Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
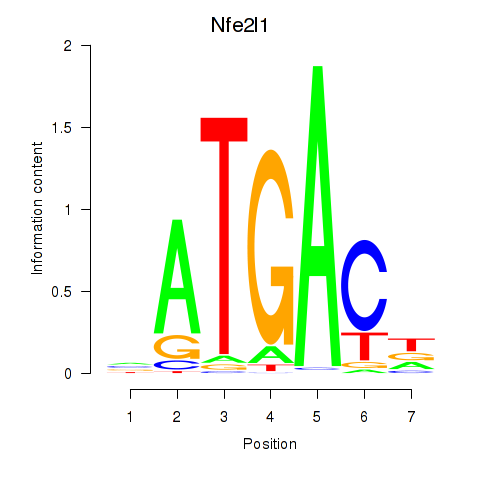
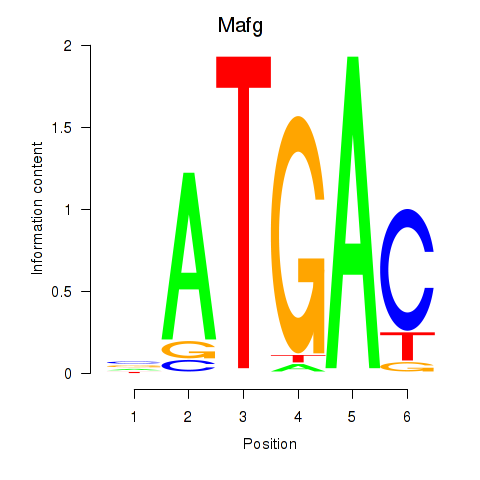
Results for Nfe2l1_Mafg
Z-value: 0.98
Transcription factors associated with Nfe2l1_Mafg
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nfe2l1
|
ENSMUSG00000038615.11 | nuclear factor, erythroid derived 2,-like 1 |
|
Mafg
|
ENSMUSG00000051510.7 | v-maf musculoaponeurotic fibrosarcoma oncogene family, protein G (avian) |
|
Mafg
|
ENSMUSG00000053906.4 | v-maf musculoaponeurotic fibrosarcoma oncogene family, protein G (avian) |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr11_120632589_120632750 | Mafg | 887 | 0.260451 | -0.97 | 1.1e-03 | Click! |
| chr11_120629221_120629376 | Mafg | 850 | 0.270332 | -0.97 | 1.2e-03 | Click! |
| chr11_120629382_120629533 | Mafg | 691 | 0.345206 | -0.95 | 4.3e-03 | Click! |
| chr11_120633556_120633720 | Mafg | 38 | 0.910356 | -0.95 | 4.4e-03 | Click! |
| chr11_120630495_120630798 | Mafg | 10 | 0.914170 | -0.87 | 2.4e-02 | Click! |
| chr11_96824156_96824315 | Nfe2l1 | 108 | 0.933526 | -0.98 | 3.7e-04 | Click! |
| chr11_96822735_96822898 | Nfe2l1 | 91 | 0.939857 | -0.94 | 5.7e-03 | Click! |
| chr11_96826613_96826784 | Nfe2l1 | 1021 | 0.354776 | -0.92 | 1.0e-02 | Click! |
| chr11_96826021_96826172 | Nfe2l1 | 1544 | 0.232334 | 0.89 | 1.7e-02 | Click! |
| chr11_96829364_96829648 | Nfe2l1 | 4 | 0.954420 | -0.87 | 2.3e-02 | Click! |
Activity of the Nfe2l1_Mafg motif across conditions
Conditions sorted by the z-value of the Nfe2l1_Mafg motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
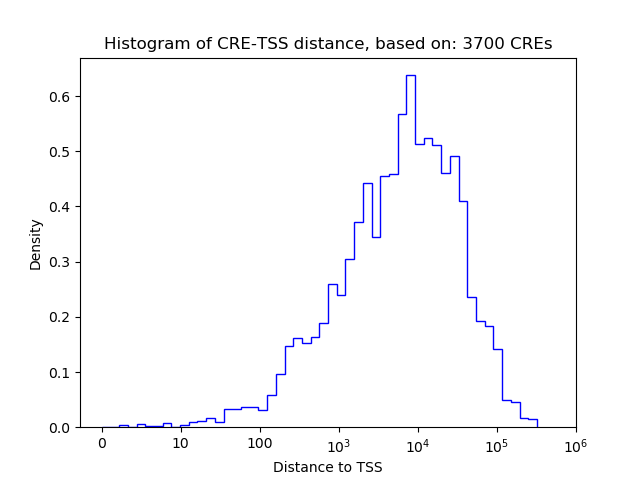
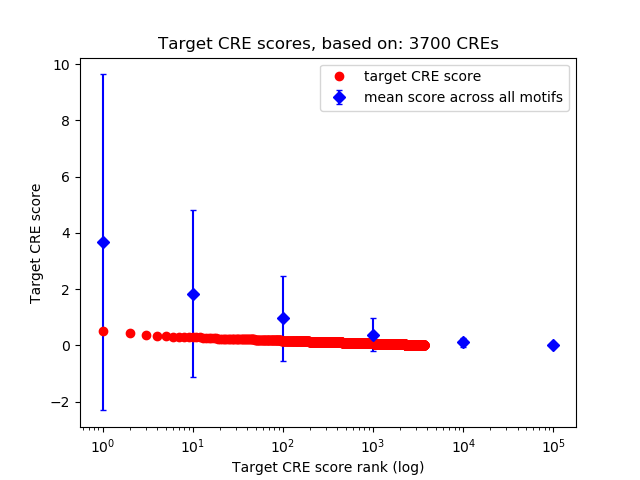
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr10_75873468_75873620 | 0.49 |
Gm47744 |
predicted gene, 47744 |
1887 |
0.18 |
| chr3_158062237_158062399 | 0.43 |
Lrrc40 |
leucine rich repeat containing 40 |
40 |
0.97 |
| chr19_40153747_40153898 | 0.36 |
Cyp2c70 |
cytochrome P450, family 2, subfamily c, polypeptide 70 |
33464 |
0.13 |
| chr12_104000672_104000833 | 0.33 |
Gm28577 |
predicted gene 28577 |
4173 |
0.12 |
| chr11_7816093_7816451 | 0.31 |
Gm27393 |
predicted gene, 27393 |
70233 |
0.13 |
| chr6_149189382_149189536 | 0.31 |
Amn1 |
antagonist of mitotic exit network 1 |
747 |
0.6 |
| chr3_138282244_138282429 | 0.30 |
Adh1 |
alcohol dehydrogenase 1 (class I) |
4685 |
0.14 |
| chr12_104082595_104082890 | 0.29 |
Serpina4-ps1 |
serine (or cysteine) peptidase inhibitor, clade A, member 4, pseudogene 1 |
2093 |
0.18 |
| chr13_82200174_82200343 | 0.29 |
Gm48155 |
predicted gene, 48155 |
110501 |
0.07 |
| chr13_4270161_4270342 | 0.28 |
Akr1c12 |
aldo-keto reductase family 1, member C12 |
9182 |
0.15 |
| chr4_141955631_141955810 | 0.28 |
Fhad1 |
forkhead-associated (FHA) phosphopeptide binding domain 1 |
1526 |
0.3 |
| chr6_22340862_22341016 | 0.28 |
Fam3c |
family with sequence similarity 3, member C |
2974 |
0.34 |
| chr19_40158328_40158479 | 0.27 |
Cyp2c70 |
cytochrome P450, family 2, subfamily c, polypeptide 70 |
28883 |
0.13 |
| chr1_162892140_162892544 | 0.27 |
Fmo2 |
flavin containing monooxygenase 2 |
5827 |
0.19 |
| chr2_58773911_58774203 | 0.25 |
Upp2 |
uridine phosphorylase 2 |
8732 |
0.21 |
| chr18_81319673_81319824 | 0.25 |
Gm30192 |
predicted gene, 30192 |
55040 |
0.14 |
| chr2_134491985_134492278 | 0.25 |
Hao1 |
hydroxyacid oxidase 1, liver |
62176 |
0.15 |
| chr5_49060180_49060366 | 0.24 |
Gm43047 |
predicted gene 43047 |
9730 |
0.1 |
| chr7_63923519_63923771 | 0.24 |
Klf13 |
Kruppel-like factor 13 |
1225 |
0.37 |
| chr4_104868512_104868743 | 0.24 |
C8a |
complement component 8, alpha polypeptide |
7756 |
0.2 |
| chr12_104084479_104084838 | 0.24 |
Serpina4-ps1 |
serine (or cysteine) peptidase inhibitor, clade A, member 4, pseudogene 1 |
4009 |
0.12 |
| chr4_76964290_76964441 | 0.24 |
Gm23159 |
predicted gene, 23159 |
5867 |
0.27 |
| chr13_45623869_45624201 | 0.24 |
Gmpr |
guanosine monophosphate reductase |
77882 |
0.1 |
| chr8_9501766_9501947 | 0.23 |
4930435N07Rik |
RIKEN cDNA 4930435N07 gene |
42764 |
0.13 |
| chr6_24049175_24049330 | 0.23 |
Slc13a1 |
solute carrier family 13 (sodium/sulfate symporters), member 1 |
59319 |
0.12 |
| chr8_93164567_93164980 | 0.23 |
Ces1d |
carboxylesterase 1D |
5202 |
0.15 |
| chr11_90321650_90321810 | 0.23 |
Hlf |
hepatic leukemia factor |
60205 |
0.12 |
| chr5_43310353_43310569 | 0.23 |
Gm43020 |
predicted gene 43020 |
44798 |
0.11 |
| chr6_135782336_135782495 | 0.22 |
Gm22892 |
predicted gene, 22892 |
41418 |
0.15 |
| chr8_40651238_40651389 | 0.22 |
Mtmr7 |
myotubularin related protein 7 |
16516 |
0.14 |
| chr3_18138170_18138321 | 0.22 |
Gm23686 |
predicted gene, 23686 |
39380 |
0.16 |
| chr2_147988721_147988917 | 0.22 |
9030622O22Rik |
RIKEN cDNA 9030622O22 gene |
12634 |
0.21 |
| chr13_24377886_24378061 | 0.22 |
Cmah |
cytidine monophospho-N-acetylneuraminic acid hydroxylase |
1890 |
0.23 |
| chr1_193926856_193927030 | 0.22 |
Gm21362 |
predicted gene, 21362 |
59934 |
0.16 |
| chr6_24610042_24610446 | 0.22 |
Lmod2 |
leiomodin 2 (cardiac) |
12482 |
0.14 |
| chr19_40156899_40157166 | 0.22 |
Cyp2c70 |
cytochrome P450, family 2, subfamily c, polypeptide 70 |
30254 |
0.13 |
| chr3_18111157_18111308 | 0.22 |
Gm23726 |
predicted gene, 23726 |
41906 |
0.14 |
| chr19_56171122_56171273 | 0.21 |
Gm31912 |
predicted gene, 31912 |
64887 |
0.11 |
| chr18_42796496_42796654 | 0.21 |
Ppp2r2b |
protein phosphatase 2, regulatory subunit B, beta |
102240 |
0.07 |
| chr7_35748679_35748830 | 0.21 |
Dpy19l3 |
dpy-19-like 3 (C. elegans) |
5601 |
0.2 |
| chr8_36265736_36265923 | 0.21 |
Lonrf1 |
LON peptidase N-terminal domain and ring finger 1 |
16313 |
0.2 |
| chr6_51657319_51657493 | 0.21 |
Gm22914 |
predicted gene, 22914 |
35774 |
0.17 |
| chr7_132967481_132967632 | 0.21 |
Zranb1 |
zinc finger, RAN-binding domain containing 1 |
13303 |
0.14 |
| chr17_10401671_10401822 | 0.21 |
A230009B12Rik |
RIKEN cDNA A230009B12 gene |
54470 |
0.13 |
| chr11_16808519_16808674 | 0.21 |
Egfros |
epidermal growth factor receptor, opposite strand |
22106 |
0.19 |
| chr6_137784539_137784690 | 0.21 |
Dera |
deoxyribose-phosphate aldolase (putative) |
3791 |
0.29 |
| chr8_105095287_105095438 | 0.21 |
Ces3b |
carboxylesterase 3B |
6743 |
0.11 |
| chr12_69758939_69759328 | 0.21 |
Mir681 |
microRNA 681 |
4811 |
0.14 |
| chr10_87865511_87865999 | 0.20 |
Igf1os |
insulin-like growth factor 1, opposite strand |
2374 |
0.3 |
| chr13_102050615_102050815 | 0.20 |
Gm38133 |
predicted gene, 38133 |
65320 |
0.13 |
| chr15_59005379_59005677 | 0.20 |
4930544F09Rik |
RIKEN cDNA 4930544F09 gene |
21392 |
0.16 |
| chr14_11132064_11132231 | 0.20 |
Fhit |
fragile histidine triad gene |
6556 |
0.25 |
| chr3_95879412_95879563 | 0.20 |
Ciart |
circadian associated repressor of transcription |
1759 |
0.16 |
| chr1_91464737_91464909 | 0.20 |
Gm28381 |
predicted gene 28381 |
5275 |
0.12 |
| chr1_21264707_21265188 | 0.20 |
Gm28836 |
predicted gene 28836 |
6646 |
0.11 |
| chr5_63837978_63838129 | 0.20 |
0610040J01Rik |
RIKEN cDNA 0610040J01 gene |
25533 |
0.17 |
| chr18_11317026_11317247 | 0.19 |
Gata6 |
GATA binding protein 6 |
258089 |
0.02 |
| chr12_57317079_57317230 | 0.19 |
Mipol1 |
mirror-image polydactyly 1 |
11071 |
0.22 |
| chr4_54954362_54954516 | 0.19 |
Zfp462 |
zinc finger protein 462 |
6463 |
0.29 |
| chr15_83202825_83203000 | 0.19 |
7530414M10Rik |
RIKEN cDNA 7530414M10 gene |
26467 |
0.1 |
| chr12_57515437_57515691 | 0.19 |
Gm2568 |
predicted gene 2568 |
5060 |
0.18 |
| chr7_25884401_25884651 | 0.19 |
Gm6434 |
predicted gene 6434 |
2137 |
0.18 |
| chr5_100408563_100408720 | 0.19 |
Sec31a |
Sec31 homolog A (S. cerevisiae) |
971 |
0.48 |
| chr17_64684263_64684445 | 0.19 |
Man2a1 |
mannosidase 2, alpha 1 |
29066 |
0.2 |
| chr16_79086922_79087073 | 0.19 |
Tmprss15 |
transmembrane protease, serine 15 |
4095 |
0.35 |
| chr16_93333790_93334136 | 0.19 |
1810053B23Rik |
RIKEN cDNA 1810053B23 gene |
19226 |
0.18 |
| chr6_147954211_147954549 | 0.19 |
Far2 |
fatty acyl CoA reductase 2 |
92879 |
0.07 |
| chr8_93191975_93192208 | 0.19 |
Gm45909 |
predicted gene 45909 |
733 |
0.57 |
| chr6_90478594_90478745 | 0.18 |
Gm44421 |
predicted gene, 44421 |
203 |
0.89 |
| chr9_122056710_122056883 | 0.18 |
Gm39465 |
predicted gene, 39465 |
5333 |
0.13 |
| chr8_93192874_93193058 | 0.18 |
Gm45909 |
predicted gene 45909 |
1608 |
0.28 |
| chr7_63899068_63899509 | 0.18 |
Gm27252 |
predicted gene 27252 |
1314 |
0.37 |
| chr2_160797768_160797919 | 0.18 |
Plcg1 |
phospholipase C, gamma 1 |
37725 |
0.11 |
| chr6_51578534_51578718 | 0.18 |
Snx10 |
sorting nexin 10 |
34060 |
0.16 |
| chr6_38769722_38769895 | 0.18 |
Hipk2 |
homeodomain interacting protein kinase 2 |
26548 |
0.2 |
| chr5_125529429_125529580 | 0.18 |
Tmem132b |
transmembrane protein 132B |
2270 |
0.26 |
| chr8_77410509_77410680 | 0.18 |
Gm45407 |
predicted gene 45407 |
38078 |
0.15 |
| chr3_51226339_51227024 | 0.18 |
Noct |
nocturnin |
2211 |
0.23 |
| chr6_67441579_67441755 | 0.18 |
Gm44083 |
predicted gene, 44083 |
23616 |
0.11 |
| chr7_99969637_99969788 | 0.18 |
Rnf169 |
ring finger protein 169 |
3978 |
0.16 |
| chr7_98358819_98359055 | 0.18 |
Tsku |
tsukushi, small leucine rich proteoglycan |
1142 |
0.47 |
| chr14_18304273_18304424 | 0.18 |
Ube2e1 |
ubiquitin-conjugating enzyme E2E 1 |
22943 |
0.14 |
| chr19_40183641_40183792 | 0.18 |
Cyp2c70 |
cytochrome P450, family 2, subfamily c, polypeptide 70 |
3570 |
0.19 |
| chr2_131452202_131452379 | 0.18 |
Gm14304 |
predicted gene 14304 |
2350 |
0.25 |
| chr7_70499142_70499303 | 0.18 |
Gm7656 |
predicted gene 7656 |
8374 |
0.15 |
| chr9_66697369_66697520 | 0.18 |
Car12 |
carbonic anhydrase 12 |
16242 |
0.19 |
| chr5_33024447_33024632 | 0.18 |
Ywhah |
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, eta polypeptide |
2748 |
0.21 |
| chr2_5839502_5839908 | 0.18 |
Cdc123 |
cell division cycle 123 |
5146 |
0.16 |
| chr13_96960473_96960682 | 0.18 |
Gm48597 |
predicted gene, 48597 |
9663 |
0.17 |
| chr14_10751127_10751283 | 0.18 |
Fhitos |
fragile histidine triad gene, opposite strand |
12652 |
0.25 |
| chr14_86444521_86444715 | 0.17 |
Gm32913 |
predicted gene, 32913 |
35519 |
0.14 |
| chr14_118008711_118008863 | 0.17 |
Dct |
dopachrome tautomerase |
18748 |
0.19 |
| chr2_31493649_31493953 | 0.17 |
Ass1 |
argininosuccinate synthetase 1 |
3971 |
0.22 |
| chr3_142862185_142862381 | 0.17 |
Pkn2 |
protein kinase N2 |
6548 |
0.15 |
| chr9_64357609_64357961 | 0.17 |
Dis3l |
DIS3 like exosome 3'-5' exoribonuclease |
16497 |
0.16 |
| chr6_128432450_128432601 | 0.17 |
Fkbp4 |
FK506 binding protein 4 |
5128 |
0.08 |
| chr15_3470533_3470724 | 0.17 |
Ghr |
growth hormone receptor |
1016 |
0.68 |
| chr4_6919330_6919515 | 0.17 |
Tox |
thymocyte selection-associated high mobility group box |
71061 |
0.12 |
| chr16_76330855_76331006 | 0.17 |
Nrip1 |
nuclear receptor interacting protein 1 |
7272 |
0.24 |
| chr19_31881164_31881315 | 0.17 |
A1cf |
APOBEC1 complementation factor |
12458 |
0.22 |
| chr2_31510334_31510882 | 0.17 |
Ass1 |
argininosuccinate synthetase 1 |
7882 |
0.18 |
| chr15_3506426_3506577 | 0.17 |
Ghr |
growth hormone receptor |
34857 |
0.21 |
| chr13_44285946_44286097 | 0.17 |
Gm29676 |
predicted gene, 29676 |
5572 |
0.22 |
| chr3_133740354_133740509 | 0.17 |
Gm6135 |
prediticted gene 6135 |
51073 |
0.13 |
| chr1_192812225_192812480 | 0.17 |
Gm38360 |
predicted gene, 38360 |
8775 |
0.14 |
| chr18_36665669_36665887 | 0.17 |
Eif4ebp3 |
eukaryotic translation initiation factor 4E binding protein 3 |
1718 |
0.17 |
| chr11_28695418_28695578 | 0.16 |
2810471M01Rik |
RIKEN cDNA 2810471M01 gene |
13934 |
0.17 |
| chr3_97637473_97637939 | 0.16 |
Fmo5 |
flavin containing monooxygenase 5 |
8823 |
0.13 |
| chr13_44250819_44251012 | 0.16 |
Gm29676 |
predicted gene, 29676 |
1755 |
0.34 |
| chr19_14736673_14736829 | 0.16 |
Gm26026 |
predicted gene, 26026 |
18271 |
0.26 |
| chr1_107939897_107940125 | 0.16 |
D830032E09Rik |
RIKEN cDNA D830032E09 gene |
3302 |
0.23 |
| chr8_114151087_114151399 | 0.16 |
Nudt7 |
nudix (nucleoside diphosphate linked moiety X)-type motif 7 |
17601 |
0.25 |
| chr3_52011173_52011324 | 0.16 |
Gm37465 |
predicted gene, 37465 |
7223 |
0.13 |
| chr3_116915063_116915225 | 0.16 |
Frrs1 |
ferric-chelate reductase 1 |
18434 |
0.13 |
| chr16_42172601_42172764 | 0.16 |
Gm49737 |
predicted gene, 49737 |
12235 |
0.2 |
| chr11_16851040_16851282 | 0.16 |
Egfros |
epidermal growth factor receptor, opposite strand |
20459 |
0.18 |
| chr5_33459431_33459801 | 0.16 |
Gm43851 |
predicted gene 43851 |
22152 |
0.16 |
| chr1_88132349_88132542 | 0.16 |
Ugt1a6a |
UDP glucuronosyltransferase 1 family, polypeptide A6A |
2364 |
0.1 |
| chr8_114142907_114143190 | 0.16 |
Nudt7 |
nudix (nucleoside diphosphate linked moiety X)-type motif 7 |
9406 |
0.28 |
| chr15_55043831_55044027 | 0.16 |
Taf2 |
TATA-box binding protein associated factor 2 |
2237 |
0.26 |
| chr2_109488302_109488457 | 0.16 |
Gm13925 |
predicted gene 13925 |
11376 |
0.23 |
| chr5_151042731_151042918 | 0.16 |
Gm8675 |
predicted gene 8675 |
33523 |
0.17 |
| chr3_51198208_51198374 | 0.16 |
Noct |
nocturnin |
26156 |
0.14 |
| chr9_72702491_72702671 | 0.16 |
Nedd4 |
neural precursor cell expressed, developmentally down-regulated 4 |
9142 |
0.1 |
| chr3_97225695_97225846 | 0.16 |
Bcl9 |
B cell CLL/lymphoma 9 |
1561 |
0.36 |
| chr11_6158649_6158802 | 0.16 |
Rps15a-ps6 |
ribosomal protein S15A, pseudogene 6 |
14311 |
0.13 |
| chr10_89471287_89471467 | 0.16 |
Gas2l3 |
growth arrest-specific 2 like 3 |
27410 |
0.18 |
| chr4_99028977_99029128 | 0.16 |
Angptl3 |
angiopoietin-like 3 |
1902 |
0.32 |
| chr11_16818044_16818409 | 0.16 |
Egfros |
epidermal growth factor receptor, opposite strand |
12476 |
0.22 |
| chr3_98738287_98738502 | 0.16 |
Hsd3b2 |
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 2 |
13851 |
0.13 |
| chr12_83685921_83686270 | 0.16 |
Psen1 |
presenilin 1 |
2057 |
0.23 |
| chr15_85766228_85766381 | 0.16 |
Ppara |
peroxisome proliferator activated receptor alpha |
5910 |
0.16 |
| chr10_14012990_14013158 | 0.16 |
Hivep2 |
human immunodeficiency virus type I enhancer binding protein 2 |
46050 |
0.14 |
| chr1_161240339_161240617 | 0.16 |
Prdx6 |
peroxiredoxin 6 |
10076 |
0.16 |
| chr9_106240931_106241179 | 0.15 |
Alas1 |
aminolevulinic acid synthase 1 |
1022 |
0.36 |
| chr6_21827909_21828060 | 0.15 |
Tspan12 |
tetraspanin 12 |
23844 |
0.17 |
| chr14_89610572_89610930 | 0.15 |
Gm25415 |
predicted gene, 25415 |
96054 |
0.09 |
| chr19_17311642_17311793 | 0.15 |
Gcnt1 |
glucosaminyl (N-acetyl) transferase 1, core 2 |
23719 |
0.17 |
| chr11_5499044_5499213 | 0.15 |
Gm11963 |
predicted gene 11963 |
288 |
0.86 |
| chr1_67230512_67230663 | 0.15 |
Gm15668 |
predicted gene 15668 |
18613 |
0.21 |
| chr17_45924651_45924805 | 0.15 |
Gm49805 |
predicted gene, 49805 |
37131 |
0.12 |
| chr7_119961192_119961374 | 0.15 |
Dnah3 |
dynein, axonemal, heavy chain 3 |
6559 |
0.16 |
| chr10_87532342_87532676 | 0.15 |
Pah |
phenylalanine hydroxylase |
10482 |
0.2 |
| chr8_29133062_29133289 | 0.15 |
Svet1 |
subventricular expressed transcript 1 |
20399 |
0.19 |
| chr3_29835547_29835722 | 0.15 |
Mannr |
Mecom adjacent non-protein coding RNA |
55380 |
0.12 |
| chr18_75498893_75499229 | 0.15 |
Gm10532 |
predicted gene 10532 |
15584 |
0.25 |
| chr9_106271509_106271729 | 0.15 |
Poc1a |
POC1 centriolar protein A |
9442 |
0.11 |
| chr14_118948462_118948868 | 0.15 |
Dnajc3 |
DnaJ heat shock protein family (Hsp40) member C3 |
10689 |
0.16 |
| chr10_127407277_127407450 | 0.15 |
Gm23819 |
predicted gene, 23819 |
2971 |
0.15 |
| chr7_72323573_72323742 | 0.15 |
Mctp2 |
multiple C2 domains, transmembrane 2 |
17049 |
0.28 |
| chrX_166678508_166678678 | 0.15 |
Gm1720 |
predicted gene 1720 |
16568 |
0.21 |
| chr6_24602762_24602931 | 0.15 |
Lmod2 |
leiomodin 2 (cardiac) |
5084 |
0.17 |
| chr13_14180280_14180431 | 0.15 |
Arid4b |
AT rich interactive domain 4B (RBP1-like) |
2372 |
0.25 |
| chr10_93075982_93076133 | 0.15 |
Cfap54 |
cilia and flagella associated protein 54 |
5539 |
0.22 |
| chr10_117437437_117437791 | 0.15 |
Gm9002 |
predicted gene 9002 |
7996 |
0.14 |
| chr11_75737051_75737202 | 0.15 |
Ywhae |
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, epsilon polypeptide |
3978 |
0.19 |
| chr1_21259661_21259812 | 0.15 |
Gsta3 |
glutathione S-transferase, alpha 3 |
6215 |
0.11 |
| chr9_48599126_48599309 | 0.14 |
Nnmt |
nicotinamide N-methyltransferase |
1768 |
0.43 |
| chr10_87536307_87536462 | 0.14 |
Pah |
phenylalanine hydroxylase |
10289 |
0.2 |
| chr10_67100073_67100416 | 0.14 |
Reep3 |
receptor accessory protein 3 |
3299 |
0.24 |
| chr1_192126218_192126369 | 0.14 |
Rcor3 |
REST corepressor 3 |
2008 |
0.23 |
| chr11_16865620_16865819 | 0.14 |
Egfr |
epidermal growth factor receptor |
12431 |
0.2 |
| chr14_21422563_21422878 | 0.14 |
Gm25864 |
predicted gene, 25864 |
27754 |
0.17 |
| chr10_99349668_99349865 | 0.14 |
B530045E10Rik |
RIKEN cDNA B530045E10 gene |
50600 |
0.1 |
| chr11_110249489_110249640 | 0.14 |
Abca6 |
ATP-binding cassette, sub-family A (ABC1), member 6 |
2212 |
0.39 |
| chr3_93578904_93579055 | 0.14 |
S100a10 |
S100 calcium binding protein A10 (calpactin) |
18252 |
0.11 |
| chr11_11845438_11845699 | 0.14 |
Ddc |
dopa decarboxylase |
9288 |
0.18 |
| chr5_43309511_43309822 | 0.14 |
6030400A10Rik |
RIKEN cDNA 6030400A10 gene |
44509 |
0.11 |
| chr9_53359307_53359484 | 0.14 |
Exph5 |
exophilin 5 |
18206 |
0.15 |
| chr13_101508264_101508415 | 0.14 |
Gm47533 |
predicted gene, 47533 |
36867 |
0.13 |
| chr7_140720541_140720872 | 0.14 |
Olfr542-ps1 |
olfactory receptor 542, pseudogene 1 |
576 |
0.58 |
| chr1_87572169_87572320 | 0.14 |
Ngef |
neuronal guanine nucleotide exchange factor |
1626 |
0.26 |
| chr9_74964080_74964349 | 0.14 |
Fam214a |
family with sequence similarity 214, member A |
11142 |
0.2 |
| chr19_10063756_10063907 | 0.14 |
Fads2 |
fatty acid desaturase 2 |
7356 |
0.13 |
| chr12_102327592_102327745 | 0.14 |
Rin3 |
Ras and Rab interactor 3 |
27077 |
0.18 |
| chr8_40270006_40270157 | 0.14 |
Fgf20 |
fibroblast growth factor 20 |
16872 |
0.2 |
| chr11_16839622_16839885 | 0.14 |
Egfros |
epidermal growth factor receptor, opposite strand |
9051 |
0.21 |
| chr1_51774175_51774474 | 0.14 |
Myo1b |
myosin IB |
2112 |
0.33 |
| chr15_81253437_81253614 | 0.14 |
8430426J06Rik |
RIKEN cDNA 8430426J06 gene |
1663 |
0.31 |
| chr2_8116897_8117048 | 0.14 |
Gm13254 |
predicted gene 13254 |
30893 |
0.26 |
| chr8_76147308_76147459 | 0.14 |
Gm45742 |
predicted gene 45742 |
30356 |
0.21 |
| chr19_12654035_12654281 | 0.14 |
Gm24521 |
predicted gene, 24521 |
11097 |
0.09 |
| chr13_37601794_37602225 | 0.14 |
Gm47753 |
predicted gene, 47753 |
19738 |
0.1 |
| chr16_26723002_26723153 | 0.14 |
Il1rap |
interleukin 1 receptor accessory protein |
643 |
0.79 |
| chr13_101746510_101746927 | 0.14 |
Gm36638 |
predicted gene, 36638 |
5170 |
0.24 |
| chr18_59170100_59170266 | 0.14 |
Chsy3 |
chondroitin sulfate synthase 3 |
5218 |
0.33 |
| chr15_59061763_59061914 | 0.14 |
Mtss1 |
MTSS I-BAR domain containing 1 |
6374 |
0.24 |
| chr9_107303087_107303371 | 0.14 |
Gm17041 |
predicted gene 17041 |
1391 |
0.24 |
| chr4_108780980_108781662 | 0.14 |
Zfyve9 |
zinc finger, FYVE domain containing 9 |
523 |
0.73 |
| chr1_74047166_74047356 | 0.14 |
Tns1 |
tensin 1 |
10104 |
0.21 |
| chr4_63232014_63232237 | 0.14 |
Col27a1 |
collagen, type XXVII, alpha 1 |
2161 |
0.28 |
| chr11_80974473_80974624 | 0.14 |
Gm11416 |
predicted gene 11416 |
72246 |
0.1 |
| chr3_138455131_138455312 | 0.14 |
Adh5 |
alcohol dehydrogenase 5 (class III), chi polypeptide |
816 |
0.51 |
| chr4_150239968_150240153 | 0.14 |
Eno1 |
enolase 1, alpha non-neuron |
2360 |
0.22 |
| chr18_67648032_67648194 | 0.14 |
Psmg2 |
proteasome (prosome, macropain) assembly chaperone 2 |
6505 |
0.16 |
| chr6_24262961_24263227 | 0.14 |
Slc13a1 |
solute carrier family 13 (sodium/sulfate symporters), member 1 |
95002 |
0.08 |
| chr4_102557092_102557428 | 0.14 |
Pde4b |
phosphodiesterase 4B, cAMP specific |
12835 |
0.29 |
| chr7_132924974_132925182 | 0.14 |
1500002F19Rik |
RIKEN cDNA 1500002F19 gene |
6025 |
0.15 |
| chr12_79481989_79482309 | 0.14 |
Rad51b |
RAD51 paralog B |
154796 |
0.04 |
| chr2_6870734_6870924 | 0.13 |
Celf2 |
CUGBP, Elav-like family member 2 |
1143 |
0.52 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.1 | 0.2 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.0 | 0.1 | GO:0006701 | progesterone biosynthetic process(GO:0006701) |
| 0.0 | 0.1 | GO:0072592 | oxygen metabolic process(GO:0072592) |
| 0.0 | 0.1 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.0 | 0.1 | GO:0045726 | positive regulation of integrin biosynthetic process(GO:0045726) |
| 0.0 | 0.1 | GO:0015959 | diadenosine polyphosphate metabolic process(GO:0015959) |
| 0.0 | 0.2 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.0 | 0.1 | GO:0009730 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 0.0 | 0.1 | GO:0060741 | prostate gland stromal morphogenesis(GO:0060741) |
| 0.0 | 0.1 | GO:0010046 | response to mycotoxin(GO:0010046) |
| 0.0 | 0.2 | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.0 | 0.2 | GO:0006549 | isoleucine metabolic process(GO:0006549) |
| 0.0 | 0.2 | GO:0006559 | L-phenylalanine catabolic process(GO:0006559) erythrose 4-phosphate/phosphoenolpyruvate family amino acid catabolic process(GO:1902222) |
| 0.0 | 0.1 | GO:0051081 | membrane disassembly(GO:0030397) nuclear envelope disassembly(GO:0051081) |
| 0.0 | 0.1 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.0 | 0.1 | GO:0034310 | primary alcohol catabolic process(GO:0034310) |
| 0.0 | 0.1 | GO:2000302 | positive regulation of synaptic vesicle exocytosis(GO:2000302) |
| 0.0 | 0.1 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.2 | GO:0021540 | corpus callosum morphogenesis(GO:0021540) |
| 0.0 | 0.1 | GO:0045719 | negative regulation of glycogen biosynthetic process(GO:0045719) |
| 0.0 | 0.0 | GO:0034770 | histone H4-K20 methylation(GO:0034770) |
| 0.0 | 0.1 | GO:0009202 | deoxyribonucleoside triphosphate biosynthetic process(GO:0009202) |
| 0.0 | 0.1 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.0 | 0.1 | GO:0030647 | polyketide metabolic process(GO:0030638) aminoglycoside antibiotic metabolic process(GO:0030647) daunorubicin metabolic process(GO:0044597) doxorubicin metabolic process(GO:0044598) |
| 0.0 | 0.1 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.0 | 0.1 | GO:0032066 | nucleolus to nucleoplasm transport(GO:0032066) |
| 0.0 | 0.3 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.1 | GO:0050883 | musculoskeletal movement, spinal reflex action(GO:0050883) |
| 0.0 | 0.1 | GO:0021847 | ventricular zone neuroblast division(GO:0021847) |
| 0.0 | 0.1 | GO:0034146 | toll-like receptor 5 signaling pathway(GO:0034146) |
| 0.0 | 0.1 | GO:0019676 | ammonia assimilation cycle(GO:0019676) |
| 0.0 | 0.1 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.0 | 0.1 | GO:0002278 | eosinophil activation involved in immune response(GO:0002278) eosinophil mediated immunity(GO:0002447) eosinophil degranulation(GO:0043308) |
| 0.0 | 0.1 | GO:0006680 | glucosylceramide catabolic process(GO:0006680) |
| 0.0 | 0.1 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 0.0 | 0.1 | GO:0097167 | circadian regulation of translation(GO:0097167) |
| 0.0 | 0.1 | GO:0015842 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.0 | 0.1 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.0 | 0.1 | GO:0018317 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.0 | 0.1 | GO:1903660 | regulation of complement-dependent cytotoxicity(GO:1903659) negative regulation of complement-dependent cytotoxicity(GO:1903660) |
| 0.0 | 0.1 | GO:0072602 | interleukin-4 secretion(GO:0072602) |
| 0.0 | 0.1 | GO:0009256 | 10-formyltetrahydrofolate metabolic process(GO:0009256) |
| 0.0 | 0.1 | GO:1900045 | negative regulation of protein K63-linked ubiquitination(GO:1900045) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.0 | 0.1 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.0 | 0.1 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.0 | GO:0070874 | negative regulation of glycogen metabolic process(GO:0070874) |
| 0.0 | 0.1 | GO:0090435 | protein localization to nuclear envelope(GO:0090435) |
| 0.0 | 0.0 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.0 | 0.0 | GO:0003289 | atrial septum primum morphogenesis(GO:0003289) |
| 0.0 | 0.1 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.1 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.0 | 0.0 | GO:0002121 | inter-male aggressive behavior(GO:0002121) |
| 0.0 | 0.0 | GO:2000807 | regulation of synaptic vesicle clustering(GO:2000807) |
| 0.0 | 0.0 | GO:0010909 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.0 | 0.0 | GO:1901078 | negative regulation of relaxation of muscle(GO:1901078) |
| 0.0 | 0.1 | GO:0019482 | beta-alanine metabolic process(GO:0019482) |
| 0.0 | 0.0 | GO:0060916 | mesenchymal cell proliferation involved in lung development(GO:0060916) |
| 0.0 | 0.0 | GO:0003162 | atrioventricular node development(GO:0003162) |
| 0.0 | 0.1 | GO:0015781 | nucleotide-sugar transport(GO:0015780) pyrimidine nucleotide-sugar transport(GO:0015781) |
| 0.0 | 0.0 | GO:0090158 | endoplasmic reticulum membrane organization(GO:0090158) |
| 0.0 | 0.0 | GO:0046271 | phenylpropanoid catabolic process(GO:0046271) |
| 0.0 | 0.0 | GO:0010040 | response to iron(II) ion(GO:0010040) |
| 0.0 | 0.0 | GO:0019065 | receptor-mediated endocytosis of virus by host cell(GO:0019065) endocytosis involved in viral entry into host cell(GO:0075509) |
| 0.0 | 0.0 | GO:0006435 | threonyl-tRNA aminoacylation(GO:0006435) |
| 0.0 | 0.0 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.0 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.0 | 0.1 | GO:0052405 | negative regulation by host of symbiont molecular function(GO:0052405) modification by host of symbiont molecular function(GO:0052428) |
| 0.0 | 0.1 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.0 | 0.0 | GO:0070836 | caveola assembly(GO:0070836) |
| 0.0 | 0.0 | GO:1990705 | cholangiocyte proliferation(GO:1990705) |
| 0.0 | 0.1 | GO:0006183 | GTP biosynthetic process(GO:0006183) |
| 0.0 | 0.1 | GO:0046130 | purine nucleoside catabolic process(GO:0006152) purine ribonucleoside catabolic process(GO:0046130) |
| 0.0 | 0.0 | GO:0016080 | synaptic vesicle targeting(GO:0016080) |
| 0.0 | 0.1 | GO:0043651 | linoleic acid metabolic process(GO:0043651) |
| 0.0 | 0.1 | GO:0097105 | presynaptic membrane assembly(GO:0097105) |
| 0.0 | 0.0 | GO:0042525 | tyrosine phosphorylation of Stat6 protein(GO:0042505) regulation of tyrosine phosphorylation of Stat6 protein(GO:0042525) |
| 0.0 | 0.0 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 0.0 | 0.0 | GO:0051660 | establishment of centrosome localization(GO:0051660) |
| 0.0 | 0.0 | GO:0061002 | negative regulation of dendritic spine morphogenesis(GO:0061002) |
| 0.0 | 0.0 | GO:1900740 | regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900739) positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900740) |
| 0.0 | 0.0 | GO:0034773 | histone H4-K20 trimethylation(GO:0034773) |
| 0.0 | 0.0 | GO:0048254 | snoRNA localization(GO:0048254) |
| 0.0 | 0.1 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.0 | 0.0 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.0 | 0.1 | GO:0033147 | negative regulation of intracellular estrogen receptor signaling pathway(GO:0033147) |
| 0.0 | 0.0 | GO:0060161 | positive regulation of dopamine receptor signaling pathway(GO:0060161) |
| 0.0 | 0.1 | GO:0006528 | asparagine metabolic process(GO:0006528) |
| 0.0 | 0.0 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.0 | 0.0 | GO:0090361 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 0.0 | 0.1 | GO:0006517 | protein deglycosylation(GO:0006517) |
| 0.0 | 0.1 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.0 | 0.1 | GO:0071397 | cellular response to cholesterol(GO:0071397) |
| 0.0 | 0.0 | GO:1902659 | regulation of glucose mediated signaling pathway(GO:1902659) positive regulation of glucose mediated signaling pathway(GO:1902661) |
| 0.0 | 0.0 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.0 | 0.1 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.0 | 0.0 | GO:0006069 | ethanol metabolic process(GO:0006067) ethanol oxidation(GO:0006069) |
| 0.0 | 0.0 | GO:1901475 | pyruvate transmembrane transport(GO:1901475) |
| 0.0 | 0.1 | GO:0052697 | xenobiotic glucuronidation(GO:0052697) |
| 0.0 | 0.1 | GO:0009404 | toxin metabolic process(GO:0009404) |
| 0.0 | 0.0 | GO:0042524 | negative regulation of tyrosine phosphorylation of Stat5 protein(GO:0042524) |
| 0.0 | 0.0 | GO:0060468 | prevention of polyspermy(GO:0060468) |
| 0.0 | 0.0 | GO:0071476 | cellular hypotonic response(GO:0071476) |
| 0.0 | 0.1 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.0 | 0.1 | GO:0006002 | fructose 6-phosphate metabolic process(GO:0006002) |
| 0.0 | 0.1 | GO:2000510 | positive regulation of dendritic cell chemotaxis(GO:2000510) |
| 0.0 | 0.1 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.0 | 0.0 | GO:0030050 | vesicle transport along actin filament(GO:0030050) |
| 0.0 | 0.0 | GO:0034140 | negative regulation of toll-like receptor 3 signaling pathway(GO:0034140) |
| 0.0 | 0.1 | GO:0010519 | negative regulation of phospholipase activity(GO:0010519) |
| 0.0 | 0.1 | GO:1900103 | positive regulation of endoplasmic reticulum unfolded protein response(GO:1900103) |
| 0.0 | 0.1 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 | 0.0 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 0.0 | 0.0 | GO:0060523 | prostate epithelial cord elongation(GO:0060523) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.0 | 0.1 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.0 | 0.1 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.0 | 0.1 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.0 | 0.1 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.0 | 0.1 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.0 | 0.1 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.1 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.0 | 0.1 | GO:0045293 | mRNA editing complex(GO:0045293) |
| 0.0 | 0.1 | GO:0042824 | MHC class I peptide loading complex(GO:0042824) |
| 0.0 | 0.0 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.1 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.0 | 0.0 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 0.1 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.0 | 0.1 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.0 | 0.1 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.1 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.0 | 0.0 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.0 | 0.0 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.0 | 0.1 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.0 | 0.0 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.0 | 0.1 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.0 | 0.0 | GO:0030478 | actin cap(GO:0030478) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.1 | 0.3 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 0.0 | 0.1 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.0 | 0.1 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 0.0 | 0.1 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.0 | 0.1 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.0 | 0.3 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.1 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.0 | 0.2 | GO:0047760 | butyrate-CoA ligase activity(GO:0047760) |
| 0.0 | 0.1 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.0 | 0.1 | GO:0071614 | linoleic acid epoxygenase activity(GO:0071614) |
| 0.0 | 0.1 | GO:0016743 | carboxyl- or carbamoyltransferase activity(GO:0016743) |
| 0.0 | 0.2 | GO:0016151 | nickel cation binding(GO:0016151) |
| 0.0 | 0.2 | GO:0051920 | peroxiredoxin activity(GO:0051920) |
| 0.0 | 0.2 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.0 | 0.1 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.0 | 0.1 | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0003829) |
| 0.0 | 0.1 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.1 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.0 | 0.1 | GO:0038181 | bile acid receptor activity(GO:0038181) |
| 0.0 | 0.1 | GO:0016899 | oxidoreductase activity, acting on the CH-OH group of donors, oxygen as acceptor(GO:0016899) |
| 0.0 | 0.1 | GO:0035663 | Toll-like receptor 2 binding(GO:0035663) |
| 0.0 | 0.1 | GO:0047023 | androsterone dehydrogenase activity(GO:0047023) |
| 0.0 | 0.0 | GO:0043125 | ErbB-3 class receptor binding(GO:0043125) |
| 0.0 | 0.2 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.1 | GO:0017098 | sulfonylurea receptor binding(GO:0017098) |
| 0.0 | 0.1 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.0 | 0.1 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 0.1 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.0 | 0.1 | GO:0043426 | MRF binding(GO:0043426) |
| 0.0 | 0.1 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.0 | 0.2 | GO:0016215 | acyl-CoA desaturase activity(GO:0016215) |
| 0.0 | 0.0 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.0 | 0.0 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) |
| 0.0 | 0.1 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.2 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.1 | GO:0000403 | Y-form DNA binding(GO:0000403) |
| 0.0 | 0.1 | GO:0047105 | aminobutyraldehyde dehydrogenase activity(GO:0019145) 4-trimethylammoniobutyraldehyde dehydrogenase activity(GO:0047105) |
| 0.0 | 0.1 | GO:1990239 | steroid hormone binding(GO:1990239) |
| 0.0 | 0.0 | GO:0004829 | threonine-tRNA ligase activity(GO:0004829) |
| 0.0 | 0.1 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.0 | 0.0 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.0 | 0.0 | GO:0000295 | adenine nucleotide transmembrane transporter activity(GO:0000295) purine ribonucleotide transmembrane transporter activity(GO:0005346) purine nucleotide transmembrane transporter activity(GO:0015216) |
| 0.0 | 0.3 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.0 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.0 | 0.1 | GO:0015377 | cation:chloride symporter activity(GO:0015377) |
| 0.0 | 0.1 | GO:0004673 | protein histidine kinase activity(GO:0004673) |
| 0.0 | 0.0 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.0 | 0.1 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.0 | 0.0 | GO:0019797 | procollagen-proline 3-dioxygenase activity(GO:0019797) |
| 0.0 | 0.1 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.0 | 0.0 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.0 | 0.0 | GO:0004487 | methenyltetrahydrofolate cyclohydrolase activity(GO:0004477) methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
| 0.0 | 0.0 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.0 | 0.1 | GO:0004075 | biotin carboxylase activity(GO:0004075) |
| 0.0 | 0.0 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.0 | 0.0 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.0 | 0.1 | GO:0047391 | alkylglycerophosphoethanolamine phosphodiesterase activity(GO:0047391) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.1 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.3 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.0 | REACTOME VIF MEDIATED DEGRADATION OF APOBEC3G | Genes involved in Vif-mediated degradation of APOBEC3G |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.1 | REACTOME ER PHAGOSOME PATHWAY | Genes involved in ER-Phagosome pathway |
| 0.0 | 0.3 | REACTOME GRB2 EVENTS IN ERBB2 SIGNALING | Genes involved in GRB2 events in ERBB2 signaling |
| 0.0 | 0.1 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.1 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.1 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.0 | 0.1 | REACTOME PROLONGED ERK ACTIVATION EVENTS | Genes involved in Prolonged ERK activation events |
| 0.0 | 0.1 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.0 | 0.0 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |