Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
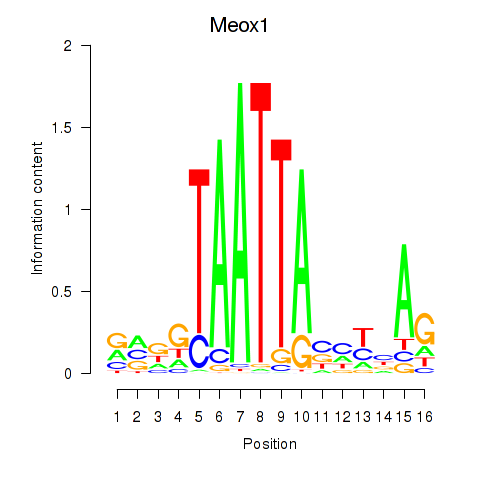
Results for Meox1
Z-value: 0.52
Transcription factors associated with Meox1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Meox1
|
ENSMUSG00000001493.9 | mesenchyme homeobox 1 |
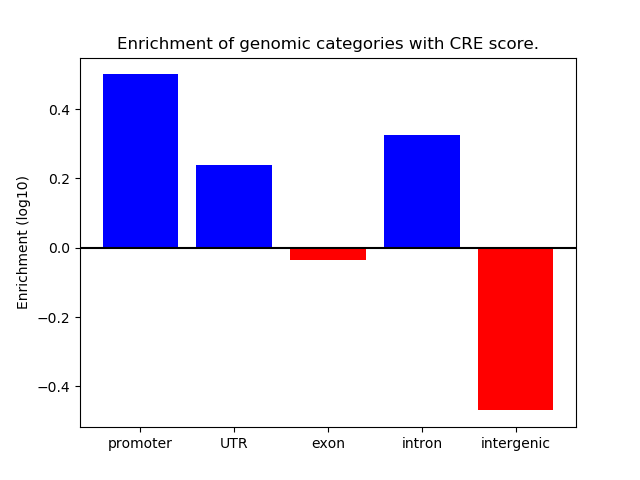
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr11_101912289_101912454 | Meox1 | 17997 | 0.101798 | 0.08 | 8.8e-01 | Click! |
| chr11_101905236_101905402 | Meox1 | 10945 | 0.115203 | 0.06 | 9.1e-01 | Click! |
| chr11_101910339_101910504 | Meox1 | 16047 | 0.105432 | 0.03 | 9.5e-01 | Click! |
Activity of the Meox1 motif across conditions
Conditions sorted by the z-value of the Meox1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
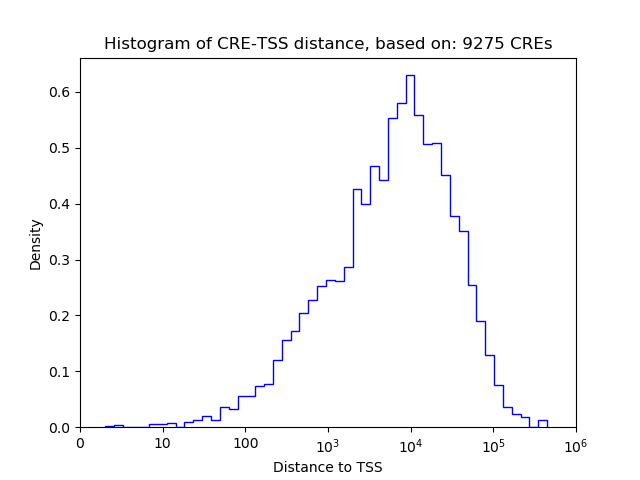
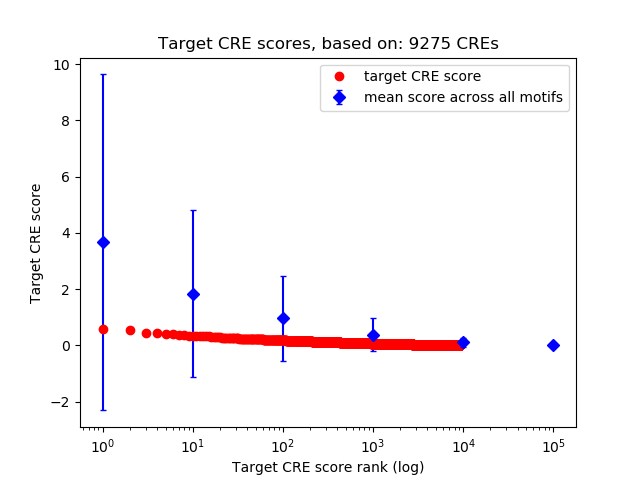
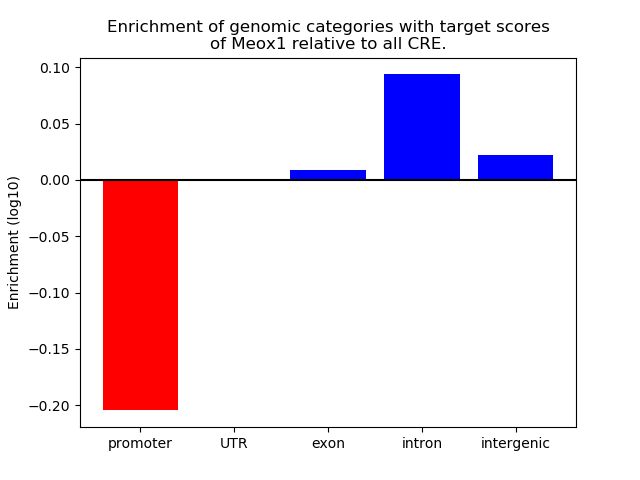
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr15_34500771_34501210 | 0.59 |
Pop1 |
processing of precursor 1, ribonuclease P/MRP family, (S. cerevisiae) |
5667 |
0.12 |
| chr19_44394517_44394772 | 0.54 |
Scd1 |
stearoyl-Coenzyme A desaturase 1 |
12046 |
0.14 |
| chr12_104083020_104083260 | 0.43 |
Serpina4-ps1 |
serine (or cysteine) peptidase inhibitor, clade A, member 4, pseudogene 1 |
2491 |
0.16 |
| chr16_41161594_41161767 | 0.42 |
Gm26381 |
predicted gene, 26381 |
56065 |
0.15 |
| chr13_4270161_4270342 | 0.41 |
Akr1c12 |
aldo-keto reductase family 1, member C12 |
9182 |
0.15 |
| chr13_44437957_44438156 | 0.40 |
1700029N11Rik |
RIKEN cDNA 1700029N11 gene |
1656 |
0.27 |
| chr19_40153747_40153898 | 0.37 |
Cyp2c70 |
cytochrome P450, family 2, subfamily c, polypeptide 70 |
33464 |
0.13 |
| chr1_162891903_162892071 | 0.36 |
Fmo2 |
flavin containing monooxygenase 2 |
5472 |
0.19 |
| chr4_135789148_135789435 | 0.34 |
Myom3 |
myomesin family, member 3 |
8899 |
0.14 |
| chr16_42947764_42948084 | 0.34 |
Ndufs5-ps |
NADH:ubiquinone oxidoreductase core subunit S5, pseudogene |
7707 |
0.21 |
| chr9_114857411_114857562 | 0.33 |
Cmtm8 |
CKLF-like MARVEL transmembrane domain containing 8 |
13330 |
0.16 |
| chr1_100298746_100298897 | 0.33 |
Gm29667 |
predicted gene 29667 |
16030 |
0.18 |
| chr7_63922331_63923024 | 0.32 |
Klf13 |
Kruppel-like factor 13 |
2193 |
0.22 |
| chr3_12608407_12608567 | 0.32 |
Gm2429 |
predicted gene 2429 |
22773 |
0.28 |
| chr2_113578462_113578613 | 0.31 |
Gm13964 |
predicted gene 13964 |
74503 |
0.09 |
| chr4_121078016_121078199 | 0.31 |
Zmpste24 |
zinc metallopeptidase, STE24 |
369 |
0.74 |
| chrX_85734642_85734808 | 0.30 |
Gk |
glycerol kinase |
2885 |
0.21 |
| chr8_35219364_35219515 | 0.29 |
Gm34474 |
predicted gene, 34474 |
801 |
0.55 |
| chr5_102009929_102010121 | 0.28 |
Wdfy3 |
WD repeat and FYVE domain containing 3 |
28467 |
0.16 |
| chr3_133759501_133760150 | 0.28 |
Gm6135 |
prediticted gene 6135 |
31679 |
0.18 |
| chr16_10555941_10556092 | 0.27 |
Clec16a |
C-type lectin domain family 16, member A |
10607 |
0.17 |
| chr11_80974682_80974867 | 0.27 |
Gm11416 |
predicted gene 11416 |
72020 |
0.1 |
| chr8_93188530_93188681 | 0.27 |
Gm45909 |
predicted gene 45909 |
2753 |
0.19 |
| chr4_105328178_105328374 | 0.26 |
Gm12722 |
predicted gene 12722 |
46670 |
0.19 |
| chr2_38823684_38823849 | 0.26 |
Nr6a1os |
nuclear receptor subfamily 6, group A, member 1, opposite strand |
425 |
0.74 |
| chr9_74888354_74888522 | 0.26 |
Onecut1 |
one cut domain, family member 1 |
21954 |
0.14 |
| chr17_28420558_28420744 | 0.25 |
Fkbp5 |
FK506 binding protein 5 |
2124 |
0.18 |
| chr6_85813280_85813431 | 0.25 |
Nat8f6 |
N-acetyltransferase 8 (GCN5-related) family member 6 |
3492 |
0.11 |
| chr6_136906482_136906715 | 0.25 |
Erp27 |
endoplasmic reticulum protein 27 |
15542 |
0.11 |
| chr19_44401346_44401497 | 0.25 |
Scd1 |
stearoyl-Coenzyme A desaturase 1 |
5269 |
0.16 |
| chr11_28668598_28668749 | 0.25 |
2810471M01Rik |
RIKEN cDNA 2810471M01 gene |
12891 |
0.2 |
| chr3_60525375_60525543 | 0.25 |
Mbnl1 |
muscleblind like splicing factor 1 |
2669 |
0.31 |
| chr1_67182326_67182537 | 0.24 |
Cps1 |
carbamoyl-phosphate synthetase 1 |
59405 |
0.11 |
| chr3_144446211_144446394 | 0.24 |
Gm5857 |
predicted gene 5857 |
18630 |
0.18 |
| chr10_121349776_121349943 | 0.24 |
Gm48435 |
predicted gene, 48435 |
601 |
0.58 |
| chr2_31519719_31520357 | 0.24 |
Ass1 |
argininosuccinate synthetase 1 |
1548 |
0.36 |
| chr3_100893002_100893153 | 0.23 |
Trim45 |
tripartite motif-containing 45 |
29125 |
0.16 |
| chrX_170020226_170020377 | 0.23 |
Erdr1 |
erythroid differentiation regulator 1 |
9557 |
0.18 |
| chr5_20858729_20858886 | 0.23 |
Phtf2 |
putative homeodomain transcription factor 2 |
3616 |
0.22 |
| chr3_58520131_58520481 | 0.23 |
Eif2a |
eukaryotic translation initiation factor 2A |
5515 |
0.16 |
| chr14_89652024_89652190 | 0.23 |
Gm25415 |
predicted gene, 25415 |
54698 |
0.17 |
| chr2_31513022_31513372 | 0.23 |
Ass1 |
argininosuccinate synthetase 1 |
5293 |
0.19 |
| chr12_100392491_100392648 | 0.22 |
Ttc7b |
tetratricopeptide repeat domain 7B |
6669 |
0.19 |
| chr7_4765364_4765545 | 0.22 |
Fam71e2 |
family with sequence similarity 71, member E2 |
5848 |
0.07 |
| chr8_35216178_35216374 | 0.22 |
Gm34474 |
predicted gene, 34474 |
2362 |
0.23 |
| chr13_58189859_58190041 | 0.22 |
Gm48357 |
predicted gene, 48357 |
1368 |
0.28 |
| chr17_28513542_28513712 | 0.22 |
Fkbp5 |
FK506 binding protein 5 |
2270 |
0.13 |
| chr3_97635501_97635936 | 0.22 |
Fmo5 |
flavin containing monooxygenase 5 |
6835 |
0.14 |
| chr8_73953761_73954148 | 0.22 |
Gm7948 |
predicted gene 7948 |
82261 |
0.11 |
| chr15_59067650_59068255 | 0.22 |
Mtss1 |
MTSS I-BAR domain containing 1 |
12488 |
0.22 |
| chr5_123090732_123091094 | 0.22 |
Tmem120b |
transmembrane protein 120B |
8988 |
0.09 |
| chr8_36290613_36290918 | 0.21 |
Lonrf1 |
LON peptidase N-terminal domain and ring finger 1 |
41249 |
0.14 |
| chr3_136048131_136048315 | 0.21 |
Gm5281 |
predicted gene 5281 |
3465 |
0.25 |
| chr17_63370765_63370929 | 0.21 |
Gm24813 |
predicted gene, 24813 |
23176 |
0.18 |
| chrX_77601507_77601673 | 0.21 |
Gm23121 |
predicted gene, 23121 |
22103 |
0.2 |
| chr13_51093099_51093344 | 0.21 |
Spin1 |
spindlin 1 |
7659 |
0.25 |
| chr9_60982764_60983106 | 0.21 |
Gm5122 |
predicted gene 5122 |
30546 |
0.14 |
| chr19_44394826_44394979 | 0.21 |
Scd1 |
stearoyl-Coenzyme A desaturase 1 |
11788 |
0.14 |
| chr1_31051542_31051713 | 0.21 |
Gm28644 |
predicted gene 28644 |
9708 |
0.17 |
| chr11_51727078_51727230 | 0.21 |
Gm25291 |
predicted gene, 25291 |
10768 |
0.14 |
| chr4_15212980_15213172 | 0.21 |
Tmem64 |
transmembrane protein 64 |
52755 |
0.13 |
| chr7_144551116_144551314 | 0.21 |
Ppfia1 |
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 1 |
1147 |
0.45 |
| chr5_43309511_43309822 | 0.21 |
6030400A10Rik |
RIKEN cDNA 6030400A10 gene |
44509 |
0.11 |
| chr17_64763760_64763987 | 0.21 |
Dreh |
down-regulated in hepatocellular carcinoma |
3238 |
0.25 |
| chr2_103028308_103028465 | 0.20 |
Pdhx |
pyruvate dehydrogenase complex, component X |
44949 |
0.13 |
| chr11_119048222_119048411 | 0.20 |
Cbx8 |
chromobox 8 |
7347 |
0.16 |
| chr12_57281727_57281906 | 0.20 |
Mipol1 |
mirror-image polydactyly 1 |
357 |
0.88 |
| chr14_17864277_17864546 | 0.20 |
Gm48320 |
predicted gene, 48320 |
93289 |
0.08 |
| chr12_51274477_51274668 | 0.20 |
Rps11-ps4 |
ribosomal protein S11, pseudogene 4 |
22915 |
0.22 |
| chr13_24360335_24360523 | 0.20 |
Gm11342 |
predicted gene 11342 |
15521 |
0.12 |
| chr14_7827742_7828144 | 0.20 |
Flnb |
filamin, beta |
9986 |
0.16 |
| chr9_79796831_79797014 | 0.19 |
4930429F24Rik |
RIKEN cDNA 4930429F24 gene |
3281 |
0.18 |
| chr9_122826708_122826859 | 0.19 |
Gm35549 |
predicted gene, 35549 |
17103 |
0.1 |
| chr11_16814706_16815122 | 0.19 |
Egfros |
epidermal growth factor receptor, opposite strand |
15788 |
0.21 |
| chr6_90499813_90499975 | 0.19 |
Gm44236 |
predicted gene, 44236 |
13408 |
0.1 |
| chr2_14596063_14596214 | 0.19 |
Cacnb2 |
calcium channel, voltage-dependent, beta 2 subunit |
6950 |
0.13 |
| chr13_109635481_109635828 | 0.19 |
Pde4d |
phosphodiesterase 4D, cAMP specific |
2874 |
0.42 |
| chr2_154405536_154405724 | 0.19 |
Snta1 |
syntrophin, acidic 1 |
2350 |
0.26 |
| chr16_43465927_43466078 | 0.19 |
Zbtb20 |
zinc finger and BTB domain containing 20 |
37612 |
0.16 |
| chr15_62121397_62121548 | 0.19 |
Pvt1 |
Pvt1 oncogene |
22443 |
0.25 |
| chr6_58611375_58611579 | 0.18 |
Abcg2 |
ATP binding cassette subfamily G member 2 (Junior blood group) |
14806 |
0.19 |
| chr13_9932808_9933042 | 0.18 |
Gm47406 |
predicted gene, 47406 |
40015 |
0.14 |
| chr11_86679364_86679596 | 0.18 |
Vmp1 |
vacuole membrane protein 1 |
4341 |
0.18 |
| chr3_102384557_102384708 | 0.18 |
Gm43245 |
predicted gene 43245 |
5375 |
0.21 |
| chr17_83838220_83838652 | 0.18 |
Haao |
3-hydroxyanthranilate 3,4-dioxygenase |
2468 |
0.26 |
| chr6_29571610_29572022 | 0.18 |
Tnpo3 |
transportin 3 |
297 |
0.87 |
| chr2_155100983_155101285 | 0.18 |
2310005A03Rik |
RIKEN cDNA 2310005A03 gene |
1053 |
0.43 |
| chr8_122898814_122898990 | 0.18 |
Ankrd11 |
ankyrin repeat domain 11 |
878 |
0.49 |
| chr6_72120521_72121047 | 0.18 |
4933431G14Rik |
RIKEN cDNA 4933431G14 gene |
2156 |
0.2 |
| chr13_101718622_101718773 | 0.18 |
Pik3r1 |
phosphoinositide-3-kinase regulatory subunit 1 |
16593 |
0.2 |
| chr11_50295296_50295447 | 0.18 |
Canx |
calnexin |
2948 |
0.16 |
| chr13_20205112_20205287 | 0.18 |
Elmo1 |
engulfment and cell motility 1 |
19992 |
0.25 |
| chr13_52979672_52979858 | 0.18 |
Nfil3 |
nuclear factor, interleukin 3, regulated |
1308 |
0.43 |
| chr9_44087034_44087185 | 0.18 |
Usp2 |
ubiquitin specific peptidase 2 |
81 |
0.91 |
| chr15_34501595_34501764 | 0.18 |
Pop1 |
processing of precursor 1, ribonuclease P/MRP family, (S. cerevisiae) |
6356 |
0.12 |
| chr11_68854403_68854915 | 0.18 |
Ndel1 |
nudE neurodevelopment protein 1 like 1 |
1524 |
0.3 |
| chr10_67176584_67176792 | 0.18 |
Jmjd1c |
jumonji domain containing 1C |
9060 |
0.22 |
| chr2_43509495_43509670 | 0.18 |
Gm13464 |
predicted gene 13464 |
9816 |
0.28 |
| chr1_50806813_50807031 | 0.18 |
Gm28321 |
predicted gene 28321 |
8453 |
0.28 |
| chr1_140421414_140421565 | 0.18 |
Kcnt2 |
potassium channel, subfamily T, member 2 |
21447 |
0.27 |
| chr14_21411001_21411208 | 0.18 |
Gm25864 |
predicted gene, 25864 |
39370 |
0.14 |
| chr2_73628966_73629117 | 0.18 |
Chn1 |
chimerin 1 |
3301 |
0.21 |
| chr9_116136676_116136901 | 0.18 |
Tgfbr2 |
transforming growth factor, beta receptor II |
38477 |
0.15 |
| chr8_35564396_35564553 | 0.17 |
Mfhas1 |
malignant fibrous histiocytoma amplified sequence 1 |
23324 |
0.16 |
| chr10_111583801_111583952 | 0.17 |
4933440J02Rik |
RIKEN cDNA 4933440J02 gene |
10397 |
0.16 |
| chr15_31250860_31251157 | 0.17 |
Dap |
death-associated protein |
17328 |
0.15 |
| chr7_66150558_66150709 | 0.17 |
Gm26827 |
predicted gene, 26827 |
22488 |
0.11 |
| chr10_88505415_88505608 | 0.17 |
Chpt1 |
choline phosphotransferase 1 |
1438 |
0.36 |
| chr8_22623689_22623868 | 0.17 |
Dkk4 |
dickkopf WNT signaling pathway inhibitor 4 |
265 |
0.89 |
| chr16_87627804_87628113 | 0.17 |
Gm22808 |
predicted gene, 22808 |
6838 |
0.21 |
| chr15_56746775_56746946 | 0.17 |
Has2os |
hyaluronan synthase 2, opposite strand |
18600 |
0.21 |
| chr13_107542484_107542646 | 0.17 |
Gm32004 |
predicted gene, 32004 |
22758 |
0.2 |
| chr13_24807889_24808047 | 0.17 |
Gm22358 |
predicted gene, 22358 |
3600 |
0.15 |
| chr9_74872523_74872674 | 0.17 |
Onecut1 |
one cut domain, family member 1 |
6114 |
0.16 |
| chr16_22076472_22076942 | 0.17 |
Igf2bp2 |
insulin-like growth factor 2 mRNA binding protein 2 |
2411 |
0.27 |
| chr5_106224232_106224439 | 0.17 |
Gm5987 |
predicted gene 5987 |
8684 |
0.21 |
| chr9_36726106_36726470 | 0.17 |
Chek1 |
checkpoint kinase 1 |
86 |
0.95 |
| chr11_11820349_11820865 | 0.17 |
Ddc |
dopa decarboxylase |
2314 |
0.29 |
| chr9_14061834_14061992 | 0.17 |
1700019J19Rik |
RIKEN cDNA 1700019J19 gene |
27707 |
0.17 |
| chr19_12697726_12697882 | 0.16 |
Keg1 |
kidney expressed gene 1 |
1990 |
0.18 |
| chr18_38176274_38176683 | 0.16 |
Pcdh1 |
protocadherin 1 |
26685 |
0.12 |
| chr7_39809662_39809816 | 0.16 |
4930558N11Rik |
RIKEN cDNA 4930558N11 gene |
11997 |
0.16 |
| chr15_62204340_62204858 | 0.16 |
Pvt1 |
Pvt1 oncogene |
13624 |
0.23 |
| chr3_79525088_79525251 | 0.16 |
Fnip2 |
folliculin interacting protein 2 |
6707 |
0.17 |
| chr17_12941646_12941826 | 0.16 |
Acat3 |
acetyl-Coenzyme A acetyltransferase 3 |
1136 |
0.27 |
| chr5_3375724_3375875 | 0.16 |
Cdk6 |
cyclin-dependent kinase 6 |
31487 |
0.16 |
| chr9_69204954_69205120 | 0.16 |
Rora |
RAR-related orphan receptor alpha |
9496 |
0.29 |
| chr4_6242659_6242849 | 0.16 |
Gm11798 |
predicted gene 11798 |
18211 |
0.19 |
| chr5_135036438_135036589 | 0.16 |
Stx1a |
syntaxin 1A (brain) |
998 |
0.3 |
| chrX_155151974_155152135 | 0.16 |
Gm4997 |
predicted gene 4997 |
743 |
0.48 |
| chr12_104341895_104342090 | 0.16 |
Serpina3k |
serine (or cysteine) peptidase inhibitor, clade A, member 3K |
3506 |
0.14 |
| chr3_133740753_133740929 | 0.16 |
Gm6135 |
prediticted gene 6135 |
50663 |
0.13 |
| chr11_111996515_111996723 | 0.16 |
Gm11679 |
predicted gene 11679 |
46959 |
0.19 |
| chr18_9504029_9504426 | 0.16 |
Gm7527 |
predicted gene 7527 |
8606 |
0.15 |
| chr4_44777019_44777170 | 0.16 |
Zcchc7 |
zinc finger, CCHC domain containing 7 |
16650 |
0.16 |
| chr18_61424158_61424549 | 0.16 |
Gm8755 |
predicted gene 8755 |
9848 |
0.12 |
| chr10_87883611_87883967 | 0.16 |
Igf1os |
insulin-like growth factor 1, opposite strand |
20408 |
0.18 |
| chr8_45362709_45362860 | 0.16 |
Fam149a |
family with sequence similarity 149, member A |
3919 |
0.19 |
| chr4_118031195_118031493 | 0.16 |
St3gal3 |
ST3 beta-galactoside alpha-2,3-sialyltransferase 3 |
538 |
0.78 |
| chr9_108729006_108729157 | 0.16 |
Gm24259 |
predicted gene, 24259 |
9777 |
0.1 |
| chr3_51252801_51253044 | 0.15 |
Elf2 |
E74-like factor 2 |
7319 |
0.13 |
| chr2_128972020_128972207 | 0.15 |
Gm10762 |
predicted gene 10762 |
4069 |
0.12 |
| chr5_33465811_33466048 | 0.15 |
Gm43851 |
predicted gene 43851 |
28465 |
0.15 |
| chr16_37591583_37591771 | 0.15 |
Gm46559 |
predicted gene, 46559 |
4513 |
0.17 |
| chr2_72206740_72207276 | 0.15 |
Rapgef4 |
Rap guanine nucleotide exchange factor (GEF) 4 |
8209 |
0.18 |
| chr7_65902304_65902456 | 0.15 |
Pcsk6 |
proprotein convertase subtilisin/kexin type 6 |
39948 |
0.14 |
| chr10_17260347_17260498 | 0.15 |
Gm47761 |
predicted gene, 47761 |
61189 |
0.13 |
| chr17_45570529_45570680 | 0.15 |
Hsp90ab1 |
heat shock protein 90 alpha (cytosolic), class B member 1 |
868 |
0.38 |
| chr5_125526040_125526191 | 0.15 |
Tmem132b |
transmembrane protein 132B |
5659 |
0.17 |
| chr17_62789514_62789722 | 0.15 |
Efna5 |
ephrin A5 |
91526 |
0.1 |
| chr2_104594636_104594787 | 0.15 |
Cstf3 |
cleavage stimulation factor, 3' pre-RNA, subunit 3 |
4093 |
0.15 |
| chr1_13561804_13561990 | 0.15 |
Tram1 |
translocating chain-associating membrane protein 1 |
17901 |
0.2 |
| chr15_34501373_34501524 | 0.15 |
Pop1 |
processing of precursor 1, ribonuclease P/MRP family, (S. cerevisiae) |
6125 |
0.12 |
| chr1_13322152_13322303 | 0.15 |
Gm23169 |
predicted gene, 23169 |
12382 |
0.11 |
| chr5_91213314_91213507 | 0.15 |
Gm23092 |
predicted gene, 23092 |
4594 |
0.26 |
| chr12_55440243_55440394 | 0.15 |
Psma6 |
proteasome subunit alpha 6 |
32935 |
0.13 |
| chr14_34339389_34339887 | 0.15 |
Glud1 |
glutamate dehydrogenase 1 |
4266 |
0.12 |
| chr18_33337551_33337772 | 0.15 |
Gm5503 |
predicted gene 5503 |
47294 |
0.17 |
| chr4_53221393_53221544 | 0.15 |
4930412L05Rik |
RIKEN cDNA 4930412L05 gene |
3611 |
0.22 |
| chr13_112314994_112315164 | 0.15 |
Ankrd55 |
ankyrin repeat domain 55 |
3375 |
0.23 |
| chr3_18165162_18165350 | 0.15 |
Gm23686 |
predicted gene, 23686 |
12369 |
0.23 |
| chr1_195223205_195223404 | 0.15 |
Gm37887 |
predicted gene, 37887 |
538 |
0.73 |
| chr9_55280024_55280330 | 0.15 |
Nrg4 |
neuregulin 4 |
3395 |
0.23 |
| chr13_75387054_75387205 | 0.15 |
Gm48234 |
predicted gene, 48234 |
92258 |
0.07 |
| chr1_23237656_23237879 | 0.15 |
Gm29506 |
predicted gene 29506 |
2094 |
0.23 |
| chr8_84841387_84841538 | 0.15 |
Rad23a |
RAD23 homolog A, nucleotide excision repair protein |
797 |
0.35 |
| chr15_34501830_34501985 | 0.15 |
Pop1 |
processing of precursor 1, ribonuclease P/MRP family, (S. cerevisiae) |
6584 |
0.12 |
| chr9_123296355_123296514 | 0.15 |
Gm47162 |
predicted gene, 47162 |
10650 |
0.15 |
| chr3_129450558_129450716 | 0.15 |
Rpl7a-ps7 |
ribosomal protein L7A, pseudogene 7 |
11335 |
0.17 |
| chr2_31489958_31490302 | 0.14 |
Ass1 |
argininosuccinate synthetase 1 |
7642 |
0.19 |
| chr14_21435070_21435221 | 0.14 |
Gm25864 |
predicted gene, 25864 |
15329 |
0.19 |
| chr12_45067249_45067400 | 0.14 |
Stxbp6 |
syntaxin binding protein 6 (amisyn) |
6788 |
0.22 |
| chr11_52374066_52374363 | 0.14 |
Vdac1 |
voltage-dependent anion channel 1 |
49 |
0.97 |
| chr1_164543147_164543444 | 0.14 |
D630023O14Rik |
RIKEN cDNA D630023O14 gene |
32160 |
0.13 |
| chr1_67215139_67215608 | 0.14 |
Gm15668 |
predicted gene 15668 |
33827 |
0.17 |
| chr3_118443694_118443845 | 0.14 |
Gm9916 |
predicted gene 9916 |
8999 |
0.15 |
| chr10_128526597_128526748 | 0.14 |
Esyt1 |
extended synaptotagmin-like protein 1 |
801 |
0.3 |
| chr6_149225033_149225369 | 0.14 |
1700003I16Rik |
RIKEN cDNA 1700003I16 gene |
10016 |
0.16 |
| chr15_58983651_58983950 | 0.14 |
4930544F09Rik |
RIKEN cDNA 4930544F09 gene |
336 |
0.87 |
| chr14_7834896_7835063 | 0.14 |
Flnb |
filamin, beta |
17022 |
0.15 |
| chr2_85021399_85021563 | 0.14 |
P2rx3 |
purinergic receptor P2X, ligand-gated ion channel, 3 |
11713 |
0.11 |
| chr4_45800098_45800462 | 0.14 |
Aldh1b1 |
aldehyde dehydrogenase 1 family, member B1 |
1159 |
0.41 |
| chr4_120465821_120465983 | 0.14 |
Scmh1 |
sex comb on midleg homolog 1 |
3822 |
0.27 |
| chrX_169974457_169974608 | 0.14 |
Mid1 |
midline 1 |
727 |
0.6 |
| chr6_58641968_58642181 | 0.14 |
Abcg2 |
ATP binding cassette subfamily G member 2 (Junior blood group) |
1492 |
0.46 |
| chr19_38683120_38683271 | 0.14 |
Gm8717 |
predicted gene 8717 |
21129 |
0.19 |
| chr6_71999696_71999905 | 0.14 |
Gm26628 |
predicted gene, 26628 |
36025 |
0.1 |
| chr18_54739961_54740121 | 0.14 |
Gm5821 |
predicted gene 5821 |
26091 |
0.21 |
| chr8_74993299_74993474 | 0.14 |
Hmgxb4 |
HMG box domain containing 4 |
30 |
0.98 |
| chr17_81386211_81386362 | 0.14 |
Gm50044 |
predicted gene, 50044 |
15453 |
0.24 |
| chr1_21247881_21248070 | 0.14 |
Gsta3 |
glutathione S-transferase, alpha 3 |
5546 |
0.12 |
| chr4_108060521_108060686 | 0.14 |
Scp2 |
sterol carrier protein 2, liver |
10760 |
0.13 |
| chr13_44445691_44446120 | 0.14 |
Gm27036 |
predicted gene, 27036 |
6092 |
0.16 |
| chr10_52420605_52420773 | 0.14 |
Nus1 |
NUS1 dehydrodolichyl diphosphate synthase subunit |
3142 |
0.15 |
| chr6_24609749_24609900 | 0.14 |
Lmod2 |
leiomodin 2 (cardiac) |
12062 |
0.14 |
| chr9_24922429_24922589 | 0.14 |
Gm48255 |
predicted gene, 48255 |
30464 |
0.13 |
| chr3_107256063_107256233 | 0.14 |
Prok1 |
prokineticin 1 |
16441 |
0.12 |
| chr2_148023249_148023719 | 0.14 |
9030622O22Rik |
RIKEN cDNA 9030622O22 gene |
14786 |
0.17 |
| chr5_115515109_115515270 | 0.14 |
Gm13840 |
predicted gene 13840 |
74 |
0.92 |
| chr10_28454512_28454940 | 0.14 |
Ptprk |
protein tyrosine phosphatase, receptor type, K |
105425 |
0.07 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0030327 | prenylated protein catabolic process(GO:0030327) |
| 0.1 | 0.2 | GO:0010046 | response to mycotoxin(GO:0010046) |
| 0.1 | 0.2 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.1 | 0.3 | GO:1903964 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.1 | 0.2 | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.1 | 0.2 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.1 | 0.2 | GO:2000211 | regulation of glutamate metabolic process(GO:2000211) |
| 0.1 | 0.2 | GO:0060741 | prostate gland stromal morphogenesis(GO:0060741) |
| 0.0 | 0.1 | GO:0035860 | glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.0 | 0.2 | GO:0019856 | 'de novo' pyrimidine nucleobase biosynthetic process(GO:0006207) pyrimidine nucleobase biosynthetic process(GO:0019856) |
| 0.0 | 0.2 | GO:0006166 | purine ribonucleoside salvage(GO:0006166) |
| 0.0 | 0.1 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.0 | 0.1 | GO:0051081 | membrane disassembly(GO:0030397) nuclear envelope disassembly(GO:0051081) |
| 0.0 | 0.1 | GO:0006059 | hexitol metabolic process(GO:0006059) |
| 0.0 | 0.1 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.0 | 0.1 | GO:0046469 | platelet activating factor biosynthetic process(GO:0006663) platelet activating factor metabolic process(GO:0046469) |
| 0.0 | 0.2 | GO:0006975 | DNA damage induced protein phosphorylation(GO:0006975) |
| 0.0 | 0.1 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.0 | 0.1 | GO:0072338 | creatinine metabolic process(GO:0046449) cellular lactam metabolic process(GO:0072338) |
| 0.0 | 0.1 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.0 | 0.1 | GO:0002309 | T cell proliferation involved in immune response(GO:0002309) |
| 0.0 | 0.1 | GO:0032383 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.0 | 0.1 | GO:0021778 | oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.0 | 0.3 | GO:0009404 | toxin metabolic process(GO:0009404) |
| 0.0 | 0.1 | GO:1902915 | negative regulation of protein K63-linked ubiquitination(GO:1900045) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.0 | 0.0 | GO:0006533 | aspartate catabolic process(GO:0006533) |
| 0.0 | 0.1 | GO:0046167 | glycerol-3-phosphate biosynthetic process(GO:0046167) |
| 0.0 | 0.1 | GO:1901162 | serotonin biosynthetic process(GO:0042427) primary amino compound biosynthetic process(GO:1901162) |
| 0.0 | 0.1 | GO:0002282 | microglial cell activation involved in immune response(GO:0002282) |
| 0.0 | 0.1 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.0 | 0.1 | GO:0051799 | negative regulation of hair follicle development(GO:0051799) |
| 0.0 | 0.1 | GO:0015959 | diadenosine polyphosphate metabolic process(GO:0015959) |
| 0.0 | 0.1 | GO:0001994 | norepinephrine-epinephrine vasoconstriction involved in regulation of systemic arterial blood pressure(GO:0001994) |
| 0.0 | 0.0 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.0 | 0.1 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.0 | 0.1 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.0 | 0.1 | GO:0006987 | activation of signaling protein activity involved in unfolded protein response(GO:0006987) |
| 0.0 | 0.1 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.1 | GO:0072107 | regulation of ureteric bud formation(GO:0072106) positive regulation of ureteric bud formation(GO:0072107) |
| 0.0 | 0.1 | GO:0034310 | primary alcohol catabolic process(GO:0034310) |
| 0.0 | 0.1 | GO:0034755 | iron ion transmembrane transport(GO:0034755) |
| 0.0 | 0.1 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.0 | 0.1 | GO:0006982 | response to lipid hydroperoxide(GO:0006982) |
| 0.0 | 0.0 | GO:0061152 | trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 0.0 | 0.1 | GO:0042695 | thelarche(GO:0042695) mammary gland branching involved in thelarche(GO:0060744) |
| 0.0 | 0.1 | GO:2001286 | regulation of caveolin-mediated endocytosis(GO:2001286) |
| 0.0 | 0.1 | GO:0016078 | tRNA catabolic process(GO:0016078) |
| 0.0 | 0.1 | GO:1901740 | negative regulation of myoblast fusion(GO:1901740) |
| 0.0 | 0.0 | GO:0044337 | canonical Wnt signaling pathway involved in positive regulation of apoptotic process(GO:0044337) |
| 0.0 | 0.1 | GO:0006538 | glutamate catabolic process(GO:0006538) |
| 0.0 | 0.1 | GO:0000459 | exonucleolytic trimming involved in rRNA processing(GO:0000459) exonucleolytic trimming to generate mature 3'-end of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000467) |
| 0.0 | 0.1 | GO:1901078 | negative regulation of relaxation of muscle(GO:1901078) |
| 0.0 | 0.2 | GO:0043651 | linoleic acid metabolic process(GO:0043651) |
| 0.0 | 0.1 | GO:1901525 | negative regulation of macromitophagy(GO:1901525) |
| 0.0 | 0.1 | GO:0006208 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) thymine metabolic process(GO:0019859) |
| 0.0 | 0.1 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.0 | 0.0 | GO:0046083 | adenine metabolic process(GO:0046083) adenine biosynthetic process(GO:0046084) |
| 0.0 | 0.1 | GO:0036112 | medium-chain fatty-acyl-CoA metabolic process(GO:0036112) |
| 0.0 | 0.1 | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 0.0 | 0.1 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.0 | 0.1 | GO:0021524 | visceral motor neuron differentiation(GO:0021524) |
| 0.0 | 0.2 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.0 | 0.2 | GO:0033147 | negative regulation of intracellular estrogen receptor signaling pathway(GO:0033147) |
| 0.0 | 0.1 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.0 | 0.1 | GO:1990086 | lens fiber cell apoptotic process(GO:1990086) |
| 0.0 | 0.1 | GO:0060750 | epithelial cell proliferation involved in mammary gland duct elongation(GO:0060750) |
| 0.0 | 0.1 | GO:0015871 | choline transport(GO:0015871) |
| 0.0 | 0.1 | GO:1902990 | mitotic telomere maintenance via semi-conservative replication(GO:1902990) |
| 0.0 | 0.1 | GO:0006203 | dGTP catabolic process(GO:0006203) |
| 0.0 | 0.1 | GO:0043102 | amino acid salvage(GO:0043102) L-methionine biosynthetic process(GO:0071265) L-methionine salvage(GO:0071267) |
| 0.0 | 0.0 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 0.1 | GO:0021943 | formation of radial glial scaffolds(GO:0021943) |
| 0.0 | 0.1 | GO:0042518 | negative regulation of tyrosine phosphorylation of Stat3 protein(GO:0042518) |
| 0.0 | 0.0 | GO:1903774 | positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 0.0 | 0.0 | GO:0048022 | negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) |
| 0.0 | 0.0 | GO:0070669 | response to interleukin-2(GO:0070669) |
| 0.0 | 0.0 | GO:0090071 | negative regulation of ribosome biogenesis(GO:0090071) |
| 0.0 | 0.0 | GO:1904017 | response to Thyroglobulin triiodothyronine(GO:1904016) cellular response to Thyroglobulin triiodothyronine(GO:1904017) |
| 0.0 | 0.0 | GO:0060125 | negative regulation of growth hormone secretion(GO:0060125) |
| 0.0 | 0.1 | GO:0006559 | L-phenylalanine catabolic process(GO:0006559) erythrose 4-phosphate/phosphoenolpyruvate family amino acid catabolic process(GO:1902222) |
| 0.0 | 0.1 | GO:0042737 | drug catabolic process(GO:0042737) |
| 0.0 | 0.0 | GO:0006106 | fumarate metabolic process(GO:0006106) |
| 0.0 | 0.1 | GO:1900103 | positive regulation of endoplasmic reticulum unfolded protein response(GO:1900103) |
| 0.0 | 0.0 | GO:0046874 | quinolinate metabolic process(GO:0046874) |
| 0.0 | 0.0 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 0.0 | 0.1 | GO:0046618 | drug export(GO:0046618) |
| 0.0 | 0.0 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 0.0 | GO:1904154 | positive regulation of retrograde protein transport, ER to cytosol(GO:1904154) |
| 0.0 | 0.0 | GO:0046098 | guanine metabolic process(GO:0046098) |
| 0.0 | 0.0 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.0 | 0.1 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.0 | 0.0 | GO:2000619 | negative regulation of histone H4-K16 acetylation(GO:2000619) |
| 0.0 | 0.1 | GO:0042997 | negative regulation of Golgi to plasma membrane protein transport(GO:0042997) |
| 0.0 | 0.1 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.0 | 0.0 | GO:0003162 | atrioventricular node development(GO:0003162) |
| 0.0 | 0.0 | GO:0009629 | response to gravity(GO:0009629) |
| 0.0 | 0.0 | GO:0034242 | negative regulation of syncytium formation by plasma membrane fusion(GO:0034242) |
| 0.0 | 0.1 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.0 | 0.0 | GO:1900126 | negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.0 | 0.1 | GO:0051561 | positive regulation of mitochondrial calcium ion concentration(GO:0051561) |
| 0.0 | 0.1 | GO:0006083 | acetate metabolic process(GO:0006083) |
| 0.0 | 0.0 | GO:0032058 | positive regulation of translational initiation in response to stress(GO:0032058) |
| 0.0 | 0.1 | GO:0006851 | mitochondrial calcium ion transport(GO:0006851) |
| 0.0 | 0.0 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.0 | 0.1 | GO:0006107 | oxaloacetate metabolic process(GO:0006107) |
| 0.0 | 0.0 | GO:0070672 | response to interleukin-15(GO:0070672) |
| 0.0 | 0.0 | GO:0006481 | C-terminal protein methylation(GO:0006481) |
| 0.0 | 0.0 | GO:1900625 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.0 | 0.0 | GO:2000574 | regulation of microtubule motor activity(GO:2000574) |
| 0.0 | 0.1 | GO:0045900 | negative regulation of translational elongation(GO:0045900) |
| 0.0 | 0.0 | GO:0000429 | carbon catabolite regulation of transcription from RNA polymerase II promoter(GO:0000429) regulation of transcription from RNA polymerase II promoter by glucose(GO:0000430) positive regulation of transcription from RNA polymerase II promoter by glucose(GO:0000432) carbon catabolite activation of transcription from RNA polymerase II promoter(GO:0000436) |
| 0.0 | 0.0 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.0 | 0.1 | GO:0060836 | lymphatic endothelial cell differentiation(GO:0060836) |
| 0.0 | 0.0 | GO:0070340 | detection of bacterial lipopeptide(GO:0070340) |
| 0.0 | 0.0 | GO:0097680 | double-strand break repair via classical nonhomologous end joining(GO:0097680) |
| 0.0 | 0.0 | GO:1990928 | response to amino acid starvation(GO:1990928) |
| 0.0 | 0.0 | GO:1903071 | positive regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903071) |
| 0.0 | 0.1 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.0 | 0.0 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.0 | GO:0006768 | biotin metabolic process(GO:0006768) |
| 0.0 | 0.0 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 0.0 | 0.1 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 0.1 | GO:2000370 | positive regulation of clathrin-mediated endocytosis(GO:2000370) |
| 0.0 | 0.1 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.0 | 0.1 | GO:0045908 | negative regulation of vasodilation(GO:0045908) |
| 0.0 | 0.0 | GO:0018211 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.0 | 0.1 | GO:0046218 | tryptophan catabolic process(GO:0006569) tryptophan catabolic process to kynurenine(GO:0019441) indole-containing compound catabolic process(GO:0042436) indolalkylamine catabolic process(GO:0046218) |
| 0.0 | 0.0 | GO:0035672 | oligopeptide transmembrane transport(GO:0035672) |
| 0.0 | 0.0 | GO:1903347 | negative regulation of bicellular tight junction assembly(GO:1903347) |
| 0.0 | 0.0 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.0 | 0.1 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.0 | 0.0 | GO:0034475 | U4 snRNA 3'-end processing(GO:0034475) polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.0 | 0.0 | GO:0019086 | late viral transcription(GO:0019086) |
| 0.0 | 0.1 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) |
| 0.0 | 0.1 | GO:0038093 | Fc receptor signaling pathway(GO:0038093) |
| 0.0 | 0.1 | GO:0036315 | cellular response to sterol(GO:0036315) |
| 0.0 | 0.0 | GO:2000664 | positive regulation of interleukin-5 secretion(GO:2000664) positive regulation of interleukin-13 secretion(GO:2000667) |
| 0.0 | 0.1 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.0 | 0.0 | GO:0086023 | adrenergic receptor signaling pathway involved in heart process(GO:0086023) |
| 0.0 | 0.0 | GO:0009957 | epidermal cell fate specification(GO:0009957) |
| 0.0 | 0.0 | GO:0032741 | positive regulation of interleukin-18 production(GO:0032741) |
| 0.0 | 0.0 | GO:0090283 | regulation of protein glycosylation in Golgi(GO:0090283) |
| 0.0 | 0.1 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.0 | 0.0 | GO:2000850 | negative regulation of steroid hormone secretion(GO:2000832) negative regulation of corticosteroid hormone secretion(GO:2000847) negative regulation of glucocorticoid secretion(GO:2000850) |
| 0.0 | 0.2 | GO:0045475 | locomotor rhythm(GO:0045475) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.0 | 0.1 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.0 | 0.1 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.0 | 0.1 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.0 | 0.1 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.0 | 0.1 | GO:0042567 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) insulin-like growth factor ternary complex(GO:0042567) |
| 0.0 | 0.1 | GO:0098536 | deuterosome(GO:0098536) |
| 0.0 | 0.1 | GO:0034666 | integrin alpha2-beta1 complex(GO:0034666) |
| 0.0 | 0.1 | GO:0097413 | Lewy body(GO:0097413) |
| 0.0 | 0.1 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.1 | GO:0009279 | cell outer membrane(GO:0009279) cell envelope(GO:0030313) external encapsulating structure part(GO:0044462) |
| 0.0 | 0.1 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.2 | GO:0046930 | pore complex(GO:0046930) |
| 0.0 | 0.1 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.0 | 0.1 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.0 | 0.1 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.0 | 0.2 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.1 | GO:0031094 | platelet dense tubular network(GO:0031094) |
| 0.0 | 0.1 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.1 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.0 | 0.1 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.0 | GO:0035355 | Toll-like receptor 2-Toll-like receptor 6 protein complex(GO:0035355) |
| 0.0 | 0.0 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.0 | 0.1 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.0 | 0.1 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.0 | 0.1 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.0 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.0 | 0.1 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.0 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0016743 | carboxyl- or carbamoyltransferase activity(GO:0016743) |
| 0.1 | 0.2 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.1 | 0.3 | GO:0032896 | palmitoyl-CoA 9-desaturase activity(GO:0032896) |
| 0.0 | 0.4 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.1 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.1 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.0 | 0.1 | GO:0038181 | bile acid receptor activity(GO:0038181) |
| 0.0 | 0.1 | GO:0016823 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.0 | 0.1 | GO:0043125 | ErbB-3 class receptor binding(GO:0043125) |
| 0.0 | 0.1 | GO:0071614 | linoleic acid epoxygenase activity(GO:0071614) |
| 0.0 | 0.2 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 0.1 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.0 | 0.1 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 0.1 | GO:0009374 | biotin binding(GO:0009374) |
| 0.0 | 0.1 | GO:0000171 | ribonuclease MRP activity(GO:0000171) |
| 0.0 | 0.1 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.0 | 0.1 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.0 | 0.1 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 0.1 | GO:0008898 | S-adenosylmethionine-homocysteine S-methyltransferase activity(GO:0008898) |
| 0.0 | 0.1 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.0 | 0.2 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.1 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.0 | 0.1 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.0 | 0.1 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.0 | 0.3 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.1 | GO:0031697 | beta-1 adrenergic receptor binding(GO:0031697) |
| 0.0 | 0.1 | GO:0004370 | glycerol kinase activity(GO:0004370) |
| 0.0 | 0.1 | GO:0047023 | androsterone dehydrogenase activity(GO:0047023) |
| 0.0 | 0.1 | GO:0055100 | adiponectin binding(GO:0055100) |
| 0.0 | 0.1 | GO:0005119 | smoothened binding(GO:0005119) |
| 0.0 | 0.1 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.0 | 0.1 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 0.0 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.0 | 0.1 | GO:0032139 | dinucleotide insertion or deletion binding(GO:0032139) |
| 0.0 | 0.1 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.0 | 0.1 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 0.0 | 0.1 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.0 | 0.1 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.0 | 0.1 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.0 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.0 | 0.1 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.0 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.1 | GO:0043426 | MRF binding(GO:0043426) |
| 0.0 | 0.1 | GO:0016453 | acetyl-CoA C-acetyltransferase activity(GO:0003985) C-acetyltransferase activity(GO:0016453) |
| 0.0 | 0.1 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 0.1 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.0 | GO:0030620 | U2 snRNA binding(GO:0030620) |
| 0.0 | 0.1 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.0 | 0.1 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.0 | 0.0 | GO:0035663 | Toll-like receptor 2 binding(GO:0035663) |
| 0.0 | 0.1 | GO:0019763 | immunoglobulin receptor activity(GO:0019763) |
| 0.0 | 0.1 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 0.0 | GO:0034648 | histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.0 | 0.1 | GO:0098518 | polynucleotide phosphatase activity(GO:0098518) |
| 0.0 | 0.0 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.0 | 0.0 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.0 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.0 | 0.1 | GO:0047760 | butyrate-CoA ligase activity(GO:0047760) |
| 0.0 | 0.0 | GO:0004075 | biotin carboxylase activity(GO:0004075) |
| 0.0 | 0.1 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 0.0 | 0.2 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.1 | GO:0003847 | 1-alkyl-2-acetylglycerophosphocholine esterase activity(GO:0003847) |
| 0.0 | 0.1 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 0.1 | GO:0016151 | nickel cation binding(GO:0016151) |
| 0.0 | 0.1 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.0 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.0 | 0.0 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.0 | 0.2 | GO:0043295 | glutathione binding(GO:0043295) oligopeptide binding(GO:1900750) |
| 0.0 | 0.0 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.0 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.0 | 0.3 | GO:0019206 | nucleoside kinase activity(GO:0019206) |
| 0.0 | 0.0 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.0 | 0.1 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.0 | 0.1 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.1 | GO:0002054 | nucleobase binding(GO:0002054) |
| 0.0 | 0.2 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.0 | GO:0016880 | acid-ammonia (or amide) ligase activity(GO:0016880) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.3 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.0 | REACTOME NRIF SIGNALS CELL DEATH FROM THE NUCLEUS | Genes involved in NRIF signals cell death from the nucleus |
| 0.0 | 0.2 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 0.2 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.0 | 0.2 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.1 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.1 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.0 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 0.1 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.1 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.1 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.0 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.1 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.1 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |