Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
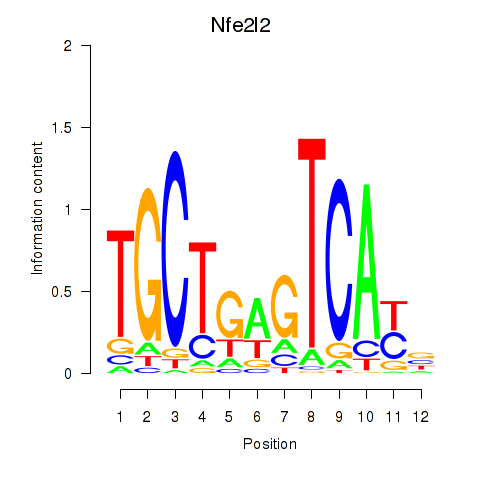
Results for Nfe2l2
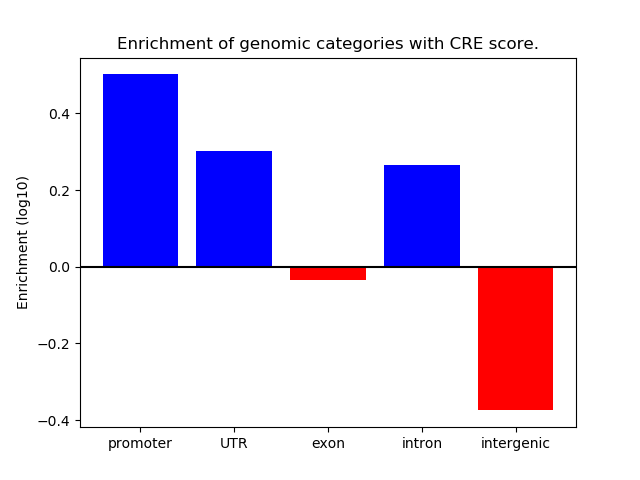
Z-value: 0.76
Transcription factors associated with Nfe2l2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nfe2l2
|
ENSMUSG00000015839.6 | nuclear factor, erythroid derived 2, like 2 |
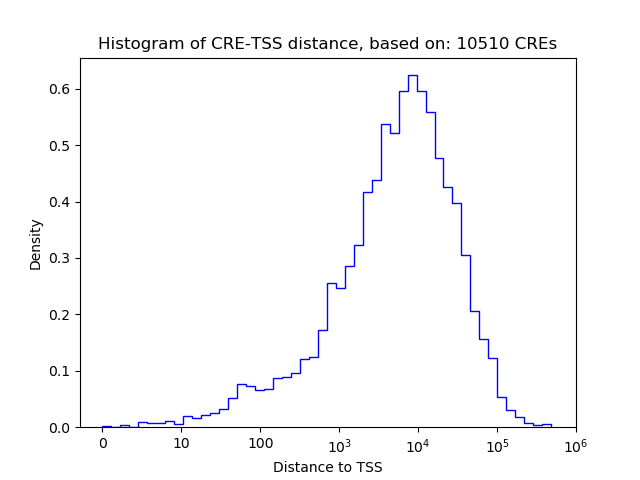
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr2_75698638_75698789 | Nfe2l2 | 5776 | 0.129144 | -0.92 | 9.5e-03 | Click! |
| chr2_75696217_75696368 | Nfe2l2 | 8197 | 0.121140 | -0.89 | 1.8e-02 | Click! |
| chr2_75699304_75699580 | Nfe2l2 | 5047 | 0.133232 | -0.89 | 1.8e-02 | Click! |
| chr2_75698077_75698279 | Nfe2l2 | 6311 | 0.126848 | -0.87 | 2.3e-02 | Click! |
| chr2_75697788_75697939 | Nfe2l2 | 6626 | 0.125689 | -0.85 | 3.1e-02 | Click! |
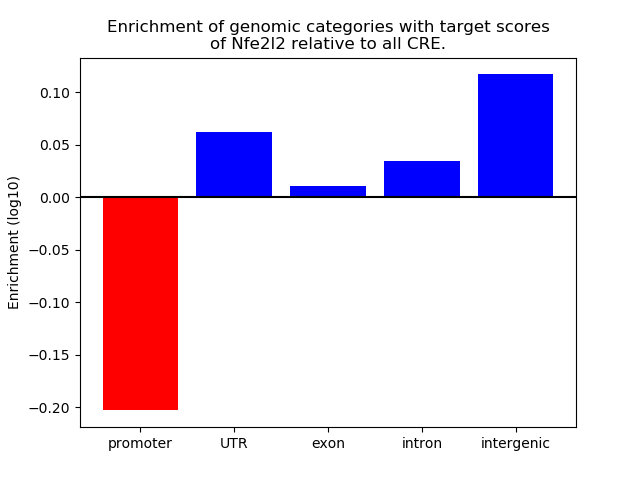
Activity of the Nfe2l2 motif across conditions
Conditions sorted by the z-value of the Nfe2l2 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
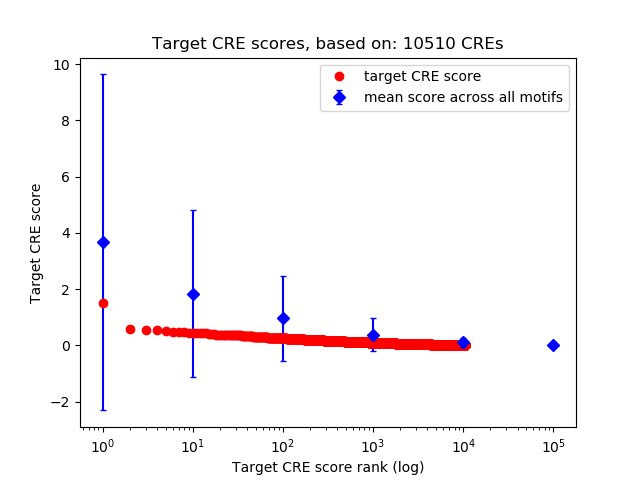
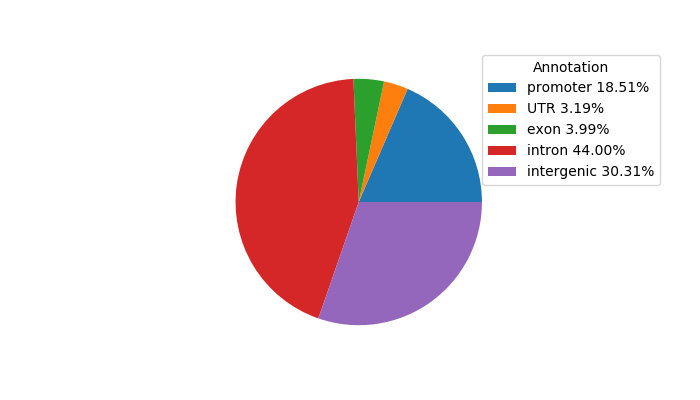
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr11_7201124_7201275 | 1.50 |
Igfbp1 |
insulin-like growth factor binding protein 1 |
3417 |
0.21 |
| chr5_139871644_139871802 | 0.58 |
Gm42423 |
predicted gene 42423 |
24086 |
0.12 |
| chr16_28991322_28991483 | 0.54 |
Gm8253 |
predicted gene 8253 |
40047 |
0.19 |
| chr16_30590365_30590575 | 0.54 |
Lsg1 |
large 60S subunit nuclear export GTPase 1 |
2878 |
0.26 |
| chr6_31066838_31067230 | 0.52 |
Gm27019 |
predicted gene, 27019 |
3539 |
0.14 |
| chr1_92683263_92683419 | 0.49 |
Gm29483 |
predicted gene 29483 |
33861 |
0.09 |
| chr5_134941937_134942100 | 0.46 |
Mettl27 |
methyltransferase like 27 |
1556 |
0.18 |
| chr19_42118558_42118858 | 0.46 |
Pi4k2a |
phosphatidylinositol 4-kinase type 2 alpha |
4030 |
0.13 |
| chr16_38711344_38711500 | 0.45 |
Arhgap31 |
Rho GTPase activating protein 31 |
1476 |
0.34 |
| chr15_102014961_102015124 | 0.44 |
Krt8 |
keratin 8 |
10560 |
0.11 |
| chr14_62465117_62465312 | 0.43 |
Gucy1b2 |
guanylate cyclase 1, soluble, beta 2 |
8925 |
0.16 |
| chr9_69356418_69356569 | 0.43 |
Gm15511 |
predicted gene 15511 |
7754 |
0.19 |
| chr10_84701256_84701556 | 0.43 |
Rfx4 |
regulatory factor X, 4 (influences HLA class II expression) |
54656 |
0.1 |
| chr8_126844756_126844953 | 0.43 |
A630001O12Rik |
RIKEN cDNA A630001O12 gene |
5621 |
0.22 |
| chr10_127915625_127915914 | 0.42 |
Gm15905 |
predicted gene 15905 |
5300 |
0.1 |
| chr9_118101918_118102083 | 0.41 |
Azi2 |
5-azacytidine induced gene 2 |
42944 |
0.13 |
| chrY_90769047_90769393 | 0.41 |
Gm21860 |
predicted gene, 21860 |
13753 |
0.16 |
| chr4_33243933_33244238 | 0.38 |
Pnrc1 |
proline-rich nuclear receptor coactivator 1 |
3490 |
0.21 |
| chr10_107943872_107944134 | 0.38 |
Gm29685 |
predicted gene, 29685 |
2642 |
0.35 |
| chr3_104531244_104531395 | 0.38 |
Lrig2 |
leucine-rich repeats and immunoglobulin-like domains 2 |
19401 |
0.12 |
| chr9_43622435_43622586 | 0.37 |
Gm29909 |
predicted gene, 29909 |
14326 |
0.17 |
| chr14_69728721_69728885 | 0.37 |
Chmp7 |
charged multivesicular body protein 7 |
2072 |
0.22 |
| chr2_132212432_132212583 | 0.37 |
Tmem230 |
transmembrane protein 230 |
35147 |
0.11 |
| chr19_37528710_37528861 | 0.37 |
Exoc6 |
exocyst complex component 6 |
7906 |
0.2 |
| chr13_64388507_64388698 | 0.36 |
Ctsl |
cathepsin L |
17712 |
0.1 |
| chr17_31874093_31874279 | 0.36 |
Sik1 |
salt inducible kinase 1 |
18382 |
0.13 |
| chr16_21828430_21828584 | 0.36 |
Map3k13 |
mitogen-activated protein kinase kinase kinase 13 |
2565 |
0.18 |
| chr10_20673746_20674078 | 0.36 |
Gm17230 |
predicted gene 17230 |
48277 |
0.14 |
| chr10_93188458_93188632 | 0.36 |
Cdk17 |
cyclin-dependent kinase 17 |
6935 |
0.21 |
| chr5_130264636_130264798 | 0.36 |
Tyw1 |
tRNA-yW synthesizing protein 1 homolog (S. cerevisiae) |
5787 |
0.13 |
| chr19_23033456_23033617 | 0.36 |
Gm50136 |
predicted gene, 50136 |
27918 |
0.18 |
| chr6_121886736_121886887 | 0.35 |
Mug1 |
murinoglobulin 1 |
1234 |
0.47 |
| chr16_76237397_76237728 | 0.35 |
Nrip1 |
nuclear receptor interacting protein 1 |
86096 |
0.08 |
| chr1_192061757_192061908 | 0.35 |
Traf5 |
TNF receptor-associated factor 5 |
2670 |
0.19 |
| chr14_47354165_47354327 | 0.34 |
Lgals3 |
lectin, galactose binding, soluble 3 |
13505 |
0.1 |
| chr16_21828238_21828402 | 0.34 |
Map3k13 |
mitogen-activated protein kinase kinase kinase 13 |
2378 |
0.19 |
| chr14_31496236_31496433 | 0.33 |
Eaf1 |
ELL associated factor 1 |
1138 |
0.32 |
| chr16_77569572_77569736 | 0.33 |
Gm37606 |
predicted gene, 37606 |
8004 |
0.11 |
| chr4_139334725_139334887 | 0.33 |
AL807811.1 |
aldo-keto reductase family 7 member A2 (AKR7A2) pseudogene |
2374 |
0.12 |
| chr1_153502177_153502328 | 0.33 |
Dhx9 |
DEAH (Asp-Glu-Ala-His) box polypeptide 9 |
14592 |
0.16 |
| chr4_46563917_46564068 | 0.32 |
Coro2a |
coronin, actin binding protein 2A |
2448 |
0.24 |
| chr2_35066631_35066785 | 0.32 |
Hc |
hemolytic complement |
5270 |
0.18 |
| chr14_78937878_78938029 | 0.32 |
Gm49016 |
predicted gene, 49016 |
9274 |
0.2 |
| chr12_84202016_84202416 | 0.32 |
Gm31513 |
predicted gene, 31513 |
6247 |
0.11 |
| chr12_54671337_54671527 | 0.32 |
Gm24971 |
predicted gene, 24971 |
4646 |
0.08 |
| chr3_67490227_67490388 | 0.32 |
Gm42568 |
predicted gene 42568 |
13329 |
0.13 |
| chr12_29100151_29100321 | 0.31 |
4833405L11Rik |
RIKEN cDNA 4833405L11 gene |
6501 |
0.24 |
| chr12_110381296_110381622 | 0.31 |
Gm47195 |
predicted gene, 47195 |
56074 |
0.08 |
| chr10_84854177_84854328 | 0.31 |
Rfx4 |
regulatory factor X, 4 (influences HLA class II expression) |
16104 |
0.21 |
| chr11_115437682_115437852 | 0.31 |
Trim80 |
tripartite motif-containing 80 |
2778 |
0.11 |
| chr11_61263826_61264208 | 0.31 |
Aldh3a2 |
aldehyde dehydrogenase family 3, subfamily A2 |
1682 |
0.33 |
| chr3_85991904_85992061 | 0.30 |
Prss48 |
protease, serine 48 |
10509 |
0.12 |
| chr16_31430067_31430241 | 0.30 |
Gm15743 |
predicted gene 15743 |
378 |
0.68 |
| chr13_34898393_34898709 | 0.30 |
Prpf4b |
pre-mRNA processing factor 4B |
3098 |
0.15 |
| chr9_94993496_94993650 | 0.30 |
Gm47168 |
predicted gene, 47168 |
889 |
0.73 |
| chr11_22269235_22269425 | 0.30 |
Ehbp1 |
EH domain binding protein 1 |
16508 |
0.26 |
| chr11_116079675_116079864 | 0.30 |
Wbp2 |
WW domain binding protein 2 |
1716 |
0.19 |
| chr2_12197838_12198004 | 0.30 |
Itga8 |
integrin alpha 8 |
4609 |
0.27 |
| chr6_8345541_8345704 | 0.30 |
Gm16055 |
predicted gene 16055 |
18514 |
0.17 |
| chr9_119405710_119405861 | 0.29 |
Gm47320 |
predicted gene, 47320 |
3231 |
0.15 |
| chr8_71405094_71405257 | 0.29 |
Ankle1 |
ankyrin repeat and LEM domain containing 1 |
835 |
0.39 |
| chr8_68908746_68908916 | 0.29 |
Lpl |
lipoprotein lipase |
18087 |
0.18 |
| chr15_100625793_100625948 | 0.29 |
Dazap2 |
DAZ associated protein 2 |
9538 |
0.08 |
| chr8_69896581_69896741 | 0.29 |
Yjefn3 |
YjeF N-terminal domain containing 3 |
2686 |
0.15 |
| chr7_35127036_35127350 | 0.28 |
Gm45091 |
predicted gene 45091 |
7588 |
0.1 |
| chr13_93627456_93627944 | 0.28 |
Gm15622 |
predicted gene 15622 |
2318 |
0.25 |
| chr5_145989518_145989698 | 0.28 |
Cyp3a25 |
cytochrome P450, family 3, subfamily a, polypeptide 25 |
2035 |
0.23 |
| chr2_101781393_101781544 | 0.28 |
Prr5l |
proline rich 5 like |
16182 |
0.23 |
| chr10_108612751_108612908 | 0.28 |
Syt1 |
synaptotagmin I |
24135 |
0.23 |
| chr12_21141056_21141213 | 0.27 |
Asap2 |
ArfGAP with SH3 domain, ankyrin repeat and PH domain 2 |
29180 |
0.17 |
| chr2_144182222_144182616 | 0.27 |
Gm5535 |
predicted gene 5535 |
3333 |
0.2 |
| chr9_107689065_107689227 | 0.27 |
Sema3f |
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3F |
771 |
0.43 |
| chr11_117965286_117965456 | 0.27 |
Socs3 |
suppressor of cytokine signaling 3 |
3815 |
0.17 |
| chr2_104130173_104130336 | 0.27 |
A930018P22Rik |
RIKEN cDNA A930018P22 gene |
7485 |
0.14 |
| chr8_45255175_45255326 | 0.27 |
F11 |
coagulation factor XI |
6781 |
0.18 |
| chr10_18399536_18399687 | 0.27 |
Nhsl1 |
NHS-like 1 |
8027 |
0.24 |
| chr12_80598944_80599110 | 0.27 |
Galnt16 |
polypeptide N-acetylgalactosaminyltransferase 16 |
924 |
0.35 |
| chr14_8356958_8357137 | 0.27 |
2610318M16Rik |
RIKEN cDNA 2610318M16 gene |
10393 |
0.18 |
| chr4_70499390_70499541 | 0.27 |
Megf9 |
multiple EGF-like-domains 9 |
35463 |
0.22 |
| chr15_3353815_3354005 | 0.27 |
Ccdc152 |
coiled-coil domain containing 152 |
50384 |
0.15 |
| chr1_63479013_63479186 | 0.27 |
Adam23 |
a disintegrin and metallopeptidase domain 23 |
8480 |
0.22 |
| chr18_20989752_20989946 | 0.27 |
Rnf138 |
ring finger protein 138 |
11492 |
0.2 |
| chr11_58931646_58931797 | 0.27 |
Rnf187 |
ring finger protein 187 |
7195 |
0.07 |
| chr15_6748493_6748644 | 0.26 |
Rictor |
RPTOR independent companion of MTOR, complex 2 |
40185 |
0.16 |
| chr15_77412067_77412222 | 0.26 |
Apol9a |
apolipoprotein L 9a |
1009 |
0.35 |
| chr16_95814866_95815042 | 0.26 |
2810404F17Rik |
RIKEN cDNA 2810404F17 gene |
6637 |
0.19 |
| chr18_20782566_20782759 | 0.26 |
B4galt6 |
UDP-Gal:betaGlcNAc beta 1,4-galactosyltransferase, polypeptide 6 |
36258 |
0.15 |
| chr18_46381013_46381189 | 0.26 |
Gm24617 |
predicted gene, 24617 |
13736 |
0.14 |
| chr11_53764208_53764367 | 0.26 |
Mir7671 |
microRNA 7671 |
633 |
0.56 |
| chr1_75660295_75660569 | 0.26 |
Gm5257 |
predicted gene 5257 |
24042 |
0.16 |
| chr10_77970592_77970747 | 0.26 |
Trpm2 |
transient receptor potential cation channel, subfamily M, member 2 |
106 |
0.94 |
| chr2_59884229_59884405 | 0.26 |
Wdsub1 |
WD repeat, SAM and U-box domain containing 1 |
1726 |
0.39 |
| chr8_89279548_89279868 | 0.26 |
Gm5356 |
predicted pseudogene 5356 |
92148 |
0.09 |
| chr13_56324320_56324471 | 0.26 |
Gm17878 |
predicted gene, 17878 |
4061 |
0.17 |
| chr11_7203353_7203538 | 0.26 |
Igfbp1 |
insulin-like growth factor binding protein 1 |
5663 |
0.18 |
| chr18_23805749_23805916 | 0.26 |
Mapre2 |
microtubule-associated protein, RP/EB family, member 2 |
1729 |
0.33 |
| chr16_37856387_37856547 | 0.26 |
Gm36028 |
predicted gene, 36028 |
39 |
0.96 |
| chr19_3850170_3850502 | 0.26 |
Chka |
choline kinase alpha |
1437 |
0.22 |
| chr6_145679594_145679775 | 0.26 |
Tuba3b |
tubulin, alpha 3B |
63927 |
0.09 |
| chr15_36904059_36904218 | 0.25 |
Gm10384 |
predicted gene 10384 |
24322 |
0.13 |
| chr8_102024599_102024751 | 0.25 |
Gm22223 |
predicted gene, 22223 |
484967 |
0.01 |
| chr4_45503959_45504122 | 0.25 |
Gm22518 |
predicted gene, 22518 |
3488 |
0.2 |
| chr1_131546654_131546962 | 0.25 |
Fam72a |
family with sequence similarity 72, member A |
18905 |
0.14 |
| chr7_24850950_24851109 | 0.25 |
Gm18207 |
predicted gene, 18207 |
9917 |
0.09 |
| chr5_145852474_145852695 | 0.25 |
Cyp3a11 |
cytochrome P450, family 3, subfamily a, polypeptide 11 |
24107 |
0.13 |
| chr5_145989114_145989402 | 0.25 |
Cyp3a25 |
cytochrome P450, family 3, subfamily a, polypeptide 25 |
2385 |
0.2 |
| chr7_25387916_25388067 | 0.25 |
Lipe |
lipase, hormone sensitive |
1233 |
0.27 |
| chr13_39452515_39452666 | 0.25 |
Gm47348 |
predicted gene, 47348 |
12025 |
0.18 |
| chr15_6882089_6882243 | 0.25 |
Osmr |
oncostatin M receptor |
7197 |
0.29 |
| chr13_10274143_10274314 | 0.24 |
Gm26861 |
predicted gene, 26861 |
83367 |
0.08 |
| chr11_64588556_64588707 | 0.24 |
Gm24275 |
predicted gene, 24275 |
2009 |
0.47 |
| chr14_73105205_73105370 | 0.24 |
Rcbtb2 |
regulator of chromosome condensation (RCC1) and BTB (POZ) domain containing protein 2 |
17750 |
0.19 |
| chr18_12882054_12882208 | 0.24 |
Osbpl1a |
oxysterol binding protein-like 1A |
2154 |
0.28 |
| chr10_24097646_24097797 | 0.24 |
Taar8c |
trace amine-associated receptor 8C |
4191 |
0.1 |
| chr4_115607059_115607329 | 0.24 |
Cyp4a32 |
cytochrome P450, family 4, subfamily a, polypeptide 32 |
6219 |
0.14 |
| chr1_180219669_180219845 | 0.24 |
Gm37336 |
predicted gene, 37336 |
7356 |
0.14 |
| chr14_51106134_51106584 | 0.24 |
Eddm3b |
epididymal protein 3B |
8067 |
0.08 |
| chr5_148674254_148674405 | 0.24 |
Gm36186 |
predicted gene, 36186 |
37137 |
0.13 |
| chr14_37134902_37135213 | 0.24 |
Ghitm |
growth hormone inducible transmembrane protein |
82 |
0.97 |
| chr1_155108763_155108914 | 0.24 |
Ier5 |
immediate early response 5 |
9202 |
0.15 |
| chr8_75089591_75089759 | 0.24 |
Hmox1 |
heme oxygenase 1 |
3946 |
0.16 |
| chr5_17854594_17854745 | 0.24 |
Cd36 |
CD36 molecule |
4877 |
0.33 |
| chr14_20135347_20135503 | 0.23 |
Saysd1 |
SAYSVFN motif domain containing 1 |
3109 |
0.21 |
| chr16_32685796_32686168 | 0.23 |
Tnk2 |
tyrosine kinase, non-receptor, 2 |
5168 |
0.16 |
| chr14_24232675_24232832 | 0.23 |
Gm17105 |
predicted gene 17105 |
10638 |
0.13 |
| chr19_6273750_6273958 | 0.23 |
Gm14963 |
predicted gene 14963 |
2751 |
0.1 |
| chr13_59875722_59876043 | 0.23 |
Gm48384 |
predicted gene, 48384 |
2240 |
0.23 |
| chr15_99814958_99815109 | 0.23 |
Gm17057 |
predicted gene 17057 |
3987 |
0.1 |
| chr4_41637175_41637517 | 0.23 |
Gm12406 |
predicted gene 12406 |
2032 |
0.17 |
| chr5_36563902_36564053 | 0.23 |
Tbc1d14 |
TBC1 domain family, member 14 |
79 |
0.97 |
| chr11_7196610_7196788 | 0.23 |
Igfbp1 |
insulin-like growth factor binding protein 1 |
1083 |
0.48 |
| chr19_6932250_6932483 | 0.23 |
Kcnk4 |
potassium channel, subfamily K, member 4 |
461 |
0.58 |
| chr8_11005778_11005929 | 0.23 |
9530052E02Rik |
RIKEN cDNA 9530052E02 gene |
1997 |
0.22 |
| chr16_16543014_16543174 | 0.23 |
Gm16139 |
predicted gene 16139 |
771 |
0.66 |
| chr2_92667342_92667493 | 0.23 |
Chst1 |
carbohydrate sulfotransferase 1 |
67710 |
0.08 |
| chr2_154591324_154591483 | 0.22 |
Pxmp4 |
peroxisomal membrane protein 4 |
12285 |
0.1 |
| chr5_90433544_90433714 | 0.22 |
Alb |
albumin |
27268 |
0.13 |
| chr8_116344914_116345106 | 0.22 |
1700018P08Rik |
RIKEN cDNA 1700018P08 gene |
12051 |
0.29 |
| chr3_41031529_41031708 | 0.22 |
Larp1b |
La ribonucleoprotein domain family, member 1B |
1897 |
0.27 |
| chr2_44142320_44142937 | 0.22 |
Arhgap15os |
Rho GTPase activating protein 15, opposite strand |
77304 |
0.11 |
| chr9_23030248_23030409 | 0.22 |
Bmper |
BMP-binding endothelial regulator |
192748 |
0.03 |
| chr16_94530638_94530789 | 0.22 |
Vps26c |
VPS26 endosomal protein sorting factor C |
3883 |
0.21 |
| chr1_170709591_170709775 | 0.22 |
Gm23523 |
predicted gene, 23523 |
42140 |
0.12 |
| chr14_73388637_73388820 | 0.22 |
Itm2b |
integral membrane protein 2B |
3439 |
0.26 |
| chr1_97672950_97673101 | 0.22 |
D1Ertd622e |
DNA segment, Chr 1, ERATO Doi 622, expressed |
10951 |
0.19 |
| chr15_59799636_59799788 | 0.22 |
Gm19510 |
predicted gene, 19510 |
4753 |
0.3 |
| chr12_111454567_111454858 | 0.22 |
Tnfaip2 |
tumor necrosis factor, alpha-induced protein 2 |
4193 |
0.14 |
| chr17_3489197_3489361 | 0.22 |
Tiam2 |
T cell lymphoma invasion and metastasis 2 |
1184 |
0.47 |
| chr19_4441877_4442205 | 0.22 |
A930001C03Rik |
RIKEN cDNA A930001C03 gene |
1697 |
0.23 |
| chr9_62435632_62436065 | 0.22 |
Coro2b |
coronin, actin binding protein, 2B |
6598 |
0.24 |
| chr16_95682151_95682417 | 0.22 |
Ets2 |
E26 avian leukemia oncogene 2, 3' domain |
19791 |
0.2 |
| chr11_12197671_12197822 | 0.22 |
Gm12001 |
predicted gene 12001 |
69748 |
0.11 |
| chr15_77727954_77728108 | 0.22 |
Apol9b |
apolipoprotein L 9b |
1008 |
0.34 |
| chr9_50590889_50591040 | 0.22 |
Sdhd |
succinate dehydrogenase complex, subunit D, integral membrane protein |
12848 |
0.09 |
| chr8_12882911_12883062 | 0.22 |
Gm15353 |
predicted gene 15353 |
730 |
0.53 |
| chr2_126591444_126591598 | 0.22 |
Hdc |
histidine decarboxylase |
6740 |
0.18 |
| chr9_31610718_31610869 | 0.22 |
Gm18820 |
predicted gene, 18820 |
32977 |
0.14 |
| chr5_131814532_131814871 | 0.21 |
4930563F08Rik |
RIKEN cDNA 4930563F08 gene |
65568 |
0.08 |
| chr17_69181315_69181466 | 0.21 |
Epb41l3 |
erythrocyte membrane protein band 4.1 like 3 |
24532 |
0.23 |
| chr4_80922189_80922362 | 0.21 |
Lurap1l |
leucine rich adaptor protein 1-like |
11629 |
0.25 |
| chr9_95549721_95549889 | 0.21 |
Gm32281 |
predicted gene, 32281 |
4692 |
0.15 |
| chr4_130075213_130075370 | 0.21 |
Col16a1 |
collagen, type XVI, alpha 1 |
1752 |
0.29 |
| chr16_32853567_32853731 | 0.21 |
Rubcn |
RUN domain and cysteine-rich domain containing, Beclin 1-interacting protein |
14689 |
0.12 |
| chr8_126686853_126687025 | 0.21 |
Gm45805 |
predicted gene 45805 |
71395 |
0.1 |
| chr8_126778914_126779065 | 0.21 |
Gm45805 |
predicted gene 45805 |
20655 |
0.21 |
| chr12_83055899_83056080 | 0.21 |
Rgs6 |
regulator of G-protein signaling 6 |
8999 |
0.21 |
| chr13_64237836_64238013 | 0.21 |
Cdc14b |
CDC14 cell division cycle 14B |
10718 |
0.1 |
| chr16_21792922_21793081 | 0.21 |
1300002E11Rik |
RIKEN cDNA 1300002E11 gene |
1317 |
0.26 |
| chr12_31913746_31913916 | 0.21 |
Cog5 |
component of oligomeric golgi complex 5 |
5842 |
0.2 |
| chr6_117604955_117605122 | 0.21 |
Gm45083 |
predicted gene 45083 |
7532 |
0.22 |
| chr16_11456411_11456595 | 0.21 |
Snx29 |
sorting nexin 29 |
9195 |
0.23 |
| chr10_81181035_81181199 | 0.21 |
Eef2 |
eukaryotic translation elongation factor 2 |
66 |
0.9 |
| chr11_62568400_62568605 | 0.20 |
Gm12280 |
predicted gene 12280 |
2119 |
0.14 |
| chr16_22108220_22108385 | 0.20 |
Igf2bp2 |
insulin-like growth factor 2 mRNA binding protein 2 |
12934 |
0.18 |
| chr11_86951820_86952000 | 0.20 |
Ypel2 |
yippee like 2 |
20114 |
0.17 |
| chr8_123162810_123162969 | 0.20 |
Gm4316 |
predicted gene 4316 |
2366 |
0.12 |
| chr11_101361469_101361620 | 0.20 |
AL590969.1 |
glucose-6-phosphatase, catalytic (G6pc) pseudogene |
5169 |
0.06 |
| chr5_100663119_100663274 | 0.20 |
Coq2 |
coenzyme Q2 4-hydroxybenzoate polyprenyltransferase |
1223 |
0.4 |
| chr7_138867340_138867520 | 0.20 |
Ppp2r2d |
protein phosphatase 2, regulatory subunit B, delta |
980 |
0.43 |
| chr18_38849992_38850159 | 0.20 |
Fgf1 |
fibroblast growth factor 1 |
8673 |
0.22 |
| chr15_43701477_43701832 | 0.20 |
Gm2140 |
predicted gene 2140 |
106807 |
0.07 |
| chr11_44172606_44172757 | 0.20 |
Gm12154 |
predicted gene 12154 |
94528 |
0.08 |
| chr9_58231879_58232044 | 0.20 |
Pml |
promyelocytic leukemia |
1424 |
0.27 |
| chr15_100227160_100227326 | 0.20 |
Atf1 |
activating transcription factor 1 |
576 |
0.67 |
| chr18_11035510_11035780 | 0.20 |
Gata6os |
GATA binding protein 6, opposite strand |
11992 |
0.21 |
| chr12_60292738_60292905 | 0.20 |
Gm32936 |
predicted gene, 32936 |
25067 |
0.2 |
| chr3_98044919_98045386 | 0.20 |
Gm42819 |
predicted gene 42819 |
14465 |
0.18 |
| chr8_10910475_10910817 | 0.20 |
Gm2814 |
predicted gene 2814 |
10088 |
0.1 |
| chr2_163661430_163661594 | 0.20 |
Pkig |
protein kinase inhibitor, gamma |
13 |
0.97 |
| chr11_101953099_101953306 | 0.20 |
4930417O22Rik |
RIKEN cDNA 4930417O22 gene |
781 |
0.45 |
| chr19_4213519_4213699 | 0.20 |
Clcf1 |
cardiotrophin-like cytokine factor 1 |
629 |
0.3 |
| chr4_115610472_115610686 | 0.20 |
Cyp4a32 |
cytochrome P450, family 4, subfamily a, polypeptide 32 |
9604 |
0.13 |
| chr5_92867642_92867801 | 0.20 |
Shroom3 |
shroom family member 3 |
11250 |
0.22 |
| chr8_94172420_94173095 | 0.20 |
Mt2 |
metallothionein 2 |
93 |
0.88 |
| chr1_191421012_191421174 | 0.20 |
Gm2272 |
predicted gene 2272 |
13427 |
0.14 |
| chr9_59487560_59487714 | 0.20 |
Arih1 |
ariadne RBR E3 ubiquitin protein ligase 1 |
1019 |
0.47 |
| chr11_103689147_103689298 | 0.20 |
Gosr2 |
golgi SNAP receptor complex member 2 |
8472 |
0.13 |
| chr6_97962459_97962619 | 0.20 |
Mitf |
melanogenesis associated transcription factor |
21621 |
0.24 |
| chr15_76552804_76552973 | 0.20 |
Slc52a2 |
solute carrier protein 52, member 2 |
13924 |
0.06 |
| chr15_85626555_85626706 | 0.20 |
Gm49539 |
predicted gene, 49539 |
19453 |
0.14 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:1903223 | positive regulation of oxidative stress-induced neuron death(GO:1903223) |
| 0.1 | 0.1 | GO:0021886 | hypothalamus gonadotrophin-releasing hormone neuron differentiation(GO:0021886) hypothalamus gonadotrophin-releasing hormone neuron development(GO:0021888) |
| 0.1 | 0.3 | GO:1903626 | positive regulation of apoptotic DNA fragmentation(GO:1902512) positive regulation of DNA catabolic process(GO:1903626) |
| 0.1 | 0.2 | GO:0060137 | maternal process involved in parturition(GO:0060137) |
| 0.1 | 0.2 | GO:2000857 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.1 | 0.1 | GO:0016095 | polyprenol catabolic process(GO:0016095) |
| 0.1 | 0.2 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.1 | 0.3 | GO:0009115 | xanthine catabolic process(GO:0009115) |
| 0.1 | 0.3 | GO:0052805 | histidine catabolic process(GO:0006548) imidazole-containing compound catabolic process(GO:0052805) |
| 0.0 | 0.1 | GO:0071502 | cellular response to temperature stimulus(GO:0071502) |
| 0.0 | 0.1 | GO:0002121 | inter-male aggressive behavior(GO:0002121) |
| 0.0 | 0.2 | GO:0021914 | negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021914) |
| 0.0 | 0.1 | GO:1902263 | apoptotic process involved in embryonic digit morphogenesis(GO:1902263) |
| 0.0 | 0.1 | GO:0034197 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.0 | 0.1 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.0 | 0.1 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.0 | 0.2 | GO:0097491 | trunk segmentation(GO:0035290) trunk neural crest cell migration(GO:0036484) ventral trunk neural crest cell migration(GO:0036486) sympathetic neuron projection extension(GO:0097490) sympathetic neuron projection guidance(GO:0097491) |
| 0.0 | 0.2 | GO:2000767 | positive regulation of cytoplasmic translation(GO:2000767) |
| 0.0 | 0.2 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 0.0 | 0.1 | GO:0061470 | T follicular helper cell differentiation(GO:0061470) |
| 0.0 | 0.2 | GO:0001712 | ectodermal cell fate commitment(GO:0001712) |
| 0.0 | 0.1 | GO:0045196 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.0 | 0.1 | GO:0046167 | glycerol-3-phosphate biosynthetic process(GO:0046167) |
| 0.0 | 0.1 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.0 | 0.1 | GO:0019747 | regulation of isoprenoid metabolic process(GO:0019747) |
| 0.0 | 0.1 | GO:0045897 | regulation of transcription during mitosis(GO:0045896) positive regulation of transcription during mitosis(GO:0045897) |
| 0.0 | 0.1 | GO:0018916 | nitrobenzene metabolic process(GO:0018916) |
| 0.0 | 0.1 | GO:2000721 | positive regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000721) |
| 0.0 | 0.1 | GO:0009093 | cysteine catabolic process(GO:0009093) L-cysteine catabolic process(GO:0019448) L-cysteine metabolic process(GO:0046439) |
| 0.0 | 0.1 | GO:0002432 | granuloma formation(GO:0002432) |
| 0.0 | 0.1 | GO:1990086 | lens fiber cell apoptotic process(GO:1990086) |
| 0.0 | 0.1 | GO:0015744 | succinate transport(GO:0015744) |
| 0.0 | 0.1 | GO:0018992 | germ-line sex determination(GO:0018992) |
| 0.0 | 0.1 | GO:1902564 | negative regulation of neutrophil activation(GO:1902564) |
| 0.0 | 0.1 | GO:0006714 | sesquiterpenoid metabolic process(GO:0006714) |
| 0.0 | 0.1 | GO:0045900 | negative regulation of translational elongation(GO:0045900) |
| 0.0 | 0.2 | GO:0071442 | positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.0 | 0.2 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.1 | GO:0070340 | detection of bacterial lipopeptide(GO:0070340) |
| 0.0 | 0.1 | GO:0003032 | detection of oxygen(GO:0003032) |
| 0.0 | 0.1 | GO:0072675 | osteoclast fusion(GO:0072675) |
| 0.0 | 0.1 | GO:2001170 | negative regulation of ATP biosynthetic process(GO:2001170) |
| 0.0 | 0.2 | GO:0070544 | histone H3-K36 demethylation(GO:0070544) |
| 0.0 | 0.1 | GO:1902744 | negative regulation of lamellipodium organization(GO:1902744) |
| 0.0 | 0.1 | GO:0046341 | CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.0 | 0.1 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.0 | 0.6 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.0 | 0.1 | GO:1902730 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.0 | 0.2 | GO:0034154 | toll-like receptor 7 signaling pathway(GO:0034154) |
| 0.0 | 0.1 | GO:0006362 | transcription elongation from RNA polymerase I promoter(GO:0006362) |
| 0.0 | 0.1 | GO:0043321 | regulation of natural killer cell degranulation(GO:0043321) |
| 0.0 | 0.1 | GO:1901679 | nucleotide transmembrane transport(GO:1901679) |
| 0.0 | 0.1 | GO:0015760 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.0 | 0.2 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.1 | GO:1990169 | detoxification of copper ion(GO:0010273) stress response to copper ion(GO:1990169) |
| 0.0 | 0.0 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.0 | 0.1 | GO:0045404 | positive regulation of interleukin-4 biosynthetic process(GO:0045404) |
| 0.0 | 0.1 | GO:0031443 | fast-twitch skeletal muscle fiber contraction(GO:0031443) |
| 0.0 | 0.1 | GO:0048295 | positive regulation of isotype switching to IgE isotypes(GO:0048295) |
| 0.0 | 0.0 | GO:0016259 | selenocysteine metabolic process(GO:0016259) |
| 0.0 | 0.2 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.0 | 0.1 | GO:0090210 | regulation of establishment of blood-brain barrier(GO:0090210) |
| 0.0 | 0.1 | GO:0010748 | negative regulation of plasma membrane long-chain fatty acid transport(GO:0010748) |
| 0.0 | 0.1 | GO:2000418 | positive regulation of eosinophil migration(GO:2000418) |
| 0.0 | 0.1 | GO:0009212 | dTTP biosynthetic process(GO:0006235) pyrimidine deoxyribonucleoside triphosphate biosynthetic process(GO:0009212) dTTP metabolic process(GO:0046075) |
| 0.0 | 0.1 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.0 | 0.1 | GO:2000468 | regulation of peroxidase activity(GO:2000468) |
| 0.0 | 0.2 | GO:0032933 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.0 | 0.1 | GO:0032792 | negative regulation of CREB transcription factor activity(GO:0032792) |
| 0.0 | 0.1 | GO:0070417 | cellular response to cold(GO:0070417) |
| 0.0 | 0.1 | GO:0019695 | choline metabolic process(GO:0019695) |
| 0.0 | 0.0 | GO:2000598 | regulation of cyclin catabolic process(GO:2000598) negative regulation of cyclin catabolic process(GO:2000599) |
| 0.0 | 0.0 | GO:0086073 | bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 0.0 | 0.1 | GO:0010886 | positive regulation of cholesterol storage(GO:0010886) |
| 0.0 | 0.1 | GO:0034093 | positive regulation of maintenance of sister chromatid cohesion(GO:0034093) positive regulation of maintenance of mitotic sister chromatid cohesion(GO:0034184) |
| 0.0 | 0.1 | GO:0060057 | apoptotic process involved in mammary gland involution(GO:0060057) positive regulation of apoptotic process involved in mammary gland involution(GO:0060058) positive regulation of apoptotic process involved in morphogenesis(GO:1902339) regulation of mammary gland involution(GO:1903519) positive regulation of mammary gland involution(GO:1903521) positive regulation of apoptotic process involved in development(GO:1904747) |
| 0.0 | 0.1 | GO:0035630 | bone mineralization involved in bone maturation(GO:0035630) |
| 0.0 | 0.1 | GO:1904417 | regulation of xenophagy(GO:1904415) positive regulation of xenophagy(GO:1904417) |
| 0.0 | 0.0 | GO:0002085 | inhibition of neuroepithelial cell differentiation(GO:0002085) negative regulation of auditory receptor cell differentiation(GO:0045608) |
| 0.0 | 0.1 | GO:0015886 | heme transport(GO:0015886) |
| 0.0 | 0.0 | GO:0071225 | cellular response to muramyl dipeptide(GO:0071225) |
| 0.0 | 0.1 | GO:0030382 | sperm mitochondrion organization(GO:0030382) |
| 0.0 | 0.1 | GO:0002414 | immunoglobulin transcytosis in epithelial cells(GO:0002414) |
| 0.0 | 0.0 | GO:0045085 | negative regulation of interleukin-2 biosynthetic process(GO:0045085) |
| 0.0 | 0.1 | GO:0006361 | transcription initiation from RNA polymerase I promoter(GO:0006361) |
| 0.0 | 0.1 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.1 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.0 | 0.2 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.0 | 0.1 | GO:0051189 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) molybdopterin cofactor biosynthetic process(GO:0032324) molybdopterin cofactor metabolic process(GO:0043545) prosthetic group metabolic process(GO:0051189) |
| 0.0 | 0.3 | GO:0035278 | miRNA mediated inhibition of translation(GO:0035278) |
| 0.0 | 0.0 | GO:0036493 | positive regulation of translation in response to endoplasmic reticulum stress(GO:0036493) |
| 0.0 | 0.1 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.0 | 0.1 | GO:0014045 | establishment of endothelial blood-brain barrier(GO:0014045) |
| 0.0 | 0.1 | GO:1990705 | cholangiocyte proliferation(GO:1990705) |
| 0.0 | 0.1 | GO:0032911 | negative regulation of transforming growth factor beta1 production(GO:0032911) |
| 0.0 | 0.0 | GO:0097296 | activation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097296) |
| 0.0 | 0.0 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.0 | 0.1 | GO:0006287 | base-excision repair, gap-filling(GO:0006287) |
| 0.0 | 0.1 | GO:0051387 | negative regulation of neurotrophin TRK receptor signaling pathway(GO:0051387) |
| 0.0 | 0.1 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.0 | 0.1 | GO:0018879 | biphenyl metabolic process(GO:0018879) |
| 0.0 | 0.1 | GO:1902510 | regulation of apoptotic DNA fragmentation(GO:1902510) |
| 0.0 | 0.1 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.0 | 0.1 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.0 | 0.1 | GO:2000850 | negative regulation of steroid hormone secretion(GO:2000832) negative regulation of corticosteroid hormone secretion(GO:2000847) negative regulation of glucocorticoid secretion(GO:2000850) |
| 0.0 | 0.1 | GO:0010424 | DNA methylation on cytosine within a CG sequence(GO:0010424) DNA methylation on cytosine(GO:0032776) |
| 0.0 | 0.1 | GO:0072718 | response to cisplatin(GO:0072718) |
| 0.0 | 0.1 | GO:0042732 | D-xylose metabolic process(GO:0042732) |
| 0.0 | 0.1 | GO:0006742 | NADP catabolic process(GO:0006742) |
| 0.0 | 0.1 | GO:0098902 | regulation of membrane depolarization during action potential(GO:0098902) |
| 0.0 | 0.1 | GO:0050812 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) regulation of acyl-CoA biosynthetic process(GO:0050812) |
| 0.0 | 0.1 | GO:0035733 | hepatic stellate cell activation(GO:0035733) regulation of hepatic stellate cell activation(GO:2000489) |
| 0.0 | 0.1 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 0.0 | 0.0 | GO:1902548 | negative regulation of cellular response to vascular endothelial growth factor stimulus(GO:1902548) |
| 0.0 | 0.0 | GO:0021593 | rhombomere morphogenesis(GO:0021593) |
| 0.0 | 0.1 | GO:0048170 | positive regulation of long-term neuronal synaptic plasticity(GO:0048170) |
| 0.0 | 0.0 | GO:0043382 | positive regulation of memory T cell differentiation(GO:0043382) |
| 0.0 | 0.1 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.0 | 0.0 | GO:0002835 | negative regulation of response to tumor cell(GO:0002835) negative regulation of immune response to tumor cell(GO:0002838) |
| 0.0 | 0.1 | GO:0010940 | positive regulation of necrotic cell death(GO:0010940) |
| 0.0 | 0.0 | GO:0072061 | inner medullary collecting duct development(GO:0072061) |
| 0.0 | 0.0 | GO:0009436 | glyoxylate catabolic process(GO:0009436) |
| 0.0 | 0.1 | GO:0072262 | metanephric glomerular mesangium development(GO:0072223) metanephric glomerular mesangial cell proliferation involved in metanephros development(GO:0072262) |
| 0.0 | 0.0 | GO:0065001 | specification of axis polarity(GO:0065001) |
| 0.0 | 0.1 | GO:0009957 | epidermal cell fate specification(GO:0009957) |
| 0.0 | 0.1 | GO:0001842 | neural fold formation(GO:0001842) |
| 0.0 | 0.1 | GO:0002291 | T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:0002291) |
| 0.0 | 0.0 | GO:1903849 | regulation of aorta morphogenesis(GO:1903847) positive regulation of aorta morphogenesis(GO:1903849) |
| 0.0 | 0.0 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.0 | 0.2 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.0 | 0.1 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.0 | 0.1 | GO:2000059 | negative regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:2000059) |
| 0.0 | 0.0 | GO:1905065 | positive regulation of vascular smooth muscle cell differentiation(GO:1905065) |
| 0.0 | 0.0 | GO:0045590 | negative regulation of regulatory T cell differentiation(GO:0045590) |
| 0.0 | 0.1 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 | 0.0 | GO:1904502 | regulation of lipophagy(GO:1904502) positive regulation of lipophagy(GO:1904504) |
| 0.0 | 0.0 | GO:1903279 | regulation of calcium:sodium antiporter activity(GO:1903279) |
| 0.0 | 0.0 | GO:0002086 | diaphragm contraction(GO:0002086) |
| 0.0 | 0.1 | GO:0051365 | cellular response to potassium ion starvation(GO:0051365) |
| 0.0 | 0.1 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.0 | 0.0 | GO:0097503 | sialylation(GO:0097503) |
| 0.0 | 0.0 | GO:1903238 | positive regulation of leukocyte tethering or rolling(GO:1903238) |
| 0.0 | 0.0 | GO:0006216 | cytidine catabolic process(GO:0006216) cytidine deamination(GO:0009972) cytidine metabolic process(GO:0046087) |
| 0.0 | 0.0 | GO:0007403 | glial cell fate determination(GO:0007403) |
| 0.0 | 0.0 | GO:0003347 | epicardial cell to mesenchymal cell transition(GO:0003347) |
| 0.0 | 0.1 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 | 0.0 | GO:0006551 | leucine metabolic process(GO:0006551) |
| 0.0 | 0.1 | GO:0002934 | desmosome organization(GO:0002934) |
| 0.0 | 0.0 | GO:0006393 | termination of mitochondrial transcription(GO:0006393) |
| 0.0 | 0.0 | GO:0006624 | vacuolar protein processing(GO:0006624) |
| 0.0 | 0.0 | GO:2000645 | negative regulation of receptor catabolic process(GO:2000645) |
| 0.0 | 0.1 | GO:0003241 | growth involved in heart morphogenesis(GO:0003241) |
| 0.0 | 0.1 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.0 | 0.1 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.0 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.0 | GO:0002901 | mature B cell apoptotic process(GO:0002901) regulation of mature B cell apoptotic process(GO:0002905) negative regulation of mature B cell apoptotic process(GO:0002906) |
| 0.0 | 0.1 | GO:2000169 | regulation of peptidyl-cysteine S-nitrosylation(GO:2000169) |
| 0.0 | 0.1 | GO:0046102 | inosine metabolic process(GO:0046102) |
| 0.0 | 0.0 | GO:0045472 | response to ether(GO:0045472) |
| 0.0 | 0.1 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.0 | 0.0 | GO:0007227 | signal transduction downstream of smoothened(GO:0007227) positive regulation of hh target transcription factor activity(GO:0007228) |
| 0.0 | 0.1 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 0.0 | GO:0045794 | negative regulation of cell volume(GO:0045794) |
| 0.0 | 0.1 | GO:0019852 | L-ascorbic acid metabolic process(GO:0019852) |
| 0.0 | 0.0 | GO:2000504 | positive regulation of blood vessel remodeling(GO:2000504) |
| 0.0 | 0.0 | GO:0051572 | negative regulation of histone H3-K4 methylation(GO:0051572) |
| 0.0 | 0.2 | GO:0007190 | activation of adenylate cyclase activity(GO:0007190) |
| 0.0 | 0.0 | GO:0071651 | positive regulation of chemokine (C-C motif) ligand 5 production(GO:0071651) |
| 0.0 | 0.0 | GO:0006933 | negative regulation of cell adhesion involved in substrate-bound cell migration(GO:0006933) |
| 0.0 | 0.2 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 0.0 | GO:0061687 | detoxification of inorganic compound(GO:0061687) |
| 0.0 | 0.1 | GO:0060298 | positive regulation of sarcomere organization(GO:0060298) |
| 0.0 | 0.0 | GO:0086042 | cardiac muscle cell-cardiac muscle cell adhesion(GO:0086042) |
| 0.0 | 0.0 | GO:0006481 | C-terminal protein methylation(GO:0006481) |
| 0.0 | 0.1 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 | 0.0 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.0 | 0.2 | GO:0043518 | negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) |
| 0.0 | 0.0 | GO:0036015 | response to interleukin-3(GO:0036015) cellular response to interleukin-3(GO:0036016) |
| 0.0 | 0.1 | GO:0042983 | amyloid precursor protein biosynthetic process(GO:0042983) regulation of amyloid precursor protein biosynthetic process(GO:0042984) |
| 0.0 | 0.1 | GO:0071267 | amino acid salvage(GO:0043102) L-methionine biosynthetic process(GO:0071265) L-methionine salvage(GO:0071267) |
| 0.0 | 0.0 | GO:0006285 | base-excision repair, AP site formation(GO:0006285) |
| 0.0 | 0.0 | GO:0060061 | Spemann organizer formation(GO:0060061) |
| 0.0 | 0.0 | GO:0034238 | macrophage fusion(GO:0034238) regulation of macrophage fusion(GO:0034239) |
| 0.0 | 0.0 | GO:2000321 | positive regulation of T-helper 17 cell differentiation(GO:2000321) |
| 0.0 | 0.0 | GO:1904479 | negative regulation of intestinal absorption(GO:1904479) |
| 0.0 | 0.0 | GO:1904707 | positive regulation of vascular smooth muscle cell proliferation(GO:1904707) |
| 0.0 | 0.1 | GO:0070561 | vitamin D receptor signaling pathway(GO:0070561) |
| 0.0 | 0.1 | GO:0060670 | branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 0.0 | 0.0 | GO:0071205 | protein localization to juxtaparanode region of axon(GO:0071205) |
| 0.0 | 0.0 | GO:0006578 | amino-acid betaine biosynthetic process(GO:0006578) |
| 0.0 | 0.1 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.0 | 0.0 | GO:0061158 | 3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.0 | 0.0 | GO:0019064 | fusion of virus membrane with host plasma membrane(GO:0019064) membrane fusion involved in viral entry into host cell(GO:0039663) multi-organism membrane fusion(GO:0044800) |
| 0.0 | 0.0 | GO:0061038 | uterus morphogenesis(GO:0061038) |
| 0.0 | 0.1 | GO:2000676 | positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.0 | 0.0 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.0 | 0.1 | GO:2001223 | negative regulation of neuron migration(GO:2001223) |
| 0.0 | 0.1 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 | 0.1 | GO:0031547 | brain-derived neurotrophic factor receptor signaling pathway(GO:0031547) |
| 0.0 | 0.0 | GO:0050713 | negative regulation of interleukin-1 beta secretion(GO:0050713) |
| 0.0 | 0.0 | GO:0034392 | negative regulation of smooth muscle cell apoptotic process(GO:0034392) |
| 0.0 | 0.1 | GO:0060484 | lung-associated mesenchyme development(GO:0060484) |
| 0.0 | 0.1 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.0 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.0 | 0.1 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.0 | 0.0 | GO:0048023 | positive regulation of melanin biosynthetic process(GO:0048023) positive regulation of secondary metabolite biosynthetic process(GO:1900378) |
| 0.0 | 0.0 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.0 | 0.0 | GO:0032348 | negative regulation of aldosterone metabolic process(GO:0032345) negative regulation of aldosterone biosynthetic process(GO:0032348) negative regulation of cortisol biosynthetic process(GO:2000065) |
| 0.0 | 0.0 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.0 | 0.0 | GO:0070782 | phosphatidylserine exposure on apoptotic cell surface(GO:0070782) |
| 0.0 | 0.0 | GO:0009794 | regulation of mitotic cell cycle, embryonic(GO:0009794) mitotic cell cycle, embryonic(GO:0045448) |
| 0.0 | 0.1 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.0 | 0.0 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.0 | 0.0 | GO:0032817 | regulation of natural killer cell proliferation(GO:0032817) positive regulation of natural killer cell proliferation(GO:0032819) |
| 0.0 | 0.0 | GO:0006553 | lysine metabolic process(GO:0006553) |
| 0.0 | 0.1 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.0 | 0.0 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.0 | 0.1 | GO:0003105 | negative regulation of glomerular filtration(GO:0003105) |
| 0.0 | 0.0 | GO:0002378 | immunoglobulin biosynthetic process(GO:0002378) |
| 0.0 | 0.0 | GO:0031508 | pericentric heterochromatin assembly(GO:0031508) |
| 0.0 | 0.1 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.0 | 0.0 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.0 | GO:0071725 | response to triacyl bacterial lipopeptide(GO:0071725) cellular response to triacyl bacterial lipopeptide(GO:0071727) |
| 0.0 | 0.0 | GO:0051599 | response to hydrostatic pressure(GO:0051599) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0036454 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) insulin-like growth factor ternary complex(GO:0042567) |
| 0.1 | 0.2 | GO:0097057 | TRAF2-GSTP1 complex(GO:0097057) |
| 0.0 | 0.1 | GO:0035838 | growing cell tip(GO:0035838) |
| 0.0 | 0.1 | GO:0035355 | Toll-like receptor 2-Toll-like receptor 6 protein complex(GO:0035355) |
| 0.0 | 0.1 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.0 | 0.2 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.1 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.0 | 0.1 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.1 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.0 | 0.1 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.0 | 0.1 | GO:1990462 | omegasome(GO:1990462) |
| 0.0 | 0.1 | GO:0005606 | laminin-1 complex(GO:0005606) |
| 0.0 | 0.2 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.2 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 0.1 | GO:0036488 | CHOP-C/EBP complex(GO:0036488) |
| 0.0 | 0.1 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.0 | 0.1 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 0.1 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.0 | 0.2 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.0 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.0 | 0.1 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.0 | 0.3 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.1 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.0 | 0.1 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.0 | 0.1 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.1 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.0 | 0.2 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.1 | GO:0005767 | secondary lysosome(GO:0005767) |
| 0.0 | 0.1 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.0 | 0.1 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.0 | GO:0031523 | Myb complex(GO:0031523) |
| 0.0 | 0.2 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.1 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.0 | 0.0 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 0.1 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.1 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.0 | 0.1 | GO:0005587 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.0 | 0.0 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 0.0 | GO:0097451 | glial limiting end-foot(GO:0097451) |
| 0.0 | 0.1 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.0 | 0.0 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.0 | 0.0 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.0 | 0.1 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 0.0 | GO:1990597 | AIP1-IRE1 complex(GO:1990597) |
| 0.0 | 0.1 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 0.1 | GO:0005683 | U7 snRNP(GO:0005683) |
| 0.0 | 0.1 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.0 | 0.1 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 0.0 | GO:0009279 | cell outer membrane(GO:0009279) cell envelope(GO:0030313) external encapsulating structure part(GO:0044462) |
| 0.0 | 0.0 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.0 | 0.0 | GO:0035363 | histone locus body(GO:0035363) |
| 0.0 | 0.0 | GO:0005745 | m-AAA complex(GO:0005745) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.1 | 0.3 | GO:0030151 | molybdenum ion binding(GO:0030151) |
| 0.1 | 0.2 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.1 | 0.3 | GO:0008390 | testosterone 16-alpha-hydroxylase activity(GO:0008390) |
| 0.1 | 0.2 | GO:0003941 | L-serine ammonia-lyase activity(GO:0003941) |
| 0.1 | 0.3 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.1 | 0.2 | GO:0097603 | temperature-gated ion channel activity(GO:0097603) |
| 0.1 | 0.2 | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0003829) |
| 0.0 | 0.1 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.0 | 0.0 | GO:0043546 | molybdopterin cofactor binding(GO:0043546) |
| 0.0 | 0.2 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.0 | 0.0 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.0 | 0.2 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.2 | GO:0071723 | lipopeptide binding(GO:0071723) |
| 0.0 | 0.1 | GO:0035651 | AP-3 adaptor complex binding(GO:0035651) |
| 0.0 | 0.1 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.1 | GO:0017153 | sodium:dicarboxylate symporter activity(GO:0017153) |
| 0.0 | 0.1 | GO:0004104 | cholinesterase activity(GO:0004104) |
| 0.0 | 0.2 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.1 | GO:0015526 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.0 | 0.1 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) |
| 0.0 | 0.1 | GO:0047751 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) cholestenone 5-alpha-reductase activity(GO:0047751) |
| 0.0 | 0.1 | GO:0004370 | glycerol kinase activity(GO:0004370) |
| 0.0 | 0.2 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.3 | GO:0038187 | signaling pattern recognition receptor activity(GO:0008329) pattern recognition receptor activity(GO:0038187) |
| 0.0 | 0.1 | GO:0050051 | leukotriene-B4 20-monooxygenase activity(GO:0050051) |
| 0.0 | 0.1 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 0.0 | 0.1 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) DNA (cytosine-5-)-methyltransferase activity, acting on CpG substrates(GO:0051718) |
| 0.0 | 0.1 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.0 | 0.1 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.1 | GO:0071535 | RING-like zinc finger domain binding(GO:0071535) |
| 0.0 | 0.1 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.0 | 0.4 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.1 | GO:0032139 | dinucleotide insertion or deletion binding(GO:0032139) |
| 0.0 | 0.2 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.0 | 0.1 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.0 | 0.3 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.1 | GO:0033592 | RNA strand annealing activity(GO:0033592) |
| 0.0 | 0.1 | GO:0050309 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.0 | 0.1 | GO:0004030 | aldehyde dehydrogenase [NAD(P)+] activity(GO:0004030) |
| 0.0 | 0.1 | GO:0002094 | polyprenyltransferase activity(GO:0002094) |
| 0.0 | 0.1 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 0.0 | GO:0016726 | xanthine dehydrogenase activity(GO:0004854) oxidoreductase activity, acting on CH or CH2 groups, NAD or NADP as acceptor(GO:0016726) |
| 0.0 | 0.1 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.0 | 0.1 | GO:0008898 | S-adenosylmethionine-homocysteine S-methyltransferase activity(GO:0008898) |
| 0.0 | 0.2 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.2 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.1 | GO:0004165 | dodecenoyl-CoA delta-isomerase activity(GO:0004165) |
| 0.0 | 0.0 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.0 | 0.1 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 0.1 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.0 | 0.1 | GO:0030984 | kininogen binding(GO:0030984) |
| 0.0 | 0.1 | GO:0004174 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.0 | 0.1 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.0 | 0.1 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.0 | 0.1 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) |
| 0.0 | 0.1 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 0.1 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.1 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.0 | 0.1 | GO:0050649 | testosterone 6-beta-hydroxylase activity(GO:0050649) |
| 0.0 | 0.1 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.2 | GO:0001013 | RNA polymerase I regulatory region DNA binding(GO:0001013) |
| 0.0 | 0.0 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.0 | 0.0 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.0 | 0.1 | GO:0047391 | alkylglycerophosphoethanolamine phosphodiesterase activity(GO:0047391) |
| 0.0 | 0.0 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.0 | 0.1 | GO:0098518 | polynucleotide phosphatase activity(GO:0098518) |
| 0.0 | 0.1 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.2 | GO:1900750 | glutathione binding(GO:0043295) oligopeptide binding(GO:1900750) |
| 0.0 | 0.0 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) |
| 0.0 | 0.1 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 0.0 | GO:0071209 | U7 snRNA binding(GO:0071209) |
| 0.0 | 0.0 | GO:0032767 | copper-dependent protein binding(GO:0032767) |
| 0.0 | 0.1 | GO:0003945 | N-acetyllactosamine synthase activity(GO:0003945) |
| 0.0 | 0.1 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.0 | 0.0 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 0.1 | GO:0004457 | lactate dehydrogenase activity(GO:0004457) |
| 0.0 | 0.1 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 0.0 | GO:0015216 | adenine nucleotide transmembrane transporter activity(GO:0000295) purine ribonucleotide transmembrane transporter activity(GO:0005346) purine nucleotide transmembrane transporter activity(GO:0015216) |
| 0.0 | 0.0 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.0 | 0.1 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.1 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.0 | 0.1 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.1 | GO:0035197 | siRNA binding(GO:0035197) |
| 0.0 | 0.1 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.0 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.0 | 0.1 | GO:0018585 | fluorene oxygenase activity(GO:0018585) |
| 0.0 | 0.1 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.0 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.0 | 0.1 | GO:0005168 | neurotrophin TRKA receptor binding(GO:0005168) |
| 0.0 | 0.1 | GO:0019763 | immunoglobulin receptor activity(GO:0019763) |
| 0.0 | 0.0 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.0 | 0.2 | GO:0016918 | retinal binding(GO:0016918) |
| 0.0 | 0.1 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.0 | 0.1 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.0 | GO:0004661 | protein geranylgeranyltransferase activity(GO:0004661) |
| 0.0 | 0.1 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.0 | 0.1 | GO:0016803 | ether hydrolase activity(GO:0016803) |
| 0.0 | 0.0 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 0.0 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 0.0 | 0.1 | GO:0016505 | peptidase activator activity involved in apoptotic process(GO:0016505) |
| 0.0 | 0.0 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.0 | 0.0 | GO:0015252 | hydrogen ion channel activity(GO:0015252) |
| 0.0 | 0.0 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.0 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 0.0 | 0.1 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) |
| 0.0 | 0.0 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.0 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.0 | 0.2 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.0 | GO:0008486 | diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) |
| 0.0 | 0.0 | GO:0034618 | arginine binding(GO:0034618) |
| 0.0 | 0.1 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.0 | 0.0 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
| 0.0 | 0.2 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.0 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 0.1 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 0.5 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.0 | 0.0 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.0 | 0.0 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 0.1 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.0 | 0.3 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.0 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.0 | 0.1 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.0 | 0.0 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.0 | 0.3 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.0 | 0.1 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.0 | PID TRAIL PATHWAY | TRAIL signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 0.1 | REACTOME RORA ACTIVATES CIRCADIAN EXPRESSION | Genes involved in RORA Activates Circadian Expression |
| 0.0 | 0.1 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.2 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.0 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.2 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.1 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.3 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 0.2 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.2 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.1 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.1 | REACTOME REGULATED PROTEOLYSIS OF P75NTR | Genes involved in Regulated proteolysis of p75NTR |
| 0.0 | 0.1 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 0.1 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.1 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 0.2 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.0 | 0.2 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.2 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.1 | REACTOME RESOLUTION OF AP SITES VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Resolution of AP sites via the single-nucleotide replacement pathway |