Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
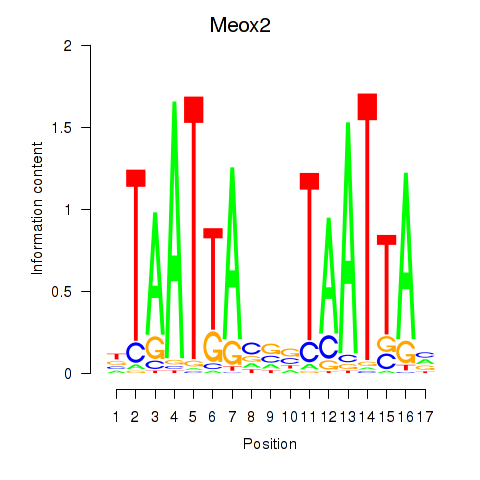
Results for Meox2
Z-value: 1.90
Transcription factors associated with Meox2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Meox2
|
ENSMUSG00000036144.5 | mesenchyme homeobox 2 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr12_37167805_37167956 | Meox2 | 59340 | 0.112105 | -0.41 | 4.2e-01 | Click! |
Activity of the Meox2 motif across conditions
Conditions sorted by the z-value of the Meox2 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr1_21250942_21251436 | 1.04 |
Gsta3 |
glutathione S-transferase, alpha 3 |
2332 |
0.18 |
| chr3_97635501_97635936 | 0.99 |
Fmo5 |
flavin containing monooxygenase 5 |
6835 |
0.14 |
| chr3_81955296_81955447 | 0.99 |
Ctso |
cathepsin O |
4434 |
0.18 |
| chr18_86383968_86384119 | 0.89 |
Neto1 |
neuropilin (NRP) and tolloid (TLL)-like 1 |
10909 |
0.24 |
| chr9_122148667_122148818 | 0.84 |
Gm47121 |
predicted gene, 47121 |
6293 |
0.13 |
| chr1_140421414_140421565 | 0.84 |
Kcnt2 |
potassium channel, subfamily T, member 2 |
21447 |
0.27 |
| chr1_80244319_80244685 | 0.84 |
Gm37932 |
predicted gene, 37932 |
11695 |
0.13 |
| chr14_30542112_30542288 | 0.76 |
Tkt |
transketolase |
6159 |
0.16 |
| chr10_69208679_69208830 | 0.76 |
Rhobtb1 |
Rho-related BTB domain containing 1 |
202 |
0.94 |
| chr1_97770980_97771325 | 0.73 |
Ppip5k2 |
diphosphoinositol pentakisphosphate kinase 2 |
741 |
0.49 |
| chr8_93185394_93185584 | 0.73 |
Gm45909 |
predicted gene 45909 |
5869 |
0.14 |
| chr8_109981175_109981347 | 0.73 |
Gm45795 |
predicted gene 45795 |
6909 |
0.13 |
| chr5_87589802_87589979 | 0.72 |
Sult1e1 |
sulfotransferase family 1E, member 1 |
1704 |
0.22 |
| chr1_23237656_23237879 | 0.70 |
Gm29506 |
predicted gene 29506 |
2094 |
0.23 |
| chr4_88893074_88893256 | 0.69 |
Ifne |
interferon epsilon |
12964 |
0.08 |
| chr6_108836766_108836917 | 0.64 |
Edem1 |
ER degradation enhancer, mannosidase alpha-like 1 |
431 |
0.86 |
| chr13_93719775_93719928 | 0.64 |
AW495222 |
expressed sequence AW495222 |
676 |
0.6 |
| chr11_28689224_28689493 | 0.64 |
2810471M01Rik |
RIKEN cDNA 2810471M01 gene |
7794 |
0.19 |
| chr1_156978437_156978634 | 0.63 |
4930439D14Rik |
RIKEN cDNA 4930439D14 gene |
38697 |
0.12 |
| chr13_104044189_104044340 | 0.63 |
Nln |
neurolysin (metallopeptidase M3 family) |
6622 |
0.22 |
| chr1_67162712_67162908 | 0.63 |
Cps1 |
carbamoyl-phosphate synthetase 1 |
39784 |
0.16 |
| chr1_67165771_67165961 | 0.62 |
Cps1 |
carbamoyl-phosphate synthetase 1 |
42840 |
0.16 |
| chr16_42998647_42998825 | 0.61 |
Ndufs5-ps |
NADH:ubiquinone oxidoreductase core subunit S5, pseudogene |
43105 |
0.16 |
| chr1_160781397_160781614 | 0.61 |
Rabgap1l |
RAB GTPase activating protein 1-like |
11433 |
0.12 |
| chr6_58611375_58611579 | 0.60 |
Abcg2 |
ATP binding cassette subfamily G member 2 (Junior blood group) |
14806 |
0.19 |
| chr3_112120839_112120990 | 0.60 |
Gm25519 |
predicted gene, 25519 |
78127 |
0.1 |
| chr7_114361745_114361949 | 0.59 |
4933406I18Rik |
RIKEN cDNA 4933406I18 gene |
53174 |
0.12 |
| chr13_4077186_4077340 | 0.59 |
Akr1c14 |
aldo-keto reductase family 1, member C14 |
991 |
0.44 |
| chr2_93821900_93822368 | 0.59 |
Ext2 |
exostosin glycosyltransferase 2 |
411 |
0.82 |
| chr4_15029409_15029632 | 0.59 |
Gm11844 |
predicted gene 11844 |
6938 |
0.27 |
| chr11_52374066_52374363 | 0.56 |
Vdac1 |
voltage-dependent anion channel 1 |
49 |
0.97 |
| chr4_108060521_108060686 | 0.56 |
Scp2 |
sterol carrier protein 2, liver |
10760 |
0.13 |
| chr17_80022007_80022158 | 0.56 |
Gm22215 |
predicted gene, 22215 |
11432 |
0.14 |
| chr11_28694681_28694832 | 0.55 |
2810471M01Rik |
RIKEN cDNA 2810471M01 gene |
13192 |
0.17 |
| chr13_93623443_93623622 | 0.54 |
Gm15622 |
predicted gene 15622 |
1850 |
0.3 |
| chr2_160805166_160805505 | 0.54 |
Gm11447 |
predicted gene 11447 |
40278 |
0.11 |
| chr16_7049906_7050057 | 0.54 |
Rbfox1 |
RNA binding protein, fox-1 homolog (C. elegans) 1 |
19865 |
0.3 |
| chr14_45659577_45659728 | 0.54 |
Ddhd1 |
DDHD domain containing 1 |
1509 |
0.23 |
| chr12_80336463_80336614 | 0.54 |
Dcaf5 |
DDB1 and CUL4 associated factor 5 |
40154 |
0.1 |
| chr6_120318643_120318978 | 0.54 |
B4galnt3 |
beta-1,4-N-acetyl-galactosaminyl transferase 3 |
24251 |
0.15 |
| chr3_107690573_107690757 | 0.54 |
Ahcyl1 |
S-adenosylhomocysteine hydrolase-like 1 |
5298 |
0.18 |
| chr18_75675824_75676322 | 0.53 |
Ctif |
CBP80/20-dependent translation initiation factor |
21481 |
0.23 |
| chr17_43672210_43672361 | 0.53 |
Cyp39a1 |
cytochrome P450, family 39, subfamily a, polypeptide 1 |
4760 |
0.18 |
| chr16_26694594_26694745 | 0.53 |
Il1rap |
interleukin 1 receptor accessory protein |
27765 |
0.23 |
| chr8_109995374_109995556 | 0.53 |
Tat |
tyrosine aminotransferase |
4959 |
0.13 |
| chr3_144110488_144110677 | 0.52 |
Gm34078 |
predicted gene, 34078 |
25172 |
0.2 |
| chr6_4820679_4820851 | 0.52 |
Gm20714 |
predicted gene 20714 |
4436 |
0.18 |
| chr19_21158185_21158336 | 0.52 |
AC106834.1 |
novel transcript |
12581 |
0.21 |
| chr11_88618341_88618520 | 0.51 |
Msi2 |
musashi RNA-binding protein 2 |
28283 |
0.2 |
| chr12_73861884_73862127 | 0.51 |
Gm15283 |
predicted gene 15283 |
7607 |
0.18 |
| chr8_56345237_56345388 | 0.51 |
Gm45540 |
predicted gene 45540 |
34008 |
0.18 |
| chr18_38176274_38176683 | 0.51 |
Pcdh1 |
protocadherin 1 |
26685 |
0.12 |
| chr3_5425056_5425268 | 0.51 |
4930555M17Rik |
RIKEN cDNA 4930555M17 gene |
90171 |
0.09 |
| chr17_45879729_45879910 | 0.50 |
4930542M03Rik |
RIKEN cDNA 4930542M03 gene |
1633 |
0.33 |
| chr13_89583759_89583911 | 0.50 |
Hapln1 |
hyaluronan and proteoglycan link protein 1 |
676 |
0.73 |
| chr8_86599602_86599753 | 0.50 |
Gm44348 |
predicted gene, 44348 |
12715 |
0.16 |
| chr2_84379041_84379217 | 0.50 |
Calcrl |
calcitonin receptor-like |
3771 |
0.25 |
| chr16_37874310_37874496 | 0.50 |
Lrrc58 |
leucine rich repeat containing 58 |
6014 |
0.14 |
| chr3_51204305_51204466 | 0.49 |
Noct |
nocturnin |
20062 |
0.14 |
| chr9_47348407_47348558 | 0.49 |
Gm31816 |
predicted gene, 31816 |
14268 |
0.26 |
| chr13_52771924_52772129 | 0.49 |
BB123696 |
expressed sequence BB123696 |
14821 |
0.27 |
| chr3_111102424_111102592 | 0.48 |
Gm43407 |
predicted gene 43407 |
5992 |
0.34 |
| chr7_118146005_118146156 | 0.48 |
Smg1 |
SMG1 homolog, phosphatidylinositol 3-kinase-related kinase (C. elegans) |
4281 |
0.18 |
| chr8_104787830_104787982 | 0.47 |
Gm45782 |
predicted gene 45782 |
2078 |
0.17 |
| chr2_64094972_64095229 | 0.47 |
Fign |
fidgetin |
2888 |
0.42 |
| chr4_139111685_139111987 | 0.47 |
Micos10 |
mitochondrial contact site and cristae organizing system subunit 10 |
593 |
0.66 |
| chr3_12608407_12608567 | 0.47 |
Gm2429 |
predicted gene 2429 |
22773 |
0.28 |
| chr11_31875210_31875374 | 0.46 |
Cpeb4 |
cytoplasmic polyadenylation element binding protein 4 |
2017 |
0.35 |
| chr4_53395599_53395871 | 0.46 |
Gm12496 |
predicted gene 12496 |
8295 |
0.23 |
| chr2_10369756_10369912 | 0.46 |
Sfmbt2 |
Scm-like with four mbt domains 2 |
676 |
0.33 |
| chr10_87040962_87041138 | 0.46 |
1700113H08Rik |
RIKEN cDNA 1700113H08 gene |
16995 |
0.16 |
| chr4_82355326_82355477 | 0.46 |
n-R5s188 |
nuclear encoded rRNA 5S 188 |
84009 |
0.1 |
| chr18_61841168_61841343 | 0.46 |
Afap1l1 |
actin filament associated protein 1-like 1 |
54553 |
0.1 |
| chr1_180179778_180180226 | 0.46 |
Coq8a |
coenzyme Q8A |
1080 |
0.44 |
| chr19_44401346_44401497 | 0.45 |
Scd1 |
stearoyl-Coenzyme A desaturase 1 |
5269 |
0.16 |
| chr17_32493567_32493723 | 0.45 |
Cyp4f39 |
cytochrome P450, family 4, subfamily f, polypeptide 39 |
1487 |
0.3 |
| chr8_100855820_100856252 | 0.45 |
Gm39232 |
predicted gene, 39232 |
129681 |
0.06 |
| chr13_108346244_108346395 | 0.45 |
Depdc1b |
DEP domain containing 1B |
11107 |
0.19 |
| chr13_93626581_93626763 | 0.45 |
Gm15622 |
predicted gene 15622 |
1290 |
0.4 |
| chr1_97603062_97603228 | 0.45 |
AC099860.1 |
proline rich protein BstNI subfamily 4 (PRB4), pseudogene |
57686 |
0.11 |
| chr10_89492629_89492900 | 0.44 |
Nr1h4 |
nuclear receptor subfamily 1, group H, member 4 |
13885 |
0.21 |
| chr7_139684440_139684595 | 0.44 |
Cfap46 |
cilia and flagella associated protein 46 |
700 |
0.66 |
| chr1_65167472_65167777 | 0.44 |
Idh1 |
isocitrate dehydrogenase 1 (NADP+), soluble |
7940 |
0.14 |
| chr9_107853667_107853838 | 0.44 |
Rbm6 |
RNA binding motif protein 6 |
2319 |
0.17 |
| chr2_19659363_19659514 | 0.44 |
Otud1 |
OTU domain containing 1 |
1686 |
0.26 |
| chr3_35169015_35169202 | 0.44 |
1700017M07Rik |
RIKEN cDNA 1700017M07 gene |
107023 |
0.07 |
| chr3_131638032_131638214 | 0.44 |
Gm42881 |
predicted gene 42881 |
24343 |
0.22 |
| chr3_51437862_51438013 | 0.44 |
Naa15 |
N(alpha)-acetyltransferase 15, NatA auxiliary subunit |
2980 |
0.14 |
| chr14_21161568_21161719 | 0.44 |
Adk |
adenosine kinase |
85491 |
0.09 |
| chr11_17745312_17745477 | 0.44 |
Gm12016 |
predicted gene 12016 |
106211 |
0.07 |
| chr18_39483316_39483539 | 0.44 |
Nr3c1 |
nuclear receptor subfamily 3, group C, member 1 |
3805 |
0.31 |
| chr10_13985883_13986077 | 0.44 |
Hivep2 |
human immunodeficiency virus type I enhancer binding protein 2 |
18956 |
0.19 |
| chr15_62121397_62121548 | 0.44 |
Pvt1 |
Pvt1 oncogene |
22443 |
0.25 |
| chr10_10843070_10843258 | 0.43 |
4930567K20Rik |
RIKEN cDNA 4930567K20 gene |
92084 |
0.07 |
| chr10_61139154_61139351 | 0.43 |
Sgpl1 |
sphingosine phosphate lyase 1 |
7646 |
0.15 |
| chr15_50404034_50404185 | 0.43 |
Gm49198 |
predicted gene, 49198 |
117211 |
0.07 |
| chr7_75852856_75853007 | 0.43 |
Klhl25 |
kelch-like 25 |
4490 |
0.26 |
| chr18_12799044_12799195 | 0.43 |
Osbpl1a |
oxysterol binding protein-like 1A |
20479 |
0.15 |
| chr6_29772335_29772499 | 0.43 |
Ahcyl2 |
S-adenosylhomocysteine hydrolase-like 2 |
3889 |
0.2 |
| chr14_8003834_8004016 | 0.43 |
Abhd6 |
abhydrolase domain containing 6 |
959 |
0.54 |
| chr3_95435996_95436155 | 0.43 |
Arnt |
aryl hydrocarbon receptor nuclear translocator |
1556 |
0.21 |
| chr1_69108268_69108432 | 0.42 |
Erbb4 |
erb-b2 receptor tyrosine kinase 4 |
291 |
0.88 |
| chr4_101198122_101198415 | 0.42 |
Gm24468 |
predicted gene, 24468 |
11504 |
0.14 |
| chr5_54000014_54000256 | 0.42 |
Stim2 |
stromal interaction molecule 2 |
1570 |
0.48 |
| chr3_133740753_133740929 | 0.42 |
Gm6135 |
prediticted gene 6135 |
50663 |
0.13 |
| chr19_12685171_12685322 | 0.42 |
Gm49772 |
predicted gene, 49772 |
3475 |
0.12 |
| chr6_51695515_51695666 | 0.42 |
Gm38811 |
predicted gene, 38811 |
15491 |
0.23 |
| chr8_24524982_24525133 | 0.42 |
Ido2 |
indoleamine 2,3-dioxygenase 2 |
15739 |
0.16 |
| chr8_72420483_72420634 | 0.42 |
Eps15l1 |
epidermal growth factor receptor pathway substrate 15-like 1 |
880 |
0.31 |
| chr16_85704612_85704964 | 0.42 |
Gm6278 |
predicted gene 6278 |
60536 |
0.13 |
| chr8_84773010_84773167 | 0.42 |
Nfix |
nuclear factor I/X |
308 |
0.82 |
| chr19_31898515_31898680 | 0.42 |
Gm19241 |
predicted gene, 19241 |
15047 |
0.2 |
| chr19_34523065_34523384 | 0.41 |
Lipa |
lysosomal acid lipase A |
4187 |
0.15 |
| chr9_46036951_46037323 | 0.41 |
Sik3 |
SIK family kinase 3 |
24137 |
0.13 |
| chr9_9228799_9229183 | 0.41 |
Gm16833 |
predicted gene, 16833 |
7297 |
0.23 |
| chr13_51093099_51093344 | 0.41 |
Spin1 |
spindlin 1 |
7659 |
0.25 |
| chr1_163817675_163817827 | 0.41 |
Kifap3 |
kinesin-associated protein 3 |
38168 |
0.16 |
| chr8_110644559_110644860 | 0.41 |
Vac14 |
Vac14 homolog (S. cerevisiae) |
871 |
0.63 |
| chr12_99351809_99352026 | 0.41 |
Foxn3 |
forkhead box N3 |
5102 |
0.14 |
| chr1_155896484_155896645 | 0.41 |
Cep350 |
centrosomal protein 350 |
1967 |
0.24 |
| chr19_44394517_44394772 | 0.41 |
Scd1 |
stearoyl-Coenzyme A desaturase 1 |
12046 |
0.14 |
| chr17_49304784_49304935 | 0.41 |
Gm17830 |
predicted gene, 17830 |
7382 |
0.19 |
| chr2_84359341_84359492 | 0.41 |
Calcrl |
calcitonin receptor-like |
15942 |
0.19 |
| chr17_15372349_15372526 | 0.40 |
Dll1 |
delta like canonical Notch ligand 1 |
606 |
0.69 |
| chr10_61135528_61135708 | 0.40 |
Gm44308 |
predicted gene, 44308 |
10383 |
0.14 |
| chr7_134192391_134192592 | 0.40 |
Adam12 |
a disintegrin and metallopeptidase domain 12 (meltrin alpha) |
19547 |
0.21 |
| chr1_48446424_48446607 | 0.40 |
Gm23240 |
predicted gene, 23240 |
15778 |
0.27 |
| chr1_191945292_191945694 | 0.40 |
Rd3 |
retinal degeneration 3 |
31877 |
0.1 |
| chr2_31513022_31513372 | 0.40 |
Ass1 |
argininosuccinate synthetase 1 |
5293 |
0.19 |
| chrX_142477976_142478290 | 0.40 |
Gm25915 |
predicted gene, 25915 |
7314 |
0.19 |
| chrX_164281566_164281717 | 0.40 |
Gm22023 |
predicted gene, 22023 |
1455 |
0.35 |
| chr16_76432143_76432294 | 0.40 |
Gm9843 |
predicted gene 9843 |
28566 |
0.17 |
| chr7_102310929_102311109 | 0.40 |
Stim1 |
stromal interaction molecule 1 |
42692 |
0.09 |
| chr15_59067650_59068255 | 0.40 |
Mtss1 |
MTSS I-BAR domain containing 1 |
12488 |
0.22 |
| chr1_67214555_67214706 | 0.40 |
Gm15668 |
predicted gene 15668 |
34570 |
0.17 |
| chr11_116211852_116212160 | 0.39 |
Ten1 |
TEN1 telomerase capping complex subunit |
1072 |
0.32 |
| chr1_67227292_67227476 | 0.39 |
Gm15668 |
predicted gene 15668 |
21816 |
0.2 |
| chr19_42540662_42540829 | 0.39 |
R3hcc1l |
R3H domain and coiled-coil containing 1 like |
21923 |
0.17 |
| chr12_100795519_100795676 | 0.39 |
Dglucy |
D-glutamate cyclase |
9455 |
0.15 |
| chr11_14186035_14186200 | 0.39 |
4930554G24Rik |
RIKEN cDNA 4930554G24 gene |
7076 |
0.27 |
| chr19_30093178_30093359 | 0.39 |
Uhrf2 |
ubiquitin-like, containing PHD and RING finger domains 2 |
1307 |
0.48 |
| chr17_84997546_84997697 | 0.39 |
Ppm1b |
protein phosphatase 1B, magnesium dependent, beta isoform |
3941 |
0.22 |
| chr17_46059620_46059782 | 0.39 |
Gm36200 |
predicted gene, 36200 |
24565 |
0.11 |
| chr3_70400102_70400273 | 0.39 |
Gm6631 |
predicted gene 6631 |
13132 |
0.29 |
| chr17_83310756_83310907 | 0.39 |
Rpl21-ps7 |
ribosomal protein L21, pseudogene 7 |
22285 |
0.18 |
| chr5_45474015_45474198 | 0.39 |
Lap3 |
leucine aminopeptidase 3 |
19268 |
0.11 |
| chr3_57743459_57743610 | 0.38 |
Rnf13 |
ring finger protein 13 |
7414 |
0.14 |
| chr6_116120008_116120203 | 0.38 |
Tmcc1 |
transmembrane and coiled coil domains 1 |
9711 |
0.15 |
| chr8_33384837_33385055 | 0.38 |
Wrn |
Werner syndrome RecQ like helicase |
526 |
0.72 |
| chr9_106245058_106245282 | 0.38 |
Alas1 |
aminolevulinic acid synthase 1 |
1536 |
0.23 |
| chr4_76942079_76942249 | 0.38 |
Gm11246 |
predicted gene 11246 |
14853 |
0.22 |
| chr6_38815502_38815672 | 0.38 |
Hipk2 |
homeodomain interacting protein kinase 2 |
2759 |
0.33 |
| chr10_24912717_24913171 | 0.38 |
Mir6905 |
microRNA 6905 |
2280 |
0.2 |
| chr5_89342174_89342864 | 0.38 |
Gc |
vitamin D binding protein |
93109 |
0.09 |
| chr16_42998396_42998547 | 0.38 |
Ndufs5-ps |
NADH:ubiquinone oxidoreductase core subunit S5, pseudogene |
42840 |
0.16 |
| chr7_128640219_128640376 | 0.38 |
Inpp5f |
inositol polyphosphate-5-phosphatase F |
18711 |
0.11 |
| chr3_5346748_5346907 | 0.38 |
Zfhx4 |
zinc finger homeodomain 4 |
105155 |
0.07 |
| chr13_81333716_81333867 | 0.38 |
Adgrv1 |
adhesion G protein-coupled receptor V1 |
9045 |
0.27 |
| chr9_48739787_48740228 | 0.37 |
Zbtb16 |
zinc finger and BTB domain containing 16 |
95938 |
0.07 |
| chr1_67227779_67227978 | 0.37 |
Gm15668 |
predicted gene 15668 |
21322 |
0.2 |
| chr4_97900129_97900314 | 0.37 |
Nfia |
nuclear factor I/A |
10812 |
0.29 |
| chr2_116075832_116075983 | 0.37 |
2810405F15Rik |
RIKEN cDNA 2810405F15 gene |
189 |
0.94 |
| chr14_17864277_17864546 | 0.37 |
Gm48320 |
predicted gene, 48320 |
93289 |
0.08 |
| chr7_98384449_98384600 | 0.37 |
Tsku |
tsukushi, small leucine rich proteoglycan |
23196 |
0.13 |
| chr8_14548995_14549164 | 0.37 |
Dlgap2 |
DLG associated protein 2 |
167083 |
0.03 |
| chr12_40016661_40016850 | 0.37 |
Arl4a |
ADP-ribosylation factor-like 4A |
2271 |
0.3 |
| chr6_50215446_50215627 | 0.37 |
Mpp6 |
membrane protein, palmitoylated 6 (MAGUK p55 subfamily member 6) |
21120 |
0.2 |
| chr9_23224794_23225147 | 0.37 |
Bmper |
BMP-binding endothelial regulator |
1684 |
0.55 |
| chr1_21297964_21298132 | 0.37 |
Gm4956 |
predicted gene 4956 |
79 |
0.95 |
| chr18_65034546_65034697 | 0.37 |
Nedd4l |
neural precursor cell expressed, developmentally down-regulated gene 4-like |
8819 |
0.23 |
| chr13_119456737_119456888 | 0.37 |
4833420G17Rik |
RIKEN cDNA 4833420G17 gene |
5956 |
0.14 |
| chr1_72009620_72009955 | 0.37 |
4933417E11Rik |
RIKEN cDNA 4933417E11 gene |
4592 |
0.18 |
| chr15_66074633_66074784 | 0.36 |
Gm27153 |
predicted gene 27153 |
156 |
0.97 |
| chr8_18713888_18714465 | 0.36 |
Angpt2 |
angiopoietin 2 |
14629 |
0.17 |
| chr15_3430435_3430588 | 0.36 |
Ghr |
growth hormone receptor |
41133 |
0.19 |
| chr10_115388269_115388460 | 0.36 |
Zfc3h1 |
zinc finger, C3H1-type containing |
3405 |
0.17 |
| chr1_67170087_67170238 | 0.36 |
Cps1 |
carbamoyl-phosphate synthetase 1 |
47136 |
0.14 |
| chr4_97025279_97025435 | 0.36 |
Gm27521 |
predicted gene, 27521 |
108337 |
0.07 |
| chr7_66724393_66724565 | 0.36 |
Gm45098 |
predicted gene 45098 |
1360 |
0.35 |
| chr9_66590494_66590818 | 0.36 |
Usp3 |
ubiquitin specific peptidase 3 |
2302 |
0.3 |
| chr3_52544102_52544286 | 0.36 |
Gm30173 |
predicted gene, 30173 |
27230 |
0.2 |
| chr2_144600133_144600679 | 0.36 |
Dtd1 |
D-tyrosyl-tRNA deacylase 1 |
509 |
0.71 |
| chr19_40141682_40141833 | 0.36 |
Cyp2c70 |
cytochrome P450, family 2, subfamily c, polypeptide 70 |
45529 |
0.11 |
| chr2_18059830_18059981 | 0.36 |
Mir7655 |
microRNA 7655 |
2059 |
0.18 |
| chr19_58421573_58421724 | 0.35 |
Gfra1 |
glial cell line derived neurotrophic factor family receptor alpha 1 |
32818 |
0.19 |
| chr5_77357793_77357964 | 0.35 |
Polr2b |
polymerase (RNA) II (DNA directed) polypeptide B |
17510 |
0.13 |
| chr12_118313299_118313450 | 0.35 |
Sp4 |
trans-acting transcription factor 4 |
11934 |
0.27 |
| chr7_98430701_98430878 | 0.35 |
Gm44980 |
predicted gene 44980 |
6730 |
0.13 |
| chr7_104995535_104995703 | 0.35 |
Olfr672 |
olfactory receptor 672 |
1283 |
0.2 |
| chr12_7816660_7817001 | 0.35 |
Gm32828 |
predicted gene, 32828 |
42861 |
0.14 |
| chr2_155587050_155587219 | 0.35 |
Gss |
glutathione synthetase |
442 |
0.63 |
| chr2_26593869_26594069 | 0.35 |
Egfl7 |
EGF-like domain 7 |
1822 |
0.16 |
| chr10_59549383_59549552 | 0.35 |
Gm10322 |
predicted gene 10322 |
66596 |
0.1 |
| chr2_118839517_118839869 | 0.35 |
Gm14091 |
predicted gene 14091 |
5571 |
0.14 |
| chr5_5514010_5514339 | 0.35 |
Cldn12 |
claudin 12 |
275 |
0.9 |
| chr16_13257585_13257940 | 0.35 |
Mrtfb |
myocardin related transcription factor B |
1256 |
0.58 |
| chr11_90184166_90184514 | 0.35 |
n-R5s72 |
nuclear encoded rRNA 5S 72 |
787 |
0.57 |
| chr19_42118053_42118204 | 0.35 |
Pi4k2a |
phosphatidylinositol 4-kinase type 2 alpha |
3450 |
0.14 |
| chr13_29774433_29774613 | 0.35 |
Cdkal1 |
CDK5 regulatory subunit associated protein 1-like 1 |
80908 |
0.11 |
| chr4_141158698_141158852 | 0.35 |
Fbxo42 |
F-box protein 42 |
10853 |
0.12 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:1904380 | endoplasmic reticulum mannose trimming(GO:1904380) |
| 0.2 | 0.6 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.2 | 1.0 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.1 | 0.4 | GO:1901078 | negative regulation of relaxation of muscle(GO:1901078) |
| 0.1 | 0.3 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.1 | 0.3 | GO:0010963 | regulation of L-arginine import(GO:0010963) |
| 0.1 | 0.5 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.1 | 0.3 | GO:0001757 | somite specification(GO:0001757) |
| 0.1 | 0.3 | GO:0032066 | nucleolus to nucleoplasm transport(GO:0032066) |
| 0.1 | 0.3 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.1 | 0.2 | GO:0015747 | urate transport(GO:0015747) |
| 0.1 | 0.2 | GO:0003431 | growth plate cartilage chondrocyte development(GO:0003431) |
| 0.1 | 0.3 | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.1 | 0.3 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.1 | 0.3 | GO:0006549 | isoleucine metabolic process(GO:0006549) |
| 0.1 | 0.3 | GO:0070858 | negative regulation of bile acid biosynthetic process(GO:0070858) negative regulation of bile acid metabolic process(GO:1904252) |
| 0.1 | 0.3 | GO:0019478 | D-amino acid catabolic process(GO:0019478) |
| 0.1 | 0.1 | GO:0048696 | regulation of collateral sprouting in absence of injury(GO:0048696) |
| 0.1 | 0.2 | GO:0071727 | response to triacyl bacterial lipopeptide(GO:0071725) cellular response to triacyl bacterial lipopeptide(GO:0071727) |
| 0.1 | 0.2 | GO:0038171 | cannabinoid signaling pathway(GO:0038171) endocannabinoid signaling pathway(GO:0071926) |
| 0.1 | 0.3 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.1 | 0.3 | GO:1904528 | regulation of microtubule binding(GO:1904526) positive regulation of microtubule binding(GO:1904528) |
| 0.1 | 0.2 | GO:1902809 | regulation of skeletal muscle fiber differentiation(GO:1902809) |
| 0.1 | 0.2 | GO:0019065 | receptor-mediated endocytosis of virus by host cell(GO:0019065) endocytosis involved in viral entry into host cell(GO:0075509) |
| 0.1 | 0.1 | GO:1900020 | regulation of protein kinase C activity(GO:1900019) positive regulation of protein kinase C activity(GO:1900020) |
| 0.1 | 0.2 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.1 | 0.3 | GO:0061073 | ciliary body morphogenesis(GO:0061073) |
| 0.1 | 0.2 | GO:2001184 | positive regulation of interleukin-12 secretion(GO:2001184) |
| 0.1 | 0.2 | GO:0090296 | regulation of mitochondrial DNA replication(GO:0090296) |
| 0.1 | 0.2 | GO:0021553 | olfactory nerve development(GO:0021553) |
| 0.1 | 0.5 | GO:0006851 | mitochondrial calcium ion transport(GO:0006851) |
| 0.1 | 0.3 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.1 | 0.4 | GO:0071265 | amino acid salvage(GO:0043102) L-methionine biosynthetic process(GO:0071265) L-methionine salvage(GO:0071267) |
| 0.1 | 0.3 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) |
| 0.1 | 0.2 | GO:0006578 | amino-acid betaine biosynthetic process(GO:0006578) |
| 0.1 | 0.1 | GO:2000809 | positive regulation of synaptic vesicle clustering(GO:2000809) |
| 0.1 | 0.4 | GO:0060136 | embryonic process involved in female pregnancy(GO:0060136) |
| 0.1 | 0.2 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.1 | 0.4 | GO:0021707 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.0 | 0.2 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.0 | 0.2 | GO:0090245 | axis elongation involved in somitogenesis(GO:0090245) |
| 0.0 | 0.1 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.0 | 0.0 | GO:1902221 | L-phenylalanine metabolic process(GO:0006558) L-phenylalanine catabolic process(GO:0006559) erythrose 4-phosphate/phosphoenolpyruvate family amino acid metabolic process(GO:1902221) erythrose 4-phosphate/phosphoenolpyruvate family amino acid catabolic process(GO:1902222) |
| 0.0 | 0.0 | GO:0046098 | guanine metabolic process(GO:0046098) |
| 0.0 | 0.2 | GO:1903964 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.0 | 0.1 | GO:1902488 | cholangiocyte apoptotic process(GO:1902488) regulation of cholangiocyte apoptotic process(GO:1904192) negative regulation of cholangiocyte apoptotic process(GO:1904193) |
| 0.0 | 0.2 | GO:0090435 | protein localization to nuclear envelope(GO:0090435) |
| 0.0 | 0.2 | GO:0006499 | N-terminal protein myristoylation(GO:0006499) |
| 0.0 | 0.1 | GO:0036493 | positive regulation of translation in response to endoplasmic reticulum stress(GO:0036493) |
| 0.0 | 0.1 | GO:0008050 | female courtship behavior(GO:0008050) |
| 0.0 | 0.5 | GO:0009404 | toxin metabolic process(GO:0009404) |
| 0.0 | 0.2 | GO:2001286 | regulation of caveolin-mediated endocytosis(GO:2001286) |
| 0.0 | 0.1 | GO:0070669 | response to interleukin-2(GO:0070669) |
| 0.0 | 0.2 | GO:0006975 | DNA damage induced protein phosphorylation(GO:0006975) |
| 0.0 | 0.2 | GO:0008295 | spermidine biosynthetic process(GO:0008295) |
| 0.0 | 0.1 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.0 | 0.1 | GO:0032788 | saturated monocarboxylic acid metabolic process(GO:0032788) unsaturated monocarboxylic acid metabolic process(GO:0032789) |
| 0.0 | 0.3 | GO:0009074 | aromatic amino acid family catabolic process(GO:0009074) |
| 0.0 | 0.0 | GO:0032741 | positive regulation of interleukin-18 production(GO:0032741) |
| 0.0 | 0.5 | GO:2000010 | positive regulation of protein localization to cell surface(GO:2000010) |
| 0.0 | 0.1 | GO:1903659 | regulation of complement-dependent cytotoxicity(GO:1903659) negative regulation of complement-dependent cytotoxicity(GO:1903660) |
| 0.0 | 0.1 | GO:0051791 | medium-chain fatty acid metabolic process(GO:0051791) |
| 0.0 | 0.1 | GO:0031087 | nuclear-transcribed mRNA catabolic process, deadenylation-independent decay(GO:0031086) deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.0 | 0.1 | GO:0030327 | prenylated protein catabolic process(GO:0030327) |
| 0.0 | 0.2 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.0 | 0.3 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.0 | 0.2 | GO:0071447 | cellular response to hydroperoxide(GO:0071447) |
| 0.0 | 0.1 | GO:0070458 | detoxification of nitrogen compound(GO:0051410) cellular detoxification of nitrogen compound(GO:0070458) |
| 0.0 | 0.1 | GO:0060523 | prostate epithelial cord elongation(GO:0060523) |
| 0.0 | 0.1 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.0 | 0.2 | GO:0007182 | common-partner SMAD protein phosphorylation(GO:0007182) |
| 0.0 | 0.1 | GO:2000409 | positive regulation of T cell extravasation(GO:2000409) |
| 0.0 | 0.3 | GO:1901407 | regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.0 | 0.3 | GO:0097105 | presynaptic membrane assembly(GO:0097105) |
| 0.0 | 0.1 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 0.0 | 0.1 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.0 | 0.1 | GO:0060447 | bud outgrowth involved in lung branching(GO:0060447) |
| 0.0 | 0.1 | GO:0046874 | quinolinate metabolic process(GO:0046874) |
| 0.0 | 0.2 | GO:1901725 | regulation of histone deacetylase activity(GO:1901725) |
| 0.0 | 0.2 | GO:0042518 | negative regulation of tyrosine phosphorylation of Stat3 protein(GO:0042518) |
| 0.0 | 0.1 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.0 | 0.1 | GO:0070857 | regulation of bile acid biosynthetic process(GO:0070857) |
| 0.0 | 0.1 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.2 | GO:0070245 | positive regulation of thymocyte apoptotic process(GO:0070245) |
| 0.0 | 0.1 | GO:0071677 | positive regulation of mononuclear cell migration(GO:0071677) |
| 0.0 | 0.1 | GO:0060125 | negative regulation of growth hormone secretion(GO:0060125) |
| 0.0 | 0.1 | GO:0003330 | regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) |
| 0.0 | 0.1 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.0 | 0.2 | GO:0015074 | DNA integration(GO:0015074) |
| 0.0 | 0.1 | GO:0036112 | medium-chain fatty-acyl-CoA metabolic process(GO:0036112) |
| 0.0 | 0.2 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.0 | 0.1 | GO:0002071 | glandular epithelial cell maturation(GO:0002071) |
| 0.0 | 0.1 | GO:0001922 | B-1 B cell homeostasis(GO:0001922) |
| 0.0 | 0.1 | GO:0033629 | negative regulation of cell adhesion mediated by integrin(GO:0033629) |
| 0.0 | 0.1 | GO:1903334 | positive regulation of protein folding(GO:1903334) |
| 0.0 | 0.1 | GO:0051081 | membrane disassembly(GO:0030397) nuclear envelope disassembly(GO:0051081) |
| 0.0 | 0.1 | GO:0006166 | purine ribonucleoside salvage(GO:0006166) |
| 0.0 | 0.2 | GO:0032488 | Cdc42 protein signal transduction(GO:0032488) |
| 0.0 | 0.1 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.0 | 0.1 | GO:1901339 | regulation of store-operated calcium channel activity(GO:1901339) |
| 0.0 | 0.1 | GO:0007228 | positive regulation of hh target transcription factor activity(GO:0007228) |
| 0.0 | 0.1 | GO:0060696 | regulation of phospholipid catabolic process(GO:0060696) |
| 0.0 | 0.2 | GO:0009052 | pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 0.0 | 0.1 | GO:0032380 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.0 | 0.2 | GO:0050916 | sensory perception of sweet taste(GO:0050916) |
| 0.0 | 0.0 | GO:0014834 | skeletal muscle satellite cell maintenance involved in skeletal muscle regeneration(GO:0014834) |
| 0.0 | 0.2 | GO:0046146 | tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.0 | 0.2 | GO:0060689 | cell differentiation involved in salivary gland development(GO:0060689) |
| 0.0 | 0.1 | GO:0003062 | regulation of heart rate by chemical signal(GO:0003062) |
| 0.0 | 0.2 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 0.1 | GO:0044789 | modulation by host of viral release from host cell(GO:0044789) positive regulation by host of viral release from host cell(GO:0044791) |
| 0.0 | 0.1 | GO:1903969 | regulation of macrophage colony-stimulating factor signaling pathway(GO:1902226) regulation of response to macrophage colony-stimulating factor(GO:1903969) regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903972) |
| 0.0 | 0.1 | GO:2001245 | regulation of phosphatidylcholine biosynthetic process(GO:2001245) |
| 0.0 | 0.1 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.0 | 0.1 | GO:0046101 | hypoxanthine metabolic process(GO:0046100) hypoxanthine biosynthetic process(GO:0046101) |
| 0.0 | 0.2 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.0 | 0.1 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 0.1 | GO:1904016 | response to Thyroglobulin triiodothyronine(GO:1904016) cellular response to Thyroglobulin triiodothyronine(GO:1904017) |
| 0.0 | 0.1 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.0 | 0.2 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.0 | 0.1 | GO:0030382 | sperm mitochondrion organization(GO:0030382) |
| 0.0 | 0.1 | GO:0021780 | oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.0 | 0.1 | GO:1990086 | lens fiber cell apoptotic process(GO:1990086) |
| 0.0 | 0.1 | GO:1902047 | polyamine transmembrane transport(GO:1902047) regulation of polyamine transmembrane transport(GO:1902267) |
| 0.0 | 0.2 | GO:0015871 | choline transport(GO:0015871) |
| 0.0 | 0.1 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.0 | 0.2 | GO:0052697 | xenobiotic glucuronidation(GO:0052697) |
| 0.0 | 0.0 | GO:0016557 | peroxisome membrane biogenesis(GO:0016557) |
| 0.0 | 0.2 | GO:0034244 | negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.0 | 0.1 | GO:1903347 | negative regulation of bicellular tight junction assembly(GO:1903347) |
| 0.0 | 0.4 | GO:0008210 | estrogen metabolic process(GO:0008210) |
| 0.0 | 0.1 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.0 | 0.3 | GO:0006699 | bile acid biosynthetic process(GO:0006699) |
| 0.0 | 0.1 | GO:2000074 | regulation of type B pancreatic cell development(GO:2000074) |
| 0.0 | 0.0 | GO:1901187 | regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.0 | 0.1 | GO:1904502 | regulation of lipophagy(GO:1904502) positive regulation of lipophagy(GO:1904504) |
| 0.0 | 0.1 | GO:0072602 | interleukin-4 secretion(GO:0072602) |
| 0.0 | 0.4 | GO:0010569 | regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.0 | 0.1 | GO:0030050 | vesicle transport along actin filament(GO:0030050) |
| 0.0 | 0.1 | GO:1900095 | regulation of dosage compensation by inactivation of X chromosome(GO:1900095) |
| 0.0 | 0.1 | GO:0002025 | vasodilation by norepinephrine-epinephrine involved in regulation of systemic arterial blood pressure(GO:0002025) |
| 0.0 | 0.2 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.0 | 0.1 | GO:0060839 | endothelial cell fate commitment(GO:0060839) |
| 0.0 | 0.0 | GO:0060375 | regulation of mast cell differentiation(GO:0060375) |
| 0.0 | 0.2 | GO:0042996 | regulation of Golgi to plasma membrane protein transport(GO:0042996) |
| 0.0 | 0.1 | GO:0045875 | negative regulation of sister chromatid cohesion(GO:0045875) |
| 0.0 | 0.3 | GO:0060294 | cilium movement involved in cell motility(GO:0060294) |
| 0.0 | 0.1 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.0 | 0.1 | GO:0034047 | regulation of protein phosphatase type 2A activity(GO:0034047) |
| 0.0 | 0.0 | GO:0021683 | cerebellar granular layer morphogenesis(GO:0021683) |
| 0.0 | 0.1 | GO:1902414 | protein localization to cell junction(GO:1902414) |
| 0.0 | 0.1 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.0 | 0.2 | GO:0090005 | negative regulation of establishment of protein localization to plasma membrane(GO:0090005) |
| 0.0 | 0.1 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.0 | 0.1 | GO:0048319 | axial mesoderm morphogenesis(GO:0048319) |
| 0.0 | 0.1 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.0 | 0.3 | GO:0045986 | negative regulation of smooth muscle contraction(GO:0045986) |
| 0.0 | 0.0 | GO:0009698 | phenylpropanoid metabolic process(GO:0009698) phenylpropanoid catabolic process(GO:0046271) |
| 0.0 | 0.1 | GO:0018317 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.0 | 0.2 | GO:0090308 | regulation of methylation-dependent chromatin silencing(GO:0090308) |
| 0.0 | 0.3 | GO:0035510 | DNA dealkylation(GO:0035510) |
| 0.0 | 0.1 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.0 | 0.1 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 0.0 | 0.1 | GO:0045583 | regulation of cytotoxic T cell differentiation(GO:0045583) positive regulation of cytotoxic T cell differentiation(GO:0045585) |
| 0.0 | 0.1 | GO:0070537 | histone H2A K63-linked deubiquitination(GO:0070537) |
| 0.0 | 0.0 | GO:0001996 | positive regulation of heart rate by epinephrine-norepinephrine(GO:0001996) |
| 0.0 | 0.1 | GO:0010764 | negative regulation of fibroblast migration(GO:0010764) |
| 0.0 | 0.2 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.0 | 0.0 | GO:0010757 | negative regulation of plasminogen activation(GO:0010757) |
| 0.0 | 0.1 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.0 | 0.1 | GO:1903071 | positive regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903071) |
| 0.0 | 0.1 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 0.1 | GO:0035356 | cellular triglyceride homeostasis(GO:0035356) |
| 0.0 | 0.1 | GO:1900060 | negative regulation of ceramide biosynthetic process(GO:1900060) |
| 0.0 | 0.1 | GO:0015772 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.0 | 0.0 | GO:1903799 | negative regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903799) |
| 0.0 | 0.1 | GO:0021943 | formation of radial glial scaffolds(GO:0021943) |
| 0.0 | 0.2 | GO:0090051 | negative regulation of cell migration involved in sprouting angiogenesis(GO:0090051) |
| 0.0 | 0.1 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.0 | 0.1 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.0 | 0.0 | GO:0021938 | smoothened signaling pathway involved in regulation of cerebellar granule cell precursor cell proliferation(GO:0021938) |
| 0.0 | 0.2 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.0 | 0.1 | GO:0032621 | interleukin-18 production(GO:0032621) |
| 0.0 | 0.0 | GO:0075136 | response to defenses of other organism involved in symbiotic interaction(GO:0052173) response to host defenses(GO:0052200) response to host(GO:0075136) |
| 0.0 | 0.0 | GO:0002155 | regulation of thyroid hormone mediated signaling pathway(GO:0002155) |
| 0.0 | 0.0 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.1 | GO:0034310 | primary alcohol catabolic process(GO:0034310) |
| 0.0 | 0.1 | GO:0060373 | regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 0.0 | 0.1 | GO:0032485 | regulation of Ral protein signal transduction(GO:0032485) |
| 0.0 | 0.0 | GO:0048296 | isotype switching to IgA isotypes(GO:0048290) regulation of isotype switching to IgA isotypes(GO:0048296) |
| 0.0 | 0.2 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.0 | 0.2 | GO:1902170 | cellular response to reactive nitrogen species(GO:1902170) |
| 0.0 | 0.0 | GO:0097680 | double-strand break repair via classical nonhomologous end joining(GO:0097680) |
| 0.0 | 0.0 | GO:0061317 | canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:0061317) |
| 0.0 | 0.2 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.0 | 0.1 | GO:0043537 | negative regulation of blood vessel endothelial cell migration(GO:0043537) |
| 0.0 | 0.1 | GO:0086046 | membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.0 | 0.1 | GO:0030638 | polyketide metabolic process(GO:0030638) aminoglycoside antibiotic metabolic process(GO:0030647) daunorubicin metabolic process(GO:0044597) doxorubicin metabolic process(GO:0044598) |
| 0.0 | 0.1 | GO:0090043 | regulation of tubulin deacetylation(GO:0090043) |
| 0.0 | 0.0 | GO:0033387 | putrescine biosynthetic process(GO:0009446) putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.0 | 0.1 | GO:0022417 | protein maturation by protein folding(GO:0022417) |
| 0.0 | 0.1 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.0 | 0.1 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.0 | 0.0 | GO:1900157 | regulation of bone mineralization involved in bone maturation(GO:1900157) |
| 0.0 | 0.2 | GO:0019184 | nonribosomal peptide biosynthetic process(GO:0019184) |
| 0.0 | 0.0 | GO:0099515 | actin filament-based transport(GO:0099515) |
| 0.0 | 0.1 | GO:1904479 | negative regulation of intestinal absorption(GO:1904479) |
| 0.0 | 0.0 | GO:1903011 | negative regulation of bone development(GO:1903011) |
| 0.0 | 0.0 | GO:0043578 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.1 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.0 | 0.1 | GO:0046473 | phosphatidic acid metabolic process(GO:0046473) |
| 0.0 | 0.1 | GO:0006072 | glycerol-3-phosphate metabolic process(GO:0006072) |
| 0.0 | 0.0 | GO:1903061 | positive regulation of protein lipidation(GO:1903061) |
| 0.0 | 0.0 | GO:0003415 | chondrocyte hypertrophy(GO:0003415) |
| 0.0 | 0.0 | GO:1901420 | negative regulation of response to alcohol(GO:1901420) |
| 0.0 | 0.0 | GO:0006546 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.0 | 0.1 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 | 0.2 | GO:2000060 | positive regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:2000060) |
| 0.0 | 0.0 | GO:0035609 | C-terminal protein deglutamylation(GO:0035609) |
| 0.0 | 0.1 | GO:0061158 | 3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.0 | 0.0 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.0 | GO:0070947 | neutrophil mediated killing of fungus(GO:0070947) |
| 0.0 | 0.0 | GO:0021898 | regulation of transcription from RNA polymerase II promoter involved in forebrain neuron fate commitment(GO:0021882) commitment of multipotent stem cells to neuronal lineage in forebrain(GO:0021898) |
| 0.0 | 0.0 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.0 | 0.0 | GO:1990036 | calcium ion import into sarcoplasmic reticulum(GO:1990036) |
| 0.0 | 0.1 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 0.0 | 0.1 | GO:0046836 | glycolipid transport(GO:0046836) |
| 0.0 | 0.0 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.0 | 0.0 | GO:0010728 | regulation of hydrogen peroxide biosynthetic process(GO:0010728) |
| 0.0 | 0.0 | GO:0006207 | 'de novo' pyrimidine nucleobase biosynthetic process(GO:0006207) pyrimidine nucleobase biosynthetic process(GO:0019856) |
| 0.0 | 0.0 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.0 | 0.1 | GO:0033313 | meiotic cell cycle checkpoint(GO:0033313) |
| 0.0 | 0.1 | GO:0032596 | protein transport into membrane raft(GO:0032596) |
| 0.0 | 0.3 | GO:0060074 | synapse maturation(GO:0060074) |
| 0.0 | 0.0 | GO:0035376 | sterol import(GO:0035376) cholesterol import(GO:0070508) |
| 0.0 | 0.1 | GO:0002072 | optic cup morphogenesis involved in camera-type eye development(GO:0002072) |
| 0.0 | 0.1 | GO:0035934 | corticosterone secretion(GO:0035934) regulation of corticosterone secretion(GO:2000852) |
| 0.0 | 0.1 | GO:0018214 | protein carboxylation(GO:0018214) |
| 0.0 | 0.0 | GO:0035283 | central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.0 | 0.0 | GO:0098904 | regulation of AV node cell action potential(GO:0098904) |
| 0.0 | 0.1 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.1 | GO:0006670 | sphingosine metabolic process(GO:0006670) |
| 0.0 | 0.1 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.0 | 0.0 | GO:0018916 | nitrobenzene metabolic process(GO:0018916) |
| 0.0 | 0.2 | GO:0043574 | protein targeting to peroxisome(GO:0006625) peroxisomal transport(GO:0043574) protein localization to peroxisome(GO:0072662) establishment of protein localization to peroxisome(GO:0072663) |
| 0.0 | 0.0 | GO:0044828 | negative regulation by host of viral genome replication(GO:0044828) |
| 0.0 | 0.0 | GO:0015755 | fructose transport(GO:0015755) |
| 0.0 | 0.0 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.0 | 0.1 | GO:0071033 | nuclear retention of pre-mRNA at the site of transcription(GO:0071033) |
| 0.0 | 0.3 | GO:2001014 | regulation of skeletal muscle cell differentiation(GO:2001014) |
| 0.0 | 0.1 | GO:0035519 | protein K29-linked ubiquitination(GO:0035519) |
| 0.0 | 0.1 | GO:0055064 | chloride ion homeostasis(GO:0055064) |
| 0.0 | 0.1 | GO:2000188 | regulation of cholesterol homeostasis(GO:2000188) |
| 0.0 | 0.0 | GO:0098700 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.0 | 0.0 | GO:0051665 | membrane raft localization(GO:0051665) |
| 0.0 | 0.0 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.0 | 0.0 | GO:0002154 | thyroid hormone mediated signaling pathway(GO:0002154) |
| 0.0 | 0.1 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.0 | 0.1 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.0 | 0.1 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.0 | 0.0 | GO:0021847 | ventricular zone neuroblast division(GO:0021847) |
| 0.0 | 0.1 | GO:2000811 | negative regulation of anoikis(GO:2000811) |
| 0.0 | 0.0 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.0 | 0.0 | GO:0001539 | cilium or flagellum-dependent cell motility(GO:0001539) cilium-dependent cell motility(GO:0060285) |
| 0.0 | 0.0 | GO:0045658 | regulation of neutrophil differentiation(GO:0045658) |
| 0.0 | 0.0 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.0 | 0.1 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.0 | 0.1 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 0.0 | 0.0 | GO:2000574 | regulation of microtubule motor activity(GO:2000574) |
| 0.0 | 0.0 | GO:0070681 | glutaminyl-tRNAGln biosynthesis via transamidation(GO:0070681) |
| 0.0 | 0.0 | GO:0002740 | negative regulation of cytokine secretion involved in immune response(GO:0002740) |
| 0.0 | 0.3 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 | 0.0 | GO:0036394 | amylase secretion(GO:0036394) |
| 0.0 | 0.1 | GO:0006702 | androgen biosynthetic process(GO:0006702) |
| 0.0 | 0.1 | GO:0050667 | homocysteine metabolic process(GO:0050667) |
| 0.0 | 0.0 | GO:0007567 | parturition(GO:0007567) |
| 0.0 | 0.0 | GO:0046005 | positive regulation of circadian sleep/wake cycle, REM sleep(GO:0046005) |
| 0.0 | 0.0 | GO:0060510 | Type II pneumocyte differentiation(GO:0060510) |
| 0.0 | 0.1 | GO:0010459 | negative regulation of heart rate(GO:0010459) |
| 0.0 | 0.0 | GO:1902969 | mitotic DNA replication(GO:1902969) |
| 0.0 | 0.0 | GO:0008054 | negative regulation of cyclin-dependent protein serine/threonine kinase by cyclin degradation(GO:0008054) |
| 0.0 | 0.0 | GO:0070827 | chromatin maintenance(GO:0070827) |
| 0.0 | 0.0 | GO:0006868 | glutamine transport(GO:0006868) |
| 0.0 | 0.0 | GO:0006551 | leucine metabolic process(GO:0006551) |
| 0.0 | 0.0 | GO:0042178 | xenobiotic catabolic process(GO:0042178) |
| 0.0 | 0.0 | GO:1902474 | positive regulation of protein localization to synapse(GO:1902474) |
| 0.0 | 0.1 | GO:1900454 | positive regulation of long term synaptic depression(GO:1900454) |
| 0.0 | 0.1 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.0 | 0.0 | GO:0046083 | adenine metabolic process(GO:0046083) adenine biosynthetic process(GO:0046084) |
| 0.0 | 0.0 | GO:0006449 | regulation of translational termination(GO:0006449) |
| 0.0 | 0.0 | GO:1903553 | positive regulation of extracellular exosome assembly(GO:1903553) |
| 0.0 | 0.0 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.0 | 0.0 | GO:0010994 | regulation of ubiquitin homeostasis(GO:0010993) free ubiquitin chain polymerization(GO:0010994) |
| 0.0 | 0.0 | GO:0015888 | thiamine transport(GO:0015888) |
| 0.0 | 0.0 | GO:0032058 | positive regulation of translational initiation in response to stress(GO:0032058) |
| 0.0 | 0.1 | GO:0008343 | adult feeding behavior(GO:0008343) |
| 0.0 | 0.1 | GO:0043619 | regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0043619) |
| 0.0 | 0.0 | GO:0033693 | neurofilament bundle assembly(GO:0033693) |
| 0.0 | 0.1 | GO:0002457 | T cell antigen processing and presentation(GO:0002457) |
| 0.0 | 0.1 | GO:0051798 | positive regulation of hair follicle development(GO:0051798) |
| 0.0 | 0.1 | GO:0042359 | vitamin D metabolic process(GO:0042359) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.1 | 0.3 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.1 | 0.2 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.1 | 0.2 | GO:0035355 | Toll-like receptor 2-Toll-like receptor 6 protein complex(GO:0035355) |
| 0.1 | 0.3 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.1 | 0.2 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.1 | 0.2 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 0.7 | GO:0046930 | pore complex(GO:0046930) |
| 0.0 | 0.2 | GO:0032021 | NELF complex(GO:0032021) |
| 0.0 | 0.3 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.0 | 0.3 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.7 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.1 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.0 | 0.1 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.0 | 0.3 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.0 | 0.3 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.1 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.0 | 0.1 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.0 | 0.0 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 0.2 | GO:0043203 | axon hillock(GO:0043203) |
| 0.0 | 0.0 | GO:0031261 | DNA replication preinitiation complex(GO:0031261) |
| 0.0 | 0.1 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.0 | 0.1 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.0 | 0.1 | GO:0000836 | ER ubiquitin ligase complex(GO:0000835) Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.0 | 0.1 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.1 | GO:0031010 | ISWI-type complex(GO:0031010) |
| 0.0 | 0.1 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.0 | 0.0 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.0 | 0.3 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 0.2 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.0 | 0.1 | GO:0016939 | kinesin II complex(GO:0016939) |
| 0.0 | 0.1 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.0 | 0.1 | GO:0031415 | NatA complex(GO:0031415) |
| 0.0 | 0.3 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 0.1 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.0 | 0.1 | GO:0044308 | axonal spine(GO:0044308) |
| 0.0 | 0.1 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.0 | 0.2 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.1 | GO:0036194 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.0 | 0.8 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.1 | GO:0005784 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.0 | 0.1 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.0 | 0.2 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 0.1 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 0.0 | 0.1 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.0 | 0.1 | GO:0045179 | apical cortex(GO:0045179) |
| 0.0 | 0.3 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.1 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.0 | 0.1 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.0 | 0.1 | GO:0055087 | Ski complex(GO:0055087) |
| 0.0 | 0.0 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 0.0 | 0.0 | GO:0000811 | GINS complex(GO:0000811) |
| 0.0 | 0.2 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.1 | GO:0044352 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.0 | 0.1 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.0 | 0.1 | GO:0030062 | mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 0.0 | 0.1 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.1 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.0 | 0.0 | GO:1990812 | growth cone filopodium(GO:1990812) |
| 0.0 | 0.5 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.2 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.0 | GO:0032010 | phagolysosome(GO:0032010) |
| 0.0 | 0.0 | GO:0070939 | Dsl1p complex(GO:0070939) |
| 0.0 | 0.1 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.0 | 0.0 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.0 | 0.0 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 0.1 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.1 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.1 | GO:0035749 | myelin sheath adaxonal region(GO:0035749) |
| 0.0 | 0.0 | GO:1990761 | growth cone lamellipodium(GO:1990761) |
| 0.0 | 0.0 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.0 | 0.0 | GO:0098536 | deuterosome(GO:0098536) |
| 0.0 | 0.0 | GO:0031467 | Cul7-RING ubiquitin ligase complex(GO:0031467) |
| 0.0 | 0.0 | GO:0030956 | glutamyl-tRNA(Gln) amidotransferase complex(GO:0030956) |
| 0.0 | 0.1 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.0 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.0 | GO:1990851 | Wnt-Frizzled-LRP5/6 complex(GO:1990851) |
| 0.0 | 0.0 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.0 | 0.1 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.0 | 0.1 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 0.4 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.0 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.1 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.1 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.0 | 0.1 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.0 | 0.2 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.0 | GO:0009279 | cell outer membrane(GO:0009279) cell envelope(GO:0030313) external encapsulating structure part(GO:0044462) |
| 0.0 | 0.1 | GO:0030132 | clathrin coat of coated pit(GO:0030132) |
| 0.0 | 0.1 | GO:0000782 | telomere cap complex(GO:0000782) nuclear telomere cap complex(GO:0000783) |
| 0.0 | 0.0 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0016743 | carboxyl- or carbamoyltransferase activity(GO:0016743) |
| 0.2 | 0.5 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.1 | 0.3 | GO:0008898 | S-adenosylmethionine-homocysteine S-methyltransferase activity(GO:0008898) |
| 0.1 | 0.6 | GO:0015288 | porin activity(GO:0015288) |
| 0.1 | 0.4 | GO:0047023 | androsterone dehydrogenase activity(GO:0047023) |
| 0.1 | 0.3 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.1 | 0.3 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.1 | 0.2 | GO:0038181 | bile acid receptor activity(GO:0038181) |
| 0.1 | 0.2 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.1 | 0.4 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.1 | 0.4 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) inositol hexakisphosphate 5-kinase activity(GO:0000832) inositol hexakisphosphate 1-kinase activity(GO:0052723) inositol hexakisphosphate 3-kinase activity(GO:0052724) |
| 0.1 | 0.6 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.1 | 0.2 | GO:0035663 | Toll-like receptor 2 binding(GO:0035663) |
| 0.1 | 0.2 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.1 | 0.3 | GO:0003846 | 2-acylglycerol O-acyltransferase activity(GO:0003846) |
| 0.1 | 0.2 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.1 | 0.4 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.1 | 0.2 | GO:0016508 | long-chain-enoyl-CoA hydratase activity(GO:0016508) |
| 0.1 | 0.2 | GO:0032551 | UTP binding(GO:0002134) pyrimidine ribonucleoside binding(GO:0032551) |
| 0.1 | 0.2 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.1 | 0.2 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.1 | 0.2 | GO:0080130 | L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.0 | 0.1 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.0 | 0.1 | GO:0030620 | U2 snRNA binding(GO:0030620) |
| 0.0 | 0.2 | GO:0032896 | palmitoyl-CoA 9-desaturase activity(GO:0032896) |
| 0.0 | 0.1 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.0 | 0.2 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 0.0 | 0.2 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 0.1 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 0.0 | 0.3 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.1 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.0 | 0.2 | GO:0015252 | hydrogen ion channel activity(GO:0015252) |
| 0.0 | 0.1 | GO:0004802 | transketolase activity(GO:0004802) |
| 0.0 | 0.3 | GO:0005167 | neurotrophin TRK receptor binding(GO:0005167) |
| 0.0 | 0.2 | GO:0002046 | opsin binding(GO:0002046) |
| 0.0 | 0.2 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.0 | 0.4 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.3 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.1 | GO:0008309 | double-stranded DNA exodeoxyribonuclease activity(GO:0008309) |
| 0.0 | 0.1 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.3 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 0.1 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.0 | 0.1 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.1 | GO:0008486 | diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) |
| 0.0 | 0.4 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 0.1 | GO:0043140 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) |
| 0.0 | 0.2 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.1 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.0 | 0.2 | GO:0033695 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 0.0 | 0.1 | GO:0070538 | oleic acid binding(GO:0070538) |
| 0.0 | 0.2 | GO:0004075 | biotin carboxylase activity(GO:0004075) |
| 0.0 | 0.3 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.1 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.0 | 0.1 | GO:0055100 | adiponectin binding(GO:0055100) |
| 0.0 | 0.1 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.0 | 0.4 | GO:0008601 | protein phosphatase type 2A regulator activity(GO:0008601) |
| 0.0 | 0.2 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.0 | 0.5 | GO:1900750 | glutathione binding(GO:0043295) oligopeptide binding(GO:1900750) |
| 0.0 | 0.1 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.0 | 0.1 | GO:0008732 | threonine aldolase activity(GO:0004793) L-allo-threonine aldolase activity(GO:0008732) |
| 0.0 | 0.1 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.0 | 0.1 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.0 | 0.1 | GO:0042979 | ornithine decarboxylase regulator activity(GO:0042979) |
| 0.0 | 0.2 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.1 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.0 | 0.1 | GO:0061513 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.0 | 0.2 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.0 | 0.2 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.1 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) |
| 0.0 | 0.1 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 0.1 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.0 | 0.1 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.0 | 0.1 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.0 | 0.1 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 0.1 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.0 | 0.1 | GO:0019959 | interleukin-8 binding(GO:0019959) |
| 0.0 | 0.1 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.0 | 0.1 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 0.1 | GO:1990239 | steroid hormone binding(GO:1990239) |
| 0.0 | 0.1 | GO:0031782 | melanocortin receptor binding(GO:0031779) type 3 melanocortin receptor binding(GO:0031781) type 4 melanocortin receptor binding(GO:0031782) |
| 0.0 | 0.1 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.0 | 0.1 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.0 | 0.1 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.1 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.0 | 0.1 | GO:0035671 | enone reductase activity(GO:0035671) |
| 0.0 | 0.1 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.0 | 0.1 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.1 | GO:0035651 | AP-3 adaptor complex binding(GO:0035651) |
| 0.0 | 0.1 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.1 | GO:0030350 | iron-responsive element binding(GO:0030350) |
| 0.0 | 0.1 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 0.1 | GO:0034191 | apolipoprotein A-I receptor binding(GO:0034191) |
| 0.0 | 0.1 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.0 | 0.1 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.2 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.0 | 0.1 | GO:0008506 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.0 | 0.5 | GO:0004118 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) cGMP-inhibited cyclic-nucleotide phosphodiesterase activity(GO:0004119) photoreceptor cyclic-nucleotide phosphodiesterase activity(GO:0004120) 7,8-dihydro-D-neopterin 2',3'-cyclic phosphate phosphodiesterase activity(GO:0044688) inositol phosphosphingolipid phospholipase activity(GO:0052712) inositol phosphorylceramide phospholipase activity(GO:0052713) mannosyl-inositol phosphorylceramide phospholipase activity(GO:0052714) mannosyl-diinositol phosphorylceramide phospholipase activity(GO:0052715) |
| 0.0 | 0.1 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.0 | 0.1 | GO:0004096 | catalase activity(GO:0004096) |
| 0.0 | 0.1 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.0 | 0.1 | GO:0004470 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) |
| 0.0 | 0.1 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.2 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.0 | 0.1 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.2 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.1 | GO:0047192 | 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 0.0 | 0.4 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.2 | GO:0005542 | folic acid binding(GO:0005542) |
| 0.0 | 0.1 | GO:0046573 | lactonohydrolase activity(GO:0046573) acyl-L-homoserine-lactone lactonohydrolase activity(GO:0102007) |
| 0.0 | 0.1 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.0 | 0.1 | GO:0050072 | m7G(5')pppN diphosphatase activity(GO:0050072) |
| 0.0 | 0.1 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.0 | 0.1 | GO:0070191 | methionine-R-sulfoxide reductase activity(GO:0070191) |
| 0.0 | 0.1 | GO:0016899 | oxidoreductase activity, acting on the CH-OH group of donors, oxygen as acceptor(GO:0016899) |
| 0.0 | 0.0 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.0 | 0.0 | GO:0004103 | choline kinase activity(GO:0004103) |
| 0.0 | 0.1 | GO:0071614 | linoleic acid epoxygenase activity(GO:0071614) |
| 0.0 | 0.1 | GO:0086007 | voltage-gated calcium channel activity involved in cardiac muscle cell action potential(GO:0086007) |
| 0.0 | 0.1 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.0 | 0.0 | GO:0035650 | AP-1 adaptor complex binding(GO:0035650) |
| 0.0 | 0.1 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.0 | 0.1 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.6 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.1 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.0 | 0.1 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.0 | 0.0 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.0 | 0.0 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 0.1 | GO:0017089 | glycolipid transporter activity(GO:0017089) |
| 0.0 | 0.1 | GO:0015173 | aromatic amino acid transmembrane transporter activity(GO:0015173) |
| 0.0 | 0.0 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.1 | GO:0005072 | transforming growth factor beta receptor, cytoplasmic mediator activity(GO:0005072) |
| 0.0 | 0.1 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.0 | 0.1 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.0 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 0.0 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.0 | 0.0 | GO:0035175 | histone kinase activity (H3-S10 specific)(GO:0035175) |
| 0.0 | 0.0 | GO:0008865 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.0 | 0.1 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.0 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 0.0 | 0.0 | GO:0008147 | structural constituent of bone(GO:0008147) |
| 0.0 | 0.1 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.2 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.2 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.0 | 0.1 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.0 | 0.3 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.3 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 0.1 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.1 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.0 | GO:0031697 | beta-1 adrenergic receptor binding(GO:0031697) |
| 0.0 | 0.1 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.0 | 0.0 | GO:0000104 | succinate dehydrogenase activity(GO:0000104) |
| 0.0 | 0.0 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.1 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.0 | 0.2 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.0 | 0.1 | GO:0043743 | cobinamide kinase activity(GO:0008819) phytol kinase activity(GO:0010276) phenol kinase activity(GO:0018720) cyclin-dependent protein kinase activating kinase regulator activity(GO:0019914) inositol tetrakisphosphate 2-kinase activity(GO:0032942) heptose 7-phosphate kinase activity(GO:0033785) aminoglycoside phosphotransferase activity(GO:0034071) eukaryotic elongation factor-2 kinase regulator activity(GO:0042556) eukaryotic elongation factor-2 kinase activator activity(GO:0042557) LPPG:FO 2-phospho-L-lactate transferase activity(GO:0043743) cytidine kinase activity(GO:0043771) glycerate 2-kinase activity(GO:0043798) (S)-lactate 2-kinase activity(GO:0043841) phosphoserine:homoserine phosphotransferase activity(GO:0043899) L-seryl-tRNA(Sec) kinase activity(GO:0043915) phosphocholine transferase activity(GO:0044605) GTP-dependent polynucleotide kinase activity(GO:0051735) farnesol kinase activity(GO:0052668) CTP:2-trans,-6-trans-farnesol kinase activity(GO:0052669) geraniol kinase activity(GO:0052670) geranylgeraniol kinase activity(GO:0052671) CTP:geranylgeraniol kinase activity(GO:0052672) prenol kinase activity(GO:0052673) 1-phosphatidylinositol-5-kinase activity(GO:0052810) 1-phosphatidylinositol-3-phosphate 4-kinase activity(GO:0052811) phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) inositol-3,4,6-trisphosphate 1-kinase activity(GO:0052835) inositol 5-diphosphate pentakisphosphate 5-kinase activity(GO:0052836) inositol diphosphate tetrakisphosphate kinase activity(GO:0052839) |
| 0.0 | 0.0 | GO:0050567 | glutaminyl-tRNA synthase (glutamine-hydrolyzing) activity(GO:0050567) |
| 0.0 | 0.0 | GO:1990932 | 5.8S rRNA binding(GO:1990932) |
| 0.0 | 0.1 | GO:0043184 | vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 0.0 | 0.1 | GO:0070513 | death domain binding(GO:0070513) |
| 0.0 | 0.0 | GO:0003958 | NADPH-hemoprotein reductase activity(GO:0003958) |
| 0.0 | 0.1 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.0 | GO:0098518 | polynucleotide phosphatase activity(GO:0098518) |
| 0.0 | 0.1 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.0 | 0.2 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.0 | 0.2 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.1 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.0 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.1 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.1 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) |
| 0.0 | 0.1 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.0 | 0.1 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.0 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 0.0 | 0.1 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.0 | 0.1 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.0 | GO:0052658 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) |
| 0.0 | 0.0 | GO:0070698 | type I activin receptor binding(GO:0070698) |
| 0.0 | 0.0 | GO:0051032 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.0 | 0.0 | GO:0004370 | glycerol kinase activity(GO:0004370) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.0 | 0.2 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.4 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 0.6 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.3 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.2 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.0 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.7 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 0.1 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.0 | 0.7 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.2 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.0 | 0.1 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 0.5 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.2 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 0.1 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 0.1 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.4 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.2 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 0.2 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.0 | 0.1 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.1 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.0 | 0.2 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.0 | 0.1 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 0.1 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 0.4 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.2 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.0 | 0.4 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.4 | REACTOME SIGNALING BY NOTCH4 | Genes involved in Signaling by NOTCH4 |
| 0.0 | 0.5 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.0 | 0.0 | REACTOME DEADENYLATION DEPENDENT MRNA DECAY | Genes involved in Deadenylation-dependent mRNA decay |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS | Genes involved in Synthesis of bile acids and bile salts |
| 0.0 | 0.8 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 0.5 | REACTOME FATTY ACYL COA BIOSYNTHESIS | Genes involved in Fatty Acyl-CoA Biosynthesis |
| 0.0 | 0.2 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.5 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.0 | REACTOME VIF MEDIATED DEGRADATION OF APOBEC3G | Genes involved in Vif-mediated degradation of APOBEC3G |
| 0.0 | 0.2 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.5 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.2 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.2 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.0 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.0 | 0.2 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 0.0 | REACTOME P53 INDEPENDENT G1 S DNA DAMAGE CHECKPOINT | Genes involved in p53-Independent G1/S DNA damage checkpoint |
| 0.0 | 0.1 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.3 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.0 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.2 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.2 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.2 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 0.1 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.2 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.4 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.1 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.0 | 0.1 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 0.4 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.0 | 0.3 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.3 | REACTOME RORA ACTIVATES CIRCADIAN EXPRESSION | Genes involved in RORA Activates Circadian Expression |
| 0.0 | 1.9 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.4 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.6 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.2 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.0 | 0.2 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 0.2 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.1 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 0.1 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.4 | REACTOME TRANSPORT OF MATURE MRNA DERIVED FROM AN INTRONLESS TRANSCRIPT | Genes involved in Transport of Mature mRNA Derived from an Intronless Transcript |
| 0.0 | 0.2 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.0 | REACTOME LIPOPROTEIN METABOLISM | Genes involved in Lipoprotein metabolism |
| 0.0 | 0.3 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.4 | REACTOME TRANSCRIPTIONAL ACTIVITY OF SMAD2 SMAD3 SMAD4 HETEROTRIMER | Genes involved in Transcriptional activity of SMAD2/SMAD3:SMAD4 heterotrimer |
| 0.0 | 0.0 | REACTOME TRIGLYCERIDE BIOSYNTHESIS | Genes involved in Triglyceride Biosynthesis |
| 0.0 | 0.3 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.0 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.0 | 0.0 | REACTOME ER PHAGOSOME PATHWAY | Genes involved in ER-Phagosome pathway |
| 0.0 | 0.1 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.0 | 0.1 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |