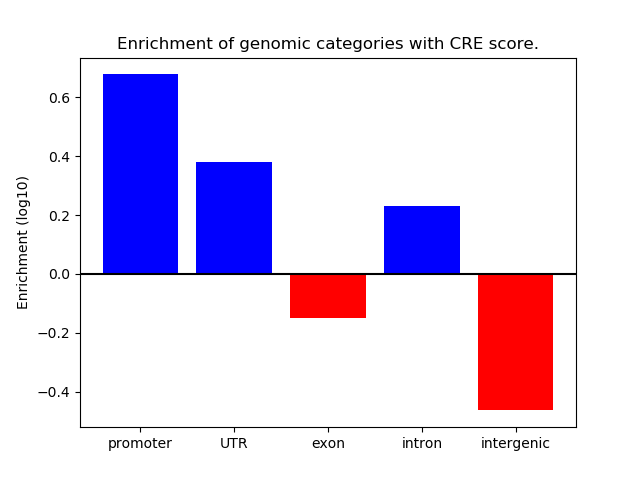
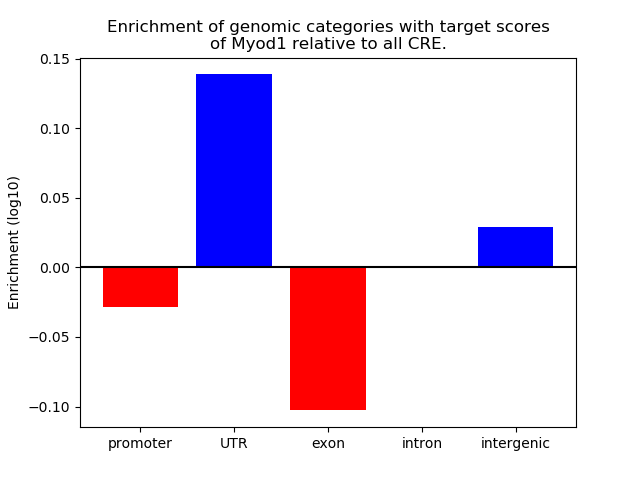
Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
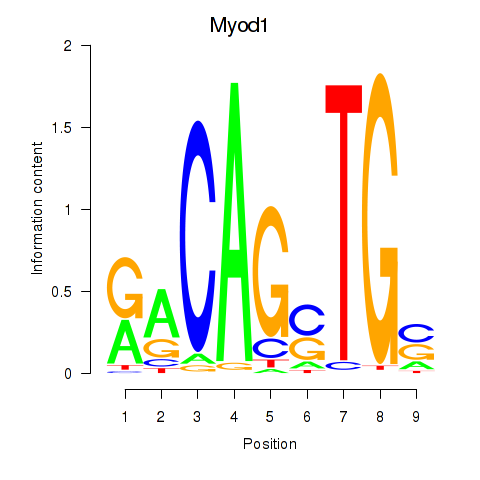
Results for Myod1
Z-value: 0.88
Transcription factors associated with Myod1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Myod1
|
ENSMUSG00000009471.3 | myogenic differentiation 1 |
Activity of the Myod1 motif across conditions
Conditions sorted by the z-value of the Myod1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
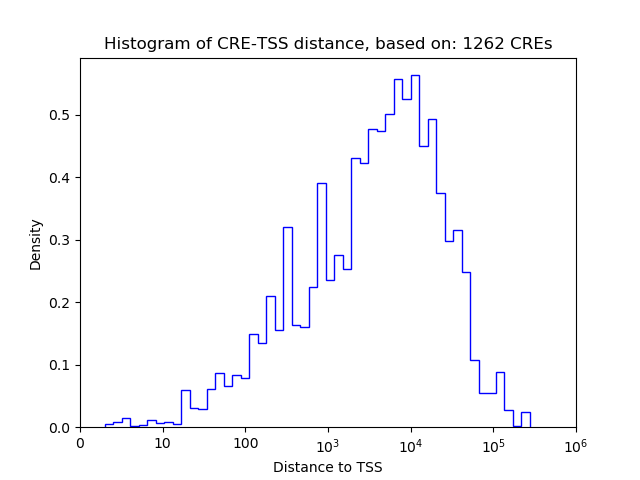
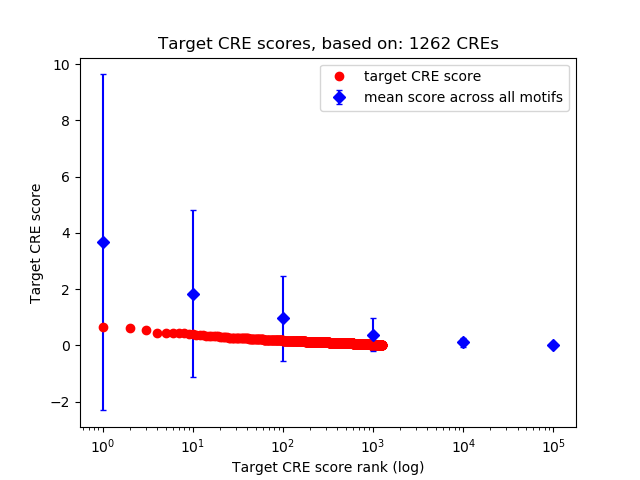
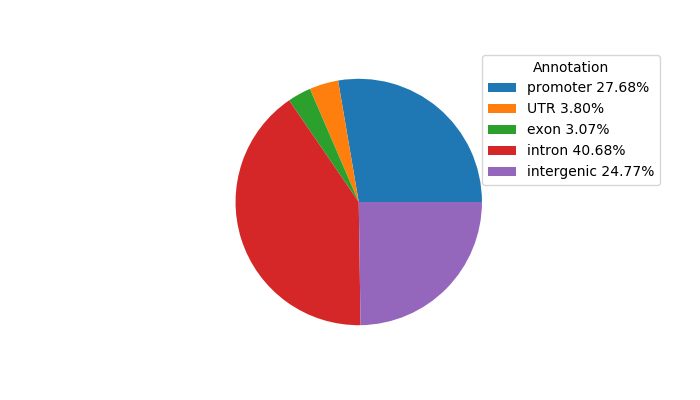
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr19_47222639_47222810 | 0.65 |
Gm50339 |
predicted gene, 50339 |
815 |
0.48 |
| chr19_46141233_46141384 | 0.61 |
Pitx3 |
paired-like homeodomain transcription factor 3 |
325 |
0.82 |
| chr9_63702505_63702656 | 0.54 |
Smad3 |
SMAD family member 3 |
9389 |
0.23 |
| chr2_167686580_167686731 | 0.45 |
Cebpb |
CCAAT/enhancer binding protein (C/EBP), beta |
2260 |
0.17 |
| chr5_8961694_8961940 | 0.44 |
Abcb4 |
ATP-binding cassette, sub-family B (MDR/TAP), member 4 |
3265 |
0.17 |
| chr12_26367487_26367659 | 0.43 |
4930549C15Rik |
RIKEN cDNA 4930549C15 gene |
20856 |
0.16 |
| chr6_121884163_121884602 | 0.43 |
Mug1 |
murinoglobulin 1 |
1195 |
0.48 |
| chr7_44810745_44811033 | 0.42 |
Atf5 |
activating transcription factor 5 |
4769 |
0.08 |
| chr6_86193078_86193250 | 0.41 |
Tgfa |
transforming growth factor alpha |
2059 |
0.28 |
| chr12_21141829_21142079 | 0.40 |
Asap2 |
ArfGAP with SH3 domain, ankyrin repeat and PH domain 2 |
30000 |
0.17 |
| chr15_25948371_25948535 | 0.38 |
Mir7212 |
microRNA 7212 |
226 |
0.93 |
| chr10_22361100_22361262 | 0.37 |
Raet1d |
retinoic acid early transcript delta |
629 |
0.64 |
| chr19_44930703_44930912 | 0.36 |
Slf2 |
SMC5-SMC6 complex localization factor 2 |
344 |
0.83 |
| chr10_8089529_8089695 | 0.35 |
Gm48614 |
predicted gene, 48614 |
68320 |
0.11 |
| chr19_61057184_61057369 | 0.35 |
Gm22520 |
predicted gene, 22520 |
43731 |
0.12 |
| chr2_26487761_26487912 | 0.34 |
Notch1 |
notch 1 |
15986 |
0.09 |
| chr8_121573324_121573508 | 0.34 |
Fbxo31 |
F-box protein 31 |
5331 |
0.12 |
| chr1_165617606_165617906 | 0.34 |
Mpzl1 |
myelin protein zero-like 1 |
3777 |
0.14 |
| chr12_21112874_21113025 | 0.33 |
Asap2 |
ArfGAP with SH3 domain, ankyrin repeat and PH domain 2 |
995 |
0.59 |
| chr10_63410268_63410432 | 0.31 |
Gm7530 |
predicted gene 7530 |
2649 |
0.19 |
| chr11_75435252_75435403 | 0.29 |
Serpinf2 |
serine (or cysteine) peptidase inhibitor, clade F, member 2 |
3573 |
0.1 |
| chr12_28910370_28910535 | 0.29 |
Gm31508 |
predicted gene, 31508 |
223 |
0.94 |
| chr1_119163374_119163565 | 0.28 |
Gm8321 |
predicted gene 8321 |
5883 |
0.26 |
| chr5_145984724_145985158 | 0.28 |
Cyp3a25 |
cytochrome P450, family 3, subfamily a, polypeptide 25 |
6702 |
0.13 |
| chr18_35887757_35887908 | 0.28 |
Gm36315 |
predicted gene, 36315 |
2505 |
0.16 |
| chr6_129542042_129542193 | 0.27 |
Gm44243 |
predicted gene, 44243 |
1896 |
0.16 |
| chr4_149978025_149978189 | 0.27 |
Spsb1 |
splA/ryanodine receptor domain and SOCS box containing 1 |
23064 |
0.13 |
| chr8_70182055_70182225 | 0.26 |
Tmem161a |
transmembrane protein 161A |
1601 |
0.22 |
| chr19_3362437_3362608 | 0.26 |
Cpt1a |
carnitine palmitoyltransferase 1a, liver |
3808 |
0.17 |
| chr2_164054518_164054669 | 0.26 |
Pabpc1l |
poly(A) binding protein, cytoplasmic 1-like |
6583 |
0.14 |
| chr7_80069741_80069903 | 0.26 |
Gm21057 |
predicted gene, 21057 |
5425 |
0.12 |
| chr4_142007387_142007541 | 0.26 |
Fhad1 |
forkhead-associated (FHA) phosphopeptide binding domain 1 |
4173 |
0.15 |
| chr5_43219459_43219610 | 0.26 |
Cpeb2 |
cytoplasmic polyadenylation element binding protein 2 |
13636 |
0.15 |
| chr13_37509524_37509684 | 0.26 |
Gm29590 |
predicted gene 29590 |
114 |
0.93 |
| chr1_40221808_40222119 | 0.25 |
Il1r1 |
interleukin 1 receptor, type I |
3117 |
0.26 |
| chr15_36471981_36472313 | 0.25 |
Ankrd46 |
ankyrin repeat domain 46 |
24568 |
0.12 |
| chr8_128465325_128465813 | 0.25 |
Nrp1 |
neuropilin 1 |
106172 |
0.06 |
| chr13_29944599_29944759 | 0.25 |
Gm11365 |
predicted gene 11365 |
4396 |
0.26 |
| chr15_27560020_27560329 | 0.25 |
Ank |
progressive ankylosis |
11273 |
0.16 |
| chr10_127659568_127659719 | 0.25 |
Stat6 |
signal transducer and activator of transcription 6 |
243 |
0.82 |
| chr18_32239390_32239541 | 0.24 |
Ercc3 |
excision repair cross-complementing rodent repair deficiency, complementation group 3 |
835 |
0.63 |
| chr19_3340116_3340320 | 0.24 |
Cpt1a |
carnitine palmitoyltransferase 1a, liver |
1409 |
0.32 |
| chr10_93495910_93496061 | 0.23 |
Hal |
histidine ammonia lyase |
420 |
0.79 |
| chr18_60737126_60737516 | 0.23 |
Ndst1 |
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1 |
4171 |
0.18 |
| chr10_119010946_119011297 | 0.23 |
Gm47461 |
predicted gene, 47461 |
10549 |
0.17 |
| chr6_72961723_72961891 | 0.23 |
Tmsb10 |
thymosin, beta 10 |
3059 |
0.2 |
| chr8_120445765_120445934 | 0.23 |
Gm22715 |
predicted gene, 22715 |
2300 |
0.29 |
| chr4_135265584_135265749 | 0.23 |
Clic4 |
chloride intracellular channel 4 (mitochondrial) |
7148 |
0.15 |
| chr15_96621805_96622214 | 0.22 |
Slc38a1 |
solute carrier family 38, member 1 |
19953 |
0.19 |
| chr10_8195740_8196032 | 0.22 |
Gm30906 |
predicted gene, 30906 |
19755 |
0.25 |
| chr1_93136180_93136798 | 0.21 |
Agxt |
alanine-glyoxylate aminotransferase |
921 |
0.46 |
| chr5_102783650_102783814 | 0.21 |
Arhgap24 |
Rho GTPase activating protein 24 |
14961 |
0.29 |
| chr13_105300041_105300198 | 0.21 |
Rnf180 |
ring finger protein 180 |
5304 |
0.3 |
| chr2_93433097_93433274 | 0.21 |
Mir7001 |
microRNA 7001 |
11178 |
0.15 |
| chr7_135609185_135609546 | 0.21 |
Ptpre |
protein tyrosine phosphatase, receptor type, E |
3537 |
0.23 |
| chr2_26500647_26500810 | 0.21 |
Notch1 |
notch 1 |
3094 |
0.15 |
| chr5_121612399_121612564 | 0.21 |
Acad12 |
acyl-Coenzyme A dehydrogenase family, member 12 |
6382 |
0.11 |
| chr9_43545212_43545661 | 0.21 |
Gm36855 |
predicted gene, 36855 |
24432 |
0.17 |
| chr16_32685796_32686168 | 0.21 |
Tnk2 |
tyrosine kinase, non-receptor, 2 |
5168 |
0.16 |
| chr12_3806542_3806693 | 0.21 |
Dnmt3a |
DNA methyltransferase 3A |
90 |
0.97 |
| chr13_24949534_24949740 | 0.20 |
Gpld1 |
glycosylphosphatidylinositol specific phospholipase D1 |
6217 |
0.14 |
| chr4_49099577_49099740 | 0.20 |
Plppr1 |
phospholipid phosphatase related 1 |
40202 |
0.18 |
| chr5_139835784_139835950 | 0.20 |
Psmg3 |
proteasome (prosome, macropain) assembly chaperone 3 |
8982 |
0.13 |
| chr12_111449466_111449652 | 0.20 |
Tnfaip2 |
tumor necrosis factor, alpha-induced protein 2 |
41 |
0.96 |
| chr4_120067294_120067548 | 0.20 |
AL607142.1 |
novel protein |
2425 |
0.34 |
| chr1_24693656_24693810 | 0.20 |
Lmbrd1 |
LMBR1 domain containing 1 |
14807 |
0.17 |
| chr15_97253131_97253396 | 0.20 |
Pced1b |
PC-esterase domain containing 1B |
4190 |
0.23 |
| chr9_108794998_108795174 | 0.20 |
Gm23034 |
predicted gene, 23034 |
302 |
0.72 |
| chr15_97019476_97019938 | 0.19 |
Slc38a4 |
solute carrier family 38, member 4 |
244 |
0.95 |
| chr15_98484617_98484776 | 0.19 |
Lalba |
lactalbumin, alpha |
1975 |
0.14 |
| chr2_145748022_145748197 | 0.19 |
Rin2 |
Ras and Rab interactor 2 |
36625 |
0.17 |
| chr17_12418655_12418810 | 0.19 |
Plg |
plasminogen |
40073 |
0.12 |
| chr7_113223800_113224075 | 0.19 |
Arntl |
aryl hydrocarbon receptor nuclear translocator-like |
10686 |
0.21 |
| chr14_14345872_14346046 | 0.19 |
Il3ra |
interleukin 3 receptor, alpha chain |
316 |
0.78 |
| chr2_32673636_32673820 | 0.19 |
Eng |
endoglin |
362 |
0.62 |
| chr8_120487540_120487709 | 0.19 |
Gse1 |
genetic suppressor element 1, coiled-coil protein |
823 |
0.56 |
| chr4_137764393_137764571 | 0.19 |
Alpl |
alkaline phosphatase, liver/bone/kidney |
1993 |
0.33 |
| chr10_80260218_80260407 | 0.19 |
Gamt |
guanidinoacetate methyltransferase |
606 |
0.42 |
| chr17_56174394_56174545 | 0.19 |
Gm44397 |
predicted gene, 44397 |
2344 |
0.15 |
| chr12_99374413_99374572 | 0.19 |
Gm47132 |
predicted gene, 47132 |
2756 |
0.19 |
| chr8_123737238_123737441 | 0.18 |
Gm45781 |
predicted gene 45781 |
2197 |
0.13 |
| chr4_11615049_11615228 | 0.18 |
Gm11832 |
predicted gene 11832 |
131 |
0.95 |
| chr1_190044023_190044190 | 0.18 |
Smyd2 |
SET and MYND domain containing 2 |
121743 |
0.05 |
| chr3_100466870_100467200 | 0.18 |
Tent5c |
terminal nucleotidyltransferase 5C |
22159 |
0.12 |
| chr5_138998044_138998247 | 0.18 |
Pdgfa |
platelet derived growth factor, alpha |
775 |
0.61 |
| chr6_126034330_126034486 | 0.18 |
Ntf3 |
neurotrophin 3 |
130552 |
0.05 |
| chr8_33892250_33892401 | 0.18 |
Rbpms |
RNA binding protein gene with multiple splicing |
561 |
0.74 |
| chr4_149799592_149799754 | 0.18 |
Gm13065 |
predicted gene 13065 |
6254 |
0.1 |
| chr15_68399262_68399413 | 0.18 |
Gm20732 |
predicted gene 20732 |
36161 |
0.13 |
| chr3_138463193_138463467 | 0.18 |
Metap1 |
methionyl aminopeptidase 1 |
5624 |
0.13 |
| chr10_81644706_81644857 | 0.18 |
Ankrd24 |
ankyrin repeat domain 24 |
978 |
0.32 |
| chr19_38223407_38224029 | 0.18 |
Fra10ac1 |
FRA10AC1 homolog (human) |
420 |
0.83 |
| chr18_20936854_20937178 | 0.18 |
Rnf125 |
ring finger protein 125 |
7609 |
0.22 |
| chr6_29117593_29117759 | 0.18 |
Gm25589 |
predicted gene, 25589 |
4070 |
0.17 |
| chr19_3840335_3840486 | 0.18 |
Chka |
choline kinase alpha |
11363 |
0.09 |
| chr15_67445781_67445973 | 0.18 |
1700012I11Rik |
RIKEN cDNA 1700012I11 gene |
219108 |
0.02 |
| chr4_118137948_118138112 | 0.17 |
St3gal3 |
ST3 beta-galactoside alpha-2,3-sialyltransferase 3 |
3116 |
0.21 |
| chr9_107689065_107689227 | 0.17 |
Sema3f |
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3F |
771 |
0.43 |
| chr14_41114673_41114824 | 0.17 |
Mat1a |
methionine adenosyltransferase I, alpha |
9367 |
0.12 |
| chr18_39498437_39498588 | 0.17 |
Nr3c1 |
nuclear receptor subfamily 3, group C, member 1 |
7211 |
0.27 |
| chr13_113536421_113536595 | 0.17 |
Gm34471 |
predicted gene, 34471 |
31259 |
0.12 |
| chr17_29138051_29138219 | 0.17 |
Rpl35a-ps3 |
ribosomal protein L35A, pseudogene 3 |
831 |
0.4 |
| chr12_103853006_103853292 | 0.17 |
Serpina1a |
serine (or cysteine) peptidase inhibitor, clade A, member 1A |
10402 |
0.09 |
| chr6_31188267_31188418 | 0.17 |
Lncpint |
long non-protein coding RNA, Trp53 induced transcript |
21826 |
0.13 |
| chr9_69306275_69306692 | 0.17 |
Rora |
RAR-related orphan receptor alpha |
16801 |
0.21 |
| chr1_72837329_72837510 | 0.16 |
Igfbp2 |
insulin-like growth factor binding protein 2 |
11280 |
0.21 |
| chr8_14257136_14257302 | 0.16 |
Gm26184 |
predicted gene, 26184 |
16092 |
0.22 |
| chr2_126585323_126585610 | 0.16 |
Hdc |
histidine decarboxylase |
12795 |
0.17 |
| chr12_39992836_39993213 | 0.16 |
Arl4a |
ADP-ribosylation factor-like 4A |
21460 |
0.17 |
| chr13_93940938_93941096 | 0.16 |
Gm24737 |
predicted gene, 24737 |
15293 |
0.17 |
| chr4_141124895_141125054 | 0.16 |
4921514A10Rik |
RIKEN cDNA 4921514A10 gene |
9314 |
0.11 |
| chr19_46623154_46623332 | 0.16 |
Wbp1l |
WW domain binding protein 1 like |
158 |
0.94 |
| chr19_61049084_61049293 | 0.16 |
Gm22520 |
predicted gene, 22520 |
35643 |
0.14 |
| chr7_18957606_18957786 | 0.16 |
Nanos2 |
nanos C2HC-type zinc finger 2 |
29704 |
0.06 |
| chr18_64332314_64332480 | 0.16 |
St8sia3os |
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 3, opposite strand |
1309 |
0.4 |
| chr1_180219669_180219845 | 0.16 |
Gm37336 |
predicted gene, 37336 |
7356 |
0.14 |
| chr15_98733475_98733629 | 0.16 |
Fkbp11 |
FK506 binding protein 11 |
5354 |
0.1 |
| chr6_50890819_50891330 | 0.16 |
G930045G22Rik |
RIKEN cDNA G930045G22 gene |
12997 |
0.17 |
| chr14_30922623_30922783 | 0.16 |
Itih3 |
inter-alpha trypsin inhibitor, heavy chain 3 |
846 |
0.45 |
| chr7_141475336_141475516 | 0.16 |
Tspan4 |
tetraspanin 4 |
84 |
0.73 |
| chr2_33178304_33178466 | 0.16 |
Gm13528 |
predicted gene 13528 |
483 |
0.76 |
| chr6_126034904_126035055 | 0.16 |
Ntf3 |
neurotrophin 3 |
129981 |
0.05 |
| chr11_51918030_51918186 | 0.15 |
Gm39822 |
predicted gene, 39822 |
19142 |
0.14 |
| chr4_148614270_148614447 | 0.15 |
Tardbp |
TAR DNA binding protein |
1780 |
0.22 |
| chr5_63897329_63897492 | 0.15 |
0610040J01Rik |
RIKEN cDNA 0610040J01 gene |
19934 |
0.16 |
| chr19_47137759_47137924 | 0.15 |
Calhm2 |
calcium homeostasis modulator family member 2 |
453 |
0.68 |
| chr6_137612326_137612520 | 0.15 |
Gm44106 |
predicted gene, 44106 |
18848 |
0.21 |
| chr7_27129962_27130129 | 0.15 |
Gm15883 |
predicted gene 15883 |
629 |
0.5 |
| chr4_120072218_120072438 | 0.15 |
AL607142.1 |
novel protein |
7332 |
0.23 |
| chr1_169916758_169916917 | 0.15 |
Ccdc190 |
coiled-coil domain containing 190 |
11811 |
0.16 |
| chr5_35973954_35974105 | 0.15 |
Afap1 |
actin filament associated protein 1 |
10159 |
0.25 |
| chr11_75431233_75431481 | 0.15 |
Gm12337 |
predicted gene 12337 |
3604 |
0.1 |
| chr7_141429450_141429618 | 0.15 |
Cend1 |
cell cycle exit and neuronal differentiation 1 |
43 |
0.91 |
| chr11_119976291_119976457 | 0.15 |
Baiap2 |
brain-specific angiogenesis inhibitor 1-associated protein 2 |
16353 |
0.1 |
| chr15_27564431_27564740 | 0.15 |
Ank |
progressive ankylosis |
6862 |
0.17 |
| chr11_32364448_32364599 | 0.15 |
Sh3pxd2b |
SH3 and PX domains 2B |
16683 |
0.16 |
| chr12_21209715_21209893 | 0.15 |
AC156032.1 |
|
37519 |
0.12 |
| chr1_75215946_75216124 | 0.15 |
Tuba4a |
tubulin, alpha 4A |
1379 |
0.16 |
| chr16_22436162_22436319 | 0.15 |
Etv5 |
ets variant 5 |
274 |
0.91 |
| chr19_6954083_6954250 | 0.15 |
Plcb3 |
phospholipase C, beta 3 |
681 |
0.4 |
| chr5_25006886_25007052 | 0.15 |
Prkag2 |
protein kinase, AMP-activated, gamma 2 non-catalytic subunit |
11216 |
0.18 |
| chr2_164862516_164862709 | 0.15 |
Pltp |
phospholipid transfer protein |
4901 |
0.1 |
| chr10_80618275_80618470 | 0.15 |
Csnk1g2 |
casein kinase 1, gamma 2 |
4466 |
0.08 |
| chr7_6727518_6727687 | 0.14 |
Peg3 |
paternally expressed 3 |
2817 |
0.13 |
| chr19_10556208_10556399 | 0.14 |
Tmem216 |
transmembrane protein 216 |
65 |
0.94 |
| chr4_134485980_134486138 | 0.14 |
Gm13250 |
predicted gene 13250 |
7163 |
0.1 |
| chr12_111051690_111051903 | 0.14 |
Rcor1 |
REST corepressor 1 |
11998 |
0.12 |
| chr5_147880438_147880628 | 0.14 |
Slc46a3 |
solute carrier family 46, member 3 |
664 |
0.66 |
| chr4_124701480_124701631 | 0.14 |
Fhl3 |
four and a half LIM domains 3 |
835 |
0.38 |
| chr14_57747701_57747852 | 0.14 |
Lats2 |
large tumor suppressor 2 |
1198 |
0.32 |
| chr19_32388450_32388613 | 0.14 |
Sgms1 |
sphingomyelin synthase 1 |
19 |
0.93 |
| chr19_47412733_47413051 | 0.14 |
Sh3pxd2a |
SH3 and PX domains 2A |
2533 |
0.32 |
| chr6_89008983_89009134 | 0.14 |
4933427D06Rik |
RIKEN cDNA 4933427D06 gene |
58375 |
0.1 |
| chr14_59550998_59551166 | 0.14 |
Cdadc1 |
cytidine and dCMP deaminase domain containing 1 |
46754 |
0.12 |
| chr4_117737643_117737808 | 0.14 |
Gm12844 |
predicted gene 12844 |
8426 |
0.13 |
| chr19_34553030_34553190 | 0.14 |
Ifit2 |
interferon-induced protein with tetratricopeptide repeats 2 |
2400 |
0.18 |
| chr13_23914881_23915050 | 0.14 |
Slc17a4 |
solute carrier family 17 (sodium phosphate), member 4 |
3 |
0.95 |
| chr7_84135593_84135937 | 0.14 |
Abhd17c |
abhydrolase domain containing 17C |
12050 |
0.15 |
| chr19_3691348_3691719 | 0.14 |
Lrp5 |
low density lipoprotein receptor-related protein 5 |
4969 |
0.12 |
| chr11_67585142_67585306 | 0.14 |
Gas7 |
growth arrest specific 7 |
1276 |
0.49 |
| chr2_32167931_32168112 | 0.14 |
Gm27805 |
predicted gene, 27805 |
5150 |
0.13 |
| chr8_106574781_106574955 | 0.14 |
Gm10073 |
predicted pseudogene 10073 |
1518 |
0.35 |
| chr17_29438491_29438658 | 0.14 |
Gm36486 |
predicted gene, 36486 |
1126 |
0.36 |
| chr8_110931698_110931872 | 0.14 |
St3gal2 |
ST3 beta-galactoside alpha-2,3-sialyltransferase 2 |
136 |
0.93 |
| chr19_37814514_37814675 | 0.14 |
Gm9067 |
predicted gene 9067 |
30328 |
0.16 |
| chr4_120073207_120073378 | 0.14 |
AL607142.1 |
novel protein |
8296 |
0.23 |
| chr15_82147771_82148080 | 0.14 |
Srebf2 |
sterol regulatory element binding factor 2 |
88 |
0.94 |
| chr12_36207522_36207673 | 0.14 |
Lrrc72 |
leucine rich repeat containing 72 |
10605 |
0.14 |
| chr2_179290486_179290655 | 0.14 |
Gm14293 |
predicted gene 14293 |
50080 |
0.15 |
| chr1_36528420_36528588 | 0.14 |
Gm38033 |
predicted gene, 38033 |
267 |
0.81 |
| chr7_16974127_16974283 | 0.14 |
Gm42372 |
predicted gene, 42372 |
8881 |
0.09 |
| chr12_103817168_103817505 | 0.14 |
Gm8893 |
predicted gene 8893 |
4907 |
0.11 |
| chr4_132063346_132063505 | 0.14 |
Epb41 |
erythrocyte membrane protein band 4.1 |
8672 |
0.11 |
| chr13_64178656_64179049 | 0.14 |
Habp4 |
hyaluronic acid binding protein 4 |
4752 |
0.13 |
| chr4_141513828_141514048 | 0.14 |
Spen |
spen family transcription repressor |
2971 |
0.19 |
| chr14_46016661_46017121 | 0.14 |
Gm6580 |
predicted gene 6580 |
10537 |
0.17 |
| chr11_40690363_40690782 | 0.14 |
Mat2b |
methionine adenosyltransferase II, beta |
2049 |
0.29 |
| chr10_71252490_71252774 | 0.14 |
Ube2d1 |
ubiquitin-conjugating enzyme E2D 1 |
4256 |
0.15 |
| chr2_164053751_164053902 | 0.14 |
Pabpc1l |
poly(A) binding protein, cytoplasmic 1-like |
5816 |
0.14 |
| chr5_111469920_111470194 | 0.13 |
Gm43119 |
predicted gene 43119 |
46468 |
0.12 |
| chr2_34407224_34407388 | 0.13 |
Mapkap1 |
mitogen-activated protein kinase associated protein 1 |
123 |
0.96 |
| chr19_7155448_7155628 | 0.13 |
Otub1 |
OTU domain, ubiquitin aldehyde binding 1 |
44926 |
0.08 |
| chr10_127326379_127326530 | 0.13 |
Arhgap9 |
Rho GTPase activating protein 9 |
2131 |
0.13 |
| chr5_28071886_28072263 | 0.13 |
Insig1 |
insulin induced gene 1 |
711 |
0.49 |
| chr12_111756775_111756948 | 0.13 |
Klc1 |
kinesin light chain 1 |
1988 |
0.2 |
| chr2_170345551_170345760 | 0.13 |
Bcas1 |
breast carcinoma amplified sequence 1 |
4085 |
0.22 |
| chr14_51101004_51101293 | 0.13 |
Rnase4 |
ribonuclease, RNase A family 4 |
4789 |
0.09 |
| chr11_79267281_79267540 | 0.13 |
Gm44787 |
predicted gene 44787 |
6434 |
0.16 |
| chr14_66147441_66147613 | 0.13 |
Chrna2 |
cholinergic receptor, nicotinic, alpha polypeptide 2 (neuronal) |
6594 |
0.17 |
| chr11_86973771_86973954 | 0.13 |
Ypel2 |
yippee like 2 |
1838 |
0.33 |
| chr19_7155121_7155413 | 0.13 |
Otub1 |
OTU domain, ubiquitin aldehyde binding 1 |
45197 |
0.08 |
| chr15_85646987_85647149 | 0.13 |
Gm49539 |
predicted gene, 49539 |
985 |
0.49 |
| chr7_117616401_117616570 | 0.13 |
Xylt1 |
xylosyltransferase 1 |
140993 |
0.05 |
| chr17_31323497_31323678 | 0.13 |
Slc37a1 |
solute carrier family 37 (glycerol-3-phosphate transporter), member 1 |
1333 |
0.36 |
| chr3_10310720_10310871 | 0.13 |
Gm38364 |
predicted gene, 38364 |
1747 |
0.2 |
| chr7_80245940_80246186 | 0.13 |
Ttll13 |
tubulin tyrosine ligase-like family, member 13 |
313 |
0.78 |
| chr19_41290060_41290220 | 0.13 |
Tm9sf3 |
transmembrane 9 superfamily member 3 |
26143 |
0.21 |
| chr1_58183291_58183442 | 0.13 |
Aox3 |
aldehyde oxidase 3 |
13742 |
0.17 |
| chr2_163481751_163481907 | 0.13 |
Fitm2 |
fat storage-inducing transmembrane protein 2 |
9200 |
0.11 |
| chr6_145703763_145703927 | 0.13 |
Gm15704 |
predicted gene 15704 |
42428 |
0.14 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0046439 | cysteine catabolic process(GO:0009093) L-cysteine catabolic process(GO:0019448) L-cysteine metabolic process(GO:0046439) |
| 0.1 | 0.2 | GO:0035771 | interleukin-4-mediated signaling pathway(GO:0035771) |
| 0.0 | 0.2 | GO:0046499 | S-adenosylmethioninamine metabolic process(GO:0046499) |
| 0.0 | 0.1 | GO:0021699 | cerebellar cortex maturation(GO:0021699) |
| 0.0 | 0.2 | GO:0006548 | histidine catabolic process(GO:0006548) imidazole-containing compound catabolic process(GO:0052805) |
| 0.0 | 0.1 | GO:0097296 | activation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097296) |
| 0.0 | 0.2 | GO:0003273 | cell migration involved in endocardial cushion formation(GO:0003273) |
| 0.0 | 0.1 | GO:0045626 | negative regulation of T-helper 1 cell differentiation(GO:0045626) |
| 0.0 | 0.1 | GO:2000591 | positive regulation of metanephric mesenchymal cell migration by platelet-derived growth factor receptor-beta signaling pathway(GO:0035793) regulation of metanephric mesenchymal cell migration by platelet-derived growth factor receptor-beta signaling pathway(GO:1900238) positive regulation of metanephric mesenchymal cell migration(GO:2000591) |
| 0.0 | 0.1 | GO:0033499 | galactose catabolic process via UDP-galactose(GO:0033499) glycolytic process from galactose(GO:0061623) |
| 0.0 | 0.2 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.1 | GO:0072367 | regulation of lipid transport by regulation of transcription from RNA polymerase II promoter(GO:0072367) |
| 0.0 | 0.1 | GO:0002432 | granuloma formation(GO:0002432) |
| 0.0 | 0.1 | GO:0002337 | B-1a B cell differentiation(GO:0002337) |
| 0.0 | 0.1 | GO:0002121 | inter-male aggressive behavior(GO:0002121) |
| 0.0 | 0.1 | GO:0048133 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.0 | 0.1 | GO:0045472 | response to ether(GO:0045472) |
| 0.0 | 0.1 | GO:0061144 | alveolar secondary septum development(GO:0061144) |
| 0.0 | 0.1 | GO:0097490 | trunk segmentation(GO:0035290) trunk neural crest cell migration(GO:0036484) ventral trunk neural crest cell migration(GO:0036486) sympathetic neuron projection extension(GO:0097490) sympathetic neuron projection guidance(GO:0097491) |
| 0.0 | 0.1 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.0 | 0.1 | GO:0010520 | regulation of reciprocal meiotic recombination(GO:0010520) |
| 0.0 | 0.0 | GO:0010248 | establishment or maintenance of transmembrane electrochemical gradient(GO:0010248) |
| 0.0 | 0.1 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.0 | 0.1 | GO:0072675 | osteoclast fusion(GO:0072675) |
| 0.0 | 0.1 | GO:0006868 | glutamine transport(GO:0006868) |
| 0.0 | 0.1 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.0 | 0.0 | GO:0007403 | glial cell fate determination(GO:0007403) |
| 0.0 | 0.1 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.2 | GO:0033683 | nucleotide-excision repair, DNA incision(GO:0033683) |
| 0.0 | 0.1 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.0 | 0.1 | GO:0006553 | lysine metabolic process(GO:0006553) |
| 0.0 | 0.1 | GO:0003176 | aortic valve development(GO:0003176) aortic valve morphogenesis(GO:0003180) |
| 0.0 | 0.1 | GO:0043321 | regulation of natural killer cell degranulation(GO:0043321) |
| 0.0 | 0.1 | GO:2001269 | positive regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001269) |
| 0.0 | 0.1 | GO:1903525 | regulation of membrane tubulation(GO:1903525) |
| 0.0 | 0.5 | GO:0070306 | lens fiber cell differentiation(GO:0070306) |
| 0.0 | 0.2 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.0 | 0.1 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.0 | 0.1 | GO:0061470 | T follicular helper cell differentiation(GO:0061470) |
| 0.0 | 0.1 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.0 | 0.1 | GO:2000659 | regulation of interleukin-1-mediated signaling pathway(GO:2000659) |
| 0.0 | 0.2 | GO:0032000 | positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.0 | 0.0 | GO:0001828 | inner cell mass cellular morphogenesis(GO:0001828) |
| 0.0 | 0.1 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.0 | 0.1 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.0 | 0.1 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.0 | 0.0 | GO:0060137 | maternal process involved in parturition(GO:0060137) |
| 0.0 | 0.0 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.0 | 0.0 | GO:0071332 | cellular response to fructose stimulus(GO:0071332) |
| 0.0 | 0.0 | GO:1901420 | negative regulation of response to alcohol(GO:1901420) |
| 0.0 | 0.0 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.0 | 0.1 | GO:0031508 | pericentric heterochromatin assembly(GO:0031508) |
| 0.0 | 0.2 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 0.0 | GO:0044805 | lysosomal microautophagy(GO:0016237) piecemeal microautophagy of nucleus(GO:0034727) late nucleophagy(GO:0044805) |
| 0.0 | 0.1 | GO:0060391 | positive regulation of SMAD protein import into nucleus(GO:0060391) |
| 0.0 | 0.0 | GO:0009448 | gamma-aminobutyric acid metabolic process(GO:0009448) |
| 0.0 | 0.0 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.0 | 0.1 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.0 | 0.1 | GO:1902262 | apoptotic process involved in patterning of blood vessels(GO:1902262) |
| 0.0 | 0.0 | GO:1990086 | lens fiber cell apoptotic process(GO:1990086) |
| 0.0 | 0.1 | GO:0051593 | response to folic acid(GO:0051593) |
| 0.0 | 0.0 | GO:0021914 | negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021914) |
| 0.0 | 0.0 | GO:0051791 | medium-chain fatty acid metabolic process(GO:0051791) |
| 0.0 | 0.0 | GO:0051795 | positive regulation of catagen(GO:0051795) |
| 0.0 | 0.0 | GO:2000468 | regulation of peroxidase activity(GO:2000468) |
| 0.0 | 0.0 | GO:1902075 | cellular response to salt(GO:1902075) |
| 0.0 | 0.1 | GO:0090051 | negative regulation of cell migration involved in sprouting angiogenesis(GO:0090051) |
| 0.0 | 0.0 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.0 | 0.1 | GO:0048069 | eye pigmentation(GO:0048069) |
| 0.0 | 0.0 | GO:0018992 | germ-line sex determination(GO:0018992) |
| 0.0 | 0.0 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.0 | 0.1 | GO:0048702 | embryonic neurocranium morphogenesis(GO:0048702) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.0 | 0.2 | GO:0044214 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.0 | 0.1 | GO:0036488 | CHOP-C/EBP complex(GO:0036488) |
| 0.0 | 0.1 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.0 | 0.2 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.0 | 0.2 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.0 | 0.1 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.0 | 0.0 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 0.0 | 0.1 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.0 | 0.0 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 0.2 | GO:0038201 | TOR complex(GO:0038201) |
| 0.0 | 0.0 | GO:0097057 | TRAF2-GSTP1 complex(GO:0097057) |
| 0.0 | 0.2 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.2 | GO:0000145 | exocyst(GO:0000145) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) DNA (cytosine-5-)-methyltransferase activity, acting on CpG substrates(GO:0051718) |
| 0.0 | 0.2 | GO:0005534 | galactose binding(GO:0005534) |
| 0.0 | 0.2 | GO:0032564 | dATP binding(GO:0032564) |
| 0.0 | 0.1 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) |
| 0.0 | 0.1 | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0003829) |
| 0.0 | 0.1 | GO:0008525 | phosphatidylcholine transporter activity(GO:0008525) |
| 0.0 | 0.1 | GO:0005072 | transforming growth factor beta receptor, cytoplasmic mediator activity(GO:0005072) |
| 0.0 | 0.1 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.0 | 0.3 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.0 | 0.1 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.1 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.2 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.0 | 0.1 | GO:0018631 | 3-(3-hydroxyphenyl)propionate hydroxylase activity(GO:0008688) 4-chlorobenzaldehyde oxidase activity(GO:0018471) 3,5-xylenol methylhydroxylase activity(GO:0018630) phenylacetate hydroxylase activity(GO:0018631) 4-nitrophenol 4-monooxygenase activity(GO:0018632) dimethyl sulfide monooxygenase activity(GO:0018633) alpha-pinene monooxygenase [NADH] activity(GO:0018634) 1-hydroxy-2-naphthoate hydroxylase activity(GO:0018637) toluene 4-monooxygenase activity(GO:0018638) xylene monooxygenase activity(GO:0018639) dibenzothiophene monooxygenase activity(GO:0018640) 6-hydroxy-3-methyl-2-oxo-1,2-dihydroquinoline 6-monooxygenase activity(GO:0018641) chlorophenol 4-monooxygenase activity(GO:0018642) carbon disulfide oxygenase activity(GO:0018643) toluene 2-monooxygenase activity(GO:0018644) 1-hydroxy-2-oxolimonene 1,2-monooxygenase activity(GO:0018646) phenanthrene 1,2-monooxygenase activity(GO:0018647) tetrahydrofuran hydroxylase activity(GO:0018649) styrene monooxygenase activity(GO:0018650) toluene-4-sulfonate monooxygenase activity(GO:0018651) toluene-sulfonate methyl-monooxygenase activity(GO:0018652) 3-methyl-2-oxo-1,2-dihydroquinoline 6-monooxygenase activity(GO:0018653) 2-hydroxy-phenylacetate hydroxylase activity(GO:0018654) 2-oxo-delta3-4,5,5-trimethylcyclopentenylacetyl-CoA 1,2-monooxygenase activity(GO:0018655) phenanthrene 3,4-monooxygenase activity(GO:0018656) toluene 3-monooxygenase activity(GO:0018657) 4-hydroxyphenylacetate,NADH:oxygen oxidoreductase (3-hydroxylating) activity(GO:0018660) limonene monooxygenase activity(GO:0019113) 2-methylnaphthalene hydroxylase activity(GO:0034526) 1-methylnaphthalene hydroxylase activity(GO:0034534) bisphenol A hydroxylase A activity(GO:0034560) salicylate 5-hydroxylase activity(GO:0034785) isobutylamine N-hydroxylase activity(GO:0034791) branched-chain dodecylbenzene sulfonate monooxygenase activity(GO:0034802) 3-HSA hydroxylase activity(GO:0034819) 4-hydroxypyridine-3-hydroxylase activity(GO:0034894) 2-octaprenyl-3-methyl-6-methoxy-1,4-benzoquinol hydroxylase activity(GO:0043719) 6-hydroxynicotinate 3-monooxygenase activity(GO:0043731) thalianol hydroxylase activity(GO:0080014) |
| 0.0 | 0.0 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.0 | 0.2 | GO:0015114 | phosphate ion transmembrane transporter activity(GO:0015114) |
| 0.0 | 0.1 | GO:0018741 | alkyl sulfatase activity(GO:0018741) endosulfan hemisulfate sulfatase activity(GO:0034889) endosulfan sulfate hydrolase activity(GO:0034902) |
| 0.0 | 0.0 | GO:0032557 | pyrimidine ribonucleotide binding(GO:0032557) |
| 0.0 | 0.1 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.0 | GO:0015182 | L-asparagine transmembrane transporter activity(GO:0015182) L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.0 | 0.0 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.0 | 0.0 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.0 | 0.0 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.0 | 0.1 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.1 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.1 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.0 | 0.1 | GO:1990239 | steroid hormone binding(GO:1990239) |
| 0.0 | 0.0 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.1 | GO:0038191 | neuropilin binding(GO:0038191) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.0 | 0.1 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.0 | 0.1 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.2 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.2 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |