Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
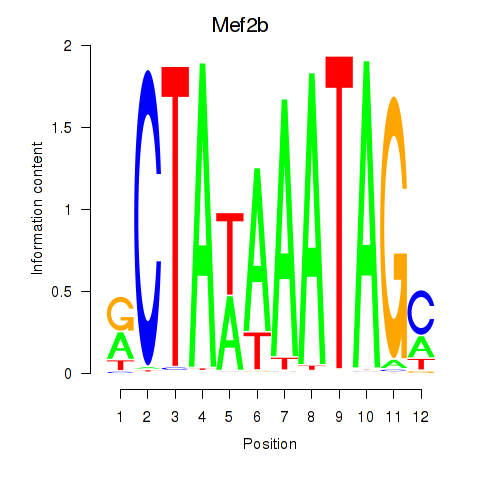
Results for Mef2b
Z-value: 2.20
Transcription factors associated with Mef2b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Mef2b
|
ENSMUSG00000079033.3 | myocyte enhancer factor 2B |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr8_70152603_70152770 | Mef2b | 92 | 0.927902 | -0.63 | 1.8e-01 | Click! |
| chr8_70158443_70158608 | Mef2b | 5744 | 0.092534 | -0.41 | 4.2e-01 | Click! |
Activity of the Mef2b motif across conditions
Conditions sorted by the z-value of the Mef2b motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
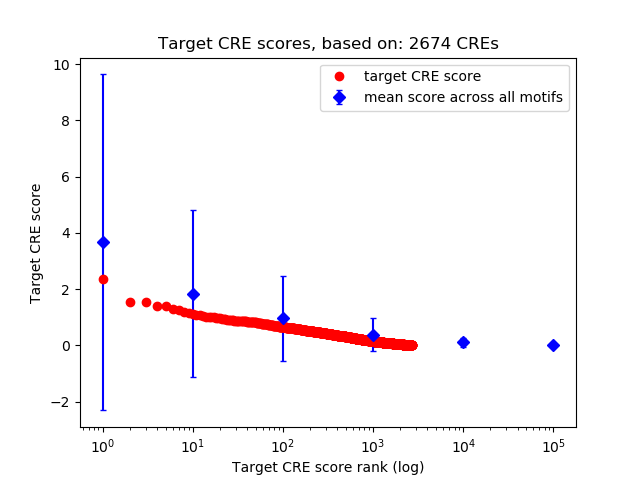
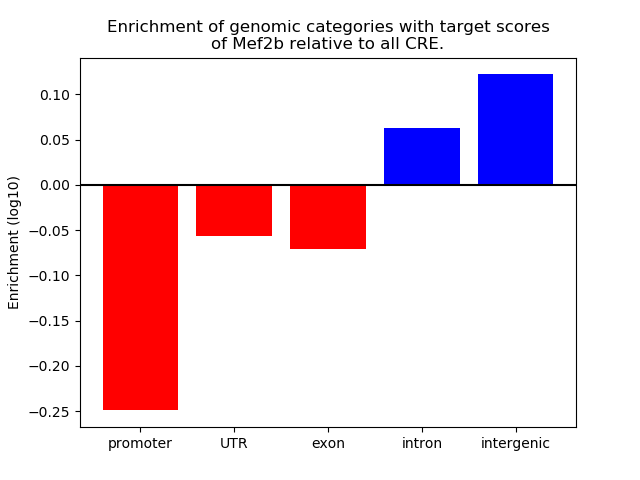
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr9_14045065_14045224 | 2.36 |
1700019J19Rik |
RIKEN cDNA 1700019J19 gene |
44476 |
0.14 |
| chr12_109521275_109521426 | 1.56 |
Gm34081 |
predicted gene, 34081 |
4610 |
0.06 |
| chr7_140776294_140776445 | 1.54 |
Gm29799 |
predicted gene, 29799 |
8146 |
0.09 |
| chrX_38422575_38423072 | 1.41 |
Lamp2 |
lysosomal-associated membrane protein 2 |
20327 |
0.15 |
| chr4_131939568_131939730 | 1.39 |
Epb41 |
erythrocyte membrane protein band 4.1 |
2262 |
0.18 |
| chr16_41161594_41161767 | 1.31 |
Gm26381 |
predicted gene, 26381 |
56065 |
0.15 |
| chr12_104346091_104346278 | 1.27 |
Serpina3k |
serine (or cysteine) peptidase inhibitor, clade A, member 3K |
7698 |
0.12 |
| chr3_148981923_148982092 | 1.20 |
Gm43573 |
predicted gene 43573 |
7145 |
0.19 |
| chr17_29323966_29324117 | 1.15 |
Gm46603 |
predicted gene, 46603 |
1492 |
0.24 |
| chr17_8283278_8283446 | 1.10 |
Mpc1 |
mitochondrial pyruvate carrier 1 |
400 |
0.78 |
| chr5_134275072_134275253 | 1.09 |
Gtf2i |
general transcription factor II I |
119 |
0.94 |
| chr5_86078864_86079015 | 1.08 |
Gm24524 |
predicted gene, 24524 |
827 |
0.55 |
| chr13_69029464_69029651 | 1.06 |
Gm37122 |
predicted gene, 37122 |
12935 |
0.17 |
| chr3_18131799_18132081 | 1.02 |
Gm23686 |
predicted gene, 23686 |
45685 |
0.14 |
| chr9_74889692_74889843 | 1.02 |
Onecut1 |
one cut domain, family member 1 |
23283 |
0.14 |
| chr15_97720259_97720503 | 1.01 |
Endou |
endonuclease, polyU-specific |
769 |
0.57 |
| chr5_75490980_75491142 | 0.99 |
Gm24502 |
predicted gene, 24502 |
70089 |
0.09 |
| chr19_40241408_40241559 | 0.97 |
Pdlim1 |
PDZ and LIM domain 1 (elfin) |
10678 |
0.15 |
| chr6_145745302_145745496 | 0.96 |
Gm15704 |
predicted gene 15704 |
874 |
0.51 |
| chr7_123405689_123405849 | 0.95 |
Lcmt1 |
leucine carboxyl methyltransferase 1 |
2411 |
0.28 |
| chr12_51848413_51848587 | 0.94 |
Gm19309 |
predicted gene, 19309 |
7879 |
0.19 |
| chr9_118646031_118646185 | 0.92 |
Itga9 |
integrin alpha 9 |
39418 |
0.14 |
| chr7_71872986_71873347 | 0.92 |
4930441H08Rik |
RIKEN cDNA 4930441H08 gene |
36527 |
0.18 |
| chr16_95439908_95440073 | 0.90 |
Erg |
ETS transcription factor |
19255 |
0.24 |
| chr16_8843944_8844115 | 0.90 |
1810013L24Rik |
RIKEN cDNA 1810013L24 gene |
12708 |
0.17 |
| chr16_52071915_52072078 | 0.90 |
Cblb |
Casitas B-lineage lymphoma b |
22343 |
0.21 |
| chr18_67738121_67738272 | 0.89 |
Ptpn2 |
protein tyrosine phosphatase, non-receptor type 2 |
13601 |
0.16 |
| chr14_105803498_105803761 | 0.89 |
Gm22406 |
predicted gene, 22406 |
13279 |
0.23 |
| chrX_102187134_102187366 | 0.88 |
Rps4x |
ribosomal protein S4, X-linked |
513 |
0.68 |
| chr15_88839321_88839531 | 0.88 |
Gm23144 |
predicted gene, 23144 |
4126 |
0.16 |
| chr7_116200646_116200810 | 0.88 |
Plekha7 |
pleckstrin homology domain containing, family A member 7 |
2167 |
0.34 |
| chr10_80260218_80260407 | 0.88 |
Gamt |
guanidinoacetate methyltransferase |
606 |
0.42 |
| chr11_110551681_110551869 | 0.88 |
Gm11682 |
predicted gene 11682 |
7268 |
0.3 |
| chr6_122826334_122826673 | 0.87 |
Foxj2 |
forkhead box J2 |
172 |
0.9 |
| chr7_145111866_145112053 | 0.86 |
Gm45181 |
predicted gene 45181 |
51037 |
0.14 |
| chr5_66067292_66067466 | 0.86 |
Gm43775 |
predicted gene 43775 |
5516 |
0.14 |
| chr7_145025998_145026178 | 0.86 |
Ccnd1 |
cyclin D1 |
86163 |
0.07 |
| chr3_116801818_116802023 | 0.85 |
Agl |
amylo-1,6-glucosidase, 4-alpha-glucanotransferase |
5497 |
0.14 |
| chr10_34513055_34513206 | 0.85 |
Frk |
fyn-related kinase |
29598 |
0.2 |
| chr14_100750255_100750445 | 0.85 |
Gm23538 |
predicted gene, 23538 |
9918 |
0.23 |
| chr13_101718024_101718189 | 0.84 |
Pik3r1 |
phosphoinositide-3-kinase regulatory subunit 1 |
16002 |
0.21 |
| chr8_126807290_126807464 | 0.83 |
A630001O12Rik |
RIKEN cDNA A630001O12 gene |
31856 |
0.17 |
| chr9_33001470_33001748 | 0.83 |
Gm27166 |
predicted gene 27166 |
30182 |
0.17 |
| chr10_25365835_25365986 | 0.83 |
Epb41l2 |
erythrocyte membrane protein band 4.1 like 2 |
6054 |
0.21 |
| chr8_80811587_80811961 | 0.83 |
Gab1 |
growth factor receptor bound protein 2-associated protein 1 |
19110 |
0.18 |
| chr10_57727739_57727910 | 0.83 |
Pkib |
protein kinase inhibitor beta, cAMP dependent, testis specific |
157 |
0.96 |
| chr5_98992026_98992185 | 0.83 |
Prkg2 |
protein kinase, cGMP-dependent, type II |
25349 |
0.2 |
| chr9_74327099_74327334 | 0.81 |
Gm24141 |
predicted gene, 24141 |
35394 |
0.17 |
| chr9_94615362_94615542 | 0.81 |
Gm39404 |
predicted gene, 39404 |
23256 |
0.15 |
| chr2_163685840_163686012 | 0.81 |
Gm16316 |
predicted gene 16316 |
6353 |
0.15 |
| chr6_31544399_31544550 | 0.81 |
Podxl |
podocalyxin-like |
19507 |
0.17 |
| chr4_63229865_63230144 | 0.81 |
Col27a1 |
collagen, type XXVII, alpha 1 |
4282 |
0.2 |
| chr15_97462722_97462873 | 0.81 |
Gm49506 |
predicted gene, 49506 |
95493 |
0.07 |
| chr15_25940308_25940999 | 0.81 |
Retreg1 |
reticulophagy regulator 1 |
49 |
0.98 |
| chr6_116004145_116004296 | 0.78 |
Plxnd1 |
plexin D1 |
9215 |
0.16 |
| chr13_93989307_93989458 | 0.78 |
Gm47216 |
predicted gene, 47216 |
2386 |
0.28 |
| chr18_46273693_46273856 | 0.78 |
Pggt1b |
protein geranylgeranyltransferase type I, beta subunit |
2155 |
0.27 |
| chr13_85127287_85127439 | 0.77 |
Gm4076 |
predicted gene 4076 |
151 |
0.96 |
| chrX_168832355_168832554 | 0.77 |
Arhgap6 |
Rho GTPase activating protein 6 |
37355 |
0.19 |
| chr11_79718866_79719052 | 0.77 |
Mir193a |
microRNA 193a |
6990 |
0.13 |
| chr1_170623064_170623228 | 0.77 |
Gm7299 |
predicted gene 7299 |
7775 |
0.19 |
| chr2_51716857_51717035 | 0.77 |
Gm13490 |
predicted gene 13490 |
15063 |
0.23 |
| chr8_47461739_47461890 | 0.76 |
Stox2 |
storkhead box 2 |
15452 |
0.16 |
| chr7_80451948_80452113 | 0.75 |
Blm |
Bloom syndrome, RecQ like helicase |
5342 |
0.14 |
| chr16_48765285_48765461 | 0.75 |
Trat1 |
T cell receptor associated transmembrane adaptor 1 |
6583 |
0.2 |
| chr3_72659248_72659399 | 0.75 |
Gm37901 |
predicted gene, 37901 |
54975 |
0.16 |
| chr16_95995234_95995402 | 0.74 |
Psmg1 |
proteasome (prosome, macropain) assembly chaperone 1 |
4358 |
0.17 |
| chr11_63827631_63827816 | 0.74 |
Gm12288 |
predicted gene 12288 |
9423 |
0.22 |
| chr14_117750473_117750636 | 0.73 |
Mir6239 |
microRNA 6239 |
203293 |
0.03 |
| chr14_33687746_33687969 | 0.73 |
Gm26228 |
predicted gene, 26228 |
43362 |
0.14 |
| chr19_27178438_27178739 | 0.73 |
Gm35438 |
predicted gene, 35438 |
13421 |
0.22 |
| chr4_149868069_149868231 | 0.73 |
Gm13070 |
predicted gene 13070 |
35856 |
0.09 |
| chr7_19952685_19952933 | 0.73 |
Rpl7a-ps8 |
ribosomal protein L7A, pseudogene 8 |
974 |
0.29 |
| chr3_98067419_98067570 | 0.72 |
Gm42819 |
predicted gene 42819 |
36807 |
0.13 |
| chr5_121612131_121612311 | 0.71 |
Acad12 |
acyl-Coenzyme A dehydrogenase family, member 12 |
6592 |
0.11 |
| chr2_121193827_121193979 | 0.71 |
Tubgcp4 |
tubulin, gamma complex associated protein 4 |
1956 |
0.22 |
| chr3_116799201_116799358 | 0.70 |
Agl |
amylo-1,6-glucosidase, 4-alpha-glucanotransferase |
8138 |
0.13 |
| chr10_84968958_84969109 | 0.70 |
Ric8b |
RIC8 guanine nucleotide exchange factor B |
31640 |
0.19 |
| chr9_42512108_42512263 | 0.70 |
Tbcel |
tubulin folding cofactor E-like |
4376 |
0.25 |
| chr5_39219850_39220015 | 0.70 |
Gm40293 |
predicted gene, 40293 |
87152 |
0.08 |
| chr12_24615310_24615477 | 0.70 |
Gm6969 |
predicted pseudogene 6969 |
23617 |
0.13 |
| chr4_9657208_9657376 | 0.69 |
Asph |
aspartate-beta-hydroxylase |
11794 |
0.19 |
| chr8_71312491_71313043 | 0.69 |
Myo9b |
myosin IXb |
21050 |
0.11 |
| chr4_37093076_37093227 | 0.69 |
Taf9-ps |
TATA-box binding protein associated factor 9, pseudogene |
46723 |
0.17 |
| chr2_158702228_158702379 | 0.69 |
Ppp1r16b |
protein phosphatase 1, regulatory subunit 16B |
6168 |
0.17 |
| chr12_79598601_79598900 | 0.68 |
Rad51b |
RAD51 paralog B |
271397 |
0.01 |
| chr11_117574902_117575083 | 0.68 |
2900041M22Rik |
RIKEN cDNA 2900041M22 gene |
36255 |
0.13 |
| chr2_134872444_134872595 | 0.68 |
Gm14036 |
predicted gene 14036 |
68570 |
0.12 |
| chr9_32906282_32906466 | 0.68 |
Gm27162 |
predicted gene 27162 |
22592 |
0.19 |
| chr5_75961858_75962064 | 0.67 |
Kdr |
kinase insert domain protein receptor |
16497 |
0.17 |
| chr18_39855902_39856245 | 0.67 |
Gm41708 |
predicted gene, 41708 |
73301 |
0.1 |
| chr7_120151008_120151184 | 0.66 |
4930505K13Rik |
RIKEN cDNA 4930505K13 gene |
5019 |
0.14 |
| chr17_86302890_86303043 | 0.66 |
2010106C02Rik |
RIKEN cDNA 2010106C02 gene |
15788 |
0.26 |
| chr4_55042640_55042808 | 0.65 |
Zfp462 |
zinc finger protein 462 |
31244 |
0.19 |
| chr4_105261100_105261481 | 0.65 |
Plpp3 |
phospholipid phosphatase 3 |
103943 |
0.07 |
| chr4_141324590_141325156 | 0.65 |
Epha2 |
Eph receptor A2 |
7960 |
0.1 |
| chr4_95212952_95213103 | 0.65 |
Gm12708 |
predicted gene 12708 |
19665 |
0.2 |
| chr11_79453041_79453205 | 0.65 |
Gm11199 |
predicted gene 11199 |
676 |
0.61 |
| chr9_32874453_32874604 | 0.65 |
Gm37167 |
predicted gene, 37167 |
34606 |
0.16 |
| chr19_29470771_29470965 | 0.64 |
Pdcd1lg2 |
programmed cell death 1 ligand 2 |
48809 |
0.1 |
| chr3_18343267_18343427 | 0.64 |
Gm43834 |
predicted gene 43834 |
44037 |
0.16 |
| chr5_66745601_66745788 | 0.64 |
Limch1 |
LIM and calponin homology domains 1 |
133 |
0.96 |
| chr19_40187264_40187454 | 0.64 |
Cyp2c70 |
cytochrome P450, family 2, subfamily c, polypeptide 70 |
26 |
0.97 |
| chr19_24251878_24252123 | 0.64 |
Fxn |
frataxin |
26171 |
0.14 |
| chr11_68956181_68956371 | 0.64 |
Arhgef15 |
Rho guanine nucleotide exchange factor (GEF) 15 |
1202 |
0.27 |
| chr16_49533485_49533653 | 0.64 |
Gm6931 |
predicted gene 6931 |
108763 |
0.06 |
| chr2_20445457_20445703 | 0.64 |
Gm13335 |
predicted gene 13335 |
12646 |
0.25 |
| chr2_124741001_124741334 | 0.63 |
Gm13994 |
predicted gene 13994 |
32512 |
0.23 |
| chr2_38917032_38917193 | 0.63 |
Nr6a1 |
nuclear receptor subfamily 6, group A, member 1 |
9105 |
0.11 |
| chr13_52067119_52067403 | 0.63 |
Gm48190 |
predicted gene, 48190 |
29381 |
0.16 |
| chr18_69690243_69690422 | 0.63 |
Tcf4 |
transcription factor 4 |
7952 |
0.3 |
| chr3_32668496_32668692 | 0.63 |
Actl6a |
actin-like 6A |
37704 |
0.11 |
| chr2_5867590_5867753 | 0.63 |
Gm13199 |
predicted gene 13199 |
4737 |
0.15 |
| chr10_42082409_42082576 | 0.63 |
Tdg-ps2 |
thymine DNA glycosylase, pseudogene 2 |
27186 |
0.19 |
| chr16_10689840_10690031 | 0.63 |
Gm15558 |
predicted gene 15558 |
6396 |
0.18 |
| chr18_12295901_12296052 | 0.63 |
Ankrd29 |
ankyrin repeat domain 29 |
15 |
0.97 |
| chr17_25067483_25067858 | 0.62 |
Tmem204 |
transmembrane protein 204 |
10554 |
0.11 |
| chr8_106718179_106718330 | 0.62 |
Tango6 |
transport and golgi organization 6 |
35108 |
0.16 |
| chr13_77045502_77045657 | 0.62 |
Slf1 |
SMC5-SMC6 complex localization factor 1 |
1818 |
0.46 |
| chr14_26441614_26441800 | 0.62 |
Slmap |
sarcolemma associated protein |
948 |
0.52 |
| chr15_59051293_59051444 | 0.62 |
Mtss1 |
MTSS I-BAR domain containing 1 |
4096 |
0.26 |
| chr13_81368031_81368195 | 0.62 |
Adgrv1 |
adhesion G protein-coupled receptor V1 |
16487 |
0.26 |
| chr11_21225073_21225224 | 0.62 |
Gm12041 |
predicted gene 12041 |
2507 |
0.2 |
| chr7_71708283_71708442 | 0.61 |
Gm44688 |
predicted gene 44688 |
2286 |
0.26 |
| chr11_77776189_77776355 | 0.61 |
Myo18a |
myosin XVIIIA |
993 |
0.46 |
| chr17_32935365_32935709 | 0.61 |
Cyp4f13 |
cytochrome P450, family 4, subfamily f, polypeptide 13 |
4529 |
0.11 |
| chr2_92424812_92425012 | 0.61 |
Cry2 |
cryptochrome 2 (photolyase-like) |
7480 |
0.11 |
| chr5_124989850_124990034 | 0.61 |
Rflna |
refilin A |
13279 |
0.18 |
| chr2_145232861_145233023 | 0.61 |
Slc24a3 |
solute carrier family 24 (sodium/potassium/calcium exchanger), member 3 |
9669 |
0.28 |
| chr8_24546361_24546522 | 0.60 |
Ido2 |
indoleamine 2,3-dioxygenase 2 |
5645 |
0.19 |
| chr4_142179591_142179879 | 0.60 |
Gm13062 |
predicted gene 13062 |
867 |
0.62 |
| chr7_107574446_107574681 | 0.60 |
Olfml1 |
olfactomedin-like 1 |
6797 |
0.16 |
| chr7_49393023_49393262 | 0.60 |
Nav2 |
neuron navigator 2 |
28421 |
0.19 |
| chr4_130715456_130715632 | 0.60 |
Snord85 |
small nucleolar RNA, C/D box 85 |
34090 |
0.11 |
| chr4_129572692_129572886 | 0.60 |
Lck |
lymphocyte protein tyrosine kinase |
852 |
0.37 |
| chr17_5324481_5324860 | 0.60 |
Gm29050 |
predicted gene 29050 |
64093 |
0.11 |
| chr19_34553030_34553190 | 0.60 |
Ifit2 |
interferon-induced protein with tetratricopeptide repeats 2 |
2400 |
0.18 |
| chr11_107264737_107265072 | 0.60 |
Gm11719 |
predicted gene 11719 |
65804 |
0.07 |
| chr9_65196417_65196590 | 0.59 |
Gm25313 |
predicted gene, 25313 |
184 |
0.88 |
| chr7_70972135_70972318 | 0.59 |
Gm34783 |
predicted gene, 34783 |
59406 |
0.13 |
| chr17_10040936_10041349 | 0.59 |
Gm49809 |
predicted gene, 49809 |
93305 |
0.08 |
| chr9_121435320_121435493 | 0.59 |
Trak1 |
trafficking protein, kinesin binding 1 |
18890 |
0.17 |
| chr8_94550090_94550250 | 0.58 |
Cpne2 |
copine II |
5344 |
0.14 |
| chr15_41447157_41447458 | 0.58 |
Oxr1 |
oxidation resistance 1 |
175 |
0.97 |
| chr7_90146413_90146603 | 0.58 |
Gm45222 |
predicted gene 45222 |
560 |
0.64 |
| chr5_147226195_147226362 | 0.58 |
Gm24556 |
predicted gene, 24556 |
26025 |
0.11 |
| chr10_116261258_116261409 | 0.58 |
Ptprb |
protein tyrosine phosphatase, receptor type, B |
14190 |
0.19 |
| chr17_73067512_73067678 | 0.58 |
Gm30420 |
predicted gene, 30420 |
38517 |
0.15 |
| chr4_136466236_136466405 | 0.58 |
Luzp1 |
leucine zipper protein 1 |
3441 |
0.19 |
| chr11_88201344_88201495 | 0.57 |
Mrps23 |
mitochondrial ribosomal protein S23 |
2969 |
0.23 |
| chr4_102486880_102487056 | 0.57 |
Pde4b |
phosphodiesterase 4B, cAMP specific |
16571 |
0.29 |
| chr7_38225087_38225250 | 0.57 |
Plekhf1 |
pleckstrin homology domain containing, family F (with FYVE domain) member 1 |
2848 |
0.19 |
| chr14_32861212_32861363 | 0.57 |
Vstm4 |
V-set and transmembrane domain containing 4 |
4531 |
0.21 |
| chr2_84387502_84387656 | 0.57 |
Calcrl |
calcitonin receptor-like |
12221 |
0.2 |
| chr5_32446427_32446736 | 0.57 |
Ppp1cb |
protein phosphatase 1 catalytic subunit beta |
12262 |
0.11 |
| chr19_37501431_37501810 | 0.57 |
Exoc6 |
exocyst complex component 6 |
19259 |
0.14 |
| chr2_32969446_32969597 | 0.56 |
Snora65 |
small nucleolar RNA, H/ACA box 65 |
6230 |
0.11 |
| chr19_8987926_8988110 | 0.56 |
Ahnak |
AHNAK nucleoprotein (desmoyokin) |
1266 |
0.26 |
| chr2_71867523_71867680 | 0.56 |
Pdk1 |
pyruvate dehydrogenase kinase, isoenzyme 1 |
5623 |
0.2 |
| chr1_68501656_68501817 | 0.55 |
Gm19335 |
predicted gene, 19335 |
91950 |
0.08 |
| chr14_26162589_26162740 | 0.55 |
Gm2274 |
predicted gene 2274 |
6628 |
0.13 |
| chr5_32328252_32328418 | 0.55 |
Gm15615 |
predicted gene 15615 |
16216 |
0.14 |
| chr2_134825894_134826247 | 0.55 |
Gm14036 |
predicted gene 14036 |
22121 |
0.2 |
| chr13_9445564_9445785 | 0.55 |
Gm48871 |
predicted gene, 48871 |
226 |
0.93 |
| chr16_89830199_89830569 | 0.55 |
Tiam1 |
T cell lymphoma invasion and metastasis 1 |
7491 |
0.3 |
| chr4_52815809_52815985 | 0.55 |
Olfr275 |
olfactory receptor 275 |
1293 |
0.37 |
| chr1_21829923_21830095 | 0.55 |
Gm38243 |
predicted gene, 38243 |
10505 |
0.24 |
| chr14_25883204_25883355 | 0.55 |
Anxa11os |
annexin A11, opposite strand |
13013 |
0.12 |
| chr18_11327208_11327359 | 0.55 |
Gata6 |
GATA binding protein 6 |
268236 |
0.01 |
| chr1_35759768_35759937 | 0.55 |
Gm25634 |
predicted gene, 25634 |
1750 |
0.42 |
| chr19_41837838_41838023 | 0.55 |
Frat1 |
frequently rearranged in advanced T cell lymphomas |
7960 |
0.14 |
| chr13_99794055_99794213 | 0.55 |
Gm24471 |
predicted gene, 24471 |
101605 |
0.06 |
| chr1_13384478_13384660 | 0.54 |
Gm29570 |
predicted gene 29570 |
8499 |
0.13 |
| chr10_84774729_84774903 | 0.54 |
Rfx4 |
regulatory factor X, 4 (influences HLA class II expression) |
12578 |
0.21 |
| chr12_85825843_85826036 | 0.54 |
Ttll5 |
tubulin tyrosine ligase-like family, member 5 |
545 |
0.68 |
| chr1_61143547_61143729 | 0.54 |
Gm11590 |
predicted gene 11590 |
19634 |
0.19 |
| chr3_101804015_101804166 | 0.54 |
Mab21l3 |
mab-21-like 3 |
32133 |
0.17 |
| chr16_23976333_23976484 | 0.53 |
Bcl6 |
B cell leukemia/lymphoma 6 |
4028 |
0.2 |
| chr13_101316368_101316712 | 0.53 |
Gm2534 |
predicted gene 2534 |
32212 |
0.18 |
| chr1_176823956_176824133 | 0.53 |
Sdccag8 |
serologically defined colon cancer antigen 8 |
9384 |
0.11 |
| chr11_87423004_87423313 | 0.53 |
Rnu1a1 |
U1a1 small nuclear RNA |
291 |
0.79 |
| chr5_28060446_28060778 | 0.53 |
Gm26608 |
predicted gene, 26608 |
5153 |
0.18 |
| chr4_67124974_67125163 | 0.52 |
Gm11403 |
predicted gene 11403 |
40411 |
0.23 |
| chr5_36620793_36620967 | 0.52 |
D5Ertd579e |
DNA segment, Chr 5, ERATO Doi 579, expressed |
375 |
0.78 |
| chr19_21274268_21274440 | 0.52 |
Zfand5 |
zinc finger, AN1-type domain 5 |
1108 |
0.47 |
| chr7_132786130_132786291 | 0.52 |
Fam53b |
family with sequence similarity 53, member B |
709 |
0.69 |
| chr9_7858246_7858397 | 0.52 |
Birc3 |
baculoviral IAP repeat-containing 3 |
2005 |
0.3 |
| chr10_68168093_68168385 | 0.52 |
Arid5b |
AT rich interactive domain 5B (MRF1-like) |
31613 |
0.21 |
| chr4_65947314_65947465 | 0.52 |
Trim32 |
tripartite motif-containing 32 |
342140 |
0.01 |
| chr18_34868711_34868905 | 0.52 |
Egr1 |
early growth response 1 |
7601 |
0.14 |
| chr1_131090290_131090446 | 0.52 |
Gm37084 |
predicted gene, 37084 |
1781 |
0.26 |
| chr15_101266552_101266884 | 0.52 |
Nr4a1 |
nuclear receptor subfamily 4, group A, member 1 |
128 |
0.93 |
| chr13_44409176_44409378 | 0.52 |
Gm33958 |
predicted gene, 33958 |
18160 |
0.15 |
| chr7_71411542_71411776 | 0.52 |
Gm29328 |
predicted gene 29328 |
41326 |
0.15 |
| chr7_29179325_29179528 | 0.52 |
Psmd8 |
proteasome (prosome, macropain) 26S subunit, non-ATPase, 8 |
405 |
0.65 |
| chr10_68842447_68842629 | 0.52 |
1700048P04Rik |
RIKEN cDNA 1700048P04 gene |
61962 |
0.13 |
| chr5_143492763_143492920 | 0.51 |
Daglb |
diacylglycerol lipase, beta |
6163 |
0.13 |
| chr10_99314465_99314820 | 0.51 |
B530045E10Rik |
RIKEN cDNA B530045E10 gene |
15476 |
0.14 |
| chr1_67201067_67201218 | 0.51 |
Gm15668 |
predicted gene 15668 |
48058 |
0.14 |
| chr8_33709174_33709335 | 0.51 |
Gm24727 |
predicted gene, 24727 |
17453 |
0.12 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:1901475 | pyruvate transmembrane transport(GO:1901475) |
| 0.2 | 0.5 | GO:1902988 | neurofibrillary tangle assembly(GO:1902988) regulation of neurofibrillary tangle assembly(GO:1902996) |
| 0.1 | 0.4 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 0.1 | 0.5 | GO:0090427 | activation of meiosis(GO:0090427) |
| 0.1 | 0.5 | GO:0035864 | response to potassium ion(GO:0035864) cellular response to potassium ion(GO:0035865) |
| 0.1 | 0.9 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.1 | 0.3 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.1 | 0.3 | GO:0006481 | C-terminal protein methylation(GO:0006481) |
| 0.1 | 0.3 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.1 | 0.3 | GO:0071288 | cellular response to mercury ion(GO:0071288) |
| 0.1 | 0.3 | GO:0034093 | positive regulation of maintenance of sister chromatid cohesion(GO:0034093) positive regulation of maintenance of mitotic sister chromatid cohesion(GO:0034184) |
| 0.1 | 0.4 | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.1 | 0.4 | GO:0071376 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.1 | 0.3 | GO:1903984 | positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) |
| 0.1 | 0.3 | GO:1903969 | regulation of macrophage colony-stimulating factor signaling pathway(GO:1902226) regulation of response to macrophage colony-stimulating factor(GO:1903969) regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903972) |
| 0.1 | 0.3 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.1 | 0.3 | GO:0050883 | musculoskeletal movement, spinal reflex action(GO:0050883) |
| 0.1 | 0.6 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.1 | 0.4 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.1 | 0.4 | GO:0003105 | negative regulation of glomerular filtration(GO:0003105) |
| 0.1 | 0.4 | GO:0071447 | cellular response to hydroperoxide(GO:0071447) |
| 0.1 | 0.1 | GO:0018197 | peptidyl-aspartic acid modification(GO:0018197) |
| 0.1 | 0.2 | GO:0003431 | growth plate cartilage chondrocyte development(GO:0003431) |
| 0.1 | 0.2 | GO:0007039 | protein catabolic process in the vacuole(GO:0007039) |
| 0.1 | 0.3 | GO:1900454 | positive regulation of long term synaptic depression(GO:1900454) |
| 0.1 | 0.2 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.1 | 1.0 | GO:0014733 | regulation of skeletal muscle adaptation(GO:0014733) |
| 0.1 | 0.3 | GO:1902897 | regulation of postsynaptic density protein 95 clustering(GO:1902897) |
| 0.1 | 0.3 | GO:2000504 | positive regulation of blood vessel remodeling(GO:2000504) |
| 0.1 | 0.1 | GO:1900825 | regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900825) |
| 0.1 | 0.2 | GO:0006578 | amino-acid betaine biosynthetic process(GO:0006578) |
| 0.1 | 0.5 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.1 | 0.2 | GO:0019287 | isopentenyl diphosphate biosynthetic process(GO:0009240) isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.1 | 0.3 | GO:0010891 | negative regulation of sequestering of triglyceride(GO:0010891) |
| 0.1 | 0.2 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 0.1 | 0.3 | GO:0061684 | chaperone-mediated autophagy(GO:0061684) |
| 0.1 | 0.1 | GO:0003162 | atrioventricular node development(GO:0003162) |
| 0.1 | 0.2 | GO:0001692 | histamine metabolic process(GO:0001692) |
| 0.1 | 0.2 | GO:0016344 | meiotic chromosome movement towards spindle pole(GO:0016344) |
| 0.1 | 0.2 | GO:1903977 | positive regulation of glial cell migration(GO:1903977) |
| 0.0 | 0.1 | GO:0071394 | cellular response to testosterone stimulus(GO:0071394) |
| 0.0 | 0.1 | GO:0060468 | prevention of polyspermy(GO:0060468) |
| 0.0 | 0.1 | GO:0034436 | glycoprotein transport(GO:0034436) |
| 0.0 | 0.0 | GO:0035793 | positive regulation of metanephric mesenchymal cell migration by platelet-derived growth factor receptor-beta signaling pathway(GO:0035793) regulation of metanephric mesenchymal cell migration by platelet-derived growth factor receptor-beta signaling pathway(GO:1900238) positive regulation of metanephric mesenchymal cell migration(GO:2000591) |
| 0.0 | 0.2 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.0 | 0.1 | GO:1902309 | negative regulation of peptidyl-serine dephosphorylation(GO:1902309) |
| 0.0 | 0.1 | GO:0002913 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.0 | 0.0 | GO:0046084 | adenine metabolic process(GO:0046083) adenine biosynthetic process(GO:0046084) |
| 0.0 | 0.2 | GO:0051771 | negative regulation of nitric-oxide synthase biosynthetic process(GO:0051771) |
| 0.0 | 0.5 | GO:0046855 | inositol phosphate dephosphorylation(GO:0046855) |
| 0.0 | 0.5 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.3 | GO:1901841 | regulation of high voltage-gated calcium channel activity(GO:1901841) |
| 0.0 | 0.2 | GO:1903553 | positive regulation of extracellular exosome assembly(GO:1903553) |
| 0.0 | 0.1 | GO:1904017 | response to Thyroglobulin triiodothyronine(GO:1904016) cellular response to Thyroglobulin triiodothyronine(GO:1904017) |
| 0.0 | 0.1 | GO:0048861 | leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.0 | 0.1 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.0 | 0.1 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.2 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.0 | 0.1 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) positive regulation of voltage-gated potassium channel activity(GO:1903818) |
| 0.0 | 0.2 | GO:0014022 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.0 | 0.3 | GO:0060613 | fat pad development(GO:0060613) |
| 0.0 | 0.1 | GO:0048294 | negative regulation of isotype switching to IgE isotypes(GO:0048294) |
| 0.0 | 0.1 | GO:0006001 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) glycolytic process through fructose-1-phosphate(GO:0061625) |
| 0.0 | 0.2 | GO:0044027 | hypermethylation of CpG island(GO:0044027) |
| 0.0 | 0.2 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.0 | 1.1 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.0 | 0.0 | GO:0072108 | positive regulation of mesenchymal to epithelial transition involved in metanephros morphogenesis(GO:0072108) |
| 0.0 | 0.1 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.0 | 0.1 | GO:0034197 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.0 | 0.1 | GO:1990705 | cholangiocyte proliferation(GO:1990705) |
| 0.0 | 0.3 | GO:0097341 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) inhibition of cysteine-type endopeptidase activity involved in apoptotic process(GO:1990001) |
| 0.0 | 0.1 | GO:0048320 | axial mesoderm formation(GO:0048320) |
| 0.0 | 0.1 | GO:0061052 | negative regulation of cell growth involved in cardiac muscle cell development(GO:0061052) |
| 0.0 | 0.1 | GO:0003221 | right ventricular cardiac muscle tissue morphogenesis(GO:0003221) |
| 0.0 | 0.1 | GO:0048597 | post-embryonic camera-type eye morphogenesis(GO:0048597) |
| 0.0 | 0.1 | GO:0031086 | nuclear-transcribed mRNA catabolic process, deadenylation-independent decay(GO:0031086) deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.0 | 0.1 | GO:0045719 | negative regulation of glycogen biosynthetic process(GO:0045719) |
| 0.0 | 0.2 | GO:1902455 | negative regulation of stem cell population maintenance(GO:1902455) |
| 0.0 | 0.1 | GO:1904263 | positive regulation of TORC1 signaling(GO:1904263) |
| 0.0 | 0.1 | GO:0006166 | purine ribonucleoside salvage(GO:0006166) |
| 0.0 | 0.3 | GO:0008300 | isoprenoid catabolic process(GO:0008300) |
| 0.0 | 0.1 | GO:0045085 | negative regulation of interleukin-2 biosynthetic process(GO:0045085) |
| 0.0 | 0.1 | GO:1900126 | negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.0 | 0.1 | GO:0072203 | cell proliferation involved in metanephros development(GO:0072203) |
| 0.0 | 0.1 | GO:0030327 | prenylated protein catabolic process(GO:0030327) |
| 0.0 | 0.1 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.0 | 0.1 | GO:1905065 | positive regulation of vascular smooth muscle cell differentiation(GO:1905065) |
| 0.0 | 0.1 | GO:0055005 | ventricular cardiac myofibril assembly(GO:0055005) |
| 0.0 | 0.0 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.1 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.0 | 0.1 | GO:0046881 | positive regulation of follicle-stimulating hormone secretion(GO:0046881) |
| 0.0 | 0.1 | GO:0008355 | olfactory learning(GO:0008355) |
| 0.0 | 0.1 | GO:0006533 | aspartate catabolic process(GO:0006533) |
| 0.0 | 0.1 | GO:0051890 | regulation of cardioblast differentiation(GO:0051890) |
| 0.0 | 0.0 | GO:0021888 | cerebral cortex tangential migration using cell-axon interactions(GO:0021824) gonadotrophin-releasing hormone neuronal migration to the hypothalamus(GO:0021828) hypothalamic tangential migration using cell-axon interactions(GO:0021856) hypothalamus gonadotrophin-releasing hormone neuron differentiation(GO:0021886) hypothalamus gonadotrophin-releasing hormone neuron development(GO:0021888) sensory neuron axon guidance(GO:0097374) facioacoustic ganglion development(GO:1903375) |
| 0.0 | 0.1 | GO:1900377 | negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) |
| 0.0 | 0.1 | GO:0090009 | primitive streak formation(GO:0090009) |
| 0.0 | 0.2 | GO:1900103 | positive regulation of endoplasmic reticulum unfolded protein response(GO:1900103) |
| 0.0 | 0.1 | GO:0002249 | lymphocyte anergy(GO:0002249) regulation of T cell anergy(GO:0002667) T cell anergy(GO:0002870) regulation of lymphocyte anergy(GO:0002911) |
| 0.0 | 0.2 | GO:0051001 | negative regulation of nitric-oxide synthase activity(GO:0051001) |
| 0.0 | 0.1 | GO:0007182 | common-partner SMAD protein phosphorylation(GO:0007182) |
| 0.0 | 0.1 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.0 | 0.1 | GO:1903336 | negative regulation of vacuolar transport(GO:1903336) |
| 0.0 | 0.1 | GO:1904970 | brush border assembly(GO:1904970) |
| 0.0 | 0.2 | GO:0036289 | peptidyl-serine autophosphorylation(GO:0036289) |
| 0.0 | 0.1 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.1 | GO:0061577 | calcium ion transmembrane transport via high voltage-gated calcium channel(GO:0061577) |
| 0.0 | 0.4 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.0 | 0.1 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.0 | 0.2 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.0 | 0.1 | GO:1903626 | positive regulation of apoptotic DNA fragmentation(GO:1902512) positive regulation of DNA catabolic process(GO:1903626) |
| 0.0 | 0.2 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.0 | 0.2 | GO:2000344 | positive regulation of acrosome reaction(GO:2000344) |
| 0.0 | 0.1 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.0 | 0.2 | GO:0061042 | vascular wound healing(GO:0061042) |
| 0.0 | 0.1 | GO:0014042 | positive regulation of neuron maturation(GO:0014042) |
| 0.0 | 0.2 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.0 | 0.2 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.0 | 0.6 | GO:0048745 | smooth muscle tissue development(GO:0048745) |
| 0.0 | 0.1 | GO:0002568 | somatic diversification of T cell receptor genes(GO:0002568) somatic recombination of T cell receptor gene segments(GO:0002681) T cell receptor V(D)J recombination(GO:0033153) |
| 0.0 | 0.2 | GO:0051418 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.0 | 0.0 | GO:0061346 | non-canonical Wnt signaling pathway involved in heart development(GO:0061341) planar cell polarity pathway involved in heart morphogenesis(GO:0061346) |
| 0.0 | 0.1 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.0 | 0.1 | GO:0060058 | apoptotic process involved in mammary gland involution(GO:0060057) positive regulation of apoptotic process involved in mammary gland involution(GO:0060058) positive regulation of apoptotic process involved in morphogenesis(GO:1902339) regulation of mammary gland involution(GO:1903519) positive regulation of mammary gland involution(GO:1903521) positive regulation of apoptotic process involved in development(GO:1904747) |
| 0.0 | 0.1 | GO:0030578 | PML body organization(GO:0030578) |
| 0.0 | 0.1 | GO:0006624 | vacuolar protein processing(GO:0006624) |
| 0.0 | 0.1 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.0 | 0.1 | GO:0072223 | metanephric glomerular mesangium development(GO:0072223) metanephric glomerular mesangial cell proliferation involved in metanephros development(GO:0072262) |
| 0.0 | 0.1 | GO:0051594 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 0.0 | 0.1 | GO:0090365 | regulation of mRNA modification(GO:0090365) |
| 0.0 | 0.1 | GO:0002636 | positive regulation of germinal center formation(GO:0002636) |
| 0.0 | 0.1 | GO:0043031 | negative regulation of macrophage activation(GO:0043031) |
| 0.0 | 0.0 | GO:0097680 | double-strand break repair via classical nonhomologous end joining(GO:0097680) |
| 0.0 | 0.1 | GO:0021960 | anterior commissure morphogenesis(GO:0021960) |
| 0.0 | 0.0 | GO:1990314 | cellular response to insulin-like growth factor stimulus(GO:1990314) |
| 0.0 | 0.1 | GO:0015755 | fructose transport(GO:0015755) |
| 0.0 | 0.1 | GO:0051365 | cellular response to potassium ion starvation(GO:0051365) |
| 0.0 | 0.2 | GO:0070474 | positive regulation of uterine smooth muscle contraction(GO:0070474) |
| 0.0 | 0.0 | GO:1903416 | response to glycoside(GO:1903416) |
| 0.0 | 0.1 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 0.0 | GO:0002666 | positive regulation of T cell tolerance induction(GO:0002666) |
| 0.0 | 0.1 | GO:0071847 | TNFSF11-mediated signaling pathway(GO:0071847) |
| 0.0 | 0.0 | GO:0021764 | amygdala development(GO:0021764) |
| 0.0 | 0.1 | GO:0070842 | aggresome assembly(GO:0070842) |
| 0.0 | 0.1 | GO:0045915 | positive regulation of catecholamine metabolic process(GO:0045915) positive regulation of dopamine metabolic process(GO:0045964) |
| 0.0 | 0.1 | GO:0003289 | septum primum development(GO:0003284) atrial septum primum morphogenesis(GO:0003289) |
| 0.0 | 0.0 | GO:0014057 | positive regulation of acetylcholine secretion, neurotransmission(GO:0014057) |
| 0.0 | 0.1 | GO:0019230 | proprioception(GO:0019230) |
| 0.0 | 0.1 | GO:0010961 | cellular magnesium ion homeostasis(GO:0010961) |
| 0.0 | 0.0 | GO:0009756 | carbohydrate mediated signaling(GO:0009756) hexose mediated signaling(GO:0009757) sugar mediated signaling pathway(GO:0010182) glucose mediated signaling pathway(GO:0010255) |
| 0.0 | 0.1 | GO:0015705 | iodide transport(GO:0015705) |
| 0.0 | 0.1 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 0.1 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.0 | 0.1 | GO:1901894 | regulation of calcium-transporting ATPase activity(GO:1901894) |
| 0.0 | 0.1 | GO:1903799 | negative regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903799) |
| 0.0 | 0.1 | GO:1902373 | negative regulation of mRNA catabolic process(GO:1902373) |
| 0.0 | 0.2 | GO:0051657 | maintenance of organelle location(GO:0051657) |
| 0.0 | 0.0 | GO:0002645 | positive regulation of tolerance induction(GO:0002645) |
| 0.0 | 0.1 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.0 | 0.1 | GO:0015817 | histidine transport(GO:0015817) |
| 0.0 | 0.1 | GO:0002071 | glandular epithelial cell maturation(GO:0002071) |
| 0.0 | 0.1 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.5 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.0 | GO:2000645 | negative regulation of receptor catabolic process(GO:2000645) |
| 0.0 | 0.1 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.0 | 0.1 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.0 | 0.1 | GO:0060178 | regulation of exocyst localization(GO:0060178) |
| 0.0 | 0.1 | GO:0001842 | neural fold formation(GO:0001842) |
| 0.0 | 0.2 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.0 | 0.2 | GO:1902400 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) signal transduction involved in mitotic G1 DNA damage checkpoint(GO:0072431) intracellular signal transduction involved in G1 DNA damage checkpoint(GO:1902400) |
| 0.0 | 0.1 | GO:1904251 | regulation of bile acid metabolic process(GO:1904251) |
| 0.0 | 0.1 | GO:0009597 | detection of virus(GO:0009597) |
| 0.0 | 0.1 | GO:0000320 | re-entry into mitotic cell cycle(GO:0000320) |
| 0.0 | 0.0 | GO:0002337 | B-1a B cell differentiation(GO:0002337) |
| 0.0 | 0.1 | GO:1901838 | positive regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901838) |
| 0.0 | 0.1 | GO:0035878 | nail development(GO:0035878) |
| 0.0 | 0.1 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.0 | 0.2 | GO:1901409 | regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.0 | 0.0 | GO:0046103 | inosine biosynthetic process(GO:0046103) |
| 0.0 | 0.1 | GO:1901341 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 0.0 | GO:0019660 | fermentation(GO:0006113) glucose catabolic process to lactate(GO:0019659) glycolytic fermentation(GO:0019660) glucose catabolic process to lactate via pyruvate(GO:0019661) |
| 0.0 | 0.1 | GO:1903279 | regulation of calcium:sodium antiporter activity(GO:1903279) |
| 0.0 | 0.0 | GO:1902202 | regulation of hepatocyte growth factor receptor signaling pathway(GO:1902202) |
| 0.0 | 0.1 | GO:0032308 | positive regulation of prostaglandin secretion(GO:0032308) |
| 0.0 | 0.0 | GO:0055099 | response to high density lipoprotein particle(GO:0055099) |
| 0.0 | 0.1 | GO:0051541 | elastin metabolic process(GO:0051541) |
| 0.0 | 0.1 | GO:0035166 | post-embryonic hemopoiesis(GO:0035166) |
| 0.0 | 0.0 | GO:0071866 | regulation of apoptotic process in bone marrow(GO:0071865) negative regulation of apoptotic process in bone marrow(GO:0071866) |
| 0.0 | 0.3 | GO:0043981 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.0 | 0.1 | GO:0097411 | hypoxia-inducible factor-1alpha signaling pathway(GO:0097411) |
| 0.0 | 0.1 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.0 | 0.1 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.0 | 0.1 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.0 | 0.0 | GO:0060447 | bud outgrowth involved in lung branching(GO:0060447) |
| 0.0 | 0.1 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.0 | 0.2 | GO:0015812 | gamma-aminobutyric acid transport(GO:0015812) |
| 0.0 | 0.1 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.1 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.0 | 0.1 | GO:1904322 | response to forskolin(GO:1904321) cellular response to forskolin(GO:1904322) |
| 0.0 | 0.1 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.1 | GO:0060339 | negative regulation of type I interferon-mediated signaling pathway(GO:0060339) |
| 0.0 | 0.1 | GO:0009436 | glyoxylate catabolic process(GO:0009436) |
| 0.0 | 0.1 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.0 | 0.1 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.0 | 0.1 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.0 | 0.1 | GO:0048007 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.0 | 0.1 | GO:1900044 | regulation of protein K63-linked ubiquitination(GO:1900044) |
| 0.0 | 0.0 | GO:0070343 | white fat cell proliferation(GO:0070343) regulation of white fat cell proliferation(GO:0070350) |
| 0.0 | 0.1 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.0 | 0.1 | GO:0021914 | negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021914) |
| 0.0 | 0.1 | GO:0030836 | positive regulation of actin filament depolymerization(GO:0030836) |
| 0.0 | 0.0 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.0 | 0.1 | GO:0034214 | protein hexamerization(GO:0034214) |
| 0.0 | 0.0 | GO:1904751 | regulation of protein localization to nucleolus(GO:1904749) positive regulation of protein localization to nucleolus(GO:1904751) |
| 0.0 | 0.1 | GO:0002934 | desmosome organization(GO:0002934) |
| 0.0 | 0.0 | GO:0014827 | intestine smooth muscle contraction(GO:0014827) |
| 0.0 | 0.0 | GO:0070858 | negative regulation of bile acid biosynthetic process(GO:0070858) negative regulation of bile acid metabolic process(GO:1904252) |
| 0.0 | 0.1 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.0 | 0.0 | GO:1902956 | regulation of mitochondrial electron transport, NADH to ubiquinone(GO:1902956) |
| 0.0 | 0.0 | GO:0010847 | regulation of chromatin assembly(GO:0010847) |
| 0.0 | 0.1 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.0 | 0.0 | GO:0060916 | mesenchymal cell proliferation involved in lung development(GO:0060916) |
| 0.0 | 0.0 | GO:0009804 | coumarin metabolic process(GO:0009804) |
| 0.0 | 0.1 | GO:0060449 | bud elongation involved in lung branching(GO:0060449) |
| 0.0 | 0.1 | GO:0010815 | bradykinin catabolic process(GO:0010815) |
| 0.0 | 0.0 | GO:1902953 | positive regulation of ER to Golgi vesicle-mediated transport(GO:1902953) |
| 0.0 | 0.0 | GO:0010725 | regulation of primitive erythrocyte differentiation(GO:0010725) |
| 0.0 | 0.0 | GO:0046208 | spermine catabolic process(GO:0046208) |
| 0.0 | 0.0 | GO:0032417 | positive regulation of sodium:proton antiporter activity(GO:0032417) |
| 0.0 | 0.0 | GO:2000095 | regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000095) |
| 0.0 | 0.0 | GO:0009233 | menaquinone metabolic process(GO:0009233) |
| 0.0 | 0.1 | GO:0072592 | oxygen metabolic process(GO:0072592) |
| 0.0 | 0.1 | GO:0015886 | heme transport(GO:0015886) |
| 0.0 | 0.1 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.1 | GO:0002322 | B cell proliferation involved in immune response(GO:0002322) |
| 0.0 | 0.1 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.0 | 0.0 | GO:0052203 | modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) |
| 0.0 | 0.0 | GO:0021912 | regulation of transcription from RNA polymerase II promoter involved in spinal cord motor neuron fate specification(GO:0021912) |
| 0.0 | 0.1 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.0 | 0.1 | GO:0072602 | interleukin-4 secretion(GO:0072602) |
| 0.0 | 0.1 | GO:0055059 | asymmetric neuroblast division(GO:0055059) |
| 0.0 | 0.0 | GO:0032286 | central nervous system myelin maintenance(GO:0032286) |
| 0.0 | 0.1 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.0 | 0.0 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.0 | 0.1 | GO:0055089 | fatty acid homeostasis(GO:0055089) |
| 0.0 | 0.0 | GO:1902369 | negative regulation of RNA catabolic process(GO:1902369) |
| 0.0 | 0.1 | GO:0019852 | L-ascorbic acid metabolic process(GO:0019852) |
| 0.0 | 0.1 | GO:0006570 | tyrosine metabolic process(GO:0006570) |
| 0.0 | 0.3 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.0 | GO:0032278 | positive regulation of gonadotropin secretion(GO:0032278) positive regulation of luteinizing hormone secretion(GO:0033686) |
| 0.0 | 0.1 | GO:0021785 | branchiomotor neuron axon guidance(GO:0021785) |
| 0.0 | 0.1 | GO:0046599 | regulation of centriole replication(GO:0046599) |
| 0.0 | 0.1 | GO:0042985 | negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 0.0 | 0.1 | GO:0060020 | Bergmann glial cell differentiation(GO:0060020) |
| 0.0 | 0.0 | GO:0006041 | glucosamine metabolic process(GO:0006041) |
| 0.0 | 0.0 | GO:1901525 | negative regulation of macromitophagy(GO:1901525) |
| 0.0 | 0.0 | GO:0042374 | phylloquinone metabolic process(GO:0042374) phylloquinone catabolic process(GO:0042376) quinone catabolic process(GO:1901662) |
| 0.0 | 0.0 | GO:0032988 | ribonucleoprotein complex disassembly(GO:0032988) |
| 0.0 | 0.1 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.0 | GO:0065001 | specification of axis polarity(GO:0065001) |
| 0.0 | 0.0 | GO:1903599 | positive regulation of mitophagy(GO:1903599) |
| 0.0 | 0.1 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 | 0.0 | GO:0035795 | negative regulation of mitochondrial membrane permeability(GO:0035795) |
| 0.0 | 0.1 | GO:0014049 | positive regulation of glutamate secretion(GO:0014049) |
| 0.0 | 0.1 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.0 | 0.1 | GO:0044351 | macropinocytosis(GO:0044351) |
| 0.0 | 0.4 | GO:0009250 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.0 | 0.1 | GO:1904948 | midbrain dopaminergic neuron differentiation(GO:1904948) |
| 0.0 | 0.1 | GO:0032596 | protein transport into membrane raft(GO:0032596) |
| 0.0 | 0.0 | GO:0090069 | regulation of ribosome biogenesis(GO:0090069) negative regulation of ribosome biogenesis(GO:0090071) |
| 0.0 | 0.0 | GO:0072137 | condensed mesenchymal cell proliferation(GO:0072137) |
| 0.0 | 0.0 | GO:0061198 | fungiform papilla formation(GO:0061198) |
| 0.0 | 0.0 | GO:1900736 | regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900736) |
| 0.0 | 0.1 | GO:0032000 | positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.0 | 0.0 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.0 | 0.1 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 0.0 | 0.0 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.0 | 0.1 | GO:0045046 | peroxisomal membrane transport(GO:0015919) protein import into peroxisome membrane(GO:0045046) |
| 0.0 | 0.0 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.0 | 0.0 | GO:0032377 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.0 | 0.1 | GO:0060211 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) |
| 0.0 | 0.0 | GO:0015884 | folic acid transport(GO:0015884) |
| 0.0 | 0.0 | GO:0031547 | brain-derived neurotrophic factor receptor signaling pathway(GO:0031547) |
| 0.0 | 0.0 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.0 | 0.1 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
| 0.0 | 0.0 | GO:0072718 | response to cisplatin(GO:0072718) |
| 0.0 | 0.0 | GO:0060066 | oviduct development(GO:0060066) |
| 0.0 | 0.0 | GO:0046341 | CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.0 | 0.1 | GO:0033182 | regulation of histone ubiquitination(GO:0033182) |
| 0.0 | 0.0 | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 0.0 | 0.0 | GO:0000087 | mitotic M phase(GO:0000087) |
| 0.0 | 0.0 | GO:0010572 | positive regulation of platelet activation(GO:0010572) |
| 0.0 | 0.0 | GO:0002426 | immunoglobulin production in mucosal tissue(GO:0002426) |
| 0.0 | 0.0 | GO:0070093 | negative regulation of glucagon secretion(GO:0070093) |
| 0.0 | 0.0 | GO:0035995 | detection of muscle stretch(GO:0035995) |
| 0.0 | 0.1 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.0 | 0.1 | GO:0008298 | intracellular mRNA localization(GO:0008298) |
| 0.0 | 0.0 | GO:0031585 | regulation of inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0031585) |
| 0.0 | 0.0 | GO:1900095 | regulation of dosage compensation by inactivation of X chromosome(GO:1900095) |
| 0.0 | 0.1 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.1 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.0 | 0.1 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.0 | 0.0 | GO:2001181 | positive regulation of interleukin-10 secretion(GO:2001181) |
| 0.0 | 0.0 | GO:0009177 | deoxyribonucleoside monophosphate biosynthetic process(GO:0009157) pyrimidine deoxyribonucleoside monophosphate biosynthetic process(GO:0009177) |
| 0.0 | 0.0 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.0 | 0.1 | GO:0043305 | negative regulation of mast cell degranulation(GO:0043305) |
| 0.0 | 0.1 | GO:1903358 | regulation of Golgi organization(GO:1903358) |
| 0.0 | 0.0 | GO:0036135 | Schwann cell migration(GO:0036135) regulation of Schwann cell migration(GO:1900147) |
| 0.0 | 0.1 | GO:0009256 | 10-formyltetrahydrofolate metabolic process(GO:0009256) |
| 0.0 | 0.1 | GO:0006528 | asparagine metabolic process(GO:0006528) |
| 0.0 | 0.1 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.0 | 0.0 | GO:0033602 | negative regulation of dopamine secretion(GO:0033602) |
| 0.0 | 0.1 | GO:0098764 | meiotic prophase I(GO:0007128) prophase(GO:0051324) meiotic cell cycle phase(GO:0098762) meiosis I cell cycle phase(GO:0098764) |
| 0.0 | 0.0 | GO:2000619 | negative regulation of histone H4-K16 acetylation(GO:2000619) |
| 0.0 | 0.0 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.0 | 0.1 | GO:0098739 | import across plasma membrane(GO:0098739) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0097418 | neurofibrillary tangle(GO:0097418) |
| 0.1 | 0.3 | GO:0032127 | dense core granule membrane(GO:0032127) |
| 0.1 | 0.3 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.1 | 0.2 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.1 | 0.4 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.1 | 0.3 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.1 | 0.1 | GO:0034679 | integrin alpha9-beta1 complex(GO:0034679) |
| 0.1 | 0.2 | GO:1990696 | USH2 complex(GO:1990696) |
| 0.1 | 0.4 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.1 | 0.3 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.0 | 0.1 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 0.1 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.0 | 0.1 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 0.1 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.0 | 0.2 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.2 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 0.4 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.1 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 0.2 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.0 | 0.1 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 0.2 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.2 | GO:0000923 | equatorial microtubule organizing center(GO:0000923) |
| 0.0 | 0.2 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.8 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.1 | GO:0097425 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.0 | 0.1 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 0.2 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.1 | GO:0046691 | intracellular canaliculus(GO:0046691) |
| 0.0 | 0.2 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 0.1 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.1 | GO:0031523 | Myb complex(GO:0031523) |
| 0.0 | 0.1 | GO:0010009 | cytoplasmic side of endosome membrane(GO:0010009) |
| 0.0 | 0.1 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 0.1 | GO:0070688 | MLL5-L complex(GO:0070688) |
| 0.0 | 0.2 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 0.1 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 0.2 | GO:0060091 | kinocilium(GO:0060091) |
| 0.0 | 0.3 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.1 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.0 | 0.1 | GO:0005775 | vacuolar lumen(GO:0005775) |
| 0.0 | 0.4 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.1 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.0 | 0.3 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.0 | 0.1 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.0 | 0.1 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.0 | 0.1 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.0 | 0.0 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.0 | 0.0 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.0 | 0.1 | GO:0000801 | central element(GO:0000801) |
| 0.0 | 0.1 | GO:0002081 | outer acrosomal membrane(GO:0002081) |
| 0.0 | 0.1 | GO:0000938 | GARP complex(GO:0000938) |
| 0.0 | 0.1 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.1 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.4 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.0 | 0.0 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.0 | 0.1 | GO:0071006 | U2-type catalytic step 1 spliceosome(GO:0071006) |
| 0.0 | 0.1 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.0 | 0.2 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 0.2 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 0.2 | GO:0001741 | XY body(GO:0001741) |
| 0.0 | 0.3 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 0.1 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 0.1 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.0 | 0.1 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.0 | 0.6 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.0 | 0.1 | GO:0031415 | NatA complex(GO:0031415) |
| 0.0 | 0.0 | GO:0031094 | platelet dense tubular network(GO:0031094) |
| 0.0 | 0.0 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.0 | 0.0 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.0 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.0 | 0.1 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.0 | 0.5 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 0.1 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.1 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.0 | 0.1 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 0.0 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.0 | 0.1 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.1 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.0 | 0.1 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 0.0 | GO:0000811 | GINS complex(GO:0000811) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.1 | 0.4 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.1 | 0.9 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.1 | 0.3 | GO:0015182 | L-asparagine transmembrane transporter activity(GO:0015182) L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.1 | 0.3 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.1 | 0.3 | GO:0016899 | oxidoreductase activity, acting on the CH-OH group of donors, oxygen as acceptor(GO:0016899) |
| 0.1 | 0.2 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.1 | 0.4 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.1 | 0.1 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.1 | 0.2 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.1 | 0.1 | GO:0005290 | L-histidine transmembrane transporter activity(GO:0005290) |
| 0.1 | 0.2 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.1 | 0.4 | GO:0018654 | 3-(3-hydroxyphenyl)propionate hydroxylase activity(GO:0008688) 4-chlorobenzaldehyde oxidase activity(GO:0018471) 3,5-xylenol methylhydroxylase activity(GO:0018630) phenylacetate hydroxylase activity(GO:0018631) 4-nitrophenol 4-monooxygenase activity(GO:0018632) dimethyl sulfide monooxygenase activity(GO:0018633) alpha-pinene monooxygenase [NADH] activity(GO:0018634) 1-hydroxy-2-naphthoate hydroxylase activity(GO:0018637) toluene 4-monooxygenase activity(GO:0018638) xylene monooxygenase activity(GO:0018639) dibenzothiophene monooxygenase activity(GO:0018640) 6-hydroxy-3-methyl-2-oxo-1,2-dihydroquinoline 6-monooxygenase activity(GO:0018641) chlorophenol 4-monooxygenase activity(GO:0018642) carbon disulfide oxygenase activity(GO:0018643) toluene 2-monooxygenase activity(GO:0018644) 1-hydroxy-2-oxolimonene 1,2-monooxygenase activity(GO:0018646) phenanthrene 1,2-monooxygenase activity(GO:0018647) tetrahydrofuran hydroxylase activity(GO:0018649) styrene monooxygenase activity(GO:0018650) toluene-4-sulfonate monooxygenase activity(GO:0018651) toluene-sulfonate methyl-monooxygenase activity(GO:0018652) 3-methyl-2-oxo-1,2-dihydroquinoline 6-monooxygenase activity(GO:0018653) 2-hydroxy-phenylacetate hydroxylase activity(GO:0018654) 2-oxo-delta3-4,5,5-trimethylcyclopentenylacetyl-CoA 1,2-monooxygenase activity(GO:0018655) phenanthrene 3,4-monooxygenase activity(GO:0018656) toluene 3-monooxygenase activity(GO:0018657) 4-hydroxyphenylacetate,NADH:oxygen oxidoreductase (3-hydroxylating) activity(GO:0018660) limonene monooxygenase activity(GO:0019113) 2-methylnaphthalene hydroxylase activity(GO:0034526) 1-methylnaphthalene hydroxylase activity(GO:0034534) bisphenol A hydroxylase A activity(GO:0034560) salicylate 5-hydroxylase activity(GO:0034785) isobutylamine N-hydroxylase activity(GO:0034791) branched-chain dodecylbenzene sulfonate monooxygenase activity(GO:0034802) 3-HSA hydroxylase activity(GO:0034819) 4-hydroxypyridine-3-hydroxylase activity(GO:0034894) 2-octaprenyl-3-methyl-6-methoxy-1,4-benzoquinol hydroxylase activity(GO:0043719) 6-hydroxynicotinate 3-monooxygenase activity(GO:0043731) thalianol hydroxylase activity(GO:0080014) |
| 0.1 | 0.2 | GO:0043125 | ErbB-3 class receptor binding(GO:0043125) |
| 0.1 | 0.2 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.1 | 0.2 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.1 | 0.4 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.1 | 0.4 | GO:0005223 | intracellular cGMP activated cation channel activity(GO:0005223) |
| 0.0 | 0.3 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.0 | 0.1 | GO:0055100 | adiponectin binding(GO:0055100) |
| 0.0 | 0.1 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.0 | 0.3 | GO:1990459 | transferrin receptor binding(GO:1990459) |
| 0.0 | 0.5 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.2 | GO:0004439 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) |
| 0.0 | 0.1 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.0 | 0.1 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.0 | 0.2 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.0 | 0.3 | GO:0004661 | protein geranylgeranyltransferase activity(GO:0004661) |
| 0.0 | 0.1 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 0.1 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.0 | 0.7 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.1 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 0.0 | 0.1 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 0.1 | GO:0033142 | progesterone receptor binding(GO:0033142) |
| 0.0 | 0.1 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.0 | 0.4 | GO:0005536 | glucose binding(GO:0005536) |
| 0.0 | 0.2 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) |
| 0.0 | 0.1 | GO:0015119 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.0 | 0.2 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.1 | GO:0102007 | lactonohydrolase activity(GO:0046573) acyl-L-homoserine-lactone lactonohydrolase activity(GO:0102007) |
| 0.0 | 0.2 | GO:1990239 | steroid hormone binding(GO:1990239) |
| 0.0 | 0.3 | GO:0046972 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.0 | 0.2 | GO:0050051 | leukotriene-B4 20-monooxygenase activity(GO:0050051) |
| 0.0 | 0.3 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 0.5 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.1 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.1 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.0 | 0.3 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.1 | GO:0043559 | insulin binding(GO:0043559) |
| 0.0 | 0.1 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.1 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.0 | 0.2 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 0.1 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.0 | 0.7 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.0 | 0.1 | GO:0070905 | serine binding(GO:0070905) |
| 0.0 | 0.2 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 0.1 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.1 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.0 | 0.1 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.1 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.0 | 0.2 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.2 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.0 | GO:0015185 | gamma-aminobutyric acid transmembrane transporter activity(GO:0015185) |
| 0.0 | 0.1 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.0 | 0.1 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 0.1 | GO:0031686 | A1 adenosine receptor binding(GO:0031686) |
| 0.0 | 0.0 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.0 | 0.1 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.0 | 0.1 | GO:0036435 | K48-linked polyubiquitin binding(GO:0036435) |
| 0.0 | 0.0 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.0 | 0.3 | GO:0071949 | FAD binding(GO:0071949) |
| 0.0 | 0.1 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 0.0 | 0.1 | GO:0008251 | tRNA-specific adenosine deaminase activity(GO:0008251) |
| 0.0 | 0.1 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.0 | 0.1 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.0 | 0.0 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.0 | 0.1 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.0 | 0.2 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.0 | 0.1 | GO:0005534 | galactose binding(GO:0005534) |
| 0.0 | 0.1 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.0 | 0.1 | GO:0031800 | type 3 metabotropic glutamate receptor binding(GO:0031800) |
| 0.0 | 0.1 | GO:0051880 | G-quadruplex DNA binding(GO:0051880) |
| 0.0 | 0.2 | GO:0030553 | cGMP binding(GO:0030553) |
| 0.0 | 0.1 | GO:0097108 | hedgehog family protein binding(GO:0097108) |
| 0.0 | 0.1 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.0 | 0.2 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.0 | 0.5 | GO:0043531 | ADP binding(GO:0043531) |
| 0.0 | 0.1 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.0 | 0.1 | GO:0017116 | single-stranded DNA-dependent ATP-dependent DNA helicase activity(GO:0017116) |
| 0.0 | 0.1 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 0.5 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.0 | 0.2 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.1 | GO:0008486 | diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) |
| 0.0 | 0.1 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.0 | 0.2 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.0 | 0.1 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.0 | 0.2 | GO:0034713 | type I transforming growth factor beta receptor binding(GO:0034713) |
| 0.0 | 0.1 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 0.0 | GO:0070698 | type I activin receptor binding(GO:0070698) |
| 0.0 | 0.2 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.2 | GO:0031994 | insulin-like growth factor I binding(GO:0031994) |
| 0.0 | 0.1 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.0 | 0.0 | GO:0032139 | dinucleotide insertion or deletion binding(GO:0032139) |
| 0.0 | 0.2 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.1 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.0 | 0.1 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.0 | 0.4 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.0 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.0 | 0.3 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.0 | GO:0043546 | molybdopterin cofactor binding(GO:0043546) |
| 0.0 | 0.1 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.0 | GO:0030021 | extracellular matrix structural constituent conferring compression resistance(GO:0030021) structural constituent of tooth enamel(GO:0030345) |
| 0.0 | 0.0 | GO:0000171 | ribonuclease MRP activity(GO:0000171) |
| 0.0 | 0.2 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.0 | 0.1 | GO:0042979 | ornithine decarboxylase regulator activity(GO:0042979) |
| 0.0 | 0.0 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.0 | 0.3 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.0 | 0.2 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 0.0 | GO:0031893 | vasopressin receptor binding(GO:0031893) |
| 0.0 | 0.0 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 0.0 | GO:0051718 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) DNA (cytosine-5-)-methyltransferase activity, acting on CpG substrates(GO:0051718) |
| 0.0 | 0.1 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.0 | 0.1 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.0 | 0.1 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 0.0 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.0 | 0.5 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.1 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.0 | 0.0 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.0 | 0.6 | GO:0008392 | arachidonic acid epoxygenase activity(GO:0008392) |
| 0.0 | 0.1 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.0 | GO:0030375 | thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 0.1 | GO:0008599 | protein phosphatase type 1 regulator activity(GO:0008599) |
| 0.0 | 0.1 | GO:0070991 | medium-chain-acyl-CoA dehydrogenase activity(GO:0070991) |
| 0.0 | 0.1 | GO:0070513 | death domain binding(GO:0070513) |
| 0.0 | 0.1 | GO:1990932 | 5.8S rRNA binding(GO:1990932) |
| 0.0 | 0.0 | GO:0015111 | iodide transmembrane transporter activity(GO:0015111) |
| 0.0 | 0.1 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.0 | 0.0 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.0 | 0.1 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.1 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 0.1 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.3 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.1 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.1 | GO:0034847 | pinocarveol dehydrogenase activity(GO:0018446) chloral hydrate dehydrogenase activity(GO:0018447) hydroxymethylmethylsilanediol oxidase activity(GO:0018448) 1-phenylethanol dehydrogenase activity(GO:0018449) myrtenol dehydrogenase activity(GO:0018450) cis-1,2-dihydroxy-1,2-dihydro-8-carboxynaphthalene dehydrogenase activity(GO:0034522) 3-hydroxy-4-methyloctanoyl-CoA dehydrogenase activity(GO:0034582) 2-hydroxy-4-isopropenylcyclohexane-1-carboxyl-CoA dehydrogenase activity(GO:0034778) cis-9,10-dihydroanthracene-9,10-diol dehydrogenase activity(GO:0034817) citronellol dehydrogenase activity(GO:0034821) naphthyl-2-hydroxymethyl-succinyl-CoA dehydrogenase activity(GO:0034847) 2,4,4-trimethyl-1-pentanol dehydrogenase activity(GO:0034863) 2,4,4-trimethyl-3-hydroxypentanoyl-CoA dehydrogenase activity(GO:0034868) 1-hydroxy-4,4-dimethylpentan-3-one dehydrogenase activity(GO:0034871) endosulfan diol dehydrogenase activity(GO:0034891) endosulfan hydroxyether dehydrogenase activity(GO:0034901) 3-hydroxy-2-methylhexanoyl-CoA dehydrogenase activity(GO:0034918) 3-hydroxy-2,6-dimethyl-5-methylene-heptanoyl-CoA dehydrogenase activity(GO:0034944) versicolorin reductase activity(GO:0042469) ketoreductase activity(GO:0045703) |
| 0.0 | 0.1 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.0 | 0.2 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.0 | GO:0016508 | long-chain-enoyl-CoA hydratase activity(GO:0016508) |
| 0.0 | 0.0 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.1 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.0 | 0.0 | GO:0035851 | Krueppel-associated box domain binding(GO:0035851) |
| 0.0 | 0.0 | GO:0019145 | aminobutyraldehyde dehydrogenase activity(GO:0019145) 4-trimethylammoniobutyraldehyde dehydrogenase activity(GO:0047105) |
| 0.0 | 0.2 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.0 | GO:0004165 | dodecenoyl-CoA delta-isomerase activity(GO:0004165) |
| 0.0 | 0.1 | GO:0090482 | vitamin transmembrane transporter activity(GO:0090482) |
| 0.0 | 0.0 | GO:0031697 | beta-1 adrenergic receptor binding(GO:0031697) |
| 0.0 | 0.1 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.0 | 0.1 | GO:0071723 | lipopeptide binding(GO:0071723) |
| 0.0 | 0.0 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 0.1 | GO:0016215 | acyl-CoA desaturase activity(GO:0016215) |
| 0.0 | 0.0 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.0 | 0.0 | GO:0034190 | apolipoprotein receptor binding(GO:0034190) |
| 0.0 | 0.1 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.0 | 0.0 | GO:0051022 | Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.0 | 0.0 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.0 | 0.0 | GO:0050252 | retinol O-fatty-acyltransferase activity(GO:0050252) |
| 0.0 | 0.0 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 0.1 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.1 | 0.1 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.1 | 0.1 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 1.4 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.4 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.0 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.0 | 0.5 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.3 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 0.4 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 0.3 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.2 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 0.0 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.0 | 0.1 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.3 | ST ADRENERGIC | Adrenergic Pathway |
| 0.0 | 0.1 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.5 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.5 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 0.3 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.2 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.2 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 0.3 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 0.5 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.0 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.0 | 0.1 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.2 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.1 | PID S1P S1P2 PATHWAY | S1P2 pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.1 | 0.2 | REACTOME REGULATION OF INSULIN SECRETION | Genes involved in Regulation of Insulin Secretion |
| 0.0 | 0.4 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.8 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.5 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.0 | 0.2 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.0 | 0.2 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.2 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.3 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.0 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.0 | 0.3 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.0 | 0.2 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.0 | REACTOME CTLA4 INHIBITORY SIGNALING | Genes involved in CTLA4 inhibitory signaling |
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.2 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.3 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.0 | 0.1 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.0 | 0.2 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.7 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.2 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 0.3 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.0 | 0.2 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 0.2 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.1 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.1 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.1 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.3 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |
| 0.0 | 0.3 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.1 | REACTOME TCR SIGNALING | Genes involved in TCR signaling |
| 0.0 | 0.1 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.0 | 0.1 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.2 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.1 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.1 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.1 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.2 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.5 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.2 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.3 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.0 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.0 | REACTOME DESTABILIZATION OF MRNA BY TRISTETRAPROLIN TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 0.0 | 0.0 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.0 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 0.1 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.1 | REACTOME NUCLEAR EVENTS KINASE AND TRANSCRIPTION FACTOR ACTIVATION | Genes involved in Nuclear Events (kinase and transcription factor activation) |
| 0.0 | 0.1 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.4 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |