GO:0031779 | melanocortin receptor binding(GO:0031779) type 3 melanocortin receptor binding(GO:0031781) type 4 melanocortin receptor binding(GO:0031782) |
| 0.0 | 0.2 | GO:0004955 | prostaglandin receptor activity(GO:0004955) |
| 0.0 | 0.1 | GO:0019153 | protein-disulfide reductase (glutathione) activity(GO:0019153) |
| 0.0 | 0.3 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.0 | 0.0 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.1 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.0 | 0.2 | GO:0016840 | carbon-nitrogen lyase activity(GO:0016840) |
| 0.0 | 0.2 | GO:0015385 | sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.1 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.0 | 0.1 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.0 | 0.1 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.0 | 0.1 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.1 | GO:0016215 | acyl-CoA desaturase activity(GO:0016215) |
| 0.0 | 0.1 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.0 | 0.1 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.0 | 0.1 | GO:0086008 | voltage-gated potassium channel activity involved in cardiac muscle cell action potential repolarization(GO:0086008) |
| 0.0 | 0.1 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.1 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.0 | 0.1 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.1 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.0 | GO:0047045 | testosterone 17-beta-dehydrogenase (NADP+) activity(GO:0047045) |
| 0.0 | 0.1 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.0 | 0.2 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.0 | 0.1 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 0.1 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.1 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 0.1 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.0 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.0 | 0.1 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.0 | 0.1 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.1 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 0.1 | GO:0019763 | immunoglobulin receptor activity(GO:0019763) |
| 0.0 | 0.2 | GO:0008409 | 5'-3' exonuclease activity(GO:0008409) |
| 0.0 | 0.1 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 0.0 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 0.1 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.0 | 0.1 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.0 | 0.1 | GO:0031544 | peptidyl-proline 3-dioxygenase activity(GO:0031544) |
| 0.0 | 0.0 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.0 | 0.1 | GO:0048185 | activin binding(GO:0048185) |
| 0.0 | 0.1 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.0 | 0.2 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.1 | GO:0043184 | vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 0.0 | 0.1 | GO:0043762 | 3-oxo-2-(2'-pentenyl)cyclopentane-1-octanoic acid CoA ligase activity(GO:0010435) 3-isopropenyl-6-oxoheptanoyl-CoA synthetase activity(GO:0018854) 2-oxo-delta3-4,5,5-trimethylcyclopentenylacetyl-CoA synthetase activity(GO:0018855) benzoyl acetate-CoA ligase activity(GO:0018856) 2,4-dichlorobenzoate-CoA ligase activity(GO:0018857) pivalate-CoA ligase activity(GO:0034783) cyclopropanecarboxylate-CoA ligase activity(GO:0034793) adipate-CoA ligase activity(GO:0034796) citronellyl-CoA ligase activity(GO:0034823) mentha-1,3-dione-CoA ligase activity(GO:0034841) thiophene-2-carboxylate-CoA ligase activity(GO:0034842) 2,4,4-trimethylpentanoate-CoA ligase activity(GO:0034865) cis-2-methyl-5-isopropylhexa-2,5-dienoate-CoA ligase activity(GO:0034942) trans-2-methyl-5-isopropylhexa-2,5-dienoate-CoA ligase activity(GO:0034943) branched-chain acyl-CoA synthetase (ADP-forming) activity(GO:0043759) aryl-CoA synthetase (ADP-forming) activity(GO:0043762) 3-hydroxypropionyl-CoA synthetase activity(GO:0043955) perillic acid:CoA ligase (ADP-forming) activity(GO:0052685) perillic acid:CoA ligase (AMP-forming) activity(GO:0052686) (3R)-3-isopropenyl-6-oxoheptanoate:CoA ligase (ADP-forming) activity(GO:0052687) (3R)-3-isopropenyl-6-oxoheptanoate:CoA ligase (AMP-forming) activity(GO:0052688) pristanate-CoA ligase activity(GO:0070251) malonyl-CoA synthetase activity(GO:0090409) |
| 0.0 | 0.0 | GO:1990188 | euchromatin binding(GO:1990188) |
| 0.0 | 0.0 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.0 | 0.0 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.0 | 0.0 | GO:0035650 | AP-1 adaptor complex binding(GO:0035650) |
| 0.0 | 0.0 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.0 | 0.1 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.0 | 0.3 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.0 | 0.0 | GO:0000171 | ribonuclease MRP activity(GO:0000171) |
| 0.0 | 0.3 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.1 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 0.0 | GO:0051033 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.0 | 0.1 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.0 | 0.1 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.0 | 0.0 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.0 | GO:0051022 | Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.0 | 0.0 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.0 | 0.1 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 0.1 | GO:0047760 | butyrate-CoA ligase activity(GO:0047760) |
| 0.0 | 0.1 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.1 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.1 | GO:0002054 | nucleobase binding(GO:0002054) |
| 0.0 | 0.1 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.0 | 0.0 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.0 | 0.0 | GO:0004090 | carbonyl reductase (NADPH) activity(GO:0004090) |
| 0.0 | 0.0 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) |
| 0.0 | 0.1 | GO:0018636 | phenanthrene 9,10-monooxygenase activity(GO:0018636) |
| 0.0 | 0.0 | GO:0070698 | type I activin receptor binding(GO:0070698) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.1 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 0.3 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.1 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.7 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.3 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.0 | 0.1 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.5 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.0 | 0.2 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 0.4 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 0.1 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.0 | 0.2 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.0 | 0.2 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.2 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.1 | 0.8 | REACTOME FATTY ACYL COA BIOSYNTHESIS | Genes involved in Fatty Acyl-CoA Biosynthesis |
| 0.0 | 0.1 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.0 | 0.6 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.2 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 0.7 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 0.0 | REACTOME IL 2 SIGNALING | Genes involved in Interleukin-2 signaling |
| 0.0 | 0.3 | REACTOME SIGNALING BY NOTCH2 | Genes involved in Signaling by NOTCH2 |
| 0.0 | 0.6 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.3 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 0.1 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.3 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.2 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.4 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.2 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.2 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.1 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.0 | 0.2 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.4 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.4 | REACTOME TGF BETA RECEPTOR SIGNALING ACTIVATES SMADS | Genes involved in TGF-beta receptor signaling activates SMADs |
| 0.0 | 0.1 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.0 | 0.1 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.1 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 0.1 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.2 | REACTOME SHC RELATED EVENTS | Genes involved in SHC-related events |
| 0.0 | 0.4 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.1 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 0.1 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.2 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.1 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.0 | 0.4 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.0 | 0.1 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 0.3 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.2 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.1 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.1 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 0.2 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.2 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.2 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |
| 0.0 | 0.3 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |