Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
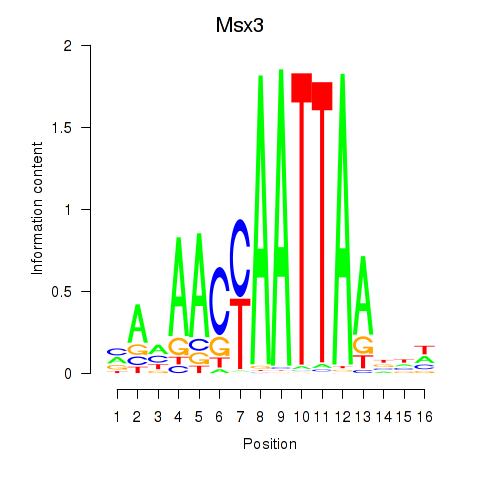
Results for Msx3
Z-value: 1.66
Transcription factors associated with Msx3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Msx3
|
ENSMUSG00000025469.9 | msh homeobox 3 |
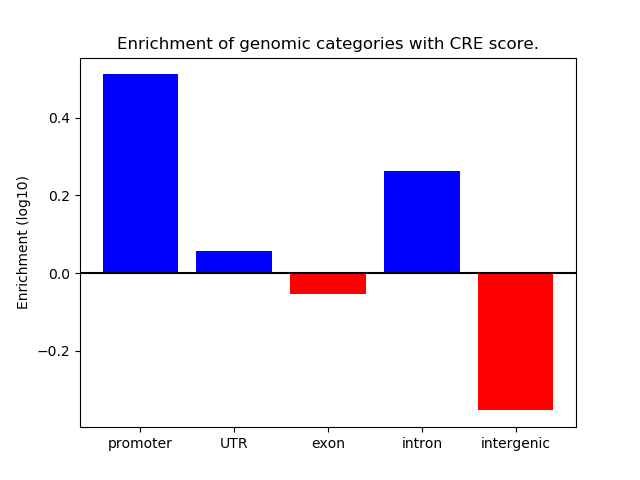
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr7_140049374_140049526 | Msx3 | 361 | 0.740301 | -0.55 | 2.5e-01 | Click! |
| chr7_140058149_140058300 | Msx3 | 9135 | 0.087534 | -0.22 | 6.8e-01 | Click! |
| chr7_140063217_140063374 | Msx3 | 14206 | 0.081201 | -0.12 | 8.2e-01 | Click! |
| chr7_140061314_140061474 | Msx3 | 12305 | 0.083391 | 0.07 | 8.9e-01 | Click! |
Activity of the Msx3 motif across conditions
Conditions sorted by the z-value of the Msx3 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
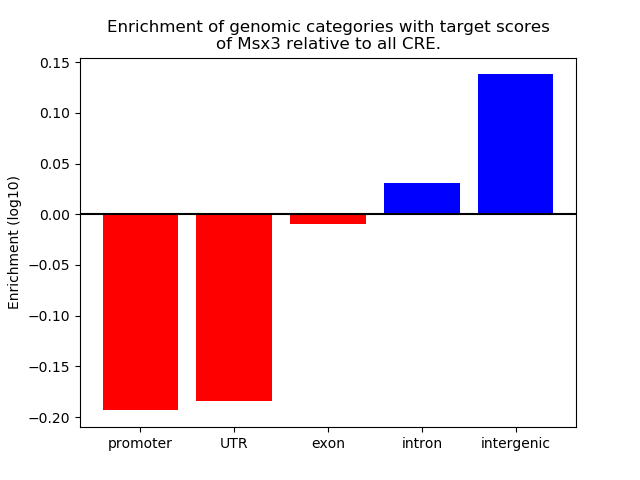
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr9_20730600_20730850 | 1.56 |
Olfm2 |
olfactomedin 2 |
2496 |
0.23 |
| chr11_83271635_83271942 | 1.52 |
Gm23444 |
predicted gene, 23444 |
290 |
0.8 |
| chr8_24445565_24445716 | 1.44 |
Gm44620 |
predicted gene 44620 |
1457 |
0.34 |
| chr11_109733399_109733703 | 1.22 |
Fam20a |
family with sequence similarity 20, member A |
11272 |
0.17 |
| chr18_7847563_7847725 | 1.17 |
Gm50027 |
predicted gene, 50027 |
2709 |
0.23 |
| chr6_35874026_35874908 | 1.10 |
Gm43442 |
predicted gene 43442 |
52244 |
0.17 |
| chr17_83938813_83938978 | 1.09 |
Gm23905 |
predicted gene, 23905 |
8693 |
0.12 |
| chr6_126033747_126033913 | 1.06 |
Ntf3 |
neurotrophin 3 |
131130 |
0.05 |
| chr18_8939044_8939213 | 1.00 |
Gm37148 |
predicted gene, 37148 |
10893 |
0.24 |
| chr10_108350464_108350766 | 0.95 |
Gm23105 |
predicted gene, 23105 |
13050 |
0.19 |
| chr19_44027305_44027461 | 0.95 |
Cyp2c23 |
cytochrome P450, family 2, subfamily c, polypeptide 23 |
1816 |
0.28 |
| chr13_106525577_106525787 | 0.94 |
Gm48216 |
predicted gene, 48216 |
6135 |
0.23 |
| chr10_52294294_52294477 | 0.92 |
Gm6627 |
predicted gene 6627 |
49272 |
0.09 |
| chr12_118887325_118887612 | 0.90 |
Abcb5 |
ATP-binding cassette, sub-family B (MDR/TAP), member 5 |
3291 |
0.3 |
| chr9_33001470_33001748 | 0.89 |
Gm27166 |
predicted gene 27166 |
30182 |
0.17 |
| chr13_101967419_101967852 | 0.87 |
Gm17832 |
predicted gene, 17832 |
47215 |
0.17 |
| chr5_43157097_43157384 | 0.84 |
Gm42552 |
predicted gene 42552 |
8913 |
0.21 |
| chr14_69628830_69628981 | 0.83 |
Gm27177 |
predicted gene 27177 |
11369 |
0.11 |
| chr4_132129267_132129447 | 0.80 |
Oprd1 |
opioid receptor, delta 1 |
15129 |
0.1 |
| chr17_5012388_5012845 | 0.79 |
Arid1b |
AT rich interactive domain 1B (SWI-like) |
16187 |
0.23 |
| chr15_38661347_38661720 | 0.79 |
Atp6v1c1 |
ATPase, H+ transporting, lysosomal V1 subunit C1 |
400 |
0.79 |
| chr2_22589828_22590052 | 0.78 |
Gm13340 |
predicted gene 13340 |
293 |
0.68 |
| chr5_110596813_110596964 | 0.77 |
Galnt9 |
polypeptide N-acetylgalactosaminyltransferase 9 |
6633 |
0.15 |
| chr11_111605518_111605669 | 0.76 |
Gm11676 |
predicted gene 11676 |
7713 |
0.32 |
| chr9_66414274_66414486 | 0.76 |
Gm28379 |
predicted gene 28379 |
6261 |
0.21 |
| chr18_64325483_64325793 | 0.75 |
St8sia3os |
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 3, opposite strand |
8068 |
0.16 |
| chr7_45264516_45264678 | 0.74 |
Slc6a16 |
solute carrier family 6, member 16 |
3553 |
0.09 |
| chr12_69759378_69759543 | 0.74 |
Mir681 |
microRNA 681 |
4484 |
0.15 |
| chr1_4859119_4859327 | 0.73 |
Tcea1 |
transcription elongation factor A (SII) 1 |
1185 |
0.36 |
| chr2_178711802_178711999 | 0.72 |
Cdh26 |
cadherin-like 26 |
251270 |
0.02 |
| chr11_108782497_108782676 | 0.71 |
Cep112 |
centrosomal protein 112 |
30381 |
0.16 |
| chr1_34439675_34439854 | 0.71 |
Imp4 |
IMP4, U3 small nucleolar ribonucleoprotein |
87 |
0.49 |
| chr8_76913299_76913495 | 0.71 |
Nr3c2 |
nuclear receptor subfamily 3, group C, member 2 |
5125 |
0.24 |
| chr15_31279226_31279377 | 0.70 |
Gm49296 |
predicted gene, 49296 |
2788 |
0.19 |
| chrX_140387482_140387633 | 0.70 |
Frmpd3 |
FERM and PDZ domain containing 3 |
5586 |
0.23 |
| chr3_116409014_116409188 | 0.70 |
Cdc14a |
CDC14 cell division cycle 14A |
3613 |
0.2 |
| chr15_99865164_99865320 | 0.70 |
Lima1 |
LIM domain and actin binding 1 |
9429 |
0.08 |
| chr13_92533605_92533774 | 0.69 |
Zfyve16 |
zinc finger, FYVE domain containing 16 |
2821 |
0.27 |
| chr6_17255791_17255960 | 0.69 |
Cav2 |
caveolin 2 |
25310 |
0.17 |
| chr7_66740464_66740653 | 0.69 |
Gm10623 |
predicted gene 10623 |
2754 |
0.19 |
| chr4_142088876_142089143 | 0.68 |
Tmem51os1 |
Tmem51 opposite strand 1 |
4153 |
0.16 |
| chr3_28736802_28736989 | 0.67 |
1700112D23Rik |
RIKEN cDNA 1700112D23 gene |
5345 |
0.16 |
| chr15_77773367_77773546 | 0.66 |
Myh9 |
myosin, heavy polypeptide 9, non-muscle |
3677 |
0.15 |
| chr15_82290409_82290560 | 0.66 |
Septin3 |
septin 3 |
6054 |
0.09 |
| chr13_58058115_58058280 | 0.65 |
Mir874 |
microRNA 874 |
34997 |
0.13 |
| chr5_105830536_105830687 | 0.65 |
Lrrc8d |
leucine rich repeat containing 8D |
6100 |
0.14 |
| chr8_81366726_81366902 | 0.64 |
Inpp4b |
inositol polyphosphate-4-phosphatase, type II |
24252 |
0.21 |
| chr3_58894660_58894811 | 0.64 |
Clrn1 |
clarin 1 |
9395 |
0.2 |
| chr19_27411429_27411595 | 0.64 |
Pum3 |
pumilio RNA-binding family member 3 |
752 |
0.7 |
| chr12_35294499_35294705 | 0.63 |
Gm44396 |
predicted gene, 44396 |
14289 |
0.25 |
| chr12_85818362_85818513 | 0.63 |
Erg28 |
ergosterol biosynthesis 28 |
3813 |
0.2 |
| chr9_90303778_90304052 | 0.63 |
Gm30849 |
predicted gene, 30849 |
115 |
0.96 |
| chr3_65357881_65358061 | 0.63 |
Kcnab1 |
potassium voltage-gated channel, shaker-related subfamily, beta member 1 |
4204 |
0.18 |
| chr2_69061493_69062160 | 0.62 |
Gm38377 |
predicted gene, 38377 |
43730 |
0.11 |
| chr11_87449212_87449363 | 0.62 |
Rnu3b3 |
U3B small nuclear RNA 3 |
660 |
0.48 |
| chr2_75202592_75202875 | 0.62 |
Gm13653 |
predicted gene 13653 |
10456 |
0.15 |
| chr2_169634592_169634925 | 0.61 |
Tshz2 |
teashirt zinc finger family member 2 |
1082 |
0.55 |
| chr5_90465676_90466047 | 0.61 |
Alb |
albumin |
3161 |
0.2 |
| chr14_61361356_61361514 | 0.61 |
Ebpl |
emopamil binding protein-like |
996 |
0.42 |
| chr4_97099326_97100044 | 0.60 |
Gm27521 |
predicted gene, 27521 |
182665 |
0.03 |
| chr1_174921018_174921617 | 0.60 |
Grem2 |
gremlin 2, DAN family BMP antagonist |
502 |
0.88 |
| chr9_123530850_123531001 | 0.60 |
Sacm1l |
SAC1 suppressor of actin mutations 1-like (yeast) |
1043 |
0.5 |
| chr1_52597389_52597548 | 0.60 |
Gm5527 |
predicted gene 5527 |
12950 |
0.13 |
| chr12_73584095_73584268 | 0.60 |
Prkch |
protein kinase C, eta |
615 |
0.73 |
| chrX_100447058_100447240 | 0.59 |
Awat2 |
acyl-CoA wax alcohol acyltransferase 2 |
4432 |
0.18 |
| chr11_86361937_86362118 | 0.59 |
Med13 |
mediator complex subunit 13 |
4425 |
0.24 |
| chr18_37949922_37950073 | 0.58 |
Hdac3 |
histone deacetylase 3 |
4433 |
0.1 |
| chr11_100330372_100330542 | 0.56 |
Gast |
gastrin |
3950 |
0.09 |
| chr4_35148195_35148566 | 0.56 |
Ifnk |
interferon kappa |
3676 |
0.22 |
| chr1_65186286_65186491 | 0.56 |
Idh1 |
isocitrate dehydrogenase 1 (NADP+), soluble |
50 |
0.79 |
| chr1_60862632_60862783 | 0.56 |
Gm38137 |
predicted gene, 38137 |
17631 |
0.12 |
| chr14_121469692_121469843 | 0.55 |
Gm33299 |
predicted gene, 33299 |
14005 |
0.17 |
| chr10_127355627_127355790 | 0.55 |
Inhbe |
inhibin beta-E |
1297 |
0.23 |
| chr17_55749906_55750061 | 0.55 |
Adgre4 |
adhesion G protein-coupled receptor E4 |
1 |
0.97 |
| chr2_132595408_132595654 | 0.54 |
AU019990 |
expressed sequence AU019990 |
2664 |
0.22 |
| chr12_26976640_26976824 | 0.54 |
Gm9866 |
predicted gene 9866 |
138263 |
0.05 |
| chr4_141718181_141718503 | 0.54 |
Ddi2 |
DNA-damage inducible protein 2 |
5077 |
0.14 |
| chr18_35681549_35681863 | 0.54 |
Dnajc18 |
DnaJ heat shock protein family (Hsp40) member C18 |
5037 |
0.1 |
| chr19_26754124_26754675 | 0.53 |
Smarca2 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
799 |
0.69 |
| chr9_61370295_61371248 | 0.53 |
Gm10655 |
predicted gene 10655 |
856 |
0.53 |
| chr5_114985285_114985436 | 0.52 |
1810017P11Rik |
RIKEN cDNA 1810017P11 gene |
9904 |
0.09 |
| chr12_106477969_106478140 | 0.52 |
Gm3191 |
predicted gene 3191 |
18904 |
0.17 |
| chr3_133765600_133766533 | 0.52 |
Gm6135 |
prediticted gene 6135 |
25438 |
0.2 |
| chr11_94370063_94370224 | 0.51 |
Abcc3 |
ATP-binding cassette, sub-family C (CFTR/MRP), member 3 |
2220 |
0.26 |
| chr14_19839858_19840045 | 0.51 |
D14Ertd670e |
DNA segment, Chr 14, ERATO Doi 670, expressed |
1026 |
0.48 |
| chr19_23024753_23025259 | 0.51 |
Gm50136 |
predicted gene, 50136 |
36448 |
0.17 |
| chr3_85550361_85550689 | 0.51 |
Gm42812 |
predicted gene 42812 |
2864 |
0.24 |
| chr8_128687947_128688299 | 0.51 |
Itgb1 |
integrin beta 1 (fibronectin receptor beta) |
2253 |
0.29 |
| chr7_145056434_145056606 | 0.50 |
Gm45181 |
predicted gene 45181 |
106476 |
0.05 |
| chr2_120170818_120170979 | 0.50 |
Ehd4 |
EH-domain containing 4 |
16292 |
0.15 |
| chr18_46336206_46336490 | 0.50 |
4930415P13Rik |
RIKEN cDNA 4930415P13 gene |
4081 |
0.17 |
| chr13_115595390_115595557 | 0.50 |
Gm6035 |
predicted gene 6035 |
65701 |
0.14 |
| chr11_70713419_70713657 | 0.49 |
Mir6925 |
microRNA 6925 |
7548 |
0.08 |
| chr1_185836743_185836921 | 0.49 |
Gm9722 |
predicted gene 9722 |
42053 |
0.18 |
| chr17_63343186_63343351 | 0.49 |
Gm17228 |
predicted gene 17228 |
4142 |
0.22 |
| chr8_93154464_93154622 | 0.48 |
Gm45910 |
predicted gene 45910 |
427 |
0.78 |
| chr14_45453135_45453314 | 0.48 |
Gm34250 |
predicted gene, 34250 |
6151 |
0.13 |
| chr1_73931861_73932066 | 0.48 |
Tns1 |
tensin 1 |
1152 |
0.54 |
| chr13_58999542_58999699 | 0.48 |
Gm34245 |
predicted gene, 34245 |
78676 |
0.08 |
| chr17_8343197_8343376 | 0.48 |
Prr18 |
proline rich 18 |
2547 |
0.17 |
| chr15_64334950_64335143 | 0.48 |
Gm10926 |
predicted gene 10926 |
6029 |
0.2 |
| chr18_20187452_20187603 | 0.48 |
Dsg1c |
desmoglein 1 gamma |
59813 |
0.12 |
| chr18_56607212_56607383 | 0.48 |
Gm50283 |
predicted gene, 50283 |
6362 |
0.14 |
| chr1_24042012_24042182 | 0.48 |
Fam135a |
family with sequence similarity 135, member A |
13011 |
0.18 |
| chr2_132134128_132134319 | 0.48 |
Gm14051 |
predicted gene 14051 |
10310 |
0.16 |
| chr13_14192509_14192814 | 0.47 |
Arid4b |
AT rich interactive domain 4B (RBP1-like) |
1484 |
0.37 |
| chr10_115337191_115337351 | 0.47 |
Tmem19 |
transmembrane protein 19 |
11793 |
0.14 |
| chr14_60635934_60636102 | 0.47 |
Spata13 |
spermatogenesis associated 13 |
1263 |
0.52 |
| chr3_149170339_149170514 | 0.47 |
Gm42647 |
predicted gene 42647 |
39845 |
0.16 |
| chr15_61562347_61562502 | 0.47 |
Gm49498 |
predicted gene, 49498 |
18241 |
0.28 |
| chr14_30351980_30352170 | 0.47 |
Cacna1d |
calcium channel, voltage-dependent, L type, alpha 1D subunit |
889 |
0.57 |
| chr2_131180305_131180466 | 0.47 |
Cenpb |
centromere protein B |
318 |
0.78 |
| chr6_4009646_4009797 | 0.47 |
Gng11 |
guanine nucleotide binding protein (G protein), gamma 11 |
5817 |
0.17 |
| chr4_149453846_149453997 | 0.47 |
Rbp7 |
retinol binding protein 7, cellular |
497 |
0.65 |
| chr9_55233053_55233204 | 0.46 |
Nrg4 |
neuregulin 4 |
9333 |
0.17 |
| chr7_46767478_46767782 | 0.46 |
Hps5 |
HPS5, biogenesis of lysosomal organelles complex 2 subunit 2 |
833 |
0.4 |
| chr3_41465297_41465458 | 0.46 |
Gm40040 |
predicted gene, 40040 |
16508 |
0.13 |
| chr9_8003099_8003325 | 0.46 |
Yap1 |
yes-associated protein 1 |
545 |
0.68 |
| chr9_119341051_119341435 | 0.46 |
Acaa1a |
acetyl-Coenzyme A acyltransferase 1A |
51 |
0.65 |
| chr7_84112155_84112947 | 0.45 |
Cemip |
cell migration inducing protein, hyaluronan binding |
26049 |
0.14 |
| chr18_10251145_10251313 | 0.44 |
Gm5686 |
predicted gene 5686 |
10978 |
0.14 |
| chr10_53753298_53753451 | 0.44 |
Fam184a |
family with sequence similarity 184, member A |
2251 |
0.35 |
| chr16_70563529_70563695 | 0.44 |
Gbe1 |
glucan (1,4-alpha-), branching enzyme 1 |
2618 |
0.36 |
| chr11_34998043_34998214 | 0.44 |
Gm25799 |
predicted gene, 25799 |
99111 |
0.07 |
| chr2_82446048_82446229 | 0.44 |
A130030D18Rik |
RIKEN cDNA A130030D18 gene |
53936 |
0.14 |
| chr13_16269739_16270144 | 0.43 |
Gm48491 |
predicted gene, 48491 |
7897 |
0.23 |
| chr6_31148929_31149080 | 0.43 |
Gm37728 |
predicted gene, 37728 |
1585 |
0.28 |
| chr2_48816834_48817005 | 0.43 |
Acvr2a |
activin receptor IIA |
2810 |
0.3 |
| chr19_43511120_43511314 | 0.43 |
Got1 |
glutamic-oxaloacetic transaminase 1, soluble |
4648 |
0.14 |
| chr16_27391982_27392165 | 0.43 |
Ccdc50 |
coiled-coil domain containing 50 |
2105 |
0.33 |
| chr16_26248335_26248499 | 0.42 |
Cldn1 |
claudin 1 |
112377 |
0.07 |
| chr17_36032663_36032990 | 0.42 |
H2-T23 |
histocompatibility 2, T region locus 23 |
29 |
0.92 |
| chr2_132143179_132143352 | 0.42 |
Gm14051 |
predicted gene 14051 |
1268 |
0.37 |
| chr10_68248229_68248529 | 0.42 |
Arid5b |
AT rich interactive domain 5B (MRF1-like) |
30342 |
0.19 |
| chr3_118507933_118508602 | 0.42 |
Gm37773 |
predicted gene, 37773 |
17884 |
0.14 |
| chr10_9902050_9902213 | 0.42 |
Gm46210 |
predicted gene, 46210 |
818 |
0.46 |
| chr13_78124273_78124441 | 0.42 |
Gm18990 |
predicted gene, 18990 |
3287 |
0.19 |
| chr10_41627379_41627538 | 0.41 |
Ccdc162 |
coiled-coil domain containing 162 |
14621 |
0.15 |
| chr15_34493251_34493646 | 0.41 |
Rida |
reactive intermediate imine deaminase A homolog |
1723 |
0.21 |
| chr9_122814663_122814835 | 0.41 |
Tcaim |
T cell activation inhibitor, mitochondrial |
9146 |
0.12 |
| chr3_138231897_138232387 | 0.41 |
Adh7 |
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
4091 |
0.14 |
| chr16_25351479_25351642 | 0.41 |
Tprg |
transformation related protein 63 regulated |
64739 |
0.14 |
| chr3_116967199_116967673 | 0.41 |
4930455H04Rik |
RIKEN cDNA 4930455H04 gene |
831 |
0.45 |
| chr10_70924491_70924642 | 0.41 |
Bicc1 |
BicC family RNA binding protein 1 |
23923 |
0.12 |
| chr5_34380236_34380602 | 0.41 |
Fam193a |
family with sequence homology 193, member A |
10486 |
0.14 |
| chr2_77563772_77563946 | 0.41 |
Zfp385b |
zinc finger protein 385B |
1879 |
0.47 |
| chr2_122081842_122082023 | 0.40 |
Spg11 |
SPG11, spatacsin vesicle trafficking associated |
36364 |
0.11 |
| chr12_3811591_3811763 | 0.40 |
Dnmt3a |
DNA methyltransferase 3A |
4517 |
0.2 |
| chr19_43767883_43768034 | 0.40 |
Cutc |
cutC copper transporter |
3085 |
0.18 |
| chr7_123124873_123125354 | 0.40 |
Tnrc6a |
trinucleotide repeat containing 6a |
857 |
0.66 |
| chr17_26024045_26024196 | 0.40 |
Rab11fip3 |
RAB11 family interacting protein 3 (class II) |
2260 |
0.16 |
| chr1_72145974_72146125 | 0.39 |
Gm4319 |
predicted gene 4319 |
24048 |
0.15 |
| chr11_16883025_16883203 | 0.39 |
Egfr |
epidermal growth factor receptor |
4964 |
0.23 |
| chr9_64457749_64457913 | 0.39 |
Megf11 |
multiple EGF-like-domains 11 |
43505 |
0.16 |
| chr19_12005901_12006348 | 0.39 |
Olfr1421-ps1 |
olfactory receptor 1421, pseudogene 1 |
14051 |
0.08 |
| chr3_94338287_94338715 | 0.39 |
Gm43743 |
predicted gene 43743 |
3169 |
0.1 |
| chr6_71823720_71824038 | 0.38 |
Mrpl35 |
mitochondrial ribosomal protein L35 |
69 |
0.95 |
| chr3_61217316_61217479 | 0.38 |
Gm37719 |
predicted gene, 37719 |
74049 |
0.1 |
| chr19_36735506_36735662 | 0.38 |
Ppp1r3c |
protein phosphatase 1, regulatory subunit 3C |
1069 |
0.56 |
| chr2_63580816_63581021 | 0.38 |
Gm23503 |
predicted gene, 23503 |
148724 |
0.05 |
| chr4_105215829_105216004 | 0.38 |
Plpp3 |
phospholipid phosphatase 3 |
58569 |
0.15 |
| chr7_111726929_111727297 | 0.38 |
Galnt18 |
polypeptide N-acetylgalactosaminyltransferase 18 |
52864 |
0.15 |
| chr12_35294940_35295100 | 0.38 |
Gm44396 |
predicted gene, 44396 |
13871 |
0.25 |
| chr17_84385788_84385950 | 0.38 |
Gm24722 |
predicted gene, 24722 |
5560 |
0.22 |
| chr1_136229022_136229181 | 0.37 |
Inava |
innate immunity activator |
918 |
0.4 |
| chr1_133251724_133252043 | 0.37 |
Gm19461 |
predicted gene, 19461 |
2409 |
0.23 |
| chr11_83703751_83703986 | 0.37 |
Wfdc17 |
WAP four-disulfide core domain 17 |
123 |
0.9 |
| chr1_43066043_43066369 | 0.37 |
Tgfbrap1 |
transforming growth factor, beta receptor associated protein 1 |
5273 |
0.14 |
| chr4_65532386_65532616 | 0.37 |
Trim32 |
tripartite motif-containing 32 |
72485 |
0.14 |
| chr6_41044443_41044618 | 0.37 |
2210010C04Rik |
RIKEN cDNA 2210010C04 gene |
9021 |
0.07 |
| chr11_102040134_102040466 | 0.37 |
Cd300lg |
CD300 molecule like family member G |
1209 |
0.28 |
| chr17_8974900_8975051 | 0.37 |
1700010I14Rik |
RIKEN cDNA 1700010I14 gene |
13358 |
0.23 |
| chr12_57514709_57514889 | 0.37 |
Gm2568 |
predicted gene 2568 |
5825 |
0.17 |
| chr2_142767278_142767429 | 0.37 |
Gm22310 |
predicted gene, 22310 |
19543 |
0.24 |
| chr3_145621208_145621546 | 0.37 |
Znhit6 |
zinc finger, HIT type 6 |
25509 |
0.15 |
| chr11_7153168_7153348 | 0.37 |
Adcy1 |
adenylate cyclase 1 |
6624 |
0.21 |
| chr4_11865116_11865283 | 0.36 |
Gm25002 |
predicted gene, 25002 |
83688 |
0.07 |
| chr7_49397440_49397604 | 0.36 |
Nav2 |
neuron navigator 2 |
32801 |
0.18 |
| chr9_79213455_79213848 | 0.36 |
Gm47499 |
predicted gene, 47499 |
11644 |
0.17 |
| chrX_74270256_74270420 | 0.36 |
Rpl10 |
ribosomal protein L10 |
474 |
0.52 |
| chr4_116689870_116690150 | 0.36 |
Prdx1 |
peroxiredoxin 1 |
2698 |
0.15 |
| chr9_77662986_77663137 | 0.36 |
Klhl31 |
kelch-like 31 |
26561 |
0.14 |
| chr11_29719333_29719653 | 0.36 |
Rtn4 |
reticulon 4 |
930 |
0.53 |
| chr10_108074024_108074345 | 0.36 |
Gm47999 |
predicted gene, 47999 |
68248 |
0.12 |
| chr15_97031052_97031203 | 0.36 |
Slc38a4 |
solute carrier family 38, member 4 |
84 |
0.98 |
| chr15_10273020_10273171 | 0.36 |
Prlr |
prolactin receptor |
23535 |
0.21 |
| chr11_60537500_60537667 | 0.36 |
Alkbh5 |
alkB homolog 5, RNA demethylase |
395 |
0.72 |
| chr12_14302401_14302552 | 0.36 |
Gm16497 |
predicted gene 16497 |
40920 |
0.19 |
| chr15_77208076_77208227 | 0.36 |
Rbfox2 |
RNA binding protein, fox-1 homolog (C. elegans) 2 |
4715 |
0.18 |
| chr3_138426007_138426180 | 0.36 |
Adh4 |
alcohol dehydrogenase 4 (class II), pi polypeptide |
10598 |
0.13 |
| chr5_9056138_9056296 | 0.36 |
Gm40264 |
predicted gene, 40264 |
21093 |
0.14 |
| chr6_50176334_50176508 | 0.36 |
Mpp6 |
membrane protein, palmitoylated 6 (MAGUK p55 subfamily member 6) |
3879 |
0.27 |
| chr15_61398964_61399115 | 0.36 |
Gm24696 |
predicted gene, 24696 |
41035 |
0.19 |
| chr6_121963215_121963471 | 0.36 |
Mug2 |
murinoglobulin 2 |
43418 |
0.12 |
| chr8_115984246_115984444 | 0.35 |
Gm45733 |
predicted gene 45733 |
12274 |
0.31 |
| chr14_64331778_64331949 | 0.35 |
Msra |
methionine sulfoxide reductase A |
85085 |
0.09 |
| chr1_24612181_24612351 | 0.35 |
Gm28439 |
predicted gene 28439 |
144 |
0.72 |
| chr6_94814990_94815159 | 0.35 |
Gm7833 |
predicted gene 7833 |
5456 |
0.15 |
| chr8_123032866_123033108 | 0.35 |
Ankrd11 |
ankyrin repeat domain 11 |
9033 |
0.11 |
| chr4_8361870_8362071 | 0.35 |
Gm37386 |
predicted gene, 37386 |
92277 |
0.08 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0060300 | regulation of cytokine activity(GO:0060300) |
| 0.1 | 0.4 | GO:2000566 | positive regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000566) |
| 0.1 | 0.4 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.1 | 0.4 | GO:0010835 | regulation of protein ADP-ribosylation(GO:0010835) |
| 0.1 | 0.3 | GO:0086046 | membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.1 | 0.5 | GO:0046487 | glyoxylate metabolic process(GO:0046487) |
| 0.1 | 0.1 | GO:0010166 | wax biosynthetic process(GO:0010025) wax metabolic process(GO:0010166) |
| 0.1 | 0.3 | GO:0035513 | oxidative RNA demethylation(GO:0035513) oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.1 | 0.2 | GO:0019065 | receptor-mediated endocytosis of virus by host cell(GO:0019065) endocytosis involved in viral entry into host cell(GO:0075509) |
| 0.1 | 0.5 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.1 | 0.2 | GO:0002414 | immunoglobulin transcytosis in epithelial cells(GO:0002414) |
| 0.1 | 0.2 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.1 | 0.2 | GO:0043134 | hindgut contraction(GO:0043133) regulation of hindgut contraction(GO:0043134) |
| 0.1 | 0.3 | GO:1902459 | positive regulation of stem cell population maintenance(GO:1902459) |
| 0.1 | 0.1 | GO:0006982 | response to lipid hydroperoxide(GO:0006982) |
| 0.1 | 0.1 | GO:1904502 | regulation of lipophagy(GO:1904502) positive regulation of lipophagy(GO:1904504) |
| 0.1 | 0.2 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.1 | 0.2 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.1 | 0.2 | GO:0007161 | calcium-independent cell-matrix adhesion(GO:0007161) |
| 0.1 | 0.3 | GO:0042663 | regulation of endodermal cell fate specification(GO:0042663) |
| 0.1 | 0.2 | GO:0072566 | chemokine (C-X-C motif) ligand 1 production(GO:0072566) regulation of chemokine (C-X-C motif) ligand 1 production(GO:2000338) |
| 0.0 | 0.0 | GO:0070447 | positive regulation of oligodendrocyte progenitor proliferation(GO:0070447) |
| 0.0 | 0.2 | GO:0003383 | apical constriction(GO:0003383) |
| 0.0 | 0.1 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.0 | 0.3 | GO:0035811 | negative regulation of urine volume(GO:0035811) |
| 0.0 | 0.2 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.0 | 0.3 | GO:0035336 | long-chain fatty-acyl-CoA metabolic process(GO:0035336) |
| 0.0 | 0.2 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.0 | 0.1 | GO:1990705 | cholangiocyte proliferation(GO:1990705) |
| 0.0 | 0.1 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.0 | 0.6 | GO:0035278 | miRNA mediated inhibition of translation(GO:0035278) |
| 0.0 | 0.3 | GO:0002591 | positive regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002591) |
| 0.0 | 0.1 | GO:0010911 | regulation of isomerase activity(GO:0010911) positive regulation of isomerase activity(GO:0010912) |
| 0.0 | 0.1 | GO:2000173 | negative regulation of branching morphogenesis of a nerve(GO:2000173) |
| 0.0 | 0.1 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.0 | 0.4 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.0 | 0.1 | GO:0034395 | regulation of transcription from RNA polymerase II promoter in response to iron(GO:0034395) |
| 0.0 | 0.2 | GO:0042713 | sperm ejaculation(GO:0042713) |
| 0.0 | 0.1 | GO:0097503 | sialylation(GO:0097503) |
| 0.0 | 0.1 | GO:0001907 | killing by symbiont of host cells(GO:0001907) disruption by symbiont of host cell(GO:0044004) |
| 0.0 | 0.1 | GO:0003213 | cardiac right atrium morphogenesis(GO:0003213) |
| 0.0 | 0.2 | GO:0071447 | cellular response to hydroperoxide(GO:0071447) |
| 0.0 | 0.1 | GO:0007403 | glial cell fate determination(GO:0007403) |
| 0.0 | 0.2 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.0 | 0.1 | GO:1901492 | positive regulation of lymphangiogenesis(GO:1901492) |
| 0.0 | 0.1 | GO:0071926 | cannabinoid signaling pathway(GO:0038171) endocannabinoid signaling pathway(GO:0071926) |
| 0.0 | 0.1 | GO:0051295 | establishment of meiotic spindle localization(GO:0051295) |
| 0.0 | 0.1 | GO:0034773 | histone H4-K20 trimethylation(GO:0034773) |
| 0.0 | 0.1 | GO:0034627 | 'de novo' NAD biosynthetic process(GO:0034627) |
| 0.0 | 0.2 | GO:0019852 | L-ascorbic acid metabolic process(GO:0019852) |
| 0.0 | 0.1 | GO:0043974 | histone H3-K27 acetylation(GO:0043974) regulation of histone H3-K27 acetylation(GO:1901674) |
| 0.0 | 0.1 | GO:2000017 | positive regulation of determination of dorsal identity(GO:2000017) |
| 0.0 | 0.1 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.0 | 0.1 | GO:1900019 | regulation of protein kinase C activity(GO:1900019) positive regulation of protein kinase C activity(GO:1900020) |
| 0.0 | 0.1 | GO:0045409 | negative regulation of interleukin-6 biosynthetic process(GO:0045409) |
| 0.0 | 0.4 | GO:0072673 | lamellipodium morphogenesis(GO:0072673) |
| 0.0 | 0.2 | GO:0000022 | mitotic spindle elongation(GO:0000022) mitotic spindle midzone assembly(GO:0051256) |
| 0.0 | 0.0 | GO:0035511 | oxidative DNA demethylation(GO:0035511) |
| 0.0 | 0.1 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.0 | 0.0 | GO:0046101 | hypoxanthine metabolic process(GO:0046100) hypoxanthine biosynthetic process(GO:0046101) |
| 0.0 | 0.1 | GO:1902809 | regulation of skeletal muscle fiber differentiation(GO:1902809) |
| 0.0 | 0.3 | GO:0046838 | phosphorylated carbohydrate dephosphorylation(GO:0046838) |
| 0.0 | 0.0 | GO:0045658 | regulation of neutrophil differentiation(GO:0045658) |
| 0.0 | 0.1 | GO:0046499 | S-adenosylmethioninamine metabolic process(GO:0046499) |
| 0.0 | 0.1 | GO:0006393 | termination of mitochondrial transcription(GO:0006393) |
| 0.0 | 0.1 | GO:0070681 | glutaminyl-tRNAGln biosynthesis via transamidation(GO:0070681) |
| 0.0 | 0.3 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 0.2 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.0 | 0.1 | GO:0042706 | eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate commitment(GO:0046552) |
| 0.0 | 0.1 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.0 | GO:0050923 | regulation of negative chemotaxis(GO:0050923) |
| 0.0 | 0.1 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.1 | GO:1902513 | regulation of organelle transport along microtubule(GO:1902513) |
| 0.0 | 0.1 | GO:1903265 | positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 0.0 | 0.1 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.0 | 0.1 | GO:0090230 | regulation of centromere complex assembly(GO:0090230) |
| 0.0 | 0.1 | GO:0001514 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.0 | 0.1 | GO:0045917 | positive regulation of complement activation(GO:0045917) positive regulation of protein activation cascade(GO:2000259) |
| 0.0 | 0.1 | GO:0061144 | alveolar secondary septum development(GO:0061144) |
| 0.0 | 0.0 | GO:0046504 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) cellular lipid biosynthetic process(GO:0097384) ether biosynthetic process(GO:1901503) |
| 0.0 | 0.1 | GO:0048254 | snoRNA localization(GO:0048254) |
| 0.0 | 0.4 | GO:0044783 | G1 DNA damage checkpoint(GO:0044783) |
| 0.0 | 0.1 | GO:0034116 | positive regulation of heterotypic cell-cell adhesion(GO:0034116) |
| 0.0 | 0.1 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.0 | 0.1 | GO:0045590 | negative regulation of regulatory T cell differentiation(GO:0045590) |
| 0.0 | 0.2 | GO:0046040 | IMP metabolic process(GO:0046040) |
| 0.0 | 0.1 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.0 | 0.1 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 0.1 | GO:1900108 | negative regulation of nodal signaling pathway(GO:1900108) |
| 0.0 | 0.1 | GO:0060398 | regulation of growth hormone receptor signaling pathway(GO:0060398) |
| 0.0 | 0.1 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.0 | 0.1 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.0 | 0.2 | GO:0071157 | negative regulation of cell cycle arrest(GO:0071157) |
| 0.0 | 0.3 | GO:0070498 | interleukin-1-mediated signaling pathway(GO:0070498) |
| 0.0 | 0.1 | GO:0061073 | ciliary body morphogenesis(GO:0061073) |
| 0.0 | 0.2 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.0 | 0.1 | GO:0006532 | aspartate biosynthetic process(GO:0006532) |
| 0.0 | 0.0 | GO:2001187 | positive regulation of CD8-positive, alpha-beta T cell activation(GO:2001187) |
| 0.0 | 0.1 | GO:0060020 | Bergmann glial cell differentiation(GO:0060020) |
| 0.0 | 0.1 | GO:0071318 | cellular response to ATP(GO:0071318) |
| 0.0 | 0.1 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 0.2 | GO:0051386 | regulation of neurotrophin TRK receptor signaling pathway(GO:0051386) |
| 0.0 | 0.1 | GO:0015817 | histidine transport(GO:0015817) |
| 0.0 | 0.1 | GO:2001274 | negative regulation of glucose import in response to insulin stimulus(GO:2001274) |
| 0.0 | 0.0 | GO:0090071 | negative regulation of ribosome biogenesis(GO:0090071) |
| 0.0 | 0.0 | GO:0045901 | positive regulation of translational elongation(GO:0045901) |
| 0.0 | 0.1 | GO:0061368 | behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 0.0 | 0.2 | GO:0071257 | cellular response to electrical stimulus(GO:0071257) |
| 0.0 | 0.1 | GO:0060613 | fat pad development(GO:0060613) |
| 0.0 | 0.1 | GO:0018214 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.0 | 0.0 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.0 | 0.3 | GO:0045780 | positive regulation of bone resorption(GO:0045780) positive regulation of bone remodeling(GO:0046852) |
| 0.0 | 0.0 | GO:0061092 | regulation of phospholipid translocation(GO:0061091) positive regulation of phospholipid translocation(GO:0061092) |
| 0.0 | 0.0 | GO:1902075 | cellular response to salt(GO:1902075) |
| 0.0 | 0.2 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.1 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) |
| 0.0 | 0.0 | GO:0061643 | chemorepulsion of axon(GO:0061643) |
| 0.0 | 0.0 | GO:0090259 | regulation of retinal ganglion cell axon guidance(GO:0090259) |
| 0.0 | 0.0 | GO:1901856 | negative regulation of cellular respiration(GO:1901856) |
| 0.0 | 0.0 | GO:0040033 | negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.0 | 0.0 | GO:0042504 | tyrosine phosphorylation of Stat4 protein(GO:0042504) regulation of tyrosine phosphorylation of Stat4 protein(GO:0042519) |
| 0.0 | 0.0 | GO:0061470 | T follicular helper cell differentiation(GO:0061470) |
| 0.0 | 0.1 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.0 | 0.1 | GO:0048935 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.0 | 0.0 | GO:0036215 | response to stem cell factor(GO:0036215) cellular response to stem cell factor stimulus(GO:0036216) Kit signaling pathway(GO:0038109) |
| 0.0 | 0.1 | GO:0060023 | soft palate development(GO:0060023) |
| 0.0 | 0.2 | GO:0010663 | positive regulation of striated muscle cell apoptotic process(GO:0010663) positive regulation of cardiac muscle cell apoptotic process(GO:0010666) |
| 0.0 | 0.0 | GO:0009826 | unidimensional cell growth(GO:0009826) |
| 0.0 | 0.1 | GO:0016188 | synaptic vesicle maturation(GO:0016188) |
| 0.0 | 0.1 | GO:0015808 | L-alanine transport(GO:0015808) |
| 0.0 | 0.1 | GO:0044789 | modulation by host of viral release from host cell(GO:0044789) positive regulation by host of viral release from host cell(GO:0044791) |
| 0.0 | 0.0 | GO:0061502 | early endosome to recycling endosome transport(GO:0061502) |
| 0.0 | 0.0 | GO:0070672 | response to interleukin-15(GO:0070672) |
| 0.0 | 0.1 | GO:0042997 | negative regulation of Golgi to plasma membrane protein transport(GO:0042997) |
| 0.0 | 0.2 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.0 | 0.0 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.0 | 0.0 | GO:0071286 | cellular response to magnesium ion(GO:0071286) |
| 0.0 | 0.0 | GO:0050847 | progesterone receptor signaling pathway(GO:0050847) |
| 0.0 | 0.1 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.0 | 0.1 | GO:0035507 | regulation of myosin-light-chain-phosphatase activity(GO:0035507) |
| 0.0 | 0.0 | GO:1903553 | positive regulation of extracellular exosome assembly(GO:1903553) |
| 0.0 | 0.0 | GO:0034140 | negative regulation of toll-like receptor 3 signaling pathway(GO:0034140) |
| 0.0 | 0.1 | GO:0048012 | hepatocyte growth factor receptor signaling pathway(GO:0048012) |
| 0.0 | 0.0 | GO:0030046 | parallel actin filament bundle assembly(GO:0030046) |
| 0.0 | 0.1 | GO:0002023 | reduction of food intake in response to dietary excess(GO:0002023) |
| 0.0 | 0.1 | GO:0099515 | actin filament-based transport(GO:0099515) |
| 0.0 | 0.1 | GO:0010815 | bradykinin catabolic process(GO:0010815) |
| 0.0 | 0.1 | GO:0071404 | cellular response to low-density lipoprotein particle stimulus(GO:0071404) |
| 0.0 | 0.1 | GO:0043031 | negative regulation of macrophage activation(GO:0043031) |
| 0.0 | 0.1 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.0 | 0.0 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 0.0 | 0.0 | GO:0072318 | clathrin coat disassembly(GO:0072318) |
| 0.0 | 0.0 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.0 | 0.0 | GO:1990168 | protein K29-linked deubiquitination(GO:0035523) protein K33-linked deubiquitination(GO:1990168) |
| 0.0 | 0.0 | GO:0048633 | positive regulation of skeletal muscle tissue growth(GO:0048633) |
| 0.0 | 0.1 | GO:0071380 | cellular response to prostaglandin E stimulus(GO:0071380) |
| 0.0 | 0.0 | GO:0032077 | positive regulation of deoxyribonuclease activity(GO:0032077) |
| 0.0 | 0.1 | GO:0009446 | putrescine biosynthetic process(GO:0009446) |
| 0.0 | 0.0 | GO:0030886 | negative regulation of myeloid dendritic cell activation(GO:0030886) |
| 0.0 | 0.0 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.0 | 0.0 | GO:0051791 | medium-chain fatty acid metabolic process(GO:0051791) |
| 0.0 | 0.0 | GO:0031296 | B cell costimulation(GO:0031296) |
| 0.0 | 0.1 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.0 | 0.1 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.0 | GO:0006041 | glucosamine metabolic process(GO:0006041) |
| 0.0 | 0.0 | GO:0001997 | positive regulation of the force of heart contraction by epinephrine-norepinephrine(GO:0001997) positive regulation of the force of heart contraction by chemical signal(GO:0003099) |
| 0.0 | 0.0 | GO:0072718 | response to cisplatin(GO:0072718) |
| 0.0 | 0.1 | GO:1903817 | negative regulation of delayed rectifier potassium channel activity(GO:1902260) negative regulation of voltage-gated potassium channel activity(GO:1903817) |
| 0.0 | 0.1 | GO:0061000 | negative regulation of dendritic spine development(GO:0061000) |
| 0.0 | 0.1 | GO:0006104 | succinyl-CoA metabolic process(GO:0006104) |
| 0.0 | 0.1 | GO:0032494 | response to peptidoglycan(GO:0032494) |
| 0.0 | 0.1 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.0 | 0.4 | GO:0009250 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.0 | 0.4 | GO:0010761 | fibroblast migration(GO:0010761) |
| 0.0 | 0.1 | GO:0014028 | notochord formation(GO:0014028) |
| 0.0 | 0.0 | GO:0006772 | thiamine metabolic process(GO:0006772) thiamine-containing compound metabolic process(GO:0042723) |
| 0.0 | 0.0 | GO:2000383 | regulation of ectoderm development(GO:2000383) negative regulation of ectoderm development(GO:2000384) |
| 0.0 | 0.0 | GO:0097296 | activation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097296) |
| 0.0 | 0.3 | GO:0042832 | defense response to protozoan(GO:0042832) |
| 0.0 | 0.0 | GO:1903525 | regulation of membrane tubulation(GO:1903525) |
| 0.0 | 0.1 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.0 | 0.0 | GO:0097477 | lateral motor column neuron migration(GO:0097477) |
| 0.0 | 0.1 | GO:0009435 | NAD biosynthetic process(GO:0009435) |
| 0.0 | 0.0 | GO:0090210 | regulation of establishment of blood-brain barrier(GO:0090210) |
| 0.0 | 0.3 | GO:0015988 | energy coupled proton transmembrane transport, against electrochemical gradient(GO:0015988) ATP hydrolysis coupled proton transport(GO:0015991) ATP hydrolysis coupled transmembrane transport(GO:0090662) |
| 0.0 | 0.1 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 | 0.0 | GO:0000966 | RNA 5'-end processing(GO:0000966) |
| 0.0 | 0.0 | GO:0010796 | regulation of multivesicular body size(GO:0010796) |
| 0.0 | 0.1 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 | 0.1 | GO:0006662 | glycerol ether metabolic process(GO:0006662) |
| 0.0 | 0.0 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.0 | 0.1 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.0 | 0.0 | GO:1904706 | negative regulation of vascular smooth muscle cell proliferation(GO:1904706) |
| 0.0 | 0.0 | GO:2001166 | regulation of histone H2B ubiquitination(GO:2001166) positive regulation of histone H2B ubiquitination(GO:2001168) |
| 0.0 | 0.0 | GO:1903116 | positive regulation of actin filament-based movement(GO:1903116) |
| 0.0 | 0.0 | GO:0015747 | urate transport(GO:0015747) |
| 0.0 | 0.1 | GO:0010801 | negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.1 | 0.6 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.1 | 0.2 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.1 | 0.2 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.1 | 0.2 | GO:0034679 | integrin alpha9-beta1 complex(GO:0034679) |
| 0.1 | 0.2 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.0 | 0.1 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.0 | 0.1 | GO:0097413 | Lewy body(GO:0097413) |
| 0.0 | 0.6 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.0 | 0.1 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.0 | 0.1 | GO:0097629 | extrinsic component of omegasome membrane(GO:0097629) |
| 0.0 | 0.3 | GO:0020005 | symbiont-containing vacuole membrane(GO:0020005) |
| 0.0 | 0.2 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.0 | 0.2 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.1 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.0 | 0.4 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.0 | 0.9 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.2 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 0.2 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.2 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.1 | GO:0010009 | cytoplasmic side of endosome membrane(GO:0010009) |
| 0.0 | 0.1 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.0 | 0.1 | GO:0030956 | glutamyl-tRNA(Gln) amidotransferase complex(GO:0030956) |
| 0.0 | 0.2 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.0 | 0.1 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 0.1 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 0.1 | GO:0045179 | apical cortex(GO:0045179) |
| 0.0 | 0.0 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 0.1 | GO:1990851 | Wnt-Frizzled-LRP5/6 complex(GO:1990851) |
| 0.0 | 0.0 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 0.0 | 0.0 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.1 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.1 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.0 | 0.3 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.1 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.1 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.3 | GO:0030137 | COPI-coated vesicle(GO:0030137) |
| 0.0 | 0.1 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.0 | 0.1 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.0 | 0.1 | GO:0030689 | Noc complex(GO:0030689) |
| 0.0 | 0.1 | GO:0034991 | nuclear meiotic cohesin complex(GO:0034991) |
| 0.0 | 0.4 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.0 | GO:0032010 | phagolysosome(GO:0032010) |
| 0.0 | 0.1 | GO:0070419 | nonhomologous end joining complex(GO:0070419) |
| 0.0 | 0.2 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.0 | 0.0 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.0 | 0.0 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 0.0 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.0 | 0.2 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 0.0 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.0 | 0.6 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 0.0 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.0 | 0.1 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 0.0 | 0.1 | GO:0060091 | kinocilium(GO:0060091) |
| 0.0 | 0.1 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.4 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.1 | 0.3 | GO:0030519 | snoRNP binding(GO:0030519) |
| 0.1 | 0.4 | GO:0016401 | palmitoyl-CoA oxidase activity(GO:0016401) |
| 0.1 | 0.3 | GO:0050252 | retinol O-fatty-acyltransferase activity(GO:0050252) |
| 0.1 | 0.4 | GO:0003696 | satellite DNA binding(GO:0003696) |
| 0.1 | 0.3 | GO:0035515 | oxidative RNA demethylase activity(GO:0035515) |
| 0.1 | 0.4 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.1 | 0.2 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.1 | 0.5 | GO:0036122 | BMP binding(GO:0036122) |
| 0.1 | 0.2 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 0.1 | 0.3 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.1 | 0.1 | GO:0035673 | oligopeptide transmembrane transporter activity(GO:0035673) |
| 0.1 | 0.2 | GO:0004346 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.1 | 0.2 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.1 | 0.7 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.1 | 0.2 | GO:0086007 | voltage-gated calcium channel activity involved in cardiac muscle cell action potential(GO:0086007) |
| 0.1 | 0.2 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.0 | 0.1 | GO:0034596 | phosphatidylinositol phosphate 4-phosphatase activity(GO:0034596) |
| 0.0 | 0.2 | GO:0090482 | vitamin transmembrane transporter activity(GO:0090482) |
| 0.0 | 0.2 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.0 | 0.3 | GO:0004438 | phosphatidylinositol-3-phosphatase activity(GO:0004438) |
| 0.0 | 0.2 | GO:0019960 | C-X3-C chemokine binding(GO:0019960) |
| 0.0 | 0.2 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.0 | 0.3 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.0 | 0.4 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.0 | 0.1 | GO:0051718 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) DNA (cytosine-5-)-methyltransferase activity, acting on CpG substrates(GO:0051718) |
| 0.0 | 0.1 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.3 | GO:0019841 | retinol binding(GO:0019841) |
| 0.0 | 0.1 | GO:0000171 | ribonuclease MRP activity(GO:0000171) |
| 0.0 | 0.1 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.0 | 0.2 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.1 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.0 | 0.0 | GO:0035514 | DNA demethylase activity(GO:0035514) |
| 0.0 | 0.1 | GO:0050897 | cobalt ion binding(GO:0050897) |
| 0.0 | 0.9 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 0.1 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) |
| 0.0 | 0.1 | GO:0050567 | glutaminyl-tRNA synthase (glutamine-hydrolyzing) activity(GO:0050567) |
| 0.0 | 0.2 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.0 | 0.1 | GO:0031800 | type 3 metabotropic glutamate receptor binding(GO:0031800) |
| 0.0 | 0.0 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.0 | 0.1 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.0 | 0.3 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.6 | GO:0051990 | (R)-2-hydroxyglutarate dehydrogenase activity(GO:0051990) |
| 0.0 | 0.1 | GO:0016823 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.0 | 0.1 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.0 | 0.1 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) |
| 0.0 | 0.1 | GO:0009374 | biotin binding(GO:0009374) |
| 0.0 | 0.1 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) |
| 0.0 | 0.1 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.0 | 0.2 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.0 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.0 | 0.3 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.0 | 0.2 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.1 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 0.1 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.0 | 0.1 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.1 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.1 | GO:0008430 | selenium binding(GO:0008430) |
| 0.0 | 0.1 | GO:0047760 | butyrate-CoA ligase activity(GO:0047760) |
| 0.0 | 0.1 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.0 | 0.1 | GO:1901474 | L-histidine transmembrane transporter activity(GO:0005290) azole transporter activity(GO:0045118) azole transmembrane transporter activity(GO:1901474) |
| 0.0 | 0.1 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.1 | GO:0031748 | D1 dopamine receptor binding(GO:0031748) |
| 0.0 | 0.1 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.0 | 0.0 | GO:0016778 | diphosphotransferase activity(GO:0016778) |
| 0.0 | 0.2 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.0 | 0.1 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.0 | 0.3 | GO:0019825 | oxygen binding(GO:0019825) |
| 0.0 | 0.1 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.0 | 0.1 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.1 | GO:0005024 | transforming growth factor beta-activated receptor activity(GO:0005024) |
| 0.0 | 0.0 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.0 | 0.1 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.0 | 0.0 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.0 | 0.1 | GO:0016892 | endoribonuclease activity, producing 3'-phosphomonoesters(GO:0016892) |
| 0.0 | 0.0 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 0.0 | 0.1 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.0 | 0.1 | GO:0002046 | opsin binding(GO:0002046) |
| 0.0 | 0.1 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.0 | 0.6 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.1 | GO:0022858 | L-alanine transmembrane transporter activity(GO:0015180) alanine transmembrane transporter activity(GO:0022858) |
| 0.0 | 0.2 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.1 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.0 | 0.1 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.0 | 0.0 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.0 | 0.4 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 0.1 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.0 | 0.1 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.0 | 0.0 | GO:0052827 | inositol-1,3,4,5,6-pentakisphosphate 3-phosphatase activity(GO:0030351) inositol-1,4,5,6-tetrakisphosphate 6-phosphatase activity(GO:0030352) inositol pentakisphosphate phosphatase activity(GO:0052827) |
| 0.0 | 0.2 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.1 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.0 | 0.1 | GO:0019534 | toxin transporter activity(GO:0019534) |
| 0.0 | 0.1 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 0.0 | GO:0032139 | dinucleotide insertion or deletion binding(GO:0032139) |
| 0.0 | 0.0 | GO:0015252 | hydrogen ion channel activity(GO:0015252) |
| 0.0 | 0.1 | GO:0001094 | TFIID-class transcription factor binding(GO:0001094) |
| 0.0 | 0.0 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.0 | 0.1 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.0 | 0.1 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) |
| 0.0 | 0.1 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.0 | GO:0034739 | histone deacetylase activity (H4-K16 specific)(GO:0034739) |
| 0.0 | 0.2 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.1 | GO:0043140 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.5 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.8 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.0 | 0.2 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.1 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 0.4 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.1 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 0.1 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.2 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.1 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.4 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 0.8 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.1 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.0 | 0.6 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 0.6 | REACTOME TRANSFERRIN ENDOCYTOSIS AND RECYCLING | Genes involved in Transferrin endocytosis and recycling |
| 0.0 | 0.2 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 0.3 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 0.2 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.2 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.3 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.0 | 0.4 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.2 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 0.2 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 0.3 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.3 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.0 | 0.1 | REACTOME P75NTR RECRUITS SIGNALLING COMPLEXES | Genes involved in p75NTR recruits signalling complexes |
| 0.0 | 0.2 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.5 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.4 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 0.0 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.1 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.0 | REACTOME SIGNALLING BY NGF | Genes involved in Signalling by NGF |
| 0.0 | 0.1 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.3 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.0 | 0.1 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 0.2 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.0 | 0.2 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.1 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 0.1 | REACTOME SHC1 EVENTS IN EGFR SIGNALING | Genes involved in SHC1 events in EGFR signaling |