Project
ENCSR904DTN: DNase-seq of mouse liver in constant darkness
Navigation
Downloads
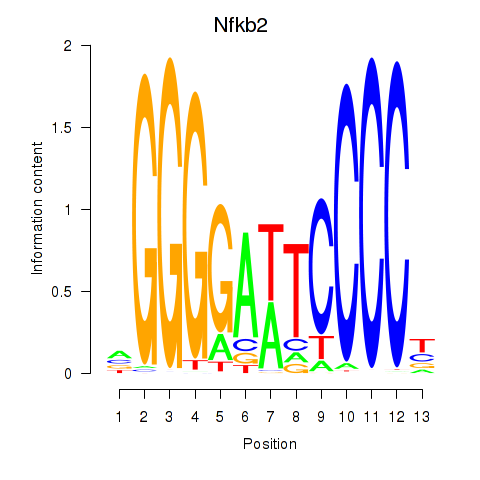
Results for Nfkb2
Z-value: 1.72
Transcription factors associated with Nfkb2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nfkb2
|
ENSMUSG00000025225.8 | nuclear factor of kappa light polypeptide gene enhancer in B cells 2, p49/p100 |
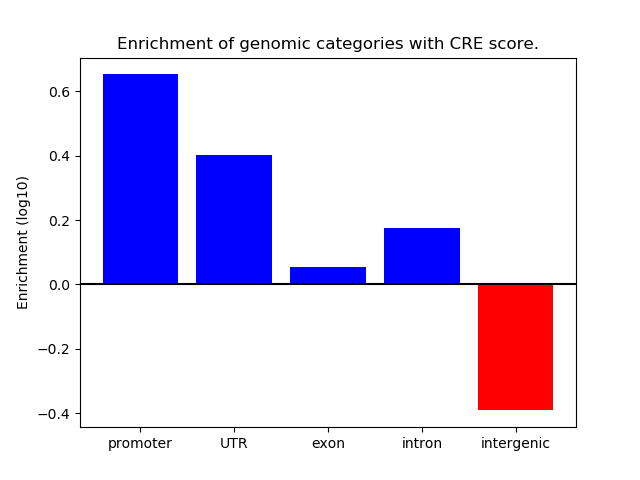
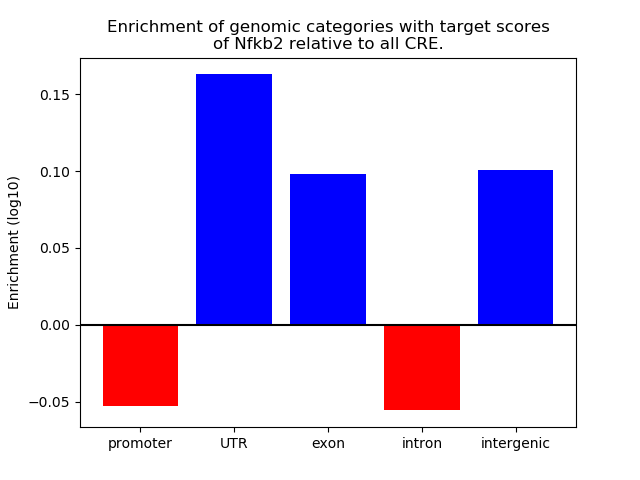
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr19_46304529_46304702 | Nfkb2 | 129 | 0.881438 | 0.66 | 1.5e-01 | Click! |
| chr19_46303966_46304121 | Nfkb2 | 277 | 0.782293 | -0.53 | 2.7e-01 | Click! |
| chr19_46305579_46305735 | Nfkb2 | 39 | 0.492610 | 0.35 | 5.0e-01 | Click! |
| chr19_46304773_46305233 | Nfkb2 | 203 | 0.758340 | -0.25 | 6.4e-01 | Click! |
| chr19_46304225_46304382 | Nfkb2 | 17 | 0.935385 | -0.13 | 8.1e-01 | Click! |
Activity of the Nfkb2 motif across conditions
Conditions sorted by the z-value of the Nfkb2 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
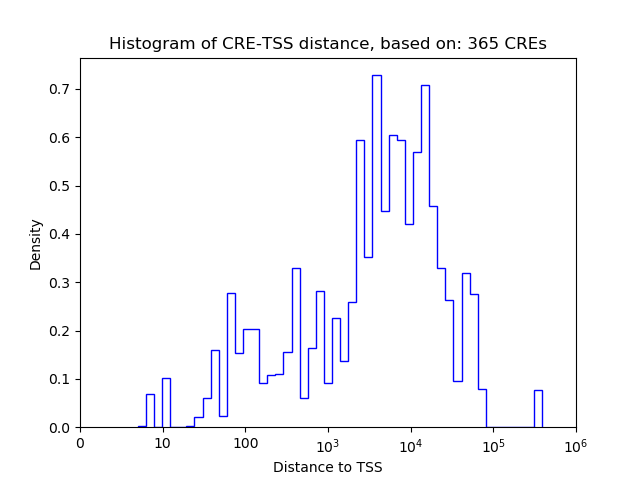
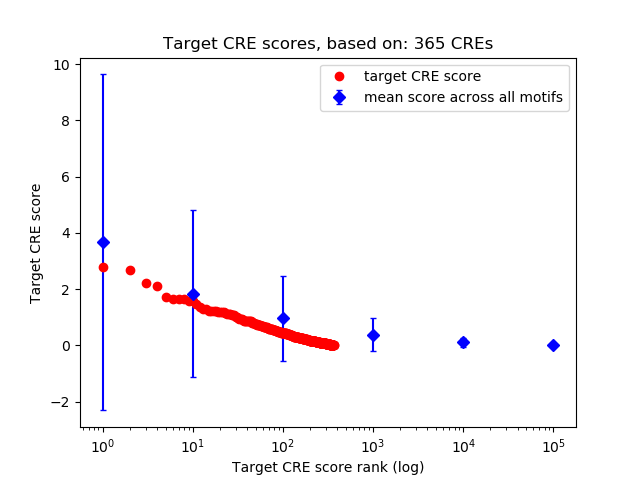
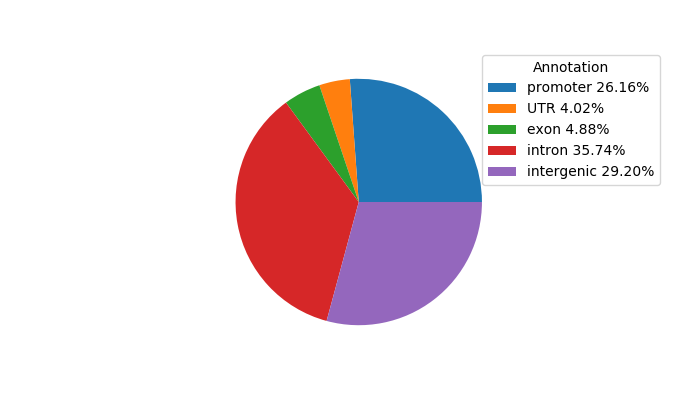
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr7_141628129_141628296 | 2.78 |
Mir7063 |
microRNA 7063 |
7511 |
0.13 |
| chr1_72829492_72829643 | 2.68 |
Igfbp2 |
insulin-like growth factor binding protein 2 |
4245 |
0.26 |
| chr18_65099329_65099521 | 2.21 |
Nedd4l |
neural precursor cell expressed, developmentally down-regulated gene 4-like |
16371 |
0.23 |
| chr6_87994497_87994648 | 2.10 |
4933412L11Rik |
RIKEN cDNA 4933412L11 gene |
393 |
0.71 |
| chr7_101302097_101302264 | 1.71 |
Atg16l2 |
autophagy related 16-like 2 (S. cerevisiae) |
71 |
0.95 |
| chr3_104796399_104796698 | 1.64 |
Rhoc |
ras homolog family member C |
4565 |
0.1 |
| chr10_81430090_81430281 | 1.64 |
Nfic |
nuclear factor I/C |
820 |
0.33 |
| chr17_29394251_29394433 | 1.63 |
Fgd2 |
FYVE, RhoGEF and PH domain containing 2 |
17841 |
0.11 |
| chr8_124343978_124344300 | 1.58 |
Gm15775 |
predicted gene 15775 |
12344 |
0.14 |
| chr12_71438664_71438829 | 1.56 |
4930404H11Rik |
RIKEN cDNA 4930404H11 gene |
49539 |
0.12 |
| chr9_21136037_21136188 | 1.45 |
Tyk2 |
tyrosine kinase 2 |
4869 |
0.1 |
| chr5_151413141_151413328 | 1.36 |
1700028E10Rik |
RIKEN cDNA 1700028E10 gene |
6708 |
0.17 |
| chr2_25095384_25095554 | 1.31 |
Noxa1 |
NADPH oxidase activator 1 |
320 |
0.75 |
| chr16_95728927_95729113 | 1.28 |
Ets2 |
E26 avian leukemia oncogene 2, 3' domain |
11286 |
0.18 |
| chr9_20804931_20805096 | 1.23 |
Col5a3 |
collagen, type V, alpha 3 |
10054 |
0.11 |
| chr7_80846015_80846166 | 1.23 |
Zscan2 |
zinc finger and SCAN domain containing 2 |
14830 |
0.13 |
| chr7_68709256_68709407 | 1.21 |
Gm44692 |
predicted gene 44692 |
17136 |
0.21 |
| chr8_70538049_70538211 | 1.20 |
Ell |
elongation factor RNA polymerase II |
1327 |
0.23 |
| chr5_139730884_139731070 | 1.19 |
Micall2 |
MICAL-like 2 |
1857 |
0.27 |
| chr7_19321614_19321765 | 1.18 |
Gm44698 |
predicted gene 44698 |
11440 |
0.07 |
| chr2_50964157_50964313 | 1.18 |
Gm13498 |
predicted gene 13498 |
54551 |
0.16 |
| chr3_83841601_83841954 | 1.17 |
Tlr2 |
toll-like receptor 2 |
10 |
0.98 |
| chr8_84786434_84786598 | 1.15 |
Nfix |
nuclear factor I/X |
12147 |
0.1 |
| chr12_110219862_110220027 | 1.12 |
Gm40576 |
predicted gene, 40576 |
16418 |
0.11 |
| chr13_93989307_93989458 | 1.11 |
Gm47216 |
predicted gene, 47216 |
2386 |
0.28 |
| chr17_23967060_23967351 | 1.11 |
Prss30 |
protease, serine 30 |
6542 |
0.07 |
| chr10_59441733_59442104 | 1.09 |
Oit3 |
oncoprotein induced transcript 3 |
140 |
0.96 |
| chr11_50213100_50213265 | 1.07 |
Mgat4b |
mannoside acetylglucosaminyltransferase 4, isoenzyme B |
2292 |
0.16 |
| chr5_8964466_8964675 | 1.07 |
Abcb4 |
ATP-binding cassette, sub-family B (MDR/TAP), member 4 |
6018 |
0.13 |
| chr9_5308705_5308876 | 1.01 |
Casp4 |
caspase 4, apoptosis-related cysteine peptidase |
38 |
0.98 |
| chr3_91483574_91483744 | 1.00 |
S100a7l2 |
S100 calcium binding protein A7 like 2 |
392856 |
0.01 |
| chr17_35218598_35218782 | 0.97 |
Gm16181 |
predicted gene 16181 |
2144 |
0.08 |
| chr12_21160412_21160576 | 0.94 |
Asap2 |
ArfGAP with SH3 domain, ankyrin repeat and PH domain 2 |
48540 |
0.12 |
| chr15_59728167_59728580 | 0.93 |
Gm20150 |
predicted gene, 20150 |
56260 |
0.12 |
| chr17_29423486_29423671 | 0.92 |
Gm36199 |
predicted gene, 36199 |
9257 |
0.12 |
| chr11_98766772_98767040 | 0.92 |
Nr1d1 |
nuclear receptor subfamily 1, group D, member 1 |
3520 |
0.12 |
| chr10_13966865_13967046 | 0.88 |
Hivep2 |
human immunodeficiency virus type I enhancer binding protein 2 |
6 |
0.98 |
| chr10_80050367_80050530 | 0.88 |
Gpx4 |
glutathione peroxidase 4 |
3040 |
0.12 |
| chr4_130829443_130829594 | 0.87 |
Gm26716 |
predicted gene, 26716 |
3205 |
0.16 |
| chr18_39800281_39800456 | 0.86 |
Pabpc2 |
poly(A) binding protein, cytoplasmic 2 |
26871 |
0.21 |
| chr8_36665428_36665609 | 0.86 |
Dlc1 |
deleted in liver cancer 1 |
51575 |
0.16 |
| chr4_57137427_57137593 | 0.86 |
Epb41l4b |
erythrocyte membrane protein band 4.1 like 4b |
682 |
0.73 |
| chr13_111490237_111490399 | 0.85 |
Gpbp1 |
GC-rich promoter binding protein 1 |
207 |
0.94 |
| chr14_51102195_51102364 | 0.85 |
Rnase4 |
ribonuclease, RNase A family 4 |
5920 |
0.08 |
| chr9_113749434_113749589 | 0.83 |
Clasp2 |
CLIP associating protein 2 |
7779 |
0.2 |
| chr9_114828742_114828949 | 0.80 |
Gm23889 |
predicted gene, 23889 |
9328 |
0.16 |
| chr6_115996180_115996399 | 0.79 |
Plxnd1 |
plexin D1 |
1284 |
0.4 |
| chr2_70870934_70871085 | 0.78 |
Gm13632 |
predicted gene 13632 |
6820 |
0.2 |
| chr18_60737126_60737516 | 0.77 |
Ndst1 |
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1 |
4171 |
0.18 |
| chr14_76698781_76698988 | 0.77 |
1700108F19Rik |
RIKEN cDNA 1700108F19 gene |
17130 |
0.2 |
| chr12_91668609_91668790 | 0.75 |
Gm16876 |
predicted gene, 16876 |
15966 |
0.15 |
| chr5_137482252_137482414 | 0.73 |
Epo |
erythropoietin |
3483 |
0.1 |
| chr2_129330671_129330856 | 0.73 |
Gm25703 |
predicted gene, 25703 |
15284 |
0.1 |
| chr2_69227808_69227986 | 0.73 |
G6pc2 |
glucose-6-phosphatase, catalytic, 2 |
7869 |
0.15 |
| chr19_5430069_5430231 | 0.72 |
AI837181 |
expressed sequence AI837181 |
4993 |
0.05 |
| chr7_90023157_90023457 | 0.72 |
E230029C05Rik |
RIKEN cDNA E230029C05 gene |
5852 |
0.14 |
| chr17_33978829_33978988 | 0.70 |
H2-K2 |
histocompatibility 2, K region locus 2 |
81 |
0.86 |
| chr2_128738093_128738244 | 0.69 |
Gm14011 |
predicted gene 14011 |
15537 |
0.15 |
| chr7_80635100_80635316 | 0.69 |
Gm15880 |
predicted gene 15880 |
809 |
0.58 |
| chr7_84212215_84212389 | 0.68 |
Gm44826 |
predicted gene 44826 |
1788 |
0.29 |
| chr7_46723990_46724178 | 0.66 |
Saa3 |
serum amyloid A 3 |
8384 |
0.09 |
| chr11_46077638_46077800 | 0.65 |
Adam19 |
a disintegrin and metallopeptidase domain 19 (meltrin beta) |
21661 |
0.11 |
| chr14_63899614_63899769 | 0.65 |
Pinx1 |
PIN2/TERF1 interacting, telomerase inhibitor 1 |
78 |
0.98 |
| chr1_82636436_82636587 | 0.64 |
n-R5s213 |
nuclear encoded rRNA 5S 213 |
1498 |
0.38 |
| chr14_8244058_8244210 | 0.64 |
Acox2 |
acyl-Coenzyme A oxidase 2, branched chain |
2269 |
0.3 |
| chr6_120579657_120579808 | 0.64 |
Gm44124 |
predicted gene, 44124 |
444 |
0.74 |
| chr4_133185342_133185538 | 0.63 |
Wasf2 |
WAS protein family, member 2 |
3578 |
0.16 |
| chr3_131463750_131463954 | 0.61 |
Sgms2 |
sphingomyelin synthase 2 |
26627 |
0.2 |
| chr8_84204114_84204277 | 0.60 |
Gm37352 |
predicted gene, 37352 |
3326 |
0.07 |
| chr2_158306303_158306481 | 0.59 |
Lbp |
lipopolysaccharide binding protein |
101 |
0.95 |
| chr1_59670213_59670380 | 0.58 |
Sumo1 |
small ubiquitin-like modifier 1 |
443 |
0.68 |
| chr15_22791886_22792091 | 0.58 |
Hnrnpa1l2-ps2 |
heterogeneous nuclear ribonucleoprotein A1-like 2, pseudogene 2 |
77158 |
0.12 |
| chr19_3359958_3360148 | 0.58 |
Cpt1a |
carnitine palmitoyltransferase 1a, liver |
6277 |
0.14 |
| chr4_133669920_133670071 | 0.57 |
Pigv |
phosphatidylinositol glycan anchor biosynthesis, class V |
2652 |
0.18 |
| chr10_77064998_77065227 | 0.57 |
Col18a1 |
collagen, type XVIII, alpha 1 |
5115 |
0.17 |
| chr15_100732715_100732889 | 0.57 |
I730030J21Rik |
RIKEN cDNA I730030J21 gene |
65 |
0.95 |
| chr3_36551644_36551818 | 0.56 |
Exosc9 |
exosome component 9 |
875 |
0.45 |
| chr7_74647398_74647580 | 0.55 |
Gm7726 |
predicted gene 7726 |
52367 |
0.15 |
| chrX_169989673_169989848 | 0.54 |
Gm15247 |
predicted gene 15247 |
2821 |
0.2 |
| chr11_50213555_50213755 | 0.54 |
Mgat4b |
mannoside acetylglucosaminyltransferase 4, isoenzyme B |
2765 |
0.14 |
| chrY_90763950_90764120 | 0.54 |
Gm21860 |
predicted gene, 21860 |
8568 |
0.17 |
| chr19_47330818_47330996 | 0.53 |
Sh3pxd2a |
SH3 and PX domains 2A |
16156 |
0.18 |
| chr10_23842034_23842208 | 0.53 |
Vnn3 |
vanin 3 |
9341 |
0.12 |
| chr6_99375099_99375287 | 0.52 |
Gm20705 |
predicted gene 20705 |
13306 |
0.23 |
| chr9_44734580_44734731 | 0.52 |
Phldb1 |
pleckstrin homology like domain, family B, member 1 |
543 |
0.54 |
| chr15_73668210_73668361 | 0.51 |
1700010B13Rik |
RIKEN cDNA 1700010B13 gene |
22411 |
0.14 |
| chr5_90891009_90891200 | 0.50 |
Cxcl1 |
chemokine (C-X-C motif) ligand 1 |
137 |
0.93 |
| chr1_43403479_43403632 | 0.50 |
Gm29041 |
predicted gene 29041 |
6029 |
0.23 |
| chr4_120924973_120925153 | 0.49 |
Exo5 |
exonuclease 5 |
46 |
0.96 |
| chr10_125930262_125930456 | 0.48 |
Lrig3 |
leucine-rich repeats and immunoglobulin-like domains 3 |
35809 |
0.22 |
| chr10_127621012_127621163 | 0.48 |
Lrp1 |
low density lipoprotein receptor-related protein 1 |
61 |
0.94 |
| chr8_64946983_64947186 | 0.47 |
Tmem192 |
transmembrane protein 192 |
101 |
0.88 |
| chr2_78868542_78868714 | 0.46 |
Ube2e3 |
ubiquitin-conjugating enzyme E2E 3 |
390 |
0.89 |
| chr8_91395852_91396020 | 0.46 |
Gm45427 |
predicted gene 45427 |
3991 |
0.18 |
| chr6_113237667_113237824 | 0.46 |
Mtmr14 |
myotubularin related protein 14 |
98 |
0.95 |
| chr9_70934414_70934936 | 0.45 |
Lipc |
lipase, hepatic |
62 |
0.98 |
| chr5_118294350_118294584 | 0.45 |
Gm25076 |
predicted gene, 25076 |
28018 |
0.15 |
| chr19_44898749_44898900 | 0.44 |
Slf2 |
SMC5-SMC6 complex localization factor 2 |
32327 |
0.11 |
| chr4_101301690_101302027 | 0.44 |
Gm12801 |
predicted gene 12801 |
17637 |
0.1 |
| chr1_166279710_166279874 | 0.44 |
Ildr2 |
immunoglobulin-like domain containing receptor 2 |
25599 |
0.14 |
| chr10_8955944_8956228 | 0.44 |
Gm48728 |
predicted gene, 48728 |
4137 |
0.24 |
| chr4_60419310_60419496 | 0.44 |
Mup9 |
major urinary protein 9 |
1181 |
0.4 |
| chrX_52988316_52988476 | 0.44 |
Hprt |
hypoxanthine guanine phosphoribosyl transferase |
259 |
0.87 |
| chr10_41489453_41489604 | 0.44 |
Smpd2 |
sphingomyelin phosphodiesterase 2, neutral |
151 |
0.88 |
| chr10_37231934_37232493 | 0.43 |
4930543K20Rik |
RIKEN cDNA 4930543K20 gene |
31535 |
0.2 |
| chr7_99441628_99441801 | 0.43 |
Gdpd5 |
glycerophosphodiester phosphodiesterase domain containing 5 |
206 |
0.91 |
| chr4_61436878_61437320 | 0.42 |
Mup15 |
major urinary protein 15 |
2644 |
0.27 |
| chr12_104247702_104247874 | 0.42 |
Serpina3h |
serine (or cysteine) peptidase inhibitor, clade A, member 3H |
117 |
0.93 |
| chr13_98595769_98595925 | 0.42 |
Gm4815 |
predicted gene 4815 |
17654 |
0.12 |
| chr7_38230449_38230651 | 0.42 |
Plekhf1 |
pleckstrin homology domain containing, family F (with FYVE domain) member 1 |
2534 |
0.2 |
| chr2_162991733_162991902 | 0.41 |
Sgk2 |
serum/glucocorticoid regulated kinase 2 |
4315 |
0.15 |
| chr4_61779591_61780024 | 0.41 |
Mup19 |
major urinary protein 19 |
2417 |
0.19 |
| chr12_25165797_25165968 | 0.41 |
Gm36287 |
predicted gene, 36287 |
14967 |
0.16 |
| chr5_36398668_36398855 | 0.41 |
Sorcs2 |
sortilin-related VPS10 domain containing receptor 2 |
622 |
0.74 |
| chr2_167375199_167375534 | 0.40 |
B4galt5 |
UDP-Gal:betaGlcNAc beta 1,4-galactosyltransferase, polypeptide 5 |
26183 |
0.15 |
| chr4_61301111_61301509 | 0.39 |
Mup14 |
major urinary protein 14 |
2055 |
0.32 |
| chr16_96944027_96944232 | 0.39 |
Gm32432 |
predicted gene, 32432 |
25372 |
0.26 |
| chr18_53279796_53279963 | 0.38 |
Snx24 |
sorting nexing 24 |
34110 |
0.18 |
| chr1_62750521_62750741 | 0.38 |
Gm29083 |
predicted gene 29083 |
32558 |
0.14 |
| chr11_98775849_98776027 | 0.38 |
Nr1d1 |
nuclear receptor subfamily 1, group D, member 1 |
605 |
0.56 |
| chr1_155533394_155533583 | 0.38 |
Gm5532 |
predicted gene 5532 |
6345 |
0.23 |
| chr14_57409253_57409404 | 0.36 |
Cryl1 |
crystallin, lambda 1 |
10799 |
0.13 |
| chr12_76555022_76555208 | 0.36 |
Plekhg3 |
pleckstrin homology domain containing, family G (with RhoGef domain) member 3 |
7123 |
0.14 |
| chr5_150595505_150595664 | 0.36 |
N4bp2l1 |
NEDD4 binding protein 2-like 1 |
1056 |
0.31 |
| chr9_116091537_116091709 | 0.35 |
Gm9385 |
predicted pseudogene 9385 |
50559 |
0.13 |
| chr7_28593760_28594110 | 0.33 |
Pak4 |
p21 (RAC1) activated kinase 4 |
4151 |
0.1 |
| chr2_52930153_52930305 | 0.33 |
Fmnl2 |
formin-like 2 |
72361 |
0.12 |
| chr11_59506517_59506712 | 0.33 |
Zkscan17 |
zinc finger with KRAB and SCAN domains 17 |
30 |
0.51 |
| chr8_83748782_83748958 | 0.32 |
Gm45778 |
predicted gene 45778 |
634 |
0.6 |
| chr4_47251261_47251412 | 0.32 |
Col15a1 |
collagen, type XV, alpha 1 |
7199 |
0.22 |
| chr17_31828262_31828413 | 0.32 |
Sik1 |
salt inducible kinase 1 |
23238 |
0.14 |
| chr15_31367525_31367697 | 0.31 |
Ankrd33b |
ankyrin repeat domain 33B |
43 |
0.93 |
| chr13_52709679_52709876 | 0.31 |
BB123696 |
expressed sequence BB123696 |
25831 |
0.24 |
| chr12_104262958_104263126 | 0.31 |
Serpina3i |
serine (or cysteine) peptidase inhibitor, clade A, member 3I |
80 |
0.94 |
| chr4_86874228_86874398 | 0.31 |
Acer2 |
alkaline ceramidase 2 |
83 |
0.97 |
| chr4_61225558_61225753 | 0.31 |
Mup13 |
major urinary protein 13 |
2616 |
0.28 |
| chr8_127196588_127196779 | 0.30 |
Pard3 |
par-3 family cell polarity regulator |
25279 |
0.24 |
| chr10_128458359_128458657 | 0.30 |
Mir8105 |
microRNA 8105 |
172 |
0.8 |
| chr5_149045840_149046001 | 0.30 |
Hmgb1 |
high mobility group box 1 |
4527 |
0.13 |
| chr3_107775838_107775989 | 0.29 |
Gm43233 |
predicted gene 43233 |
5943 |
0.15 |
| chr4_60067672_60068037 | 0.29 |
Mup7 |
major urinary protein 7 |
2557 |
0.26 |
| chrY_90763569_90763747 | 0.29 |
Gm21860 |
predicted gene, 21860 |
8191 |
0.18 |
| chrX_169989293_169989480 | 0.28 |
Gm15247 |
predicted gene 15247 |
2447 |
0.22 |
| chr1_120105769_120105951 | 0.28 |
Dbi |
diazepam binding inhibitor |
14278 |
0.16 |
| chr8_123064834_123064990 | 0.28 |
Spg7 |
SPG7, paraplegin matrix AAA peptidase subunit |
267 |
0.84 |
| chr9_24896994_24897192 | 0.28 |
Gm48255 |
predicted gene, 48255 |
55880 |
0.09 |
| chr11_106514783_106514945 | 0.28 |
Gm22711 |
predicted gene, 22711 |
13619 |
0.14 |
| chr17_29395434_29395615 | 0.28 |
Fgd2 |
FYVE, RhoGEF and PH domain containing 2 |
19023 |
0.11 |
| chr10_19087108_19087313 | 0.28 |
1700124M09Rik |
RIKEN cDNA 1700124M09 gene |
32 |
0.97 |
| chr2_26483564_26483901 | 0.28 |
Notch1 |
notch 1 |
13234 |
0.1 |
| chr13_34627445_34627664 | 0.27 |
Gm47127 |
predicted gene, 47127 |
15308 |
0.13 |
| chr7_45053037_45053201 | 0.27 |
Prr12 |
proline rich 12 |
238 |
0.75 |
| chr9_123112837_123113034 | 0.27 |
Zdhhc3 |
zinc finger, DHHC domain containing 3 |
103 |
0.72 |
| chr15_66814050_66814231 | 0.27 |
Sla |
src-like adaptor |
1547 |
0.4 |
| chr2_104688429_104688597 | 0.26 |
Tcp11l1 |
t-complex 11 like 1 |
4247 |
0.17 |
| chr4_134813997_134814173 | 0.26 |
Maco1 |
macoilin 1 |
2872 |
0.26 |
| chr4_120283294_120283461 | 0.26 |
Foxo6 |
forkhead box O6 |
3972 |
0.25 |
| chr3_149326826_149327020 | 0.26 |
Gm30382 |
predicted gene, 30382 |
3049 |
0.25 |
| chr7_19876833_19877030 | 0.26 |
Gm44659 |
predicted gene 44659 |
5493 |
0.08 |
| chr18_38277884_38278265 | 0.25 |
Pcdh12 |
protocadherin 12 |
115 |
0.93 |
| chr5_142818964_142819126 | 0.25 |
Tnrc18 |
trinucleotide repeat containing 18 |
1383 |
0.4 |
| chr2_35658400_35658598 | 0.25 |
Dab2ip |
disabled 2 interacting protein |
2851 |
0.31 |
| chr19_36375272_36375459 | 0.25 |
Pcgf5 |
polycomb group ring finger 5 |
3703 |
0.17 |
| chr2_14631725_14631884 | 0.25 |
Gm13268 |
predicted gene 13268 |
4483 |
0.13 |
| chr7_44895674_44895835 | 0.25 |
Fuz |
fuzzy planar cell polarity protein |
325 |
0.69 |
| chr2_132923749_132923930 | 0.24 |
Fermt1 |
fermitin family member 1 |
18124 |
0.14 |
| chr10_127962398_127962784 | 0.24 |
Gm47949 |
predicted gene, 47949 |
1035 |
0.33 |
| chr4_61671437_61671659 | 0.24 |
Mup18 |
major urinary protein 18 |
2588 |
0.25 |
| chr10_66631116_66631267 | 0.24 |
Gm28139 |
predicted gene 28139 |
9727 |
0.22 |
| chr8_70865758_70865918 | 0.23 |
Kcnn1 |
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 1 |
2580 |
0.11 |
| chr11_102578797_102578962 | 0.23 |
Gm11627 |
predicted gene 11627 |
240 |
0.86 |
| chr19_45114190_45114341 | 0.23 |
Gm35867 |
predicted gene, 35867 |
11024 |
0.14 |
| chr15_38518188_38518368 | 0.22 |
Azin1 |
antizyme inhibitor 1 |
57 |
0.89 |
| chr4_135312823_135313200 | 0.22 |
Gm12982 |
predicted gene 12982 |
5041 |
0.13 |
| chr8_70865457_70865628 | 0.22 |
Kcnn1 |
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 1 |
2284 |
0.12 |
| chr10_38970068_38970231 | 0.22 |
Lama4 |
laminin, alpha 4 |
4477 |
0.27 |
| chr11_98782747_98783111 | 0.21 |
Nr1d1 |
nuclear receptor subfamily 1, group D, member 1 |
7596 |
0.1 |
| chr17_35393925_35394090 | 0.21 |
H2-Q5 |
histocompatibility 2, Q region locus 5 |
119 |
0.9 |
| chr6_99172196_99172535 | 0.21 |
Foxp1 |
forkhead box P1 |
9347 |
0.29 |
| chr4_60579511_60579724 | 0.21 |
Mup10 |
major urinary protein 10 |
1480 |
0.33 |
| chr1_39102477_39102975 | 0.21 |
Gm37091 |
predicted gene, 37091 |
19388 |
0.18 |
| chr4_140983428_140983616 | 0.21 |
Atp13a2 |
ATPase type 13A2 |
3351 |
0.14 |
| chr6_88737071_88737256 | 0.21 |
Gm44005 |
predicted gene, 44005 |
1029 |
0.4 |
| chr19_46599600_46600340 | 0.20 |
Wbp1l |
WW domain binding protein 1 like |
846 |
0.52 |
| chr17_83213875_83214050 | 0.20 |
Pkdcc |
protein kinase domain containing, cytoplasmic |
1330 |
0.5 |
| chr11_98950041_98950197 | 0.20 |
Gm22061 |
predicted gene, 22061 |
3675 |
0.13 |
| chr11_88848716_88848881 | 0.20 |
Akap1 |
A kinase (PRKA) anchor protein 1 |
2705 |
0.21 |
| chr2_38185774_38185930 | 0.20 |
Dennd1a |
DENN/MADD domain containing 1A |
18033 |
0.18 |
| chr4_86545734_86545885 | 0.20 |
Saxo1 |
stabilizer of axonemal microtubules 1 |
6692 |
0.15 |
| chr1_39612023_39612366 | 0.20 |
Gm5100 |
predicted gene 5100 |
14189 |
0.12 |
| chr2_30823578_30823729 | 0.19 |
Asb6 |
ankyrin repeat and SOCS box-containing 6 |
3841 |
0.14 |
| chr7_19817545_19817767 | 0.19 |
Gm16174 |
predicted gene 16174 |
1103 |
0.24 |
| chr16_35310487_35310877 | 0.19 |
Gm49706 |
predicted gene, 49706 |
12384 |
0.15 |
| chr8_35632526_35632677 | 0.19 |
Mfhas1 |
malignant fibrous histiocytoma amplified sequence 1 |
25324 |
0.16 |
| chr6_59451195_59451399 | 0.18 |
Gprin3 |
GPRIN family member 3 |
25003 |
0.26 |
| chr11_34010872_34011034 | 0.17 |
Kcnip1 |
Kv channel-interacting protein 1 |
17801 |
0.19 |
| chr14_55899622_55899827 | 0.17 |
Sdr39u1 |
short chain dehydrogenase/reductase family 39U, member 1 |
131 |
0.91 |
| chr15_78832278_78832454 | 0.17 |
Cdc42ep1 |
CDC42 effector protein (Rho GTPase binding) 1 |
10258 |
0.09 |
| chr16_8597709_8597863 | 0.17 |
Abat |
4-aminobutyrate aminotransferase |
14159 |
0.12 |
| chr11_76677188_76677353 | 0.17 |
Trarg1 |
trafficking regulator of GLUT4 (SLC2A4) 1 |
2538 |
0.25 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.6 | GO:0039689 | negative stranded viral RNA replication(GO:0039689) multi-organism biosynthetic process(GO:0044034) |
| 0.4 | 1.2 | GO:0032741 | positive regulation of interleukin-18 production(GO:0032741) |
| 0.2 | 0.6 | GO:1904742 | regulation of telomeric DNA binding(GO:1904742) |
| 0.2 | 0.5 | GO:0070948 | regulation of neutrophil mediated cytotoxicity(GO:0070948) regulation of neutrophil mediated killing of symbiont cell(GO:0070949) |
| 0.1 | 1.6 | GO:0060263 | regulation of respiratory burst(GO:0060263) |
| 0.1 | 0.4 | GO:2000586 | regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000586) negative regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000587) |
| 0.1 | 0.4 | GO:0043096 | purine nucleobase salvage(GO:0043096) |
| 0.1 | 1.1 | GO:0035234 | ectopic germ cell programmed cell death(GO:0035234) |
| 0.1 | 0.3 | GO:0034476 | U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) |
| 0.1 | 0.3 | GO:0090283 | regulation of protein glycosylation in Golgi(GO:0090283) |
| 0.1 | 0.5 | GO:2001286 | regulation of caveolin-mediated endocytosis(GO:2001286) |
| 0.1 | 0.4 | GO:1901896 | positive regulation of calcium-transporting ATPase activity(GO:1901896) |
| 0.1 | 0.4 | GO:2001197 | regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904259) positive regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904261) basement membrane assembly involved in embryonic body morphogenesis(GO:2001197) |
| 0.1 | 0.5 | GO:0034382 | chylomicron remnant clearance(GO:0034382) triglyceride-rich lipoprotein particle clearance(GO:0071830) |
| 0.1 | 0.6 | GO:0051549 | regulation of keratinocyte migration(GO:0051547) positive regulation of keratinocyte migration(GO:0051549) |
| 0.1 | 0.3 | GO:0014734 | skeletal muscle hypertrophy(GO:0014734) |
| 0.1 | 0.2 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.1 | 0.3 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.1 | 0.4 | GO:0034144 | negative regulation of toll-like receptor 4 signaling pathway(GO:0034144) |
| 0.1 | 0.2 | GO:0001712 | ectodermal cell fate commitment(GO:0001712) |
| 0.1 | 0.2 | GO:2000313 | fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060825) regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000313) |
| 0.0 | 0.2 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.0 | 0.5 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.0 | 0.1 | GO:0061091 | regulation of phospholipid translocation(GO:0061091) positive regulation of phospholipid translocation(GO:0061092) |
| 0.0 | 0.3 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.0 | 0.0 | GO:0021828 | cerebral cortex tangential migration using cell-axon interactions(GO:0021824) gonadotrophin-releasing hormone neuronal migration to the hypothalamus(GO:0021828) hypothalamic tangential migration using cell-axon interactions(GO:0021856) facioacoustic ganglion development(GO:1903375) |
| 0.0 | 0.1 | GO:0018242 | immunoglobulin biosynthetic process(GO:0002378) protein O-linked glycosylation via serine(GO:0018242) |
| 0.0 | 0.1 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.0 | 0.1 | GO:0070350 | white fat cell proliferation(GO:0070343) regulation of white fat cell proliferation(GO:0070350) |
| 0.0 | 0.1 | GO:0061470 | T follicular helper cell differentiation(GO:0061470) |
| 0.0 | 0.1 | GO:0008050 | female courtship behavior(GO:0008050) |
| 0.0 | 0.1 | GO:0071374 | cellular response to parathyroid hormone stimulus(GO:0071374) |
| 0.0 | 0.7 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.0 | 0.1 | GO:0006768 | biotin metabolic process(GO:0006768) |
| 0.0 | 0.1 | GO:0048619 | embryonic hindgut morphogenesis(GO:0048619) |
| 0.0 | 0.2 | GO:1900086 | positive regulation of peptidyl-tyrosine autophosphorylation(GO:1900086) |
| 0.0 | 0.4 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.0 | 0.1 | GO:0035279 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.0 | 0.1 | GO:0072675 | osteoclast fusion(GO:0072675) |
| 0.0 | 0.1 | GO:0051791 | medium-chain fatty acid metabolic process(GO:0051791) |
| 0.0 | 0.1 | GO:0009223 | pyrimidine deoxyribonucleotide catabolic process(GO:0009223) |
| 0.0 | 0.4 | GO:0048505 | regulation of timing of cell differentiation(GO:0048505) |
| 0.0 | 0.1 | GO:0042997 | negative regulation of Golgi to plasma membrane protein transport(GO:0042997) |
| 0.0 | 0.1 | GO:0009414 | response to water deprivation(GO:0009414) |
| 0.0 | 0.2 | GO:0031659 | positive regulation of cyclin-dependent protein serine/threonine kinase activity involved in G1/S transition of mitotic cell cycle(GO:0031659) |
| 0.0 | 0.1 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.1 | GO:0045658 | regulation of neutrophil differentiation(GO:0045658) |
| 0.0 | 0.1 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.0 | 0.1 | GO:1903553 | positive regulation of extracellular exosome assembly(GO:1903553) |
| 0.0 | 0.1 | GO:0015772 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.0 | 0.0 | GO:0018992 | germ-line sex determination(GO:0018992) |
| 0.0 | 0.1 | GO:1900108 | negative regulation of nodal signaling pathway(GO:1900108) |
| 0.0 | 0.1 | GO:0061052 | negative regulation of cell growth involved in cardiac muscle cell development(GO:0061052) |
| 0.0 | 0.1 | GO:0002337 | B-1a B cell differentiation(GO:0002337) |
| 0.0 | 0.1 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.0 | 0.1 | GO:0008627 | intrinsic apoptotic signaling pathway in response to osmotic stress(GO:0008627) regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902218) negative regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902219) |
| 0.0 | 0.0 | GO:0061724 | lipophagy(GO:0061724) |
| 0.0 | 0.1 | GO:1903898 | negative regulation of PERK-mediated unfolded protein response(GO:1903898) |
| 0.0 | 0.1 | GO:0046618 | drug export(GO:0046618) |
| 0.0 | 0.0 | GO:0003162 | atrioventricular node development(GO:0003162) |
| 0.0 | 0.1 | GO:0043435 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.0 | 0.1 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.0 | 0.1 | GO:2000359 | regulation of binding of sperm to zona pellucida(GO:2000359) |
| 0.0 | 0.5 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.1 | GO:0003383 | apical constriction(GO:0003383) |
| 0.0 | 0.0 | GO:0002925 | positive regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002925) |
| 0.0 | 0.1 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) |
| 0.0 | 0.1 | GO:0032430 | positive regulation of phospholipase A2 activity(GO:0032430) |
| 0.0 | 0.0 | GO:0031145 | anaphase-promoting complex-dependent catabolic process(GO:0031145) |
| 0.0 | 0.1 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.0 | 0.1 | GO:1901727 | positive regulation of histone deacetylase activity(GO:1901727) |
| 0.0 | 0.2 | GO:0051895 | negative regulation of focal adhesion assembly(GO:0051895) |
| 0.0 | 0.1 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.0 | 0.0 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 0.0 | 0.1 | GO:0060334 | regulation of interferon-gamma-mediated signaling pathway(GO:0060334) |
| 0.0 | 0.1 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.0 | 0.1 | GO:2001260 | regulation of semaphorin-plexin signaling pathway(GO:2001260) |
| 0.0 | 0.1 | GO:0031665 | negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.0 | 0.0 | GO:0036151 | phosphatidylcholine acyl-chain remodeling(GO:0036151) |
| 0.0 | 0.1 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 0.1 | GO:0015705 | iodide transport(GO:0015705) |
| 0.0 | 0.1 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.0 | 0.0 | GO:0021699 | cerebellar cortex maturation(GO:0021699) |
| 0.0 | 0.1 | GO:0015884 | folic acid transport(GO:0015884) |
| 0.0 | 0.0 | GO:0007290 | spermatid nucleus elongation(GO:0007290) |
| 0.0 | 0.1 | GO:0009437 | carnitine metabolic process(GO:0009437) |
| 0.0 | 0.2 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 | 0.0 | GO:0001923 | B-1 B cell differentiation(GO:0001923) |
| 0.0 | 0.0 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.0 | GO:0048819 | regulation of hair follicle maturation(GO:0048819) |
| 0.0 | 0.0 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.1 | GO:0008298 | intracellular mRNA localization(GO:0008298) |
| 0.0 | 0.1 | GO:0019370 | leukotriene biosynthetic process(GO:0019370) |
| 0.0 | 0.2 | GO:0051443 | positive regulation of ubiquitin-protein transferase activity(GO:0051443) |
| 0.0 | 0.0 | GO:0042091 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 0.0 | 0.3 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.0 | GO:0045875 | negative regulation of sister chromatid cohesion(GO:0045875) |
| 0.0 | 0.1 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.0 | 0.0 | GO:0036016 | response to interleukin-3(GO:0036015) cellular response to interleukin-3(GO:0036016) |
| 0.0 | 0.2 | GO:0008089 | anterograde axonal transport(GO:0008089) |
| 0.0 | 0.0 | GO:0050427 | 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) |
| 0.0 | 0.0 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.0 | 0.2 | GO:2000353 | positive regulation of endothelial cell apoptotic process(GO:2000353) |
| 0.0 | 0.2 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.0 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.0 | 0.0 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.0 | 0.0 | GO:1902093 | positive regulation of sperm motility(GO:1902093) |
| 0.0 | 0.0 | GO:0097368 | establishment of Sertoli cell barrier(GO:0097368) |
| 0.0 | 0.0 | GO:0018197 | peptidyl-aspartic acid modification(GO:0018197) |
| 0.0 | 0.1 | GO:1902036 | regulation of hematopoietic stem cell differentiation(GO:1902036) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | GO:0035355 | Toll-like receptor 2-Toll-like receptor 6 protein complex(GO:0035355) |
| 0.2 | 1.0 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 0.1 | 1.0 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.1 | 1.6 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.1 | 0.2 | GO:0005745 | m-AAA complex(GO:0005745) |
| 0.1 | 0.3 | GO:0035363 | histone locus body(GO:0035363) |
| 0.1 | 0.4 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 0.1 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.2 | GO:0005767 | secondary lysosome(GO:0005767) |
| 0.0 | 0.3 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 0.1 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.0 | 0.1 | GO:0030313 | cell outer membrane(GO:0009279) cell envelope(GO:0030313) external encapsulating structure part(GO:0044462) |
| 0.0 | 0.7 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 0.5 | GO:0001741 | XY body(GO:0001741) |
| 0.0 | 0.1 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 0.4 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.1 | GO:1990597 | AIP1-IRE1 complex(GO:1990597) |
| 0.0 | 0.1 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.2 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.1 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.0 | 0.3 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 0.1 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) ERCC4-ERCC1 complex(GO:0070522) |
| 0.0 | 0.1 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.1 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 0.1 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 0.1 | GO:0098651 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.0 | 0.1 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 0.0 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.0 | 0.1 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.0 | 0.1 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.0 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 0.0 | 0.2 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.0 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.1 | GO:0090543 | Flemming body(GO:0090543) |
| 0.0 | 0.0 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.0 | 0.1 | GO:0005818 | aster(GO:0005818) |
| 0.0 | 0.0 | GO:0005879 | axonemal microtubule(GO:0005879) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.7 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.2 | 1.1 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.1 | 1.0 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.1 | 0.5 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.1 | 0.6 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.1 | 0.5 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.1 | 0.9 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.1 | 0.3 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.1 | 0.2 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.1 | 0.5 | GO:0004438 | phosphatidylinositol-3-phosphatase activity(GO:0004438) |
| 0.1 | 0.2 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.4 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.2 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.1 | GO:0008525 | phosphatidylcholine transporter activity(GO:0008525) |
| 0.0 | 0.4 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.0 | 0.4 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.2 | GO:0042979 | ornithine decarboxylase regulator activity(GO:0042979) |
| 0.0 | 0.5 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.1 | GO:0050309 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.0 | 0.2 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.0 | 0.1 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.1 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 0.0 | 0.4 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.1 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.0 | 0.2 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.0 | 0.1 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.0 | 0.3 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 0.1 | GO:0035514 | DNA demethylase activity(GO:0035514) |
| 0.0 | 0.3 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.3 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 0.1 | GO:0015157 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.0 | 0.1 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.0 | 0.0 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.0 | 0.1 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.0 | 0.1 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) |
| 0.0 | 0.1 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.0 | 1.0 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.0 | 0.1 | GO:0071535 | RING-like zinc finger domain binding(GO:0071535) |
| 0.0 | 0.0 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.0 | 0.1 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.0 | 0.2 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.1 | GO:0070326 | very-low-density lipoprotein particle receptor binding(GO:0070326) |
| 0.0 | 0.2 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.1 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 0.1 | GO:0008430 | selenium binding(GO:0008430) |
| 0.0 | 0.0 | GO:0015526 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.0 | 0.4 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) ubiquitin-like protein conjugating enzyme activity(GO:0061650) |
| 0.0 | 0.3 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.1 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.1 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.0 | 0.1 | GO:0008517 | folic acid transporter activity(GO:0008517) |
| 0.0 | 0.0 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.0 | 0.1 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.0 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.0 | 0.0 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.0 | 0.0 | GO:0051718 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) DNA (cytosine-5-)-methyltransferase activity, acting on CpG substrates(GO:0051718) |
| 0.0 | 0.1 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.0 | 0.2 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.1 | GO:0001094 | TFIID-class transcription factor binding(GO:0001094) |
| 0.0 | 0.1 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.1 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.0 | 0.1 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.0 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.0 | 0.0 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.0 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.1 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.0 | 1.2 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.4 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.2 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 0.4 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 1.2 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 0.4 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 0.7 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 0.7 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.0 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 0.8 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.4 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 0.5 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.2 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.4 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 0.2 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.2 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.0 | 0.3 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.0 | 0.6 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 1.0 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.0 | 0.4 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 0.5 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.3 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.3 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.4 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.3 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.1 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.0 | 0.3 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 0.4 | REACTOME RORA ACTIVATES CIRCADIAN EXPRESSION | Genes involved in RORA Activates Circadian Expression |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 0.0 | 0.4 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.0 | 0.2 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.1 | REACTOME TAK1 ACTIVATES NFKB BY PHOSPHORYLATION AND ACTIVATION OF IKKS COMPLEX | Genes involved in TAK1 activates NFkB by phosphorylation and activation of IKKs complex |
| 0.0 | 0.1 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.4 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.5 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.1 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 0.2 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.0 | 0.0 | REACTOME CDK MEDIATED PHOSPHORYLATION AND REMOVAL OF CDC6 | Genes involved in CDK-mediated phosphorylation and removal of Cdc6 |