Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
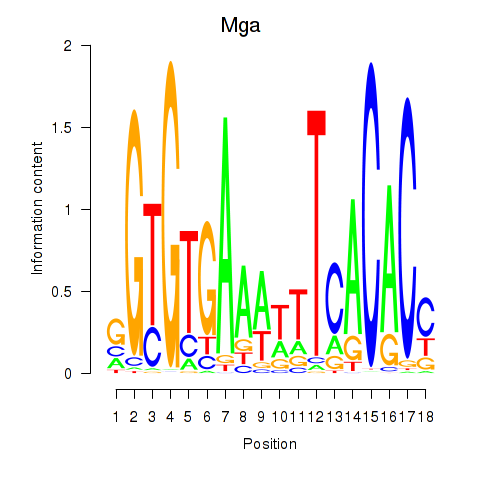
Results for Mga
Z-value: 1.21
Transcription factors associated with Mga
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Mga
|
ENSMUSG00000033943.9 | MAX gene associated |
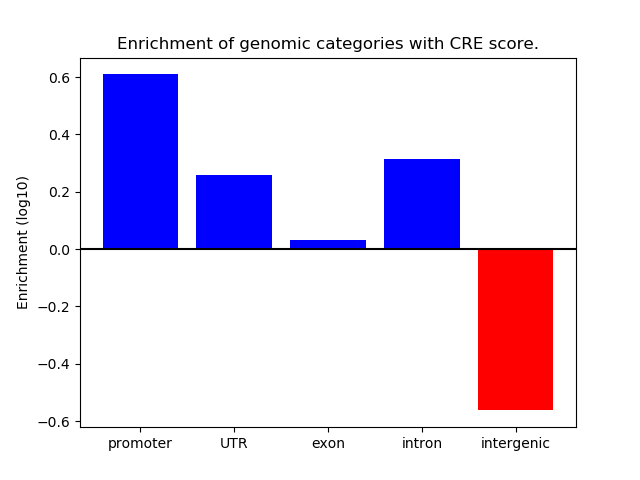
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr2_119897434_119897605 | Mga | 214 | 0.922854 | -0.88 | 2.0e-02 | Click! |
| chr2_119898035_119898287 | Mga | 856 | 0.554990 | 0.88 | 2.2e-02 | Click! |
| chr2_119897133_119897292 | Mga | 16 | 0.972614 | -0.83 | 4.2e-02 | Click! |
| chr2_119902218_119902387 | Mga | 4997 | 0.175261 | -0.80 | 5.6e-02 | Click! |
| chr2_119887854_119888033 | Mga | 9285 | 0.150912 | 0.75 | 8.6e-02 | Click! |
Activity of the Mga motif across conditions
Conditions sorted by the z-value of the Mga motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
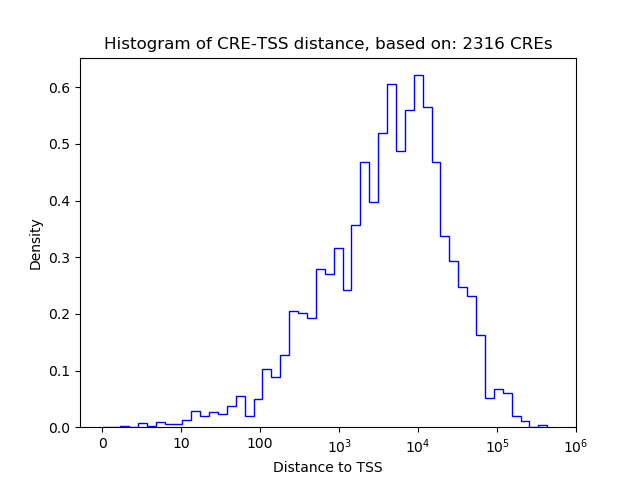
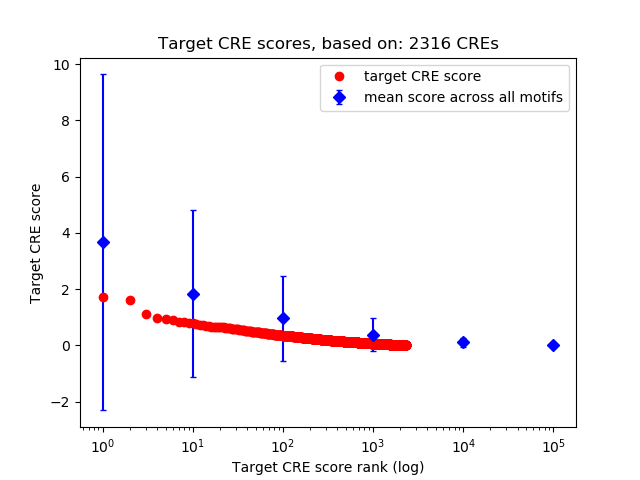
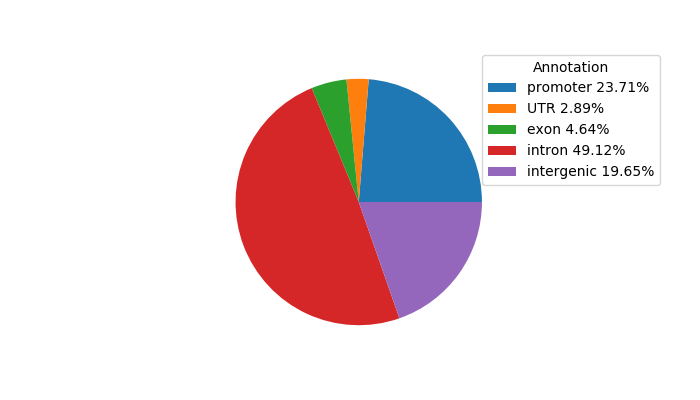
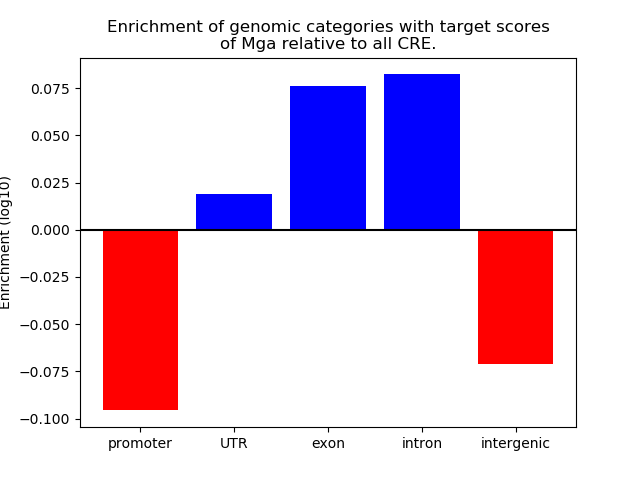
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr5_125526040_125526191 | 1.71 |
Tmem132b |
transmembrane protein 132B |
5659 |
0.17 |
| chr6_145866005_145866307 | 1.60 |
Bhlhe41 |
basic helix-loop-helix family, member e41 |
598 |
0.7 |
| chr11_7816093_7816451 | 1.11 |
Gm27393 |
predicted gene, 27393 |
70233 |
0.13 |
| chr18_38856344_38856590 | 0.98 |
Fgf1 |
fibroblast growth factor 1 |
2281 |
0.33 |
| chr14_56617767_56617918 | 0.95 |
Gm16573 |
predicted gene 16573 |
2655 |
0.22 |
| chr1_21248997_21249314 | 0.90 |
Gsta3 |
glutathione S-transferase, alpha 3 |
4366 |
0.13 |
| chr9_119299844_119299995 | 0.84 |
Gm22729 |
predicted gene, 22729 |
6838 |
0.14 |
| chr3_18146058_18146209 | 0.81 |
Gm23686 |
predicted gene, 23686 |
31492 |
0.18 |
| chr8_93193292_93193577 | 0.79 |
Gm45909 |
predicted gene 45909 |
2076 |
0.23 |
| chr9_21837693_21838006 | 0.79 |
Angptl8 |
angiopoietin-like 8 |
2339 |
0.17 |
| chr3_95864904_95865315 | 0.74 |
Mrps21 |
mitochondrial ribosomal protein S21 |
1802 |
0.16 |
| chr2_31501565_31501761 | 0.74 |
Ass1 |
argininosuccinate synthetase 1 |
937 |
0.56 |
| chr4_76942367_76942518 | 0.72 |
Gm11246 |
predicted gene 11246 |
15131 |
0.21 |
| chr10_87865511_87865999 | 0.70 |
Igf1os |
insulin-like growth factor 1, opposite strand |
2374 |
0.3 |
| chr9_106234809_106235123 | 0.67 |
Alas1 |
aminolevulinic acid synthase 1 |
2118 |
0.17 |
| chr2_70838613_70839186 | 0.67 |
Tlk1 |
tousled-like kinase 1 |
13171 |
0.18 |
| chr5_125524109_125525122 | 0.67 |
Tmem132b |
transmembrane protein 132B |
7159 |
0.16 |
| chr3_97653036_97653209 | 0.65 |
Prkab2 |
protein kinase, AMP-activated, beta 2 non-catalytic subunit |
5071 |
0.15 |
| chr2_177470070_177470297 | 0.65 |
Zfp970 |
zinc finger protein 970 |
5337 |
0.17 |
| chr15_97032998_97033149 | 0.65 |
Slc38a4 |
solute carrier family 38, member 4 |
1862 |
0.46 |
| chr1_67104802_67105196 | 0.65 |
Cps1 |
carbamoyl-phosphate synthetase 1 |
18027 |
0.21 |
| chr2_77503355_77503533 | 0.64 |
Zfp385b |
zinc finger protein 385B |
16091 |
0.23 |
| chr18_75504426_75504613 | 0.63 |
Gm10532 |
predicted gene 10532 |
10126 |
0.27 |
| chr11_16821774_16821942 | 0.63 |
Egfros |
epidermal growth factor receptor, opposite strand |
8844 |
0.22 |
| chr7_70346773_70346973 | 0.62 |
Gm44948 |
predicted gene 44948 |
823 |
0.39 |
| chr2_177901070_177901508 | 0.61 |
Gm14327 |
predicted gene 14327 |
2996 |
0.19 |
| chr3_97632897_97633210 | 0.60 |
Fmo5 |
flavin containing monooxygenase 5 |
4170 |
0.16 |
| chr6_35256147_35256298 | 0.59 |
1810058I24Rik |
RIKEN cDNA 1810058I24 gene |
3490 |
0.19 |
| chrX_142478453_142478626 | 0.59 |
Gm25915 |
predicted gene, 25915 |
6908 |
0.19 |
| chr2_148034677_148034980 | 0.59 |
9030622O22Rik |
RIKEN cDNA 9030622O22 gene |
3442 |
0.21 |
| chr2_85071327_85071510 | 0.58 |
Tnks1bp1 |
tankyrase 1 binding protein 1 |
10454 |
0.12 |
| chr9_48724021_48724519 | 0.56 |
Zbtb16 |
zinc finger and BTB domain containing 16 |
111675 |
0.06 |
| chr1_88055038_88055189 | 0.55 |
Ugt1a10 |
UDP glycosyltransferase 1 family, polypeptide A10 |
275 |
0.78 |
| chr9_106231818_106232229 | 0.54 |
Alas1 |
aminolevulinic acid synthase 1 |
5061 |
0.11 |
| chr2_158413370_158413558 | 0.54 |
Ralgapb |
Ral GTPase activating protein, beta subunit (non-catalytic) |
3362 |
0.12 |
| chr11_16797325_16797505 | 0.53 |
Egfros |
epidermal growth factor receptor, opposite strand |
33287 |
0.16 |
| chr11_60205379_60205543 | 0.53 |
Srebf1 |
sterol regulatory element binding transcription factor 1 |
651 |
0.56 |
| chr1_92944221_92944381 | 0.53 |
Capn10 |
calpain 10 |
1597 |
0.22 |
| chr1_153742082_153742457 | 0.52 |
Rgs16 |
regulator of G-protein signaling 16 |
1920 |
0.18 |
| chr7_133011265_133011604 | 0.51 |
Ctbp2 |
C-terminal binding protein 2 |
3814 |
0.2 |
| chr6_37542487_37542638 | 0.50 |
Gm7463 |
predicted gene 7463 |
1148 |
0.52 |
| chr5_122292884_122293053 | 0.50 |
Pptc7 |
PTC7 protein phosphatase homolog |
2804 |
0.16 |
| chr1_91440412_91440614 | 0.49 |
Per2 |
period circadian clock 2 |
9352 |
0.11 |
| chr4_46832518_46832720 | 0.49 |
Gabbr2 |
gamma-aminobutyric acid (GABA) B receptor, 2 |
27283 |
0.22 |
| chr6_91580636_91580787 | 0.49 |
Gm45215 |
predicted gene 45215 |
14047 |
0.13 |
| chr7_114463119_114463303 | 0.49 |
Pde3b |
phosphodiesterase 3B, cGMP-inhibited |
42967 |
0.15 |
| chr19_18148360_18148511 | 0.48 |
Gm18610 |
predicted gene, 18610 |
57360 |
0.15 |
| chr12_17450888_17451065 | 0.48 |
Gm36752 |
predicted gene, 36752 |
14358 |
0.19 |
| chr9_55125183_55125348 | 0.47 |
Ube2q2 |
ubiquitin-conjugating enzyme E2Q family member 2 |
23589 |
0.13 |
| chr8_84841387_84841538 | 0.47 |
Rad23a |
RAD23 homolog A, nucleotide excision repair protein |
797 |
0.35 |
| chr10_68129785_68130007 | 0.47 |
Arid5b |
AT rich interactive domain 5B (MRF1-like) |
6730 |
0.27 |
| chr11_98295864_98296043 | 0.46 |
Gm20644 |
predicted gene 20644 |
10571 |
0.09 |
| chr17_84734402_84734930 | 0.46 |
Lrpprc |
leucine-rich PPR-motif containing |
3482 |
0.21 |
| chr10_61138290_61138447 | 0.46 |
Sgpl1 |
sphingosine phosphate lyase 1 |
8530 |
0.15 |
| chr4_6274068_6274223 | 0.46 |
Cyp7a1 |
cytochrome P450, family 7, subfamily a, polypeptide 1 |
1486 |
0.41 |
| chr10_87891898_87892303 | 0.45 |
Igf1os |
insulin-like growth factor 1, opposite strand |
28719 |
0.16 |
| chr4_109105267_109105418 | 0.45 |
Osbpl9 |
oxysterol binding protein-like 9 |
2689 |
0.29 |
| chr3_152189220_152189460 | 0.45 |
Dnajb4 |
DnaJ heat shock protein family (Hsp40) member B4 |
4505 |
0.14 |
| chr18_56413923_56414074 | 0.45 |
Gramd3 |
GRAM domain containing 3 |
5172 |
0.22 |
| chr8_40271252_40271403 | 0.44 |
Fgf20 |
fibroblast growth factor 20 |
15626 |
0.2 |
| chr11_28689224_28689493 | 0.44 |
2810471M01Rik |
RIKEN cDNA 2810471M01 gene |
7794 |
0.19 |
| chr11_3387371_3387539 | 0.44 |
Limk2 |
LIM motif-containing protein kinase 2 |
16079 |
0.11 |
| chr14_21422563_21422878 | 0.43 |
Gm25864 |
predicted gene, 25864 |
27754 |
0.17 |
| chr7_141373770_141373967 | 0.43 |
B230206H07Rik |
RIKEN cDNA B230206H07 gene |
8758 |
0.07 |
| chr9_90166671_90167004 | 0.43 |
Adamts7 |
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 7 |
3611 |
0.19 |
| chr3_129444766_129445057 | 0.42 |
Gm5712 |
predicted gene 5712 |
11238 |
0.17 |
| chr19_10060762_10061072 | 0.42 |
Fads3 |
fatty acid desaturase 3 |
4665 |
0.15 |
| chr7_90141326_90141489 | 0.41 |
Gm45222 |
predicted gene 45222 |
4541 |
0.13 |
| chr9_30906121_30906748 | 0.41 |
Adamts15 |
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 15 |
3223 |
0.28 |
| chr12_12271220_12271371 | 0.41 |
Fam49a |
family with sequence similarity 49, member A |
9106 |
0.29 |
| chr1_51815647_51815823 | 0.41 |
Myo1b |
myosin IB |
4811 |
0.21 |
| chr3_116520495_116520697 | 0.41 |
Dbt |
dihydrolipoamide branched chain transacylase E2 |
268 |
0.83 |
| chr1_9742772_9742949 | 0.40 |
1700034P13Rik |
RIKEN cDNA 1700034P13 gene |
4788 |
0.16 |
| chr10_96107868_96108070 | 0.40 |
Gm49817 |
predicted gene, 49817 |
11964 |
0.16 |
| chr19_40223143_40223322 | 0.40 |
Pdlim1 |
PDZ and LIM domain 1 (elfin) |
124 |
0.95 |
| chr8_93173439_93173590 | 0.40 |
Ces1d |
carboxylesterase 1D |
1775 |
0.27 |
| chr10_111329724_111329924 | 0.40 |
Gm40761 |
predicted gene, 40761 |
7300 |
0.21 |
| chr8_67923947_67924108 | 0.39 |
Psd3 |
pleckstrin and Sec7 domain containing 3 |
13095 |
0.23 |
| chr5_124353121_124353272 | 0.39 |
Cdk2ap1 |
CDK2 (cyclin-dependent kinase 2)-associated protein 1 |
963 |
0.41 |
| chr16_45924599_45924776 | 0.38 |
Gm15640 |
predicted gene 15640 |
11636 |
0.17 |
| chr13_69640079_69640317 | 0.38 |
Nsun2 |
NOL1/NOP2/Sun domain family member 2 |
9458 |
0.1 |
| chr6_116056575_116057222 | 0.38 |
Tmcc1 |
transmembrane and coiled coil domains 1 |
57 |
0.97 |
| chr1_124069161_124069328 | 0.38 |
Dpp10 |
dipeptidylpeptidase 10 |
23685 |
0.27 |
| chr17_64740276_64740464 | 0.38 |
Gm17133 |
predicted gene 17133 |
13726 |
0.19 |
| chr11_98563759_98564290 | 0.37 |
Zpbp2 |
zona pellucida binding protein 2 |
8581 |
0.11 |
| chrY_90798307_90798477 | 0.37 |
Gm47283 |
predicted gene, 47283 |
7941 |
0.18 |
| chr17_5261920_5262119 | 0.37 |
Gm29050 |
predicted gene 29050 |
126744 |
0.05 |
| chr5_124234244_124234932 | 0.37 |
Gm42425 |
predicted gene 42425 |
3136 |
0.14 |
| chr5_102010547_102010959 | 0.37 |
Wdfy3 |
WD repeat and FYVE domain containing 3 |
29195 |
0.16 |
| chr6_52009095_52009246 | 0.37 |
Skap2 |
src family associated phosphoprotein 2 |
3345 |
0.2 |
| chr17_81386388_81386573 | 0.36 |
Gm50044 |
predicted gene, 50044 |
15647 |
0.24 |
| chr7_99132344_99132620 | 0.36 |
Uvrag |
UV radiation resistance associated gene |
8636 |
0.13 |
| chr11_11960671_11960874 | 0.36 |
Grb10 |
growth factor receptor bound protein 10 |
4748 |
0.24 |
| chr8_122047070_122047266 | 0.36 |
Banp |
BTG3 associated nuclear protein |
46321 |
0.11 |
| chr6_116208623_116208898 | 0.36 |
Washc2 |
WASH complex subunit 2` |
244 |
0.79 |
| chr18_39437876_39438027 | 0.36 |
Gm15337 |
predicted gene 15337 |
48526 |
0.13 |
| chr13_9057691_9057882 | 0.35 |
Gm36264 |
predicted gene, 36264 |
18665 |
0.13 |
| chr5_9108773_9108929 | 0.35 |
Tmem243 |
transmembrane protein 243, mitochondrial |
4397 |
0.19 |
| chr7_19701985_19702180 | 0.35 |
Tomm40 |
translocase of outer mitochondrial membrane 40 |
1058 |
0.26 |
| chr4_76485956_76486132 | 0.35 |
Gm42303 |
predicted gene, 42303 |
34813 |
0.18 |
| chr10_87523673_87523889 | 0.35 |
Pah |
phenylalanine hydroxylase |
1754 |
0.37 |
| chr3_152407292_152407443 | 0.34 |
Zzz3 |
zinc finger, ZZ domain containing 3 |
10673 |
0.11 |
| chr6_108290766_108291042 | 0.34 |
Itpr1 |
inositol 1,4,5-trisphosphate receptor 1 |
37946 |
0.19 |
| chr12_84328286_84328437 | 0.34 |
Zfp410 |
zinc finger protein 410 |
921 |
0.43 |
| chr15_38068899_38069087 | 0.34 |
Ubr5 |
ubiquitin protein ligase E3 component n-recognin 5 |
9432 |
0.15 |
| chr4_99046002_99046218 | 0.34 |
Dock7 |
dedicator of cytokinesis 7 |
9223 |
0.18 |
| chr9_122849425_122849576 | 0.34 |
Gm47140 |
predicted gene, 47140 |
1082 |
0.34 |
| chr2_92424553_92424704 | 0.34 |
Cry2 |
cryptochrome 2 (photolyase-like) |
7764 |
0.11 |
| chr9_65748739_65748890 | 0.34 |
Zfp609 |
zinc finger protein 609 |
47670 |
0.11 |
| chr6_24210867_24211018 | 0.33 |
Slc13a1 |
solute carrier family 13 (sodium/sulfate symporters), member 1 |
42850 |
0.16 |
| chr15_68363065_68363728 | 0.33 |
Gm20732 |
predicted gene 20732 |
220 |
0.9 |
| chr1_88024764_88024940 | 0.33 |
Gm15375 |
predicted gene 15375 |
359 |
0.71 |
| chr12_32822209_32822364 | 0.33 |
Nampt |
nicotinamide phosphoribosyltransferase |
1485 |
0.44 |
| chrX_77496822_77496989 | 0.33 |
Tbl1x |
transducin (beta)-like 1 X-linked |
14108 |
0.22 |
| chr1_190171694_190171885 | 0.33 |
Prox1 |
prospero homeobox 1 |
1075 |
0.41 |
| chr13_52979915_52980103 | 0.33 |
Nfil3 |
nuclear factor, interleukin 3, regulated |
1064 |
0.5 |
| chr1_67192197_67192413 | 0.33 |
Gm15668 |
predicted gene 15668 |
56895 |
0.12 |
| chr10_93884020_93884171 | 0.33 |
Metap2 |
methionine aminopeptidase 2 |
3391 |
0.17 |
| chr6_82053565_82053903 | 0.33 |
Gm15864 |
predicted gene 15864 |
1153 |
0.38 |
| chr16_20715133_20715551 | 0.33 |
Clcn2 |
chloride channel, voltage-sensitive 2 |
769 |
0.38 |
| chr5_36621044_36621360 | 0.32 |
D5Ertd579e |
DNA segment, Chr 5, ERATO Doi 579, expressed |
53 |
0.96 |
| chr1_9744946_9745115 | 0.32 |
1700034P13Rik |
RIKEN cDNA 1700034P13 gene |
2618 |
0.2 |
| chr17_28429327_28429723 | 0.32 |
Fkbp5 |
FK506 binding protein 5 |
896 |
0.41 |
| chr2_146555306_146555646 | 0.32 |
4933406D12Rik |
RIKEN cDNA 4933406D12 gene |
12545 |
0.25 |
| chr17_26589554_26589705 | 0.32 |
Ergic1 |
endoplasmic reticulum-golgi intermediate compartment (ERGIC) 1 |
5247 |
0.13 |
| chr3_130059270_130059430 | 0.32 |
Sec24b |
Sec24 related gene family, member B (S. cerevisiae) |
1557 |
0.34 |
| chr8_77199905_77200064 | 0.32 |
Gm23260 |
predicted gene, 23260 |
5289 |
0.24 |
| chr2_63925859_63926184 | 0.32 |
Fign |
fidgetin |
171967 |
0.04 |
| chr13_44430370_44430571 | 0.32 |
1700029N11Rik |
RIKEN cDNA 1700029N11 gene |
9242 |
0.15 |
| chr13_64205035_64205186 | 0.31 |
Cdc14b |
CDC14 cell division cycle 14B |
4072 |
0.13 |
| chr9_106235366_106235517 | 0.31 |
Alas1 |
aminolevulinic acid synthase 1 |
1643 |
0.22 |
| chr15_25714947_25715172 | 0.31 |
Myo10 |
myosin X |
1188 |
0.53 |
| chr18_64484966_64485209 | 0.31 |
Fech |
ferrochelatase |
3854 |
0.19 |
| chr3_89113405_89113607 | 0.31 |
Gm29704 |
predicted gene, 29704 |
3218 |
0.1 |
| chr11_11845100_11845416 | 0.30 |
Ddc |
dopa decarboxylase |
8978 |
0.18 |
| chr8_93258499_93258650 | 0.30 |
Ces1f |
carboxylesterase 1F |
165 |
0.94 |
| chr3_52011173_52011324 | 0.30 |
Gm37465 |
predicted gene, 37465 |
7223 |
0.13 |
| chr19_8735955_8736119 | 0.30 |
Wdr74 |
WD repeat domain 74 |
155 |
0.8 |
| chr14_48058567_48058745 | 0.30 |
Gm49306 |
predicted gene, 49306 |
11645 |
0.15 |
| chr6_51578097_51578258 | 0.30 |
Snx10 |
sorting nexin 10 |
33611 |
0.16 |
| chr14_59084522_59084678 | 0.30 |
Gm48453 |
predicted gene, 48453 |
2518 |
0.26 |
| chr5_130019586_130019737 | 0.30 |
Asl |
argininosuccinate lyase |
4639 |
0.13 |
| chr15_59032105_59032256 | 0.29 |
Mtss1 |
MTSS I-BAR domain containing 1 |
8416 |
0.21 |
| chr2_52576598_52576771 | 0.29 |
Cacnb4 |
calcium channel, voltage-dependent, beta 4 subunit |
18117 |
0.18 |
| chr13_44445691_44446120 | 0.29 |
Gm27036 |
predicted gene, 27036 |
6092 |
0.16 |
| chr2_31477347_31478122 | 0.29 |
Ass1 |
argininosuccinate synthetase 1 |
7527 |
0.19 |
| chr5_23418501_23419033 | 0.29 |
5031425E22Rik |
RIKEN cDNA 5031425E22 gene |
14542 |
0.12 |
| chr5_139362838_139363176 | 0.29 |
Mir339 |
microRNA 339 |
6738 |
0.11 |
| chr7_29247733_29248167 | 0.29 |
2200002D01Rik |
RIKEN cDNA 2200002D01 gene |
388 |
0.72 |
| chr5_125513670_125513845 | 0.29 |
Aacs |
acetoacetyl-CoA synthetase |
440 |
0.79 |
| chr4_117125610_117126073 | 0.29 |
Btbd19 |
BTB (POZ) domain containing 19 |
116 |
0.86 |
| chr18_25554784_25554942 | 0.29 |
Gm3227 |
predicted gene 3227 |
42627 |
0.17 |
| chr3_15201420_15201600 | 0.29 |
Gm33819 |
predicted gene, 33819 |
30068 |
0.23 |
| chr3_145560024_145560207 | 0.29 |
Col24a1 |
collagen, type XXIV, alpha 1 |
15142 |
0.19 |
| chr4_104846208_104846359 | 0.29 |
C8a |
complement component 8, alpha polypeptide |
10240 |
0.19 |
| chr4_116703446_116704003 | 0.28 |
Mmachc |
methylmalonic aciduria cblC type, with homocystinuria |
4564 |
0.12 |
| chr10_50657463_50657625 | 0.28 |
Ascc3 |
activating signal cointegrator 1 complex subunit 3 |
8489 |
0.23 |
| chr13_95828416_95828713 | 0.28 |
Iqgap2 |
IQ motif containing GTPase activating protein 2 |
63193 |
0.1 |
| chr14_32162147_32162298 | 0.28 |
Ncoa4 |
nuclear receptor coactivator 4 |
2241 |
0.18 |
| chr5_104471233_104471571 | 0.28 |
Pkd2 |
polycystin 2, transient receptor potential cation channel |
11832 |
0.16 |
| chr15_79074715_79074866 | 0.28 |
Eif3l |
eukaryotic translation initiation factor 3, subunit L |
389 |
0.68 |
| chr11_16819865_16820197 | 0.28 |
Egfros |
epidermal growth factor receptor, opposite strand |
10671 |
0.22 |
| chr14_11801019_11801346 | 0.28 |
Gm48602 |
predicted gene, 48602 |
33239 |
0.19 |
| chr9_64281484_64281643 | 0.28 |
Tipin |
timeless interacting protein |
18 |
0.96 |
| chr7_19664934_19665237 | 0.28 |
Clptm1 |
cleft lip and palate associated transmembrane protein 1 |
52 |
0.93 |
| chr12_71368459_71368708 | 0.27 |
Gm40438 |
predicted gene, 40438 |
13142 |
0.15 |
| chr1_30940456_30940607 | 0.27 |
Ptp4a1 |
protein tyrosine phosphatase 4a1 |
3773 |
0.2 |
| chr19_10077162_10077329 | 0.27 |
Fads2 |
fatty acid desaturase 2 |
6058 |
0.15 |
| chr11_87702293_87702667 | 0.27 |
1110028F11Rik |
RIKEN cDNA 1110028F11 gene |
357 |
0.76 |
| chr17_26719114_26719268 | 0.27 |
Crebrf |
CREB3 regulatory factor |
3467 |
0.2 |
| chr18_62095425_62095728 | 0.27 |
Gm41750 |
predicted gene, 41750 |
47042 |
0.14 |
| chr2_58785851_58786309 | 0.27 |
Upp2 |
uridine phosphorylase 2 |
20755 |
0.19 |
| chr18_74931662_74931813 | 0.27 |
Gm18786 |
predicted gene, 18786 |
4543 |
0.12 |
| chr11_77788021_77788193 | 0.27 |
Gm10277 |
predicted gene 10277 |
360 |
0.83 |
| chr1_74028158_74028331 | 0.27 |
Tns1 |
tensin 1 |
8889 |
0.23 |
| chr4_141559216_141559981 | 0.26 |
B330016D10Rik |
RIKEN cDNA B330016D10 gene |
13409 |
0.12 |
| chr4_106580992_106581506 | 0.26 |
Gm12744 |
predicted gene 12744 |
7859 |
0.11 |
| chr9_21688894_21689087 | 0.26 |
Gm26511 |
predicted gene, 26511 |
34460 |
0.09 |
| chr7_89404848_89405054 | 0.26 |
Fzd4 |
frizzled class receptor 4 |
596 |
0.47 |
| chr14_45378763_45379131 | 0.26 |
Gnpnat1 |
glucosamine-phosphate N-acetyltransferase 1 |
4496 |
0.14 |
| chr7_45615310_45615638 | 0.26 |
Fgf21 |
fibroblast growth factor 21 |
16 |
0.91 |
| chr5_24931562_24931772 | 0.26 |
Prkag2 |
protein kinase, AMP-activated, gamma 2 non-catalytic subunit |
22690 |
0.15 |
| chr17_56051343_56051500 | 0.26 |
Chaf1a |
chromatin assembly factor 1, subunit A (p150) |
4607 |
0.1 |
| chr9_121677553_121677957 | 0.26 |
Vipr1 |
vasoactive intestinal peptide receptor 1 |
9042 |
0.11 |
| chr2_3335831_3335993 | 0.25 |
Acbd7 |
acyl-Coenzyme A binding domain containing 7 |
256 |
0.87 |
| chr2_58758962_58759113 | 0.25 |
Upp2 |
uridine phosphorylase 2 |
3825 |
0.24 |
| chr2_162989615_162989976 | 0.25 |
Sgk2 |
serum/glucocorticoid regulated kinase 2 |
2293 |
0.2 |
| chr2_72157539_72157839 | 0.25 |
Rapgef4os1 |
Rap guanine nucleotide exchange factor (GEF) 4, opposite strand 1 |
8893 |
0.2 |
| chr17_28702072_28702269 | 0.25 |
Mapk14 |
mitogen-activated protein kinase 14 |
9602 |
0.12 |
| chr15_85742021_85742427 | 0.25 |
Ppara |
peroxisome proliferator activated receptor alpha |
6093 |
0.16 |
| chr19_60589967_60590189 | 0.25 |
Cacul1 |
CDK2 associated, cullin domain 1 |
9053 |
0.18 |
| chr11_43170710_43170884 | 0.25 |
Atp10b |
ATPase, class V, type 10B |
15126 |
0.2 |
| chr16_32896347_32896498 | 0.25 |
Fyttd1 |
forty-two-three domain containing 1 |
1331 |
0.34 |
| chr13_81332008_81332361 | 0.25 |
Adgrv1 |
adhesion G protein-coupled receptor V1 |
10652 |
0.27 |
| chr4_116450268_116450448 | 0.25 |
Gm12950 |
predicted gene 12950 |
1570 |
0.34 |
| chr2_30981826_30982365 | 0.25 |
BC005624 |
cDNA sequence BC005624 |
106 |
0.61 |
| chr4_102557092_102557428 | 0.25 |
Pde4b |
phosphodiesterase 4B, cAMP specific |
12835 |
0.29 |
| chr13_41199632_41199844 | 0.25 |
Elovl2 |
elongation of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 2 |
20424 |
0.11 |
| chr16_41258426_41258587 | 0.25 |
Gm47276 |
predicted gene, 47276 |
40729 |
0.18 |
| chr1_84826270_84826425 | 0.25 |
Gm37738 |
predicted gene, 37738 |
1664 |
0.29 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.2 | 0.5 | GO:0010046 | response to mycotoxin(GO:0010046) |
| 0.1 | 1.2 | GO:0010944 | negative regulation of transcription by competitive promoter binding(GO:0010944) |
| 0.1 | 0.3 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.1 | 0.3 | GO:0003062 | regulation of heart rate by chemical signal(GO:0003062) |
| 0.1 | 0.4 | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.1 | 0.2 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.1 | 0.2 | GO:0060178 | regulation of exocyst localization(GO:0060178) |
| 0.1 | 0.2 | GO:0060741 | prostate gland stromal morphogenesis(GO:0060741) |
| 0.1 | 0.3 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.1 | 0.5 | GO:0052697 | xenobiotic glucuronidation(GO:0052697) |
| 0.1 | 0.2 | GO:0060375 | regulation of mast cell differentiation(GO:0060375) |
| 0.0 | 0.4 | GO:0070857 | regulation of bile acid biosynthetic process(GO:0070857) |
| 0.0 | 0.2 | GO:0070535 | histone H2A K63-linked ubiquitination(GO:0070535) |
| 0.0 | 0.4 | GO:0055091 | phospholipid homeostasis(GO:0055091) |
| 0.0 | 0.1 | GO:0072361 | regulation of glycolytic process by regulation of transcription from RNA polymerase II promoter(GO:0072361) |
| 0.0 | 0.1 | GO:0000715 | nucleotide-excision repair, DNA damage recognition(GO:0000715) |
| 0.0 | 0.2 | GO:0070126 | mitochondrial translational termination(GO:0070126) |
| 0.0 | 0.2 | GO:0090080 | positive regulation of MAPKKK cascade by fibroblast growth factor receptor signaling pathway(GO:0090080) |
| 0.0 | 0.2 | GO:0075525 | viral translational termination-reinitiation(GO:0075525) |
| 0.0 | 0.1 | GO:0006166 | purine ribonucleoside salvage(GO:0006166) |
| 0.0 | 0.0 | GO:0019859 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) thymine metabolic process(GO:0019859) |
| 0.0 | 0.1 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.0 | 0.2 | GO:0060613 | fat pad development(GO:0060613) |
| 0.0 | 0.1 | GO:0045356 | positive regulation of interferon-alpha biosynthetic process(GO:0045356) |
| 0.0 | 0.1 | GO:0006549 | isoleucine metabolic process(GO:0006549) |
| 0.0 | 0.1 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.0 | 0.1 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.0 | 0.1 | GO:1901162 | serotonin biosynthetic process(GO:0042427) primary amino compound biosynthetic process(GO:1901162) |
| 0.0 | 0.1 | GO:0034287 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 0.0 | 0.1 | GO:2000686 | regulation of rubidium ion transmembrane transporter activity(GO:2000686) |
| 0.0 | 0.2 | GO:0019856 | 'de novo' pyrimidine nucleobase biosynthetic process(GO:0006207) pyrimidine nucleobase biosynthetic process(GO:0019856) |
| 0.0 | 0.1 | GO:0007223 | Wnt signaling pathway, calcium modulating pathway(GO:0007223) |
| 0.0 | 0.1 | GO:1902226 | regulation of macrophage colony-stimulating factor signaling pathway(GO:1902226) regulation of response to macrophage colony-stimulating factor(GO:1903969) regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903972) |
| 0.0 | 0.1 | GO:1901078 | negative regulation of relaxation of muscle(GO:1901078) |
| 0.0 | 0.1 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.0 | 0.1 | GO:0021603 | cranial nerve formation(GO:0021603) |
| 0.0 | 0.0 | GO:0032489 | regulation of Cdc42 protein signal transduction(GO:0032489) |
| 0.0 | 0.2 | GO:0021540 | corpus callosum morphogenesis(GO:0021540) |
| 0.0 | 0.1 | GO:0097680 | double-strand break repair via classical nonhomologous end joining(GO:0097680) |
| 0.0 | 0.1 | GO:0001827 | inner cell mass cell fate commitment(GO:0001827) |
| 0.0 | 0.1 | GO:0019482 | beta-alanine metabolic process(GO:0019482) |
| 0.0 | 0.1 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.0 | 0.1 | GO:1990168 | protein K29-linked deubiquitination(GO:0035523) protein K33-linked deubiquitination(GO:1990168) |
| 0.0 | 0.1 | GO:0051562 | negative regulation of mitochondrial calcium ion concentration(GO:0051562) |
| 0.0 | 0.1 | GO:1903347 | negative regulation of bicellular tight junction assembly(GO:1903347) |
| 0.0 | 0.1 | GO:1903071 | positive regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903071) |
| 0.0 | 0.1 | GO:0048769 | sarcomerogenesis(GO:0048769) |
| 0.0 | 0.2 | GO:0010603 | regulation of cytoplasmic mRNA processing body assembly(GO:0010603) |
| 0.0 | 0.1 | GO:1903689 | regulation of wound healing, spreading of epidermal cells(GO:1903689) |
| 0.0 | 0.1 | GO:1901420 | negative regulation of response to alcohol(GO:1901420) |
| 0.0 | 0.1 | GO:1903334 | positive regulation of protein folding(GO:1903334) |
| 0.0 | 0.2 | GO:0099515 | actin filament-based transport(GO:0099515) |
| 0.0 | 0.1 | GO:0042663 | regulation of endodermal cell fate specification(GO:0042663) |
| 0.0 | 0.2 | GO:0044829 | positive regulation by host of viral genome replication(GO:0044829) |
| 0.0 | 0.1 | GO:2000468 | regulation of peroxidase activity(GO:2000468) |
| 0.0 | 0.1 | GO:0000960 | mitochondrial RNA catabolic process(GO:0000957) regulation of mitochondrial RNA catabolic process(GO:0000960) |
| 0.0 | 0.1 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.0 | 0.1 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.0 | 0.1 | GO:1904479 | negative regulation of intestinal absorption(GO:1904479) |
| 0.0 | 0.1 | GO:0042723 | thiamine metabolic process(GO:0006772) thiamine-containing compound metabolic process(GO:0042723) |
| 0.0 | 0.1 | GO:0010424 | DNA methylation on cytosine within a CG sequence(GO:0010424) DNA methylation on cytosine(GO:0032776) |
| 0.0 | 0.1 | GO:0006701 | progesterone biosynthetic process(GO:0006701) |
| 0.0 | 0.1 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.0 | 0.1 | GO:0002386 | immune response in mucosal-associated lymphoid tissue(GO:0002386) |
| 0.0 | 0.1 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.0 | 0.1 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.0 | 0.0 | GO:0019676 | ammonia assimilation cycle(GO:0019676) |
| 0.0 | 0.1 | GO:0048669 | collateral sprouting in absence of injury(GO:0048669) |
| 0.0 | 0.1 | GO:0015959 | diadenosine polyphosphate metabolic process(GO:0015959) |
| 0.0 | 0.1 | GO:0045719 | negative regulation of glycogen biosynthetic process(GO:0045719) |
| 0.0 | 0.1 | GO:0003186 | tricuspid valve morphogenesis(GO:0003186) tricuspid valve formation(GO:0003195) |
| 0.0 | 0.1 | GO:2000302 | positive regulation of synaptic vesicle exocytosis(GO:2000302) |
| 0.0 | 0.1 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.0 | 0.1 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.0 | 0.1 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 0.1 | GO:0045196 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.0 | 0.1 | GO:0097343 | ripoptosome assembly(GO:0097343) ripoptosome assembly involved in necroptotic process(GO:1901026) |
| 0.0 | 0.1 | GO:2000676 | positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.0 | 0.1 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 0.0 | 0.0 | GO:0018879 | biphenyl metabolic process(GO:0018879) |
| 0.0 | 0.1 | GO:0034756 | regulation of iron ion transport(GO:0034756) |
| 0.0 | 0.1 | GO:0038171 | cannabinoid signaling pathway(GO:0038171) endocannabinoid signaling pathway(GO:0071926) |
| 0.0 | 0.2 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 0.1 | GO:0036091 | positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 0.0 | 0.1 | GO:0002904 | positive regulation of B cell apoptotic process(GO:0002904) |
| 0.0 | 0.1 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.0 | 0.1 | GO:0006105 | succinate metabolic process(GO:0006105) |
| 0.0 | 0.1 | GO:1903797 | positive regulation of inorganic anion transmembrane transport(GO:1903797) |
| 0.0 | 0.1 | GO:0045046 | peroxisomal membrane transport(GO:0015919) protein import into peroxisome membrane(GO:0045046) |
| 0.0 | 0.1 | GO:0042732 | D-xylose metabolic process(GO:0042732) |
| 0.0 | 0.1 | GO:1903223 | positive regulation of oxidative stress-induced neuron death(GO:1903223) |
| 0.0 | 0.1 | GO:0072566 | chemokine (C-X-C motif) ligand 1 production(GO:0072566) regulation of chemokine (C-X-C motif) ligand 1 production(GO:2000338) |
| 0.0 | 0.1 | GO:0034310 | primary alcohol catabolic process(GO:0034310) |
| 0.0 | 0.1 | GO:0061368 | behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 0.0 | 0.1 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.1 | GO:0042518 | negative regulation of tyrosine phosphorylation of Stat3 protein(GO:0042518) |
| 0.0 | 0.1 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.0 | 0.1 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.0 | 0.1 | GO:0060689 | cell differentiation involved in salivary gland development(GO:0060689) |
| 0.0 | 0.1 | GO:0006307 | DNA dealkylation involved in DNA repair(GO:0006307) |
| 0.0 | 0.1 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.0 | 0.6 | GO:0070542 | response to fatty acid(GO:0070542) |
| 0.0 | 0.1 | GO:0036233 | glycine import(GO:0036233) |
| 0.0 | 0.1 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.0 | 0.0 | GO:0031587 | positive regulation of inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0031587) |
| 0.0 | 0.1 | GO:0042148 | strand invasion(GO:0042148) |
| 0.0 | 0.1 | GO:0046984 | regulation of hemoglobin biosynthetic process(GO:0046984) |
| 0.0 | 0.0 | GO:0021699 | cerebellar cortex maturation(GO:0021699) |
| 0.0 | 0.1 | GO:0010838 | positive regulation of keratinocyte proliferation(GO:0010838) |
| 0.0 | 0.0 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.0 | GO:0035459 | cargo loading into vesicle(GO:0035459) |
| 0.0 | 0.3 | GO:0017144 | drug metabolic process(GO:0017144) |
| 0.0 | 0.0 | GO:0009212 | dTTP biosynthetic process(GO:0006235) pyrimidine deoxyribonucleoside triphosphate biosynthetic process(GO:0009212) dTTP metabolic process(GO:0046075) |
| 0.0 | 0.0 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 0.0 | 0.1 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.0 | 0.1 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.0 | 0.1 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.0 | 0.4 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 0.0 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.0 | 0.0 | GO:1902109 | negative regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902109) |
| 0.0 | 0.1 | GO:0061032 | visceral serous pericardium development(GO:0061032) |
| 0.0 | 0.1 | GO:1903242 | regulation of cardiac muscle adaptation(GO:0010612) regulation of cardiac muscle hypertrophy in response to stress(GO:1903242) |
| 0.0 | 0.2 | GO:0051770 | positive regulation of nitric-oxide synthase biosynthetic process(GO:0051770) |
| 0.0 | 0.2 | GO:0009404 | toxin metabolic process(GO:0009404) |
| 0.0 | 0.1 | GO:0033962 | cytoplasmic mRNA processing body assembly(GO:0033962) |
| 0.0 | 0.2 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.0 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 0.0 | 0.1 | GO:0060839 | endothelial cell fate commitment(GO:0060839) |
| 0.0 | 0.0 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.0 | 0.0 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.0 | 0.0 | GO:0071336 | regulation of hair follicle cell proliferation(GO:0071336) |
| 0.0 | 0.1 | GO:0060087 | relaxation of vascular smooth muscle(GO:0060087) |
| 0.0 | 0.1 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.0 | 0.2 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.0 | GO:0021553 | olfactory nerve development(GO:0021553) |
| 0.0 | 0.0 | GO:0006041 | glucosamine metabolic process(GO:0006041) |
| 0.0 | 0.0 | GO:0060084 | synaptic transmission involved in micturition(GO:0060084) |
| 0.0 | 0.0 | GO:0000320 | re-entry into mitotic cell cycle(GO:0000320) |
| 0.0 | 0.1 | GO:0046146 | tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.0 | 0.0 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.0 | GO:0090283 | regulation of protein glycosylation in Golgi(GO:0090283) |
| 0.0 | 0.0 | GO:1900060 | negative regulation of ceramide biosynthetic process(GO:1900060) |
| 0.0 | 0.0 | GO:0042532 | negative regulation of tyrosine phosphorylation of STAT protein(GO:0042532) |
| 0.0 | 0.0 | GO:0010891 | negative regulation of sequestering of triglyceride(GO:0010891) |
| 0.0 | 0.0 | GO:0046271 | phenylpropanoid metabolic process(GO:0009698) phenylpropanoid catabolic process(GO:0046271) |
| 0.0 | 0.1 | GO:0018206 | peptidyl-methionine modification(GO:0018206) |
| 0.0 | 0.1 | GO:0045634 | regulation of melanocyte differentiation(GO:0045634) |
| 0.0 | 0.0 | GO:0070318 | positive regulation of G0 to G1 transition(GO:0070318) |
| 0.0 | 0.0 | GO:0010751 | negative regulation of nitric oxide mediated signal transduction(GO:0010751) |
| 0.0 | 0.1 | GO:0010992 | ubiquitin homeostasis(GO:0010992) |
| 0.0 | 0.0 | GO:0048254 | snoRNA localization(GO:0048254) |
| 0.0 | 0.1 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.0 | 0.0 | GO:1900103 | positive regulation of endoplasmic reticulum unfolded protein response(GO:1900103) |
| 0.0 | 0.1 | GO:0006699 | bile acid biosynthetic process(GO:0006699) |
| 0.0 | 0.2 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.0 | 0.0 | GO:2000847 | negative regulation of steroid hormone secretion(GO:2000832) negative regulation of corticosteroid hormone secretion(GO:2000847) negative regulation of glucocorticoid secretion(GO:2000850) |
| 0.0 | 0.0 | GO:0060331 | negative regulation of response to interferon-gamma(GO:0060331) negative regulation of interferon-gamma-mediated signaling pathway(GO:0060336) |
| 0.0 | 0.1 | GO:0009256 | 10-formyltetrahydrofolate metabolic process(GO:0009256) |
| 0.0 | 0.0 | GO:0044821 | meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.0 | 0.1 | GO:0098908 | regulation of neuronal action potential(GO:0098908) |
| 0.0 | 0.0 | GO:0090135 | actin filament branching(GO:0090135) |
| 0.0 | 0.1 | GO:1901663 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.0 | 0.0 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.0 | 0.0 | GO:1990928 | response to amino acid starvation(GO:1990928) |
| 0.0 | 0.1 | GO:0052696 | flavonoid biosynthetic process(GO:0009813) flavonoid glucuronidation(GO:0052696) |
| 0.0 | 0.1 | GO:0000059 | protein import into nucleus, docking(GO:0000059) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.1 | 0.4 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.1 | 0.6 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.1 | 0.2 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.0 | 0.2 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 0.1 | GO:0071942 | XPC complex(GO:0071942) |
| 0.0 | 0.2 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.3 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.1 | GO:0097648 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor complex(GO:0097648) |
| 0.0 | 0.2 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.1 | GO:0097413 | Lewy body(GO:0097413) |
| 0.0 | 0.1 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.0 | 0.1 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.1 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.0 | 0.1 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.1 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.1 | GO:0097057 | TRAF2-GSTP1 complex(GO:0097057) |
| 0.0 | 0.2 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.0 | 0.1 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.0 | 0.1 | GO:0002142 | stereocilia ankle link complex(GO:0002142) |
| 0.0 | 0.1 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.1 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.0 | 0.1 | GO:0031417 | NatC complex(GO:0031417) |
| 0.0 | 0.2 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.1 | GO:0098536 | deuterosome(GO:0098536) |
| 0.0 | 0.1 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.0 | 0.1 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 1.3 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.0 | GO:1990597 | AIP1-IRE1 complex(GO:1990597) |
| 0.0 | 0.1 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.1 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 0.1 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.0 | 0.1 | GO:0032009 | early phagosome(GO:0032009) |
| 0.0 | 0.1 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 0.0 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.0 | 0.1 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.1 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.1 | GO:0000306 | extrinsic component of vacuolar membrane(GO:0000306) |
| 0.0 | 0.0 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 0.0 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.0 | 0.1 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.1 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.0 | 0.6 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.0 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.0 | 0.1 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.0 | 0.1 | GO:0046930 | pore complex(GO:0046930) |
| 0.0 | 0.1 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.0 | 0.2 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) |
| 0.0 | 0.1 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.0 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.0 | 0.0 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.1 | GO:0042824 | MHC class I peptide loading complex(GO:0042824) |
| 0.0 | 0.0 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.0 | 0.3 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 0.0 | GO:1990635 | proximal dendrite(GO:1990635) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.1 | GO:0043426 | MRF binding(GO:0043426) |
| 0.1 | 0.6 | GO:0047760 | butyrate-CoA ligase activity(GO:0047760) |
| 0.1 | 0.2 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.1 | 0.2 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.1 | 0.2 | GO:0016743 | carboxyl- or carbamoyltransferase activity(GO:0016743) |
| 0.0 | 0.1 | GO:0016149 | translation release factor activity, codon specific(GO:0016149) |
| 0.0 | 0.6 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.1 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.0 | 0.1 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 0.2 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.2 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 0.1 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.0 | 0.7 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.1 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 0.0 | 0.1 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.0 | 0.1 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.1 | GO:0016635 | oxidoreductase activity, acting on the CH-CH group of donors, quinone or related compound as acceptor(GO:0016635) |
| 0.0 | 0.1 | GO:0050252 | retinol O-fatty-acyltransferase activity(GO:0050252) |
| 0.0 | 0.2 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.0 | 0.1 | GO:0043398 | HLH domain binding(GO:0043398) |
| 0.0 | 0.2 | GO:0016215 | acyl-CoA desaturase activity(GO:0016215) |
| 0.0 | 0.3 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.0 | 0.1 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.2 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.0 | 0.0 | GO:0043125 | ErbB-3 class receptor binding(GO:0043125) |
| 0.0 | 0.1 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.1 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.0 | 0.7 | GO:0010296 | prenylcysteine methylesterase activity(GO:0010296) 1-oxa-2-oxocycloheptane lactonase activity(GO:0018731) sulfolactone hydrolase activity(GO:0018732) butyrolactone hydrolase activity(GO:0018734) endosulfan lactone lactonase activity(GO:0034892) L-ascorbate 6-phosphate lactonase activity(GO:0035460) Ser-tRNA(Thr) hydrolase activity(GO:0043905) Ala-tRNA(Pro) hydrolase activity(GO:0043906) Cys-tRNA(Pro) hydrolase activity(GO:0043907) Ser(Gly)-tRNA(Ala) hydrolase activity(GO:0043908) all-trans-retinyl-palmitate hydrolase, all-trans-retinol forming activity(GO:0047376) mannosyl-oligosaccharide 1,6-alpha-mannosidase activity(GO:0052767) mannosyl-oligosaccharide 1,3-alpha-mannosidase activity(GO:0052768) methyl indole-3-acetate esterase activity(GO:0080030) methyl salicylate esterase activity(GO:0080031) methyl jasmonate esterase activity(GO:0080032) |
| 0.0 | 0.1 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.0 | 0.1 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.0 | 0.1 | GO:0052658 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) |
| 0.0 | 0.1 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 0.1 | GO:0015375 | glycine:sodium symporter activity(GO:0015375) |
| 0.0 | 0.2 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.1 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.0 | 0.1 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.0 | 0.2 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.0 | 0.2 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 0.1 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.0 | 0.0 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.0 | 0.1 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 0.0 | GO:0019797 | procollagen-proline 3-dioxygenase activity(GO:0019797) |
| 0.0 | 0.1 | GO:0016401 | palmitoyl-CoA oxidase activity(GO:0016401) |
| 0.0 | 0.3 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.0 | 0.0 | GO:0004477 | methenyltetrahydrofolate cyclohydrolase activity(GO:0004477) methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
| 0.0 | 0.1 | GO:0035671 | enone reductase activity(GO:0035671) |
| 0.0 | 0.1 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.0 | 0.2 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.1 | GO:0097642 | calcitonin family receptor activity(GO:0097642) |
| 0.0 | 0.0 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 0.0 | GO:0030350 | iron-responsive element binding(GO:0030350) |
| 0.0 | 0.1 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 0.3 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.0 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.0 | 0.0 | GO:0031893 | vasopressin receptor binding(GO:0031893) |
| 0.0 | 0.0 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.0 | 0.0 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) |
| 0.0 | 0.0 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 0.0 | 0.2 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.0 | GO:0003846 | 2-acylglycerol O-acyltransferase activity(GO:0003846) |
| 0.0 | 0.0 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.0 | 0.1 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.1 | GO:0048019 | receptor antagonist activity(GO:0048019) |
| 0.0 | 0.6 | GO:0016763 | transferase activity, transferring pentosyl groups(GO:0016763) |
| 0.0 | 0.0 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 0.0 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 0.1 | GO:0005072 | transforming growth factor beta receptor, cytoplasmic mediator activity(GO:0005072) |
| 0.0 | 0.0 | GO:0038181 | bile acid receptor activity(GO:0038181) |
| 0.0 | 0.0 | GO:0008079 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.0 | 0.1 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.0 | 0.2 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.3 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.0 | GO:0035515 | oxidative RNA demethylase activity(GO:0035515) |
| 0.0 | 0.0 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.0 | 0.0 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.0 | 0.0 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.0 | 0.0 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.0 | 0.0 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.0 | 0.1 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) |
| 0.0 | 0.4 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 0.1 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.0 | 0.2 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.2 | GO:1900750 | glutathione binding(GO:0043295) oligopeptide binding(GO:1900750) |
| 0.0 | 0.0 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.3 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.2 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 0.4 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 0.7 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | REACTOME RORA ACTIVATES CIRCADIAN EXPRESSION | Genes involved in RORA Activates Circadian Expression |
| 0.0 | 1.4 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.5 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.4 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.0 | 0.2 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.4 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.1 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.0 | 0.3 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.2 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.1 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.2 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 0.0 | 0.1 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.3 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.2 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.3 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.0 | 0.1 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.0 | 0.3 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 0.1 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.1 | REACTOME P75NTR RECRUITS SIGNALLING COMPLEXES | Genes involved in p75NTR recruits signalling complexes |
| 0.0 | 0.1 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.1 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.0 | 0.3 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 0.1 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.1 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.0 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS | Genes involved in Synthesis of bile acids and bile salts |