Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
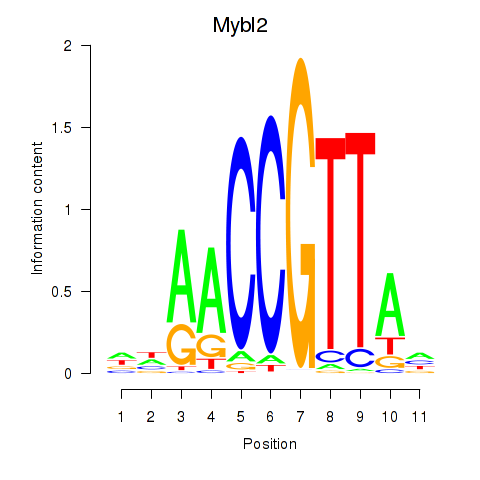
Results for Mybl2
Z-value: 1.22
Transcription factors associated with Mybl2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Mybl2
|
ENSMUSG00000017861.5 | myeloblastosis oncogene-like 2 |
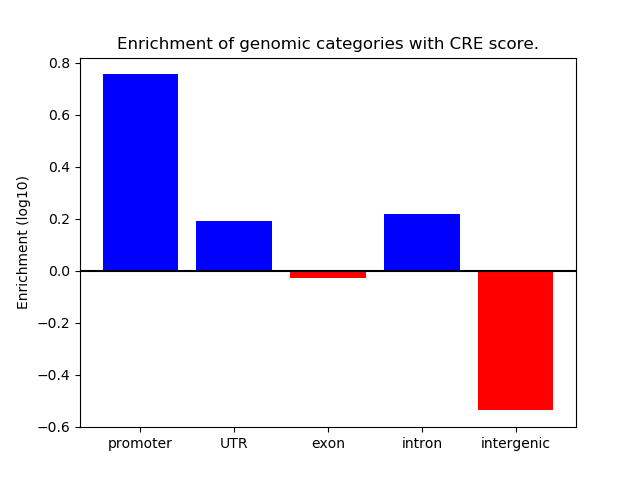
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr2_163038512_163038663 | Mybl2 | 16100 | 0.112135 | -0.77 | 7.2e-02 | Click! |
| chr2_163048181_163048359 | Mybl2 | 6417 | 0.129856 | 0.49 | 3.3e-01 | Click! |
| chr2_163054451_163054615 | Mybl2 | 154 | 0.929890 | -0.33 | 5.2e-01 | Click! |
Activity of the Mybl2 motif across conditions
Conditions sorted by the z-value of the Mybl2 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
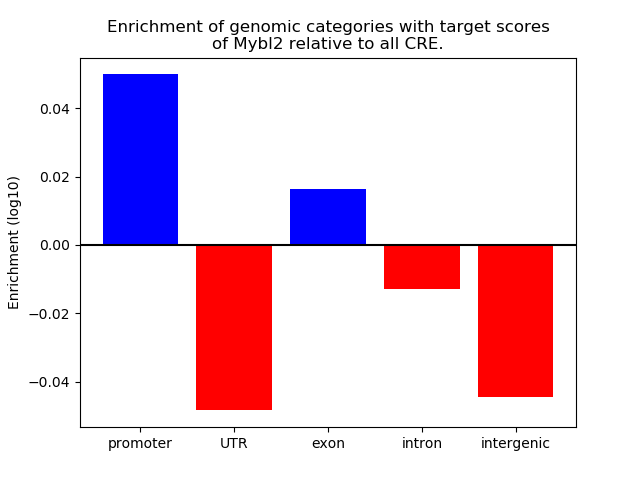
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr5_150889049_150889202 | 0.47 |
Gm43298 |
predicted gene 43298 |
6483 |
0.21 |
| chr1_66816824_66817235 | 0.38 |
Kansl1l |
KAT8 regulatory NSL complex subunit 1-like |
517 |
0.58 |
| chr9_44078711_44078862 | 0.31 |
Usp2 |
ubiquitin specific peptidase 2 |
6153 |
0.07 |
| chr13_54439539_54439690 | 0.31 |
Thoc3 |
THO complex 3 |
29174 |
0.12 |
| chr5_99533469_99533635 | 0.30 |
Gm16227 |
predicted gene 16227 |
22870 |
0.17 |
| chr16_25221904_25222082 | 0.27 |
Tprg |
transformation related protein 63 regulated |
64824 |
0.14 |
| chr9_92350909_92351360 | 0.27 |
1700057G04Rik |
RIKEN cDNA 1700057G04 gene |
49 |
0.97 |
| chr18_65223520_65223956 | 0.25 |
Nedd4l |
neural precursor cell expressed, developmentally down-regulated gene 4-like |
15854 |
0.15 |
| chr3_144266534_144266686 | 0.25 |
Gm43446 |
predicted gene 43446 |
17201 |
0.19 |
| chr4_126967080_126967454 | 0.25 |
Zmym4 |
zinc finger, MYM-type 4 |
632 |
0.66 |
| chr10_8095747_8095932 | 0.24 |
Gm48614 |
predicted gene, 48614 |
74547 |
0.1 |
| chr5_144904481_144904632 | 0.24 |
Smurf1 |
SMAD specific E3 ubiquitin protein ligase 1 |
6562 |
0.17 |
| chr19_34839173_34839483 | 0.24 |
Mir107 |
microRNA 107 |
18555 |
0.13 |
| chr10_111318102_111318256 | 0.24 |
Gm40761 |
predicted gene, 40761 |
18945 |
0.17 |
| chr19_47580113_47580699 | 0.23 |
Slk |
STE20-like kinase |
387 |
0.82 |
| chr7_12979741_12980348 | 0.22 |
Zfp446 |
zinc finger protein 446 |
524 |
0.55 |
| chr7_30361694_30362080 | 0.22 |
Lrfn3 |
leucine rich repeat and fibronectin type III domain containing 3 |
885 |
0.31 |
| chr3_84826282_84826441 | 0.22 |
Fbxw7 |
F-box and WD-40 domain protein 7 |
10595 |
0.25 |
| chr8_33834954_33835178 | 0.22 |
Rbpms |
RNA binding protein gene with multiple splicing |
8605 |
0.17 |
| chr5_120566729_120566934 | 0.22 |
2510016D11Rik |
RIKEN cDNA 2510016D11 gene |
13792 |
0.09 |
| chr13_93622998_93623302 | 0.22 |
Gm15622 |
predicted gene 15622 |
2232 |
0.26 |
| chrX_7656942_7657153 | 0.22 |
Gm14703 |
predicted gene 14703 |
211 |
0.46 |
| chr2_179261314_179261512 | 0.22 |
Gm14293 |
predicted gene 14293 |
20923 |
0.24 |
| chr14_21435070_21435221 | 0.21 |
Gm25864 |
predicted gene, 25864 |
15329 |
0.19 |
| chr2_71467270_71467440 | 0.21 |
Metap1d |
methionyl aminopeptidase type 1D (mitochondrial) |
13992 |
0.13 |
| chr18_46711768_46711921 | 0.21 |
Cdo1 |
cysteine dioxygenase 1, cytosolic |
16185 |
0.12 |
| chr13_38740448_38740614 | 0.21 |
Gm47510 |
predicted gene, 47510 |
16889 |
0.14 |
| chr5_90279563_90279724 | 0.21 |
Ankrd17 |
ankyrin repeat domain 17 |
1121 |
0.57 |
| chr16_31948717_31948903 | 0.21 |
Ncbp2 |
nuclear cap binding protein subunit 2 |
164 |
0.65 |
| chr17_12408592_12408743 | 0.21 |
Plg |
plasminogen |
30008 |
0.14 |
| chr5_90646770_90647014 | 0.21 |
Rassf6 |
Ras association (RalGDS/AF-6) domain family member 6 |
6235 |
0.18 |
| chr3_36864295_36864446 | 0.21 |
4932438A13Rik |
RIKEN cDNA 4932438A13 gene |
99 |
0.97 |
| chr9_115476457_115476622 | 0.21 |
Gm5921 |
predicted gene 5921 |
37958 |
0.14 |
| chr2_93728886_93729050 | 0.20 |
Ext2 |
exostosin glycosyltransferase 2 |
21650 |
0.2 |
| chr5_28052952_28053361 | 0.20 |
Gm26608 |
predicted gene, 26608 |
2303 |
0.25 |
| chr13_60736804_60737009 | 0.20 |
Dapk1 |
death associated protein kinase 1 |
17090 |
0.15 |
| chr4_41744512_41744692 | 0.20 |
Sigmar1 |
sigma non-opioid intracellular receptor 1 |
470 |
0.64 |
| chr13_75942831_75943082 | 0.20 |
Rhobtb3 |
Rho-related BTB domain containing 3 |
911 |
0.44 |
| chr4_94979002_94979670 | 0.20 |
Gm12694 |
predicted gene 12694 |
133 |
0.64 |
| chr11_98972114_98972272 | 0.20 |
Gjd3 |
gap junction protein, delta 3 |
10823 |
0.1 |
| chr8_35390018_35390398 | 0.20 |
Ppp1r3b |
protein phosphatase 1, regulatory subunit 3B |
13548 |
0.15 |
| chr2_10369756_10369912 | 0.20 |
Sfmbt2 |
Scm-like with four mbt domains 2 |
676 |
0.33 |
| chr1_78567590_78567756 | 0.20 |
Mogat1 |
monoacylglycerol O-acyltransferase 1 |
30045 |
0.13 |
| chr10_8086566_8086745 | 0.20 |
Gm48614 |
predicted gene, 48614 |
65363 |
0.12 |
| chr4_135789148_135789435 | 0.20 |
Myom3 |
myomesin family, member 3 |
8899 |
0.14 |
| chr3_104540522_104540806 | 0.20 |
Lrig2 |
leucine-rich repeats and immunoglobulin-like domains 2 |
28746 |
0.1 |
| chr18_60735828_60736023 | 0.19 |
Ndst1 |
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1 |
2775 |
0.22 |
| chr9_55322341_55322520 | 0.19 |
Nrg4 |
neuregulin 4 |
2757 |
0.24 |
| chr16_33464020_33464171 | 0.19 |
Slc12a8 |
solute carrier family 12 (potassium/chloride transporters), member 8 |
53233 |
0.13 |
| chr14_68304879_68305239 | 0.19 |
Gm47212 |
predicted gene, 47212 |
86806 |
0.08 |
| chr17_45701931_45702243 | 0.19 |
Mrpl14 |
mitochondrial ribosomal protein L14 |
6671 |
0.12 |
| chr7_118127356_118127544 | 0.19 |
Arl6ip1 |
ADP-ribosylation factor-like 6 interacting protein 1 |
1660 |
0.32 |
| chr6_17747909_17748076 | 0.19 |
Gm42852 |
predicted gene 42852 |
908 |
0.41 |
| chr10_8122077_8122255 | 0.19 |
Gm30906 |
predicted gene, 30906 |
53965 |
0.15 |
| chr7_132941355_132941506 | 0.19 |
Zranb1 |
zinc finger, RAN-binding domain containing 1 |
6738 |
0.15 |
| chr2_31521284_31521501 | 0.19 |
Ass1 |
argininosuccinate synthetase 1 |
2902 |
0.23 |
| chr15_36500919_36501077 | 0.19 |
Gm49246 |
predicted gene, 49246 |
2433 |
0.22 |
| chr5_130144105_130144430 | 0.19 |
Kctd7 |
potassium channel tetramerisation domain containing 7 |
594 |
0.61 |
| chr9_21367416_21367785 | 0.19 |
Ilf3 |
interleukin enhancer binding factor 3 |
271 |
0.81 |
| chr18_20933583_20933734 | 0.19 |
Rnf125 |
ring finger protein 125 |
10967 |
0.2 |
| chr3_107690573_107690757 | 0.19 |
Ahcyl1 |
S-adenosylhomocysteine hydrolase-like 1 |
5298 |
0.18 |
| chr1_155795507_155795699 | 0.19 |
Qsox1 |
quiescin Q6 sulfhydryl oxidase 1 |
186 |
0.92 |
| chr15_80684645_80684861 | 0.19 |
A430088P11Rik |
RIKEN cDNA A430088P11 gene |
12849 |
0.12 |
| chr10_45002693_45003320 | 0.19 |
Gm4795 |
predicted pseudogene 4795 |
3104 |
0.23 |
| chr4_138003808_138004018 | 0.18 |
Eif4g3 |
eukaryotic translation initiation factor 4 gamma, 3 |
839 |
0.68 |
| chr3_102383435_102383619 | 0.18 |
Gm43245 |
predicted gene 43245 |
6480 |
0.2 |
| chr1_74971297_74971448 | 0.18 |
Gm37744 |
predicted gene, 37744 |
17120 |
0.11 |
| chr13_43172983_43173301 | 0.18 |
Tbc1d7 |
TBC1 domain family, member 7 |
1641 |
0.41 |
| chr12_54453328_54453497 | 0.18 |
Gm7557 |
predicted gene 7557 |
23014 |
0.14 |
| chr6_100226321_100226472 | 0.18 |
Rybp |
RING1 and YY1 binding protein |
6735 |
0.18 |
| chr11_84016754_84017076 | 0.18 |
Synrg |
synergin, gamma |
2286 |
0.3 |
| chr19_40341503_40341709 | 0.18 |
Sorbs1 |
sorbin and SH3 domain containing 1 |
1012 |
0.56 |
| chr1_159268925_159269084 | 0.18 |
Cop1 |
COP1, E3 ubiquitin ligase |
2270 |
0.27 |
| chr12_80829408_80829559 | 0.18 |
Susd6 |
sushi domain containing 6 |
38924 |
0.1 |
| chr9_65332374_65332600 | 0.18 |
Gm39363 |
predicted gene, 39363 |
33 |
0.94 |
| chr2_167328678_167328847 | 0.18 |
B4galt5 |
UDP-Gal:betaGlcNAc beta 1,4-galactosyltransferase, polypeptide 5 |
14862 |
0.18 |
| chr11_109700431_109700593 | 0.18 |
Fam20a |
family with sequence similarity 20, member A |
17533 |
0.16 |
| chr2_103206471_103206789 | 0.18 |
Gm13869 |
predicted gene 13869 |
3727 |
0.21 |
| chr19_36362135_36362318 | 0.17 |
Gm47773 |
predicted gene, 47773 |
883 |
0.51 |
| chr2_74724645_74724796 | 0.17 |
Mir10b |
microRNA 10b |
1350 |
0.15 |
| chr5_98028317_98028767 | 0.17 |
Antxr2 |
anthrax toxin receptor 2 |
2420 |
0.22 |
| chr18_11658016_11658319 | 0.17 |
Rbbp8 |
retinoblastoma binding protein 8, endonuclease |
325 |
0.54 |
| chr2_131820393_131820556 | 0.17 |
Gm14282 |
predicted gene 14282 |
25121 |
0.12 |
| chr7_84122692_84122843 | 0.17 |
Abhd17c |
abhydrolase domain containing 17C |
25048 |
0.13 |
| chr2_69414677_69414863 | 0.17 |
Dhrs9 |
dehydrogenase/reductase (SDR family) member 9 |
34325 |
0.16 |
| chr3_107691131_107691282 | 0.17 |
Ahcyl1 |
S-adenosylhomocysteine hydrolase-like 1 |
4757 |
0.18 |
| chr8_94075372_94075662 | 0.17 |
Bbs2 |
Bardet-Biedl syndrome 2 (human) |
4239 |
0.15 |
| chr17_30590513_30590860 | 0.17 |
Gm50244 |
predicted gene, 50244 |
822 |
0.45 |
| chr19_41581597_41581748 | 0.17 |
Lcor |
ligand dependent nuclear receptor corepressor |
698 |
0.68 |
| chr9_66184918_66185081 | 0.17 |
Dapk2 |
death-associated protein kinase 2 |
26764 |
0.17 |
| chr13_55445821_55446370 | 0.17 |
Grk6 |
G protein-coupled receptor kinase 6 |
634 |
0.5 |
| chr10_93075662_93075887 | 0.17 |
Cfap54 |
cilia and flagella associated protein 54 |
5523 |
0.22 |
| chr1_59593071_59593229 | 0.17 |
Gm973 |
predicted gene 973 |
10754 |
0.14 |
| chr2_31489958_31490302 | 0.17 |
Ass1 |
argininosuccinate synthetase 1 |
7642 |
0.19 |
| chr1_90603068_90603263 | 0.17 |
Cops8 |
COP9 signalosome subunit 8 |
184 |
0.96 |
| chr17_29007079_29007233 | 0.17 |
Stk38 |
serine/threonine kinase 38 |
570 |
0.55 |
| chr19_56521472_56521623 | 0.17 |
Dclre1a |
DNA cross-link repair 1A |
15835 |
0.17 |
| chr11_30268838_30269016 | 0.17 |
Sptbn1 |
spectrin beta, non-erythrocytic 1 |
752 |
0.61 |
| chr4_76329225_76329376 | 0.16 |
Ptprd |
protein tyrosine phosphatase, receptor type, D |
4012 |
0.33 |
| chr14_51255648_51255799 | 0.16 |
Rnase2a |
ribonuclease, RNase A family, 2A (liver, eosinophil-derived neurotoxin) |
389 |
0.76 |
| chr2_165404690_165404862 | 0.16 |
Slc13a3 |
solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 3 |
4290 |
0.15 |
| chr2_32692644_32692971 | 0.16 |
Fpgs |
folylpolyglutamyl synthetase |
437 |
0.6 |
| chr15_86059275_86059609 | 0.16 |
Gramd4 |
GRAM domain containing 4 |
570 |
0.75 |
| chr10_18317869_18318162 | 0.16 |
Nhsl1 |
NHS-like 1 |
970 |
0.54 |
| chrX_140493888_140494053 | 0.16 |
Prps1 |
phosphoribosyl pyrophosphate synthetase 1 |
37357 |
0.14 |
| chr7_143296870_143297021 | 0.16 |
Kcnq1ot1 |
KCNQ1 overlapping transcript 1 |
396 |
0.65 |
| chr18_38923142_38923385 | 0.16 |
Fgf1 |
fibroblast growth factor 1 |
4492 |
0.24 |
| chr8_111721706_111721872 | 0.16 |
Bcar1 |
breast cancer anti-estrogen resistance 1 |
10316 |
0.16 |
| chr15_10890674_10891179 | 0.16 |
Gm29742 |
predicted gene, 29742 |
26511 |
0.16 |
| chr16_76330855_76331006 | 0.16 |
Nrip1 |
nuclear receptor interacting protein 1 |
7272 |
0.24 |
| chr7_39841847_39842021 | 0.16 |
4930558N11Rik |
RIKEN cDNA 4930558N11 gene |
44192 |
0.11 |
| chr3_95439022_95439237 | 0.16 |
Arnt |
aryl hydrocarbon receptor nuclear translocator |
4610 |
0.1 |
| chr3_121869502_121869683 | 0.16 |
Gm42593 |
predicted gene 42593 |
6750 |
0.18 |
| chr1_190128817_190128968 | 0.16 |
Gm28172 |
predicted gene 28172 |
39778 |
0.15 |
| chr4_133746098_133746249 | 0.16 |
Arid1a |
AT rich interactive domain 1A (SWI-like) |
7438 |
0.15 |
| chr9_35106324_35106534 | 0.16 |
St3gal4 |
ST3 beta-galactoside alpha-2,3-sialyltransferase 4 |
238 |
0.91 |
| chr1_180193628_180193822 | 0.16 |
Coq8a |
coenzyme Q8A |
63 |
0.97 |
| chr13_52155783_52156109 | 0.16 |
Gm48199 |
predicted gene, 48199 |
24465 |
0.2 |
| chr4_61857457_61857832 | 0.16 |
Mup-ps17 |
major urinary protein, pseudogene 17 |
232 |
0.85 |
| chr16_26694594_26694745 | 0.16 |
Il1rap |
interleukin 1 receptor accessory protein |
27765 |
0.23 |
| chr3_98061516_98061682 | 0.16 |
Gm42819 |
predicted gene 42819 |
30912 |
0.15 |
| chr8_79289713_79289872 | 0.16 |
Mmaa |
methylmalonic aciduria (cobalamin deficiency) type A |
4888 |
0.18 |
| chr16_93342815_93342978 | 0.15 |
1810053B23Rik |
RIKEN cDNA 1810053B23 gene |
10293 |
0.18 |
| chr5_66125192_66125593 | 0.15 |
Rbm47 |
RNA binding motif protein 47 |
25564 |
0.1 |
| chr4_125420233_125420436 | 0.15 |
Grik3 |
glutamate receptor, ionotropic, kainate 3 |
70366 |
0.1 |
| chr9_106816354_106816555 | 0.15 |
Gm22384 |
predicted gene, 22384 |
3856 |
0.16 |
| chr8_93189262_93189430 | 0.15 |
Gm45909 |
predicted gene 45909 |
2012 |
0.24 |
| chr1_136662300_136662465 | 0.15 |
Gm19705 |
predicted gene, 19705 |
8387 |
0.13 |
| chr9_48741770_48741973 | 0.15 |
Zbtb16 |
zinc finger and BTB domain containing 16 |
94074 |
0.07 |
| chr5_115543227_115543424 | 0.15 |
Pxn |
paxillin |
375 |
0.74 |
| chr6_108606639_108606790 | 0.15 |
0610040F04Rik |
RIKEN cDNA 0610040F04 gene |
7281 |
0.16 |
| chr15_89375887_89376038 | 0.15 |
Tymp |
thymidine phosphorylase |
1077 |
0.24 |
| chr1_135146355_135146506 | 0.15 |
Arl8a |
ADP-ribosylation factor-like 8A |
394 |
0.72 |
| chr2_34870625_34870806 | 0.15 |
Psmd5 |
proteasome (prosome, macropain) 26S subunit, non-ATPase, 5 |
218 |
0.88 |
| chr19_8920078_8920229 | 0.15 |
B3gat3 |
beta-1,3-glucuronyltransferase 3 (glucuronosyltransferase I) |
221 |
0.78 |
| chr5_90578056_90578210 | 0.15 |
Gm42109 |
predicted gene, 42109 |
10924 |
0.13 |
| chr1_130797794_130798113 | 0.15 |
Fcamr |
Fc receptor, IgA, IgM, high affinity |
2949 |
0.16 |
| chr8_22877841_22878072 | 0.15 |
Gm45555 |
predicted gene 45555 |
4367 |
0.18 |
| chr14_20164716_20164945 | 0.15 |
Kcnk5 |
potassium channel, subfamily K, member 5 |
16979 |
0.13 |
| chr1_24092021_24092172 | 0.15 |
Fam135a |
family with sequence similarity 135, member A |
5570 |
0.19 |
| chr9_65333109_65333260 | 0.15 |
Gm39363 |
predicted gene, 39363 |
664 |
0.49 |
| chr6_90551837_90551988 | 0.15 |
Aldh1l1 |
aldehyde dehydrogenase 1 family, member L1 |
1064 |
0.44 |
| chr1_9739631_9739800 | 0.15 |
1700034P13Rik |
RIKEN cDNA 1700034P13 gene |
7933 |
0.15 |
| chr8_36073417_36073574 | 0.15 |
Prag1 |
PEAK1 related kinase activating pseudokinase 1 |
21333 |
0.17 |
| chr16_37902046_37902197 | 0.15 |
Gpr156 |
G protein-coupled receptor 156 |
14375 |
0.14 |
| chr7_99974071_99974241 | 0.15 |
Rnf169 |
ring finger protein 169 |
6292 |
0.14 |
| chr7_123453449_123453600 | 0.15 |
Aqp8 |
aquaporin 8 |
8767 |
0.19 |
| chr3_37204278_37204436 | 0.15 |
Gm12532 |
predicted gene 12532 |
21472 |
0.1 |
| chr3_157919500_157919651 | 0.15 |
Cth |
cystathionase (cystathionine gamma-lyase) |
5476 |
0.14 |
| chr12_84225651_84225802 | 0.15 |
Elmsan1 |
ELM2 and Myb/SANT-like domain containing 1 |
6845 |
0.11 |
| chr9_55214755_55215047 | 0.15 |
Fbxo22 |
F-box protein 22 |
1238 |
0.43 |
| chr13_25019629_25019881 | 0.15 |
Mrs2 |
MRS2 magnesium transporter |
21 |
0.97 |
| chr15_38047513_38047884 | 0.15 |
Gm22353 |
predicted gene, 22353 |
10587 |
0.16 |
| chr14_66114373_66114699 | 0.15 |
Ephx2 |
epoxide hydrolase 2, cytoplasmic |
2150 |
0.27 |
| chr4_103214248_103214501 | 0.15 |
Slc35d1 |
solute carrier family 35 (UDP-glucuronic acid/UDP-N-acetylgalactosamine dual transporter), member D1 |
298 |
0.88 |
| chr19_37509643_37509826 | 0.15 |
Exoc6 |
exocyst complex component 6 |
11145 |
0.17 |
| chr11_52006220_52006371 | 0.15 |
Cdkl3 |
cyclin-dependent kinase-like 3 |
1946 |
0.21 |
| chr1_36538834_36539209 | 0.15 |
Gm42417 |
predicted gene, 42417 |
337 |
0.72 |
| chr1_39313664_39313826 | 0.14 |
Gm3617 |
predicted gene 3617 |
1154 |
0.45 |
| chr3_118606217_118606485 | 0.14 |
Dpyd |
dihydropyrimidine dehydrogenase |
44165 |
0.15 |
| chr10_128909533_128909836 | 0.14 |
Cd63 |
CD63 antigen |
15 |
0.95 |
| chr17_80814177_80814328 | 0.14 |
C230072F16Rik |
RIKEN cDNA C230072F16 gene |
82541 |
0.08 |
| chr11_102168591_102168742 | 0.14 |
Tmem101 |
transmembrane protein 101 |
12262 |
0.08 |
| chrX_166440131_166440304 | 0.14 |
Trappc2 |
trafficking protein particle complex 2 |
357 |
0.57 |
| chr9_77943161_77943330 | 0.14 |
Elovl5 |
ELOVL family member 5, elongation of long chain fatty acids (yeast) |
1954 |
0.27 |
| chr2_18080882_18081051 | 0.14 |
Mllt10 |
myeloid/lymphoid or mixed-lineage leukemia; translocated to, 10 |
16125 |
0.11 |
| chr8_22497870_22498317 | 0.14 |
Slc20a2 |
solute carrier family 20, member 2 |
8032 |
0.14 |
| chr9_43781736_43781901 | 0.14 |
Nectin1 |
nectin cell adhesion molecule 1 |
12303 |
0.17 |
| chr8_35387028_35387898 | 0.14 |
Ppp1r3b |
protein phosphatase 1, regulatory subunit 3B |
10803 |
0.16 |
| chr3_82290237_82290420 | 0.14 |
Map9 |
microtubule-associated protein 9 |
67716 |
0.12 |
| chr15_31250599_31250840 | 0.14 |
Dap |
death-associated protein |
17617 |
0.15 |
| chr6_145339191_145339494 | 0.14 |
Gm15707 |
predicted gene 15707 |
22734 |
0.11 |
| chr3_94412463_94412614 | 0.14 |
1700040D17Rik |
RIKEN cDNA 1700040D17 gene |
381 |
0.49 |
| chr11_100951927_100952102 | 0.14 |
Stat3 |
signal transducer and activator of transcription 3 |
12474 |
0.13 |
| chr6_87980501_87980658 | 0.14 |
Gm5577 |
predicted gene 5577 |
304 |
0.69 |
| chr14_70658775_70658926 | 0.14 |
Npm2 |
nucleophosmin/nucleoplasmin 2 |
394 |
0.77 |
| chr4_59006947_59007116 | 0.14 |
Gm20503 |
predicted gene 20503 |
3821 |
0.16 |
| chr6_93879123_93879274 | 0.14 |
Gm22840 |
predicted gene, 22840 |
11417 |
0.24 |
| chr4_144974231_144974382 | 0.14 |
Vps13d |
vacuolar protein sorting 13D |
9655 |
0.2 |
| chr2_5754873_5755229 | 0.14 |
Camk1d |
calcium/calmodulin-dependent protein kinase ID |
40536 |
0.14 |
| chr19_41802312_41802463 | 0.14 |
Arhgap19 |
Rho GTPase activating protein 19 |
340 |
0.86 |
| chr2_31502991_31503411 | 0.14 |
Ass1 |
argininosuccinate synthetase 1 |
2475 |
0.27 |
| chr17_83182747_83183029 | 0.14 |
Pkdcc |
protein kinase domain containing, cytoplasmic |
32404 |
0.17 |
| chr9_7988205_7988356 | 0.14 |
Yap1 |
yes-associated protein 1 |
13423 |
0.15 |
| chr13_92530147_92530703 | 0.14 |
Zfyve16 |
zinc finger, FYVE domain containing 16 |
443 |
0.84 |
| chr12_28898892_28899474 | 0.14 |
Gm31508 |
predicted gene, 31508 |
11046 |
0.17 |
| chr5_140816843_140817430 | 0.14 |
Gna12 |
guanine nucleotide binding protein, alpha 12 |
13021 |
0.21 |
| chr19_55566290_55566526 | 0.14 |
Tcf7l2 |
transcription factor 7 like 2, T cell specific, HMG box |
175412 |
0.03 |
| chr15_6445470_6445748 | 0.14 |
C9 |
complement component 9 |
237 |
0.94 |
| chr11_111362744_111362915 | 0.14 |
Gm11675 |
predicted gene 11675 |
86729 |
0.1 |
| chr10_57486792_57486966 | 0.14 |
Hsf2 |
heat shock factor 2 |
454 |
0.51 |
| chr19_36646070_36646235 | 0.14 |
Hectd2os |
Hectd2, opposite strand |
12432 |
0.22 |
| chr19_48883437_48883771 | 0.14 |
Gm50436 |
predicted gene, 50436 |
72468 |
0.11 |
| chr14_63941556_63941788 | 0.14 |
Sox7 |
SRY (sex determining region Y)-box 7 |
2001 |
0.31 |
| chr9_53291789_53291940 | 0.14 |
Exph5 |
exophilin 5 |
9806 |
0.19 |
| chr9_80167006_80167157 | 0.14 |
Myo6 |
myosin VI |
1854 |
0.34 |
| chr2_90929175_90929326 | 0.14 |
Ptpmt1 |
protein tyrosine phosphatase, mitochondrial 1 |
10992 |
0.1 |
| chr11_116165413_116165564 | 0.14 |
Fbf1 |
Fas (TNFRSF6) binding factor 1 |
395 |
0.67 |
| chr1_31033632_31033801 | 0.13 |
Gm28644 |
predicted gene 28644 |
8203 |
0.17 |
| chr2_84637297_84637592 | 0.13 |
Ctnnd1 |
catenin (cadherin associated protein), delta 1 |
661 |
0.54 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.1 | 0.2 | GO:0010046 | response to mycotoxin(GO:0010046) |
| 0.1 | 0.2 | GO:0097460 | ferrous iron import into cell(GO:0097460) |
| 0.1 | 0.2 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.1 | 0.2 | GO:0042998 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) |
| 0.0 | 0.2 | GO:2000680 | regulation of rubidium ion transport(GO:2000680) |
| 0.0 | 0.1 | GO:0010360 | negative regulation of anion channel activity(GO:0010360) |
| 0.0 | 0.1 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.0 | 0.2 | GO:0006681 | galactosylceramide metabolic process(GO:0006681) |
| 0.0 | 0.1 | GO:0009093 | cysteine catabolic process(GO:0009093) L-cysteine catabolic process(GO:0019448) L-cysteine metabolic process(GO:0046439) |
| 0.0 | 0.3 | GO:0034626 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.2 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.0 | 0.2 | GO:0010387 | COP9 signalosome assembly(GO:0010387) |
| 0.0 | 0.2 | GO:0048669 | collateral sprouting in absence of injury(GO:0048669) |
| 0.0 | 0.1 | GO:1900041 | negative regulation of interleukin-2 secretion(GO:1900041) |
| 0.0 | 0.2 | GO:0006114 | glycerol biosynthetic process(GO:0006114) |
| 0.0 | 0.3 | GO:0006991 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.0 | 0.2 | GO:0060681 | branch elongation involved in ureteric bud branching(GO:0060681) |
| 0.0 | 0.2 | GO:0060903 | positive regulation of meiosis I(GO:0060903) |
| 0.0 | 0.0 | GO:0046449 | creatinine metabolic process(GO:0046449) cellular lactam metabolic process(GO:0072338) |
| 0.0 | 0.2 | GO:0022417 | protein maturation by protein folding(GO:0022417) |
| 0.0 | 0.2 | GO:0009202 | deoxyribonucleoside triphosphate biosynthetic process(GO:0009202) |
| 0.0 | 0.1 | GO:0070827 | chromatin maintenance(GO:0070827) |
| 0.0 | 0.2 | GO:0015780 | nucleotide-sugar transport(GO:0015780) pyrimidine nucleotide-sugar transport(GO:0015781) |
| 0.0 | 0.3 | GO:0033129 | positive regulation of histone phosphorylation(GO:0033129) |
| 0.0 | 0.1 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.0 | 0.1 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.0 | 0.1 | GO:0019344 | cysteine biosynthetic process(GO:0019344) |
| 0.0 | 0.1 | GO:2000832 | negative regulation of steroid hormone secretion(GO:2000832) negative regulation of corticosteroid hormone secretion(GO:2000847) negative regulation of glucocorticoid secretion(GO:2000850) |
| 0.0 | 0.1 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.0 | 0.1 | GO:0090135 | actin filament branching(GO:0090135) |
| 0.0 | 0.2 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.0 | 0.3 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 0.2 | GO:0071267 | amino acid salvage(GO:0043102) L-methionine biosynthetic process(GO:0071265) L-methionine salvage(GO:0071267) |
| 0.0 | 0.2 | GO:0070970 | interleukin-2 secretion(GO:0070970) |
| 0.0 | 0.1 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.0 | 0.1 | GO:0055005 | ventricular cardiac myofibril assembly(GO:0055005) |
| 0.0 | 0.1 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.0 | 0.1 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.0 | 0.1 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.1 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.1 | GO:0016557 | peroxisome membrane biogenesis(GO:0016557) |
| 0.0 | 0.2 | GO:0006559 | L-phenylalanine catabolic process(GO:0006559) erythrose 4-phosphate/phosphoenolpyruvate family amino acid catabolic process(GO:1902222) |
| 0.0 | 0.1 | GO:0009256 | 10-formyltetrahydrofolate metabolic process(GO:0009256) |
| 0.0 | 0.1 | GO:0051081 | membrane disassembly(GO:0030397) nuclear envelope disassembly(GO:0051081) |
| 0.0 | 0.1 | GO:0018243 | protein O-linked glycosylation via threonine(GO:0018243) |
| 0.0 | 0.1 | GO:1903689 | regulation of wound healing, spreading of epidermal cells(GO:1903689) |
| 0.0 | 0.1 | GO:0006216 | cytidine catabolic process(GO:0006216) cytidine deamination(GO:0009972) cytidine metabolic process(GO:0046087) |
| 0.0 | 0.1 | GO:1902744 | negative regulation of lamellipodium organization(GO:1902744) |
| 0.0 | 0.2 | GO:0072102 | glomerulus morphogenesis(GO:0072102) |
| 0.0 | 0.1 | GO:0014734 | skeletal muscle hypertrophy(GO:0014734) |
| 0.0 | 0.1 | GO:1900019 | regulation of protein kinase C activity(GO:1900019) positive regulation of protein kinase C activity(GO:1900020) |
| 0.0 | 0.1 | GO:0046271 | phenylpropanoid catabolic process(GO:0046271) |
| 0.0 | 0.1 | GO:0006555 | methionine metabolic process(GO:0006555) |
| 0.0 | 0.1 | GO:1900095 | regulation of dosage compensation by inactivation of X chromosome(GO:1900095) |
| 0.0 | 0.1 | GO:0097212 | lysosomal membrane organization(GO:0097212) |
| 0.0 | 0.1 | GO:0032497 | detection of lipopolysaccharide(GO:0032497) |
| 0.0 | 0.3 | GO:0060391 | positive regulation of SMAD protein import into nucleus(GO:0060391) |
| 0.0 | 0.1 | GO:0060687 | regulation of branching involved in prostate gland morphogenesis(GO:0060687) |
| 0.0 | 0.1 | GO:1902992 | negative regulation of amyloid precursor protein catabolic process(GO:1902992) |
| 0.0 | 0.1 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.0 | 0.1 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.0 | 0.1 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.0 | 0.1 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.0 | 0.0 | GO:0051660 | establishment of centrosome localization(GO:0051660) |
| 0.0 | 0.1 | GO:0031587 | positive regulation of inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0031587) |
| 0.0 | 0.1 | GO:0046100 | hypoxanthine metabolic process(GO:0046100) hypoxanthine biosynthetic process(GO:0046101) |
| 0.0 | 0.1 | GO:0034146 | toll-like receptor 5 signaling pathway(GO:0034146) |
| 0.0 | 0.1 | GO:0015684 | ferrous iron transport(GO:0015684) |
| 0.0 | 0.1 | GO:1903223 | positive regulation of oxidative stress-induced neuron death(GO:1903223) |
| 0.0 | 0.1 | GO:0042360 | vitamin E metabolic process(GO:0042360) |
| 0.0 | 0.1 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.0 | 0.1 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 0.0 | 0.1 | GO:0035522 | monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.0 | 0.1 | GO:2001286 | regulation of caveolin-mediated endocytosis(GO:2001286) |
| 0.0 | 0.1 | GO:0009186 | deoxyribonucleoside diphosphate metabolic process(GO:0009186) |
| 0.0 | 0.1 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 0.1 | GO:0050651 | dermatan sulfate proteoglycan biosynthetic process(GO:0050651) |
| 0.0 | 0.2 | GO:0072502 | cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.0 | 0.1 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.0 | 0.1 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.1 | GO:0046618 | drug export(GO:0046618) |
| 0.0 | 0.1 | GO:0002386 | immune response in mucosal-associated lymphoid tissue(GO:0002386) |
| 0.0 | 0.1 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.0 | 0.2 | GO:0097062 | dendritic spine maintenance(GO:0097062) |
| 0.0 | 0.0 | GO:0046098 | guanine metabolic process(GO:0046098) |
| 0.0 | 0.1 | GO:0060020 | Bergmann glial cell differentiation(GO:0060020) |
| 0.0 | 0.0 | GO:0060839 | endothelial cell fate commitment(GO:0060839) |
| 0.0 | 0.0 | GO:0043101 | purine-containing compound salvage(GO:0043101) |
| 0.0 | 0.1 | GO:0072602 | interleukin-4 secretion(GO:0072602) |
| 0.0 | 0.0 | GO:0007403 | glial cell fate determination(GO:0007403) |
| 0.0 | 0.1 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.0 | 0.1 | GO:0051121 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.0 | 0.1 | GO:0019856 | 'de novo' pyrimidine nucleobase biosynthetic process(GO:0006207) pyrimidine nucleobase biosynthetic process(GO:0019856) |
| 0.0 | 0.1 | GO:0042167 | heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.0 | 0.2 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.0 | 0.1 | GO:1903677 | regulation of cap-independent translational initiation(GO:1903677) positive regulation of cap-independent translational initiation(GO:1903679) regulation of cytoplasmic translational initiation(GO:1904688) positive regulation of cytoplasmic translational initiation(GO:1904690) |
| 0.0 | 0.0 | GO:0006787 | porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) |
| 0.0 | 0.1 | GO:0030210 | heparin biosynthetic process(GO:0030210) |
| 0.0 | 0.1 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.0 | 0.1 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.1 | GO:0006113 | fermentation(GO:0006113) glucose catabolic process to lactate(GO:0019659) glycolytic fermentation(GO:0019660) glucose catabolic process to lactate via pyruvate(GO:0019661) |
| 0.0 | 0.0 | GO:0002085 | inhibition of neuroepithelial cell differentiation(GO:0002085) negative regulation of auditory receptor cell differentiation(GO:0045608) |
| 0.0 | 0.2 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.0 | 0.1 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.0 | 0.0 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) |
| 0.0 | 0.1 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.0 | 0.1 | GO:0060971 | embryonic heart tube left/right pattern formation(GO:0060971) |
| 0.0 | 0.0 | GO:0032782 | bile acid secretion(GO:0032782) |
| 0.0 | 0.0 | GO:0006166 | purine ribonucleoside salvage(GO:0006166) |
| 0.0 | 0.1 | GO:0071569 | protein ufmylation(GO:0071569) |
| 0.0 | 0.1 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.0 | 0.1 | GO:1903553 | positive regulation of extracellular exosome assembly(GO:1903553) |
| 0.0 | 0.1 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.0 | 0.0 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.0 | 0.0 | GO:1901535 | regulation of DNA demethylation(GO:1901535) negative regulation of DNA demethylation(GO:1901536) |
| 0.0 | 0.0 | GO:0003221 | right ventricular cardiac muscle tissue morphogenesis(GO:0003221) |
| 0.0 | 0.1 | GO:1904380 | endoplasmic reticulum mannose trimming(GO:1904380) |
| 0.0 | 0.1 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.0 | 0.1 | GO:0042997 | negative regulation of Golgi to plasma membrane protein transport(GO:0042997) |
| 0.0 | 0.1 | GO:0022007 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.0 | 0.1 | GO:0090005 | negative regulation of establishment of protein localization to plasma membrane(GO:0090005) |
| 0.0 | 0.2 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 0.1 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.0 | 0.2 | GO:2000060 | positive regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:2000060) |
| 0.0 | 0.0 | GO:0015766 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.0 | 0.0 | GO:0032513 | negative regulation of protein phosphatase type 2B activity(GO:0032513) |
| 0.0 | 0.0 | GO:0036023 | limb joint morphogenesis(GO:0036022) embryonic skeletal limb joint morphogenesis(GO:0036023) |
| 0.0 | 0.0 | GO:0018199 | peptidyl-glutamine modification(GO:0018199) |
| 0.0 | 0.1 | GO:0044027 | hypermethylation of CpG island(GO:0044027) |
| 0.0 | 0.0 | GO:0072361 | regulation of glycolytic process by regulation of transcription from RNA polymerase II promoter(GO:0072361) |
| 0.0 | 0.0 | GO:0090310 | negative regulation of methylation-dependent chromatin silencing(GO:0090310) |
| 0.0 | 0.0 | GO:2000313 | fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060825) regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000313) |
| 0.0 | 0.1 | GO:0090435 | protein localization to nuclear envelope(GO:0090435) |
| 0.0 | 0.0 | GO:1902564 | negative regulation of neutrophil activation(GO:1902564) |
| 0.0 | 0.0 | GO:0038095 | Fc-epsilon receptor signaling pathway(GO:0038095) |
| 0.0 | 0.1 | GO:0046726 | positive regulation by virus of viral protein levels in host cell(GO:0046726) |
| 0.0 | 0.1 | GO:0009838 | abscission(GO:0009838) |
| 0.0 | 0.0 | GO:0030997 | regulation of centriole-centriole cohesion(GO:0030997) |
| 0.0 | 0.0 | GO:0010982 | regulation of high-density lipoprotein particle clearance(GO:0010982) |
| 0.0 | 0.1 | GO:0071043 | CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) |
| 0.0 | 0.1 | GO:1903964 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.0 | 0.1 | GO:1902410 | mitotic cytokinetic process(GO:1902410) |
| 0.0 | 0.1 | GO:0042761 | very long-chain fatty acid biosynthetic process(GO:0042761) |
| 0.0 | 0.1 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.0 | 0.2 | GO:1904293 | negative regulation of ERAD pathway(GO:1904293) |
| 0.0 | 0.0 | GO:0021524 | visceral motor neuron differentiation(GO:0021524) |
| 0.0 | 0.0 | GO:0071727 | response to triacyl bacterial lipopeptide(GO:0071725) cellular response to triacyl bacterial lipopeptide(GO:0071727) |
| 0.0 | 0.1 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.1 | GO:0042364 | water-soluble vitamin biosynthetic process(GO:0042364) |
| 0.0 | 0.2 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.2 | GO:0032516 | positive regulation of phosphoprotein phosphatase activity(GO:0032516) |
| 0.0 | 0.0 | GO:0060842 | arterial endothelial cell differentiation(GO:0060842) |
| 0.0 | 0.1 | GO:0071494 | cellular response to UV-C(GO:0071494) |
| 0.0 | 0.1 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.0 | GO:0006553 | lysine metabolic process(GO:0006553) |
| 0.0 | 0.1 | GO:1903142 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.0 | 0.2 | GO:0060009 | Sertoli cell development(GO:0060009) |
| 0.0 | 0.1 | GO:1901096 | regulation of autophagosome maturation(GO:1901096) |
| 0.0 | 0.0 | GO:0036295 | cellular response to increased oxygen levels(GO:0036295) cellular response to hyperoxia(GO:0071455) |
| 0.0 | 0.1 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.0 | 0.0 | GO:0060178 | regulation of exocyst localization(GO:0060178) |
| 0.0 | 0.1 | GO:1902036 | regulation of hematopoietic stem cell differentiation(GO:1902036) |
| 0.0 | 0.2 | GO:0007020 | microtubule nucleation(GO:0007020) |
| 0.0 | 0.1 | GO:2000561 | regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000561) |
| 0.0 | 0.1 | GO:2000394 | positive regulation of lamellipodium morphogenesis(GO:2000394) |
| 0.0 | 0.1 | GO:0033578 | protein glycosylation in Golgi(GO:0033578) |
| 0.0 | 0.1 | GO:0060355 | positive regulation of cell adhesion molecule production(GO:0060355) |
| 0.0 | 0.0 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.0 | 0.1 | GO:0018406 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.0 | 0.0 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.0 | 0.0 | GO:1900094 | nodal signaling pathway involved in determination of left/right asymmetry(GO:0038107) regulation of transcription from RNA polymerase II promoter involved in determination of left/right symmetry(GO:1900094) nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900164) |
| 0.0 | 0.1 | GO:0039689 | negative stranded viral RNA replication(GO:0039689) multi-organism biosynthetic process(GO:0044034) |
| 0.0 | 0.1 | GO:0034239 | macrophage fusion(GO:0034238) regulation of macrophage fusion(GO:0034239) |
| 0.0 | 0.1 | GO:1903054 | negative regulation of extracellular matrix organization(GO:1903054) |
| 0.0 | 0.0 | GO:0032463 | negative regulation of protein homooligomerization(GO:0032463) |
| 0.0 | 0.0 | GO:0009158 | ribonucleoside monophosphate catabolic process(GO:0009158) purine ribonucleoside monophosphate catabolic process(GO:0009169) |
| 0.0 | 0.0 | GO:0019676 | ammonia assimilation cycle(GO:0019676) |
| 0.0 | 0.1 | GO:2001012 | mesenchymal cell differentiation involved in kidney development(GO:0072161) mesenchymal cell differentiation involved in renal system development(GO:2001012) |
| 0.0 | 0.1 | GO:0009052 | pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 0.0 | 0.2 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.0 | GO:0032911 | negative regulation of transforming growth factor beta1 production(GO:0032911) |
| 0.0 | 0.0 | GO:1901421 | positive regulation of response to alcohol(GO:1901421) |
| 0.0 | 0.0 | GO:0010963 | regulation of L-arginine import(GO:0010963) |
| 0.0 | 0.1 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.0 | 0.0 | GO:0035928 | rRNA import into mitochondrion(GO:0035928) |
| 0.0 | 0.1 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.0 | GO:0051599 | response to hydrostatic pressure(GO:0051599) |
| 0.0 | 0.0 | GO:0030576 | Cajal body organization(GO:0030576) |
| 0.0 | 0.1 | GO:0090160 | Golgi to lysosome transport(GO:0090160) |
| 0.0 | 0.0 | GO:1900122 | positive regulation of receptor binding(GO:1900122) |
| 0.0 | 0.0 | GO:2000370 | positive regulation of clathrin-mediated endocytosis(GO:2000370) |
| 0.0 | 0.0 | GO:1901668 | regulation of superoxide dismutase activity(GO:1901668) |
| 0.0 | 0.0 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.0 | 0.0 | GO:0033121 | regulation of purine nucleotide catabolic process(GO:0033121) negative regulation of purine nucleotide catabolic process(GO:0033122) negative regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051344) |
| 0.0 | 0.0 | GO:1900194 | negative regulation of oocyte maturation(GO:1900194) |
| 0.0 | 0.0 | GO:0042996 | regulation of Golgi to plasma membrane protein transport(GO:0042996) |
| 0.0 | 0.0 | GO:0007161 | calcium-independent cell-matrix adhesion(GO:0007161) |
| 0.0 | 0.1 | GO:0046040 | IMP metabolic process(GO:0046040) |
| 0.0 | 0.0 | GO:1902075 | cellular response to salt(GO:1902075) |
| 0.0 | 0.1 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.0 | 0.0 | GO:0061073 | ciliary body morphogenesis(GO:0061073) |
| 0.0 | 0.1 | GO:0046473 | phosphatidic acid metabolic process(GO:0046473) |
| 0.0 | 0.0 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.0 | 0.1 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.0 | 0.0 | GO:0043985 | histone H4-R3 methylation(GO:0043985) |
| 0.0 | 0.0 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.0 | 0.1 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.0 | 0.0 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.0 | 0.0 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.0 | 0.1 | GO:0060294 | cilium movement involved in cell motility(GO:0060294) |
| 0.0 | 0.0 | GO:0019346 | homoserine metabolic process(GO:0009092) transsulfuration(GO:0019346) |
| 0.0 | 0.1 | GO:0060396 | growth hormone receptor signaling pathway(GO:0060396) cellular response to growth hormone stimulus(GO:0071378) |
| 0.0 | 0.1 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.0 | 0.0 | GO:0070126 | mitochondrial translational termination(GO:0070126) |
| 0.0 | 0.0 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.0 | 0.2 | GO:0042347 | negative regulation of NF-kappaB import into nucleus(GO:0042347) |
| 0.0 | 0.0 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.0 | 0.1 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.0 | 0.1 | GO:2001138 | regulation of phospholipid transport(GO:2001138) positive regulation of phospholipid transport(GO:2001140) |
| 0.0 | 0.0 | GO:0097466 | glycoprotein ERAD pathway(GO:0097466) response to glycoprotein(GO:1904587) |
| 0.0 | 0.1 | GO:0052697 | xenobiotic glucuronidation(GO:0052697) |
| 0.0 | 0.0 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.0 | 0.0 | GO:0061156 | pulmonary artery morphogenesis(GO:0061156) |
| 0.0 | 0.0 | GO:0032849 | positive regulation of cellular pH reduction(GO:0032849) |
| 0.0 | 0.0 | GO:0070535 | histone H2A K63-linked ubiquitination(GO:0070535) |
| 0.0 | 0.0 | GO:0060008 | Sertoli cell differentiation(GO:0060008) |
| 0.0 | 0.0 | GO:0000087 | mitotic M phase(GO:0000087) |
| 0.0 | 0.0 | GO:0006344 | maintenance of chromatin silencing(GO:0006344) |
| 0.0 | 0.0 | GO:0032460 | negative regulation of protein oligomerization(GO:0032460) |
| 0.0 | 0.1 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.0 | 0.1 | GO:0006071 | glycerol metabolic process(GO:0006071) |
| 0.0 | 0.0 | GO:0071281 | cellular response to iron ion(GO:0071281) |
| 0.0 | 0.1 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.0 | 0.0 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.0 | 0.0 | GO:0030241 | skeletal muscle myosin thick filament assembly(GO:0030241) striated muscle myosin thick filament assembly(GO:0071688) |
| 0.0 | 0.1 | GO:0061318 | renal filtration cell differentiation(GO:0061318) glomerular epithelium development(GO:0072010) glomerular visceral epithelial cell differentiation(GO:0072112) glomerular epithelial cell differentiation(GO:0072311) |
| 0.0 | 0.0 | GO:0016560 | protein import into peroxisome matrix, docking(GO:0016560) |
| 0.0 | 0.0 | GO:0002924 | negative regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002924) |
| 0.0 | 0.0 | GO:1902109 | negative regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902109) |
| 0.0 | 0.1 | GO:0033133 | positive regulation of glucokinase activity(GO:0033133) positive regulation of hexokinase activity(GO:1903301) |
| 0.0 | 0.0 | GO:0090666 | scaRNA localization to Cajal body(GO:0090666) |
| 0.0 | 0.0 | GO:1903423 | positive regulation of synaptic vesicle recycling(GO:1903423) |
| 0.0 | 0.0 | GO:0033566 | gamma-tubulin complex localization(GO:0033566) |
| 0.0 | 0.1 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) |
| 0.0 | 0.1 | GO:2000774 | positive regulation of cell aging(GO:0090343) positive regulation of cellular senescence(GO:2000774) |
| 0.0 | 0.0 | GO:2001279 | regulation of prostaglandin biosynthetic process(GO:0031392) regulation of unsaturated fatty acid biosynthetic process(GO:2001279) |
| 0.0 | 0.0 | GO:0042780 | tRNA 3'-end processing(GO:0042780) |
| 0.0 | 0.0 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.0 | 0.0 | GO:0042268 | regulation of cytolysis(GO:0042268) |
| 0.0 | 0.0 | GO:0098904 | regulation of AV node cell action potential(GO:0098904) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.1 | 0.2 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.0 | 0.1 | GO:1990812 | growth cone filopodium(GO:1990812) |
| 0.0 | 0.1 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.0 | 0.2 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.2 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.1 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.0 | 0.1 | GO:0097418 | neurofibrillary tangle(GO:0097418) |
| 0.0 | 0.1 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.0 | 0.1 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.0 | 0.1 | GO:0033503 | HULC complex(GO:0033503) |
| 0.0 | 0.1 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.0 | 0.1 | GO:0090498 | extrinsic component of Golgi membrane(GO:0090498) |
| 0.0 | 0.2 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.1 | GO:0046691 | intracellular canaliculus(GO:0046691) |
| 0.0 | 0.1 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.0 | 0.1 | GO:0034715 | pICln-Sm protein complex(GO:0034715) |
| 0.0 | 0.1 | GO:0071008 | U2-type post-mRNA release spliceosomal complex(GO:0071008) |
| 0.0 | 0.1 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.0 | 0.1 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.0 | 0.1 | GO:0010009 | cytoplasmic side of endosome membrane(GO:0010009) |
| 0.0 | 0.1 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 0.0 | 0.1 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.0 | 0.1 | GO:0070688 | MLL5-L complex(GO:0070688) |
| 0.0 | 0.1 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.0 | 0.1 | GO:0005784 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.0 | 0.1 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.0 | 0.1 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 0.1 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 0.0 | GO:0005683 | U7 snRNP(GO:0005683) |
| 0.0 | 0.1 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 0.2 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 0.0 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.0 | 0.1 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.0 | 0.0 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.0 | 0.0 | GO:0032280 | symmetric synapse(GO:0032280) |
| 0.0 | 0.1 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 0.1 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.0 | 0.1 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.1 | GO:0005845 | mRNA cap binding complex(GO:0005845) RNA cap binding complex(GO:0034518) |
| 0.0 | 0.0 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 0.1 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.0 | 0.0 | GO:0035355 | Toll-like receptor 2-Toll-like receptor 6 protein complex(GO:0035355) |
| 0.0 | 0.1 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.0 | GO:0031417 | NatC complex(GO:0031417) |
| 0.0 | 0.1 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.0 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.0 | 0.1 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.1 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.0 | 0.1 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.0 | 0.0 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.0 | 0.2 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.1 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.1 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 0.1 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.1 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.1 | GO:0034464 | BBSome(GO:0034464) |
| 0.0 | 0.1 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.1 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.1 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.0 | 0.0 | GO:0097543 | ciliary inversin compartment(GO:0097543) |
| 0.0 | 0.0 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.0 | 0.1 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 0.1 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.0 | 0.1 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.0 | 0.0 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.0 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.0 | 0.0 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.0 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 0.2 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.0 | GO:0034448 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.0 | 0.5 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 0.1 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.1 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.3 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 0.1 | GO:0061617 | MICOS complex(GO:0061617) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.1 | 0.2 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.0 | 0.1 | GO:0008898 | S-adenosylmethionine-homocysteine S-methyltransferase activity(GO:0008898) |
| 0.0 | 0.2 | GO:0004528 | phosphodiesterase I activity(GO:0004528) |
| 0.0 | 0.1 | GO:0033592 | RNA strand annealing activity(GO:0033592) |
| 0.0 | 0.2 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.0 | 0.1 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.0 | 0.3 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.1 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.1 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.1 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.0 | 0.2 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.0 | 0.1 | GO:0047105 | aminobutyraldehyde dehydrogenase activity(GO:0019145) 4-trimethylammoniobutyraldehyde dehydrogenase activity(GO:0047105) |
| 0.0 | 0.1 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 0.1 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.0 | 0.2 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.0 | 0.2 | GO:0051920 | peroxiredoxin activity(GO:0051920) |
| 0.0 | 0.1 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.0 | 0.1 | GO:0036435 | K48-linked polyubiquitin binding(GO:0036435) |
| 0.0 | 0.1 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.0 | 0.1 | GO:0016743 | carboxyl- or carbamoyltransferase activity(GO:0016743) |
| 0.0 | 0.1 | GO:0061731 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.0 | 0.2 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.0 | GO:0035851 | Krueppel-associated box domain binding(GO:0035851) |
| 0.0 | 0.1 | GO:0035663 | Toll-like receptor 2 binding(GO:0035663) |
| 0.0 | 0.1 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.0 | 0.2 | GO:0070191 | methionine-R-sulfoxide reductase activity(GO:0070191) |
| 0.0 | 0.1 | GO:0080130 | L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.0 | 0.1 | GO:0071209 | U7 snRNA binding(GO:0071209) |
| 0.0 | 0.1 | GO:0015173 | aromatic amino acid transmembrane transporter activity(GO:0015173) |
| 0.0 | 0.1 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.0 | 0.1 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.0 | 0.1 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.0 | 0.1 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.0 | 0.1 | GO:0016149 | translation release factor activity, codon specific(GO:0016149) |
| 0.0 | 0.1 | GO:0004611 | phosphoenolpyruvate carboxykinase activity(GO:0004611) |
| 0.0 | 0.0 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.0 | 0.1 | GO:0000702 | oxidized base lesion DNA N-glycosylase activity(GO:0000702) |
| 0.0 | 0.3 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.0 | GO:0030620 | U2 snRNA binding(GO:0030620) |
| 0.0 | 0.1 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.0 | 0.1 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.0 | 0.0 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.0 | 0.0 | GO:0008525 | phosphatidylcholine transporter activity(GO:0008525) |
| 0.0 | 0.2 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) |
| 0.0 | 0.1 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.0 | 0.1 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.0 | 0.1 | GO:0071253 | connexin binding(GO:0071253) |
| 0.0 | 0.0 | GO:0004676 | 3-phosphoinositide-dependent protein kinase activity(GO:0004676) |
| 0.0 | 0.1 | GO:0004802 | transketolase activity(GO:0004802) |
| 0.0 | 0.1 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.0 | 0.1 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.0 | 0.1 | GO:0005345 | purine nucleobase transmembrane transporter activity(GO:0005345) pyrimidine nucleobase transmembrane transporter activity(GO:0005350) nucleobase transmembrane transporter activity(GO:0015205) |
| 0.0 | 0.1 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.0 | 0.1 | GO:0016882 | cyclo-ligase activity(GO:0016882) |
| 0.0 | 0.1 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.1 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.0 | 0.1 | GO:0003988 | acetyl-CoA C-acyltransferase activity(GO:0003988) |
| 0.0 | 0.1 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.0 | 0.0 | GO:0015157 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.0 | 0.1 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.0 | 0.3 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.0 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 0.0 | 0.1 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.0 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.0 | 0.0 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.0 | 0.0 | GO:0051733 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) |
| 0.0 | 0.0 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 0.0 | 0.0 | GO:0102007 | lactonohydrolase activity(GO:0046573) acyl-L-homoserine-lactone lactonohydrolase activity(GO:0102007) |
| 0.0 | 0.0 | GO:0001224 | RNA polymerase II transcription cofactor binding(GO:0001224) |
| 0.0 | 0.1 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.0 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.1 | GO:0016742 | hydroxymethyl-, formyl- and related transferase activity(GO:0016742) |
| 0.0 | 0.1 | GO:0032896 | palmitoyl-CoA 9-desaturase activity(GO:0032896) |
| 0.0 | 0.0 | GO:0008732 | threonine aldolase activity(GO:0004793) L-allo-threonine aldolase activity(GO:0008732) |
| 0.0 | 0.0 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.0 | GO:0035651 | AP-3 adaptor complex binding(GO:0035651) |
| 0.0 | 0.1 | GO:0070990 | snRNP binding(GO:0070990) |
| 0.0 | 0.1 | GO:0004792 | thiosulfate sulfurtransferase activity(GO:0004792) |
| 0.0 | 0.1 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.0 | 0.0 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.0 | 0.2 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.2 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.1 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.0 | 0.2 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.0 | 0.1 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.0 | 0.1 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.0 | 0.1 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.1 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.0 | 0.0 | GO:0005338 | nucleotide-sugar transmembrane transporter activity(GO:0005338) |
| 0.0 | 0.1 | GO:0016997 | exo-alpha-sialidase activity(GO:0004308) alpha-sialidase activity(GO:0016997) exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.0 | 0.1 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.2 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.0 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.0 | 0.0 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.0 | 0.1 | GO:0019211 | phosphatase activator activity(GO:0019211) |
| 0.0 | 0.1 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.0 | 0.1 | GO:0019763 | immunoglobulin receptor activity(GO:0019763) |
| 0.0 | 0.0 | GO:0016401 | palmitoyl-CoA oxidase activity(GO:0016401) |
| 0.0 | 0.1 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.2 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.0 | 0.3 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.1 | GO:0016421 | CoA carboxylase activity(GO:0016421) |
| 0.0 | 0.0 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 0.1 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.0 | 0.0 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.0 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.0 | 0.0 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) |
| 0.0 | 0.2 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.1 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 0.0 | GO:0031800 | type 3 metabotropic glutamate receptor binding(GO:0031800) |
| 0.0 | 0.0 | GO:0003846 | 2-acylglycerol O-acyltransferase activity(GO:0003846) |
| 0.0 | 0.0 | GO:0045134 | uridine-diphosphatase activity(GO:0045134) |
| 0.0 | 0.1 | GO:0018630 | 3-(3-hydroxyphenyl)propionate hydroxylase activity(GO:0008688) 4-chlorobenzaldehyde oxidase activity(GO:0018471) 3,5-xylenol methylhydroxylase activity(GO:0018630) phenylacetate hydroxylase activity(GO:0018631) 4-nitrophenol 4-monooxygenase activity(GO:0018632) dimethyl sulfide monooxygenase activity(GO:0018633) alpha-pinene monooxygenase [NADH] activity(GO:0018634) 1-hydroxy-2-naphthoate hydroxylase activity(GO:0018637) toluene 4-monooxygenase activity(GO:0018638) xylene monooxygenase activity(GO:0018639) dibenzothiophene monooxygenase activity(GO:0018640) 6-hydroxy-3-methyl-2-oxo-1,2-dihydroquinoline 6-monooxygenase activity(GO:0018641) chlorophenol 4-monooxygenase activity(GO:0018642) carbon disulfide oxygenase activity(GO:0018643) toluene 2-monooxygenase activity(GO:0018644) 1-hydroxy-2-oxolimonene 1,2-monooxygenase activity(GO:0018646) phenanthrene 1,2-monooxygenase activity(GO:0018647) tetrahydrofuran hydroxylase activity(GO:0018649) styrene monooxygenase activity(GO:0018650) toluene-4-sulfonate monooxygenase activity(GO:0018651) toluene-sulfonate methyl-monooxygenase activity(GO:0018652) 3-methyl-2-oxo-1,2-dihydroquinoline 6-monooxygenase activity(GO:0018653) 2-hydroxy-phenylacetate hydroxylase activity(GO:0018654) 2-oxo-delta3-4,5,5-trimethylcyclopentenylacetyl-CoA 1,2-monooxygenase activity(GO:0018655) phenanthrene 3,4-monooxygenase activity(GO:0018656) toluene 3-monooxygenase activity(GO:0018657) 4-hydroxyphenylacetate,NADH:oxygen oxidoreductase (3-hydroxylating) activity(GO:0018660) limonene monooxygenase activity(GO:0019113) 2-methylnaphthalene hydroxylase activity(GO:0034526) 1-methylnaphthalene hydroxylase activity(GO:0034534) bisphenol A hydroxylase A activity(GO:0034560) salicylate 5-hydroxylase activity(GO:0034785) isobutylamine N-hydroxylase activity(GO:0034791) branched-chain dodecylbenzene sulfonate monooxygenase activity(GO:0034802) 3-HSA hydroxylase activity(GO:0034819) 4-hydroxypyridine-3-hydroxylase activity(GO:0034894) 2-octaprenyl-3-methyl-6-methoxy-1,4-benzoquinol hydroxylase activity(GO:0043719) 6-hydroxynicotinate 3-monooxygenase activity(GO:0043731) thalianol hydroxylase activity(GO:0080014) |
| 0.0 | 0.2 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 0.0 | GO:0034547 | N-cyclopropylmelamine deaminase activity(GO:0034547) N-cyclopropylammeline deaminase activity(GO:0034548) N-cyclopropylammelide alkylamino hydrolase activity(GO:0034549) 2,5-diamino-6-ribitylamino-4(3H)-pyrimidinone 5'-phosphate deaminase activity(GO:0043723) tRNA-specific adenosine-37 deaminase activity(GO:0043829) archaeal-specific GTP cyclohydrolase activity(GO:0044682) tRNA-specific adenosine-34 deaminase activity(GO:0052717) |
| 0.0 | 0.1 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.0 | 0.0 | GO:0070546 | L-phenylalanine aminotransferase activity(GO:0070546) |
| 0.0 | 0.0 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.0 | 0.0 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.0 | 0.2 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.1 | GO:0004331 | fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.0 | 0.3 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 0.1 | GO:0003847 | 1-alkyl-2-acetylglycerophosphocholine esterase activity(GO:0003847) |
| 0.0 | 0.1 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.1 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.0 | GO:0031544 | peptidyl-proline 3-dioxygenase activity(GO:0031544) |
| 0.0 | 0.0 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.0 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.0 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 0.1 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 0.1 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 0.0 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 0.0 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.0 | 0.6 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.2 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.2 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.5 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.2 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 0.2 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.0 | 0.0 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.2 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.0 | 0.1 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 0.1 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.2 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.1 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 0.3 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.1 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.0 | 0.1 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.0 | REACTOME REGULATION OF APOPTOSIS | Genes involved in Regulation of Apoptosis |
| 0.0 | 0.2 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |