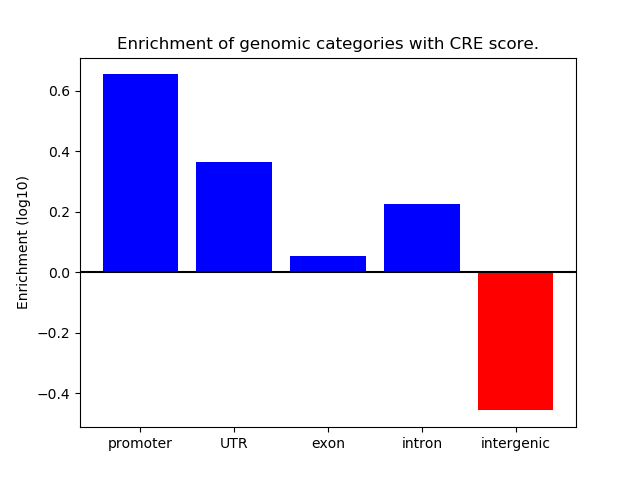
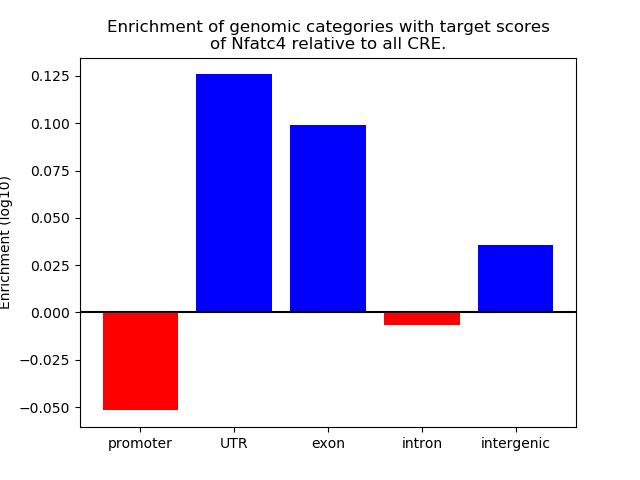
Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
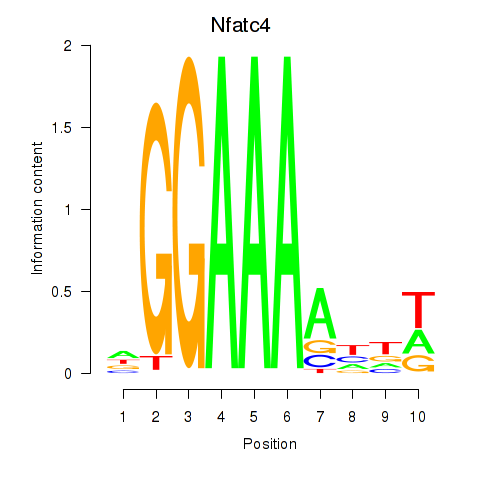
Results for Nfatc4
Z-value: 0.34
Transcription factors associated with Nfatc4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nfatc4
|
ENSMUSG00000023411.5 | nuclear factor of activated T cells, cytoplasmic, calcineurin dependent 4 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr14_55838433_55838623 | Nfatc4 | 8259 | 0.090845 | 0.78 | 6.6e-02 | Click! |
| chr14_55822593_55822771 | Nfatc4 | 462 | 0.651782 | -0.46 | 3.6e-01 | Click! |
| chr14_55822818_55822969 | Nfatc4 | 251 | 0.833439 | 0.24 | 6.5e-01 | Click! |
| chr14_55829023_55829187 | Nfatc4 | 364 | 0.740165 | -0.11 | 8.3e-01 | Click! |
Activity of the Nfatc4 motif across conditions
Conditions sorted by the z-value of the Nfatc4 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
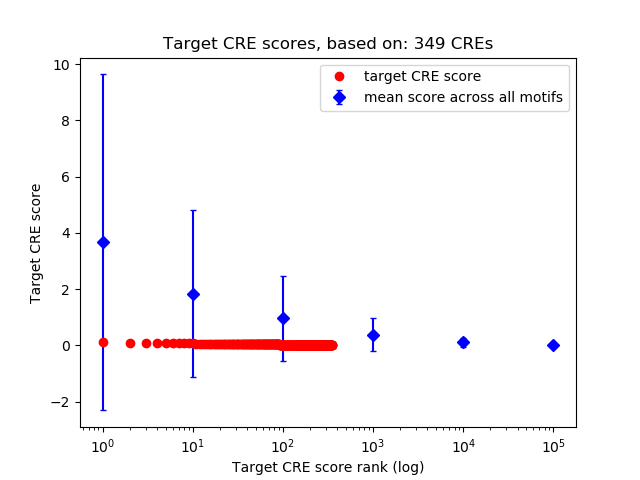
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr13_93624235_93624413 | 0.10 |
Gm15622 |
predicted gene 15622 |
1058 |
0.48 |
| chr3_83006212_83006467 | 0.09 |
Fgg |
fibrinogen gamma chain |
1385 |
0.32 |
| chr6_118478658_118478809 | 0.09 |
Zfp9 |
zinc finger protein 9 |
587 |
0.69 |
| chr4_97008802_97008953 | 0.07 |
Gm27521 |
predicted gene, 27521 |
91857 |
0.09 |
| chr7_80559132_80559287 | 0.07 |
Blm |
Bloom syndrome, RecQ like helicase |
24090 |
0.13 |
| chr5_145863264_145863516 | 0.07 |
Cyp3a11 |
cytochrome P450, family 3, subfamily a, polypeptide 11 |
13301 |
0.16 |
| chr3_18463182_18463725 | 0.07 |
Gm30667 |
predicted gene, 30667 |
2199 |
0.33 |
| chr18_64334411_64334587 | 0.07 |
St8sia3os |
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 3, opposite strand |
793 |
0.6 |
| chr12_8506611_8506851 | 0.07 |
5033421B08Rik |
RIKEN cDNA 5033421B08 gene |
4470 |
0.17 |
| chr5_128522783_128522934 | 0.07 |
Gm42499 |
predicted gene 42499 |
2637 |
0.23 |
| chr1_121350895_121351046 | 0.06 |
2210011K15Rik |
RIKEN cDNA 2210011K15 gene |
8930 |
0.16 |
| chr1_72173165_72173339 | 0.06 |
Mreg |
melanoregulin |
39055 |
0.11 |
| chr2_67974374_67974909 | 0.06 |
Gm37964 |
predicted gene, 37964 |
76045 |
0.1 |
| chr11_54587035_54587213 | 0.06 |
Rapgef6 |
Rap guanine nucleotide exchange factor (GEF) 6 |
9022 |
0.22 |
| chr1_43827020_43827171 | 0.06 |
Uxs1 |
UDP-glucuronate decarboxylase 1 |
79 |
0.97 |
| chr3_5235852_5236031 | 0.05 |
Zfhx4 |
zinc finger homeodomain 4 |
5731 |
0.22 |
| chr10_14705733_14705924 | 0.05 |
Nmbr |
neuromedin B receptor |
201 |
0.57 |
| chr9_65308324_65308479 | 0.05 |
Clpx |
caseinolytic mitochondrial matrix peptidase chaperone subunit |
1751 |
0.2 |
| chr6_52577798_52578135 | 0.05 |
Gm44445 |
predicted gene, 44445 |
17414 |
0.13 |
| chr1_162549290_162549454 | 0.05 |
Eef1aknmt |
EEF1A lysine and N-terminal methyltransferase |
821 |
0.61 |
| chr6_27671661_27671812 | 0.05 |
Gm26310 |
predicted gene, 26310 |
3649 |
0.38 |
| chr16_23102640_23102802 | 0.05 |
Eif4a2 |
eukaryotic translation initiation factor 4A2 |
4723 |
0.07 |
| chr9_62335758_62335958 | 0.05 |
Anp32a |
acidic (leucine-rich) nuclear phosphoprotein 32 family, member A |
5435 |
0.21 |
| chr18_34374027_34374186 | 0.05 |
Reep5 |
receptor accessory protein 5 |
36 |
0.97 |
| chr6_123539742_123540032 | 0.05 |
Gm22722 |
predicted gene, 22722 |
145 |
0.94 |
| chr4_53198055_53198206 | 0.05 |
4930412L05Rik |
RIKEN cDNA 4930412L05 gene |
19431 |
0.17 |
| chr4_97838379_97838530 | 0.05 |
Nfia |
nuclear factor I/A |
41665 |
0.17 |
| chr2_177839201_177839672 | 0.05 |
Gm14325 |
predicted gene 14325 |
882 |
0.56 |
| chr6_123424383_123424683 | 0.05 |
Vmn2r-ps31 |
vomeronasal 2, receptor, pseudogene 31 |
91 |
0.59 |
| chr4_148160291_148160511 | 0.05 |
Fbxo44 |
F-box protein 44 |
81 |
0.68 |
| chr2_177498894_177499115 | 0.04 |
Gm14403 |
predicted gene 14403 |
689 |
0.65 |
| chr5_25227061_25227222 | 0.04 |
E130116L18Rik |
RIKEN cDNA E130116L18 gene |
3988 |
0.18 |
| chr9_70890743_70891063 | 0.04 |
Gm3436 |
predicted pseudogene 3436 |
38327 |
0.14 |
| chr2_155091972_155092151 | 0.04 |
Gm45609 |
predicted gene 45609 |
3683 |
0.17 |
| chr13_23298661_23298830 | 0.04 |
Gm11333 |
predicted gene 11333 |
696 |
0.46 |
| chr11_83913932_83914083 | 0.04 |
Ddx52 |
DEAD (Asp-Glu-Ala-Asp) box polypeptide 52 |
28055 |
0.14 |
| chr11_87991621_87991895 | 0.04 |
Dynll2 |
dynein light chain LC8-type 2 |
4225 |
0.16 |
| chr6_38144181_38144332 | 0.04 |
D630045J12Rik |
RIKEN cDNA D630045J12 gene |
3118 |
0.22 |
| chr12_71042601_71042779 | 0.04 |
Arid4a |
AT rich interactive domain 4A (RBP1-like) |
5651 |
0.18 |
| chr5_37952218_37952372 | 0.04 |
Gm40289 |
predicted gene, 40289 |
31398 |
0.16 |
| chr19_55266477_55266642 | 0.04 |
Acsl5 |
acyl-CoA synthetase long-chain family member 5 |
6311 |
0.18 |
| chr12_26444164_26444328 | 0.04 |
Gm47799 |
predicted gene, 47799 |
7427 |
0.14 |
| chr6_31253045_31253249 | 0.04 |
2210408F21Rik |
RIKEN cDNA 2210408F21 gene |
24701 |
0.14 |
| chr9_76273523_76273711 | 0.04 |
Gm27234 |
predicted gene 27234 |
45960 |
0.14 |
| chr19_4786093_4786325 | 0.04 |
Rbm4 |
RNA binding motif protein 4 |
7642 |
0.08 |
| chr3_79849200_79849377 | 0.04 |
Tmem144 |
transmembrane protein 144 |
3480 |
0.21 |
| chr1_195014918_195015345 | 0.04 |
9630010A21Rik |
RIKEN cDNA 9630010A21 gene |
1385 |
0.22 |
| chr14_62883124_62883275 | 0.04 |
n-R5s47 |
nuclear encoded rRNA 5S 47 |
13485 |
0.13 |
| chr18_35278082_35278245 | 0.04 |
Gm50146 |
predicted gene, 50146 |
14134 |
0.16 |
| chr9_95568845_95568996 | 0.04 |
Paqr9 |
progestin and adipoQ receptor family member IX |
9263 |
0.13 |
| chr4_33245946_33246487 | 0.04 |
Pnrc1 |
proline-rich nuclear receptor coactivator 1 |
1359 |
0.39 |
| chr7_136820396_136820744 | 0.04 |
Gm9347 |
predicted gene 9347 |
9661 |
0.26 |
| chr2_166972459_166972619 | 0.04 |
Stau1 |
staufen double-stranded RNA binding protein 1 |
4232 |
0.15 |
| chr15_57468422_57468573 | 0.04 |
Slc22a22 |
solute carrier family 22 (organic cation transporter), member 22 |
9128 |
0.21 |
| chr1_172988114_172988265 | 0.04 |
Olfr16 |
olfactory receptor 16 |
31421 |
0.13 |
| chr11_3144349_3144521 | 0.04 |
Sfi1 |
Sfi1 homolog, spindle assembly associated (yeast) |
1921 |
0.21 |
| chr17_81375771_81376164 | 0.04 |
Gm50044 |
predicted gene, 50044 |
5134 |
0.28 |
| chr11_31785587_31785738 | 0.04 |
D630024D03Rik |
RIKEN cDNA D630024D03 gene |
38811 |
0.15 |
| chr14_59442931_59443277 | 0.04 |
Cab39l |
calcium binding protein 39-like |
2123 |
0.24 |
| chr8_59870347_59870498 | 0.04 |
4930470O06Rik |
RIKEN cDNA 4930470O06 gene |
44773 |
0.2 |
| chr18_56756491_56756863 | 0.04 |
Gm15345 |
predicted gene 15345 |
13040 |
0.18 |
| chr6_48677382_48677533 | 0.03 |
Gimap9 |
GTPase, IMAP family member 9 |
54 |
0.93 |
| chr3_116934960_116935115 | 0.03 |
Gm42892 |
predicted gene 42892 |
9293 |
0.14 |
| chr6_5256423_5256653 | 0.03 |
Pon3 |
paraoxonase 3 |
252 |
0.91 |
| chr5_104374044_104374195 | 0.03 |
Mepe |
matrix extracellular phosphoglycoprotein with ASARM motif (bone) |
48790 |
0.1 |
| chr7_63887410_63887577 | 0.03 |
Gm27252 |
predicted gene 27252 |
9040 |
0.15 |
| chr7_16591375_16591549 | 0.03 |
Gm29443 |
predicted gene 29443 |
22362 |
0.09 |
| chr15_6237986_6238213 | 0.03 |
Dab2 |
disabled 2, mitogen-responsive phosphoprotein |
61689 |
0.11 |
| chr10_94199177_94199540 | 0.03 |
Ndufa12 |
NADH:ubiquinone oxidoreductase subunit A12 |
321 |
0.87 |
| chr4_49407628_49408150 | 0.03 |
Acnat2 |
acyl-coenzyme A amino acid N-acyltransferase 2 |
262 |
0.88 |
| chr4_135728255_135728588 | 0.03 |
Il22ra1 |
interleukin 22 receptor, alpha 1 |
249 |
0.89 |
| chr1_67162712_67162908 | 0.03 |
Cps1 |
carbamoyl-phosphate synthetase 1 |
39784 |
0.16 |
| chr13_81315073_81315331 | 0.03 |
Adgrv1 |
adhesion G protein-coupled receptor V1 |
27634 |
0.22 |
| chr5_88693031_88693182 | 0.03 |
Grsf1 |
G-rich RNA sequence binding factor 1 |
16935 |
0.14 |
| chr1_67190099_67190250 | 0.03 |
Gm15668 |
predicted gene 15668 |
59026 |
0.11 |
| chr11_86347511_86347911 | 0.03 |
Med13 |
mediator complex subunit 13 |
9891 |
0.21 |
| chr9_86355672_86355847 | 0.03 |
Gm16202 |
predicted gene 16202 |
39075 |
0.18 |
| chr4_95586493_95586698 | 0.03 |
Fggy |
FGGY carbohydrate kinase domain containing |
320 |
0.9 |
| chr7_114066134_114066326 | 0.03 |
Gm45615 |
predicted gene 45615 |
20668 |
0.22 |
| chr2_128377797_128377975 | 0.03 |
Morrbid |
myeloid RNA regulator of BCL2L11 induced cell death |
25079 |
0.18 |
| chr6_128523521_128524414 | 0.03 |
Pzp |
PZP, alpha-2-macroglobulin like |
2736 |
0.13 |
| chr4_53011559_53011710 | 0.03 |
Nipsnap3b |
nipsnap homolog 3B |
246 |
0.9 |
| chr13_105088917_105089088 | 0.03 |
Nt5el |
5' nucleotidase, ecto-like |
6880 |
0.25 |
| chr10_44002186_44002407 | 0.03 |
Gm10812 |
predicted gene 10812 |
1858 |
0.29 |
| chr18_63689863_63690014 | 0.03 |
Txnl1 |
thioredoxin-like 1 |
2384 |
0.27 |
| chr14_109874316_109874480 | 0.03 |
Gm25901 |
predicted gene, 25901 |
8785 |
0.3 |
| chr5_36557509_36557955 | 0.03 |
Tbc1d14 |
TBC1 domain family, member 14 |
6324 |
0.17 |
| chr2_27626035_27626261 | 0.03 |
Rxra |
retinoid X receptor alpha |
50292 |
0.12 |
| chr18_16797289_16797482 | 0.03 |
Cdh2 |
cadherin 2 |
11077 |
0.2 |
| chr11_4822321_4822472 | 0.03 |
Nf2 |
neurofibromin 2 |
2000 |
0.29 |
| chr10_128918530_128918929 | 0.03 |
Rdh5 |
retinol dehydrogenase 5 |
24 |
0.94 |
| chr19_12598432_12598680 | 0.03 |
Gm4952 |
predicted gene 4952 |
1418 |
0.23 |
| chr10_21158162_21158313 | 0.03 |
Myb |
myeloblastosis oncogene |
1592 |
0.3 |
| chr19_55260197_55260377 | 0.03 |
Acsl5 |
acyl-CoA synthetase long-chain family member 5 |
6918 |
0.18 |
| chr17_27805546_27805714 | 0.03 |
Ilrun |
inflammation and lipid regulator with UBA-like and NBR1-like domains |
14822 |
0.11 |
| chr14_57998520_57998724 | 0.03 |
Micu2 |
mitochondrial calcium uptake 2 |
576 |
0.5 |
| chr18_12761990_12762141 | 0.03 |
Osbpl1a |
oxysterol binding protein-like 1A |
5072 |
0.16 |
| chr12_111819707_111819858 | 0.03 |
Zfyve21 |
zinc finger, FYVE domain containing 21 |
3526 |
0.14 |
| chr10_54046292_54046454 | 0.03 |
Gm47917 |
predicted gene, 47917 |
17438 |
0.19 |
| chr6_13280975_13281131 | 0.03 |
AC121573.1 |
predicted 725 |
17308 |
0.23 |
| chr6_14897753_14897997 | 0.03 |
Foxp2 |
forkhead box P2 |
3474 |
0.38 |
| chr4_147426052_147426209 | 0.03 |
Gm13161 |
predicted gene 13161 |
17341 |
0.12 |
| chr7_59230441_59230601 | 0.03 |
Ube3a |
ubiquitin protein ligase E3A |
1085 |
0.24 |
| chr16_20099366_20099741 | 0.03 |
Klhl24 |
kelch-like 24 |
1961 |
0.34 |
| chr11_61956665_61956823 | 0.03 |
Specc1 |
sperm antigen with calponin homology and coiled-coil domains 1 |
19 |
0.97 |
| chr1_153065207_153065385 | 0.03 |
Gm28960 |
predicted gene 28960 |
24211 |
0.16 |
| chr19_40090171_40090346 | 0.03 |
Cyp2c50 |
cytochrome P450, family 2, subfamily c, polypeptide 50 |
523 |
0.79 |
| chr3_95675100_95675253 | 0.03 |
Adamtsl4 |
ADAMTS-like 4 |
1975 |
0.19 |
| chr4_105122583_105122738 | 0.03 |
Prkaa2 |
protein kinase, AMP-activated, alpha 2 catalytic subunit |
12770 |
0.24 |
| chr12_91752583_91752734 | 0.03 |
Ston2 |
stonin 2 |
6574 |
0.19 |
| chr8_101351773_101352122 | 0.03 |
Gm22223 |
predicted gene, 22223 |
187761 |
0.03 |
| chr6_90749634_90749799 | 0.03 |
Iqsec1 |
IQ motif and Sec7 domain 1 |
7834 |
0.18 |
| chr15_97016628_97016815 | 0.03 |
Slc38a4 |
solute carrier family 38, member 4 |
3230 |
0.35 |
| chr5_34761188_34761405 | 0.02 |
Htt |
huntingtin |
444 |
0.79 |
| chr10_93454727_93454893 | 0.02 |
Lta4h |
leukotriene A4 hydrolase |
1359 |
0.37 |
| chr13_95845056_95845225 | 0.02 |
Iqgap2 |
IQ motif containing GTPase activating protein 2 |
46617 |
0.13 |
| chr3_90466862_90467019 | 0.02 |
Npr1 |
natriuretic peptide receptor 1 |
1074 |
0.28 |
| chr17_32637044_32637195 | 0.02 |
Cyp4f37 |
cytochrome P450, family 4, subfamily f, polypeptide 37 |
2417 |
0.17 |
| chr4_3937763_3937914 | 0.02 |
Plag1 |
pleiomorphic adenoma gene 1 |
544 |
0.59 |
| chr17_24139636_24139851 | 0.02 |
Pdpk1 |
3-phosphoinositide dependent protein kinase 1 |
1064 |
0.31 |
| chr4_15092272_15092451 | 0.02 |
Necab1 |
N-terminal EF-hand calcium binding protein 1 |
56600 |
0.12 |
| chr4_65917251_65917420 | 0.02 |
Trim32 |
tripartite motif-containing 32 |
312086 |
0.01 |
| chr6_138140741_138140961 | 0.02 |
Mgst1 |
microsomal glutathione S-transferase 1 |
252 |
0.95 |
| chr17_5178904_5179070 | 0.02 |
Gm15599 |
predicted gene 15599 |
66877 |
0.12 |
| chr10_128773852_128774003 | 0.02 |
Rpsa-ps2 |
ribosomal protein SA, pseudogene 2 |
3323 |
0.1 |
| chr5_36740721_36740872 | 0.02 |
Gm43701 |
predicted gene 43701 |
7822 |
0.13 |
| chr15_96284427_96284951 | 0.02 |
2610037D02Rik |
RIKEN cDNA 2610037D02 gene |
194 |
0.94 |
| chr13_60175154_60175557 | 0.02 |
Gm48488 |
predicted gene, 48488 |
1327 |
0.33 |
| chr8_26998828_26999051 | 0.02 |
Gm45370 |
predicted gene 45370 |
2689 |
0.14 |
| chr17_47694436_47694638 | 0.02 |
Gm21981 |
predicted gene 21981 |
82 |
0.62 |
| chr12_118337129_118337535 | 0.02 |
Sp4 |
trans-acting transcription factor 4 |
35892 |
0.2 |
| chr9_43742856_43743217 | 0.02 |
Nectin1 |
nectin cell adhesion molecule 1 |
948 |
0.41 |
| chr2_10098747_10098908 | 0.02 |
Kin |
Kin17 DNA and RNA binding protein |
8530 |
0.11 |
| chr11_107376639_107376790 | 0.02 |
Gm11712 |
predicted gene 11712 |
10269 |
0.13 |
| chr9_77744627_77744823 | 0.02 |
Gclc |
glutamate-cysteine ligase, catalytic subunit |
9810 |
0.13 |
| chr3_146612054_146612933 | 0.02 |
Uox |
urate oxidase |
2508 |
0.19 |
| chr4_43561781_43561932 | 0.02 |
Tln1 |
talin 1 |
301 |
0.56 |
| chrX_166509851_166510002 | 0.02 |
Tceanc |
transcription elongation factor A (SII) N-terminal and central domain containing |
552 |
0.71 |
| chr7_127769507_127769666 | 0.02 |
Orai3 |
ORAI calcium release-activated calcium modulator 3 |
229 |
0.82 |
| chr3_151884312_151884485 | 0.02 |
Gm43617 |
predicted gene 43617 |
2905 |
0.23 |
| chr13_38959459_38959624 | 0.02 |
Slc35b3 |
solute carrier family 35, member B3 |
939 |
0.42 |
| chr8_107151582_107151800 | 0.02 |
Cyb5b |
cytochrome b5 type B |
1051 |
0.46 |
| chr7_122970929_122971159 | 0.02 |
Rbbp6 |
retinoblastoma binding protein 6, ubiquitin ligase |
126 |
0.92 |
| chr8_26131886_26132040 | 0.02 |
Gm45251 |
predicted gene 45251 |
679 |
0.52 |
| chr12_69127321_69127487 | 0.02 |
Gm31447 |
predicted gene, 31447 |
2432 |
0.12 |
| chr1_58587381_58587553 | 0.02 |
Ndufb3 |
NADH:ubiquinone oxidoreductase subunit B3 |
1083 |
0.31 |
| chr8_68008523_68008836 | 0.02 |
Gm22018 |
predicted gene, 22018 |
1661 |
0.43 |
| chr18_12251815_12252003 | 0.02 |
Npc1 |
NPC intracellular cholesterol transporter 1 |
15509 |
0.14 |
| chr10_96107868_96108070 | 0.02 |
Gm49817 |
predicted gene, 49817 |
11964 |
0.16 |
| chr11_98836582_98836875 | 0.02 |
Rapgefl1 |
Rap guanine nucleotide exchange factor (GEF)-like 1 |
57 |
0.95 |
| chr16_56528453_56528608 | 0.02 |
Abi3bp |
ABI gene family, member 3 (NESH) binding protein |
50630 |
0.16 |
| chr2_80268198_80268374 | 0.02 |
Prdx6b |
peroxiredoxin 6B |
24186 |
0.2 |
| chr11_85170747_85170928 | 0.02 |
1700125H20Rik |
RIKEN cDNA 1700125H20 gene |
259 |
0.89 |
| chr7_105651071_105651233 | 0.02 |
Dnhd1 |
dynein heavy chain domain 1 |
325 |
0.75 |
| chr11_72343979_72344154 | 0.02 |
Tekt1 |
tektin 1 |
17230 |
0.11 |
| chr9_70889261_70889499 | 0.02 |
Gm3436 |
predicted pseudogene 3436 |
36804 |
0.14 |
| chr3_115619931_115620082 | 0.02 |
S1pr1 |
sphingosine-1-phosphate receptor 1 |
95066 |
0.07 |
| chr2_174532818_174532987 | 0.02 |
Gm14385 |
predicted gene 14385 |
47326 |
0.1 |
| chr7_99827785_99828169 | 0.02 |
Neu3 |
neuraminidase 3 |
440 |
0.74 |
| chr10_13007954_13008179 | 0.02 |
Sf3b5 |
splicing factor 3b, subunit 5 |
2723 |
0.26 |
| chr2_35692024_35692382 | 0.02 |
Dab2ip |
disabled 2 interacting protein |
130 |
0.97 |
| chr14_27337841_27338102 | 0.02 |
Arhgef3 |
Rho guanine nucleotide exchange factor (GEF) 3 |
1905 |
0.38 |
| chr8_127335297_127335467 | 0.02 |
Pard3 |
par-3 family cell polarity regulator |
21033 |
0.28 |
| chr3_142348416_142348578 | 0.02 |
Pdlim5 |
PDZ and LIM domain 5 |
43311 |
0.17 |
| chr6_22363116_22363267 | 0.02 |
Fam3c |
family with sequence similarity 3, member C |
6948 |
0.28 |
| chr11_48815924_48816180 | 0.02 |
Trim41 |
tripartite motif-containing 41 |
179 |
0.88 |
| chr6_72787755_72787940 | 0.02 |
Tcf7l1 |
transcription factor 7 like 1 (T cell specific, HMG box) |
751 |
0.63 |
| chr1_182409523_182409985 | 0.02 |
Trp53bp2 |
transformation related protein 53 binding protein 2 |
582 |
0.49 |
| chr9_103349628_103349779 | 0.02 |
Topbp1 |
topoisomerase (DNA) II binding protein 1 |
3475 |
0.19 |
| chr4_131888523_131888751 | 0.02 |
Srsf4 |
serine and arginine-rich splicing factor 4 |
3429 |
0.13 |
| chr4_134767914_134768168 | 0.02 |
Ldlrap1 |
low density lipoprotein receptor adaptor protein 1 |
17 |
0.98 |
| chr1_31018387_31018720 | 0.02 |
4931428L18Rik |
RIKEN cDNA 4931428L18 gene |
18649 |
0.14 |
| chr3_82857448_82857713 | 0.02 |
Rbm46 |
RNA binding motif protein 46 |
9067 |
0.17 |
| chr9_22686906_22687072 | 0.02 |
Bbs9 |
Bardet-Biedl syndrome 9 (human) |
8239 |
0.18 |
| chr2_60046169_60046353 | 0.02 |
Gm13572 |
predicted gene 13572 |
9120 |
0.18 |
| chr2_19919291_19919460 | 0.02 |
Etl4 |
enhancer trap locus 4 |
9595 |
0.17 |
| chr2_80561396_80561547 | 0.02 |
Gm24461 |
predicted gene, 24461 |
2786 |
0.18 |
| chr12_83618299_83618462 | 0.02 |
Gm6722 |
predicted gene 6722 |
2689 |
0.19 |
| chr8_121583597_121583942 | 0.02 |
Gm17786 |
predicted gene, 17786 |
2004 |
0.19 |
| chr13_23808562_23808721 | 0.02 |
Slc17a2 |
solute carrier family 17 (sodium phosphate), member 2 |
1593 |
0.16 |
| chr19_44363419_44363596 | 0.02 |
Scd4 |
stearoyl-coenzyme A desaturase 4 |
30415 |
0.1 |
| chr11_116099131_116099411 | 0.02 |
Trim47 |
tripartite motif-containing 47 |
7812 |
0.1 |
| chr8_93328194_93328378 | 0.02 |
Ces1g |
carboxylesterase 1G |
9022 |
0.15 |
| chr19_59939631_59939782 | 0.02 |
Rab11fip2 |
RAB11 family interacting protein 2 (class I) |
3294 |
0.24 |
| chr11_106802839_106802992 | 0.02 |
Cep95 |
centrosomal protein 95 |
9411 |
0.09 |
| chr18_10913556_10913835 | 0.02 |
Gm7575 |
predicted gene 7575 |
15189 |
0.2 |
| chr10_37261190_37261352 | 0.02 |
4930543K20Rik |
RIKEN cDNA 4930543K20 gene |
2477 |
0.38 |
| chr6_143219492_143219655 | 0.02 |
1700126G02Rik |
RIKEN cDNA 1700126G02 gene |
1875 |
0.33 |
| chr2_3672979_3673130 | 0.02 |
Gm13183 |
predicted gene 13183 |
7536 |
0.17 |
| chr16_78969415_78969589 | 0.02 |
Gm15692 |
predicted gene 15692 |
5406 |
0.27 |
| chr9_30427149_30427538 | 0.02 |
Snx19 |
sorting nexin 19 |
235 |
0.64 |
| chr1_135787910_135788065 | 0.02 |
Tnni1 |
troponin I, skeletal, slow 1 |
4922 |
0.16 |
| chr8_61574352_61574983 | 0.01 |
Palld |
palladin, cytoskeletal associated protein |
16472 |
0.25 |
| chr19_3568557_3568708 | 0.01 |
Ppp6r3 |
protein phosphatase 6, regulatory subunit 3 |
7064 |
0.16 |
| chr16_30945152_30945303 | 0.01 |
Gm46565 |
predicted gene, 46565 |
9598 |
0.17 |
| chr2_121441237_121441474 | 0.01 |
Ell3 |
elongation factor RNA polymerase II-like 3 |
204 |
0.84 |
| chr10_127745122_127745273 | 0.01 |
Zbtb39 |
zinc finger and BTB domain containing 39 |
5659 |
0.09 |
| chr15_84674989_84675329 | 0.01 |
Prr5 |
proline rich 5 (renal) |
5539 |
0.21 |
| chr1_91095443_91095660 | 0.01 |
Lrrfip1 |
leucine rich repeat (in FLII) interacting protein 1 |
15663 |
0.18 |
| chr2_103204688_103204993 | 0.01 |
Gm13869 |
predicted gene 13869 |
5517 |
0.19 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | GO:0046271 | coumarin metabolic process(GO:0009804) phenylpropanoid catabolic process(GO:0046271) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | GO:0102007 | lactonohydrolase activity(GO:0046573) acyl-L-homoserine-lactone lactonohydrolase activity(GO:0102007) |
| 0.0 | 0.0 | GO:0004946 | bombesin receptor activity(GO:0004946) |